Abstract
The oncogenic transcription factor Runx1 is required for the specification of definitive hematopoietic stem cells (HSC) in the developing embryo. The activity of this master regulator is tightly controlled during development. The transcription factors that upregulate the expression of Runx1 also upregulate the expression of Smad6, the inhibitory Smad, which controls Runx1 activity by targeting it to the proteasome. Here we show that Runx1, in conjunction with Fli1, Gata2, and Scl, directly regulates the expression of Smad6 in the aorta-gonad-mesonephros (AGM) region in the developing embryo, where HSCs originate. Runx1 regulates Smad6 activity via a novel upstream enhancer, and Runx1 null embryos show reduced Smad6 transcripts in the yolk-sac and c-Kit-positive fetal liver cells. By directly regulating the expression of Smad6, Runx1 sets up a functional rheostat to control its own activity. The perturbation of this rheostat, using a proteasomal inhibitor, results in an increase in Runx1 and Smad6 levels that can be directly attributed to increased Runx1 binding to tissue-specific regulatory elements of these genes. Taken together, we describe a scenario in which a key hematopoietic transcription factor controls its own expression levels by transcriptionally controlling its controller.
INTRODUCTION
Cell fate decisions in the developing embryo are coordinated by complex gene regulatory networks. These networks are composed primarily of transcription factors (TFs) and cis-regulatory modules, which together control the rates of gene expression. As a consequence, transcriptional regulatory networks control cell function and identity by controlling the types and amounts of protein that are produced in a cell (7). Largely through the systematic analysis of transcription networks in bacteria and yeast, it is now known that information flow through complex networks is controlled by deploying a recurrent set of regulatory patterns called network motifs (1). Foremost among these are feed-forward and feed-back loops that regulate the temporal sequence and on/off kinetics of protein expression, which is essential for the coordinated growth of cells in metazoans (2, 29).
Feed-forward loops are commonly deployed in transcriptional networks to bring about the robust expression of specific genes in response to a biological stimulus, and they function to distinguish the authentic expression of individual genes from transcriptional noise. Feed-back loops, which can have either a positive or negative effect, are used in development to lock in the expression of critical proteins or equally to exclude the expression of proteins in a cell that, if expressed, could push it toward an alternate cell fate (8). Feed-back loops that act as rheostats, such as TF↔ microRNA (miRNA) rheostats, where the regulator is itself controlled by the product, are increasingly recognized (9). TF↔ miRNA rheostats that have been mapped on a genome scale show these to be highly interconnected with miRNAs and TFs in such loops regulating multiple targets (19).
Runx1/AML1 is a key regulator of definitive hematopoiesis (24, 37) and is required for the generation of hematopoietic stem cells (HSCs) from hemogenic endothelium (21). Runx1 belongs to the runt domain family of TFs, which also includes Runx2 and Runx3. All Runx proteins recognize the same DNA motif, so their distinct functional identities are maintained by tightly regulating their expression patterns (17). The deletion of Runx1 in the germ line or in the endothelium results in the absence of HSCs (5, 6). On the other hand, haploinsufficiency results in the early emergence of HSCs from the aorta-gonad-mesonephros (AGM) (5, 22). Interestingly, the deletion of Runx1 in adult HSCs per se leads to the initial expansion and subsequent exhaustion of HSCs (15). The partial or complete loss of RUNX1 function is a common underlying first hit in leukemia, although it is not sufficient per se to cause full-blown leukemia (35). Taken together, controlling the dose of Runx1 is important, and having too little or too much Runx1 at specific developmental time points can perturb HSC development and homeostasis.
The control of Runx1 levels within a narrow range during hematopoiesis is achieved by transcriptional, posttranscriptional, and posttranslational mechanisms. At a transcriptional level, the Runx1+23 enhancer mediates expression in the developing hematopoietic system in conjunction with the distal and/or proximal Runx1 promoter (3). Feed-forward loops involving upstream TFs, such as Gata2, Fli1, and Scl, upregulate Runx1 expression in cells during hematopoietic commitment (16, 23). Gata2, Fli1, and Scl form a fully connected triad, with each TF regulating itself and the other two in the AGM (30). At a posttranscriptional stage, Runx1 levels can be controlled by miR-27a in a feed-back loop, which involves the positive regulation of miR-27a by Runx1 (4). At a posttranslational stage, we have reported previously that Runx1 levels are controlled by Smad6/Smurf1-mediated proteasome degradation (28), but how this control mechanism is integrated within the hematopoietic transcriptional network is not clear.
Smad6 is an inhibitor of Bmp4 signaling (14), a key driver of HSC development (18). Both Runx1 and its inhibitor, Smad6, are transcriptional targets of Bmp4 signaling (28). The activity of Runx2, a key regulator of bone development, is also controlled by Smad6 targeting of Runx2 to the proteasome (33). Interestingly, Runx2 recently has been shown to transcriptionally regulate Smad6 expression by binding an atypical Runx binding element (RBE) in the Smad6 promoter (38). This is suggestive of an integrated Runx2↔Smad6 regulatory loop. Given the requirement for the exquisite control of Runx1 levels in embryonic hematopoiesis, these data led us to question whether a Runx1↔Smad6 rheostat fine-tunes Runx1 levels during normal blood development.
MATERIALS AND METHODS
Cell culture.
COS7 cells were maintained in Dulbecco's modified essential medium (DMEM) and 416B and K562 cells in RPMI medium, each supplemented with 10% fetal calf serum (FCS) and 1:100 penicillin-streptomycin. HPC-7 cells were maintained in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% FCS, 1.5 × 10−4M monothioglycerol (MTG), and stem cell factor as previously described (31).
Stable transfection, transactivation, and luciferase reporter assay.
416B and K562 cells were electroporated with 1 μg of linearized pGK Neo and 10 μg of linearized vector and selected at 24 h with G418, and luciferase activity was assayed at ∼2 weeks as previously described (26). Site-directed mutagenesis was performed using the Stratagene QuikChange IIXL kit according to the manufacturer's instructions. The −2006/+45-pGL3b, −1191/+45-pGL3b, mut-1191/+45-pGL3b, −829/+45-pGL3b, and 6OSE2 (RBE)-pGL3b vectors were a kind gift from D. Chen and Q. Wang. Transactivation assays were performed in Cos7 cells to test Smad6 promoter and Smad6-57 enhancer responsiveness to Runx1. For Smad6 promoter responsiveness, 5 × 105 cells were transfected with luciferase constructs (pGL3b, 6RBE-pGL3b, Smad6P-pGL3b, and mRBESmad6P-pGL3b) alone or in combination with either pCDNA3 or pCDNA3-Runx1. For Smad6 enhancer responsiveness, pGL2p, Smad6-57-pGL2p, and mutant Smad6-57-pGL2p luciferase constructs were transfected alone or in combination with pMSCV-Gata2, pCDNA3-Runx1, and pEFBOS-Cbfb. An equivalent quantity of DNA was transfected using the empty vectors pcDNA3, pMSCV, and pEFBOS as controls when necessary. Each transfection and transactivation assay was performed on at least two different days in quadruplicate (transfection) or triplicate (transactivation).
Cloning primers (mouse) were the following: Smad6-57 forward, ATAACGCGTGCTAGCTGTTCTCTCAGGCTGCTTGC; Smad6-57 reverse, ATACTCGAGACGCGTCACGGTTTGGTTGTTTGTCC. Cloning primers for human samples were the following: SMAD6-57 forward, ATAGCTAGCCTGACCCCAGACACATGCTC; SMAD6-57 reverse, ATACTCGAGGAAAGAGCGGCAGAATCCTC.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed in HPC-7, 416B, and K562 cells and primary cells from embryonic day 8.5 (E8.5) yolk sacs (YSs) and E11.5 DAs and FLs. For cell lines, 2 × 107 cells per antibody were treated with 0.4% formaldehyde, and the cross-linked chromatin was sonicated to obtain fragments of 300 to 500 bp in size. For primary tissue ChIP, 100 YSs and 120 dorsal aortas were dissected from day 8.5 and day 11.5 embryos, respectively, and treated with collagenase, whereas 120 fetal livers dissected from day 11.5 embryos were disrupted by being pipetted before being cross-linked, and the sonicated chromatin was distributed evenly for immunoprecipitation with up to six antibodies. In experiments with MG132, 416B cells were treated either with the active compound or dimethylsulfoxide (DMSO) for up to 16 h prior to formaldehyde cross-linking. Enrichment was measured by real-time PCR using SYBR green (Stratagene) with primer sets designed using Beacon Designer software (Premier Biosoft International, CA) or processed for high-throughput sequencing as described below. For ChIP-reverse transcription (RT), dissociation curves were run to establish that single products were amplified in each reaction. The levels of enrichment were normalized to that obtained with a control IgG antibody and calculated as the fold increase above that measured at a control region (an intronic region of the LMO2 locus [human cells] or the AFP promoter [mouse cells]). For high-throughput sequencing, each sample was amplified and sequenced using an Illumina GAII to obtain ∼20 million raw reads per lane, of which ∼70% was aligned to either the human (K562) or mouse (HPC-7) genomes using model-based analysis of ChIP-Seq (MACS) (41), converted into density plots, and displayed as UCSC genome browser custom tracks (4, 40). Processed and unprocessed data are available at the NCBI Gene Expression Omnibus portal (accession number GSE22178). The following antibodies were used: AcH3 (Millipore 06-599), Fli1 (Abcam ab15289), Gata2 (sc9008x; Santa Cruz), Scl (sc12984X; Santa Cruz), Runx1 (PC284; Calbiochem), and Runx1 (ab23980-100; Abcam). Human ChIP primers were the following: AFP forward, GGCAAATGTCCCATTTTCAA; AFP reverse, TTCTTTTATACTCTTTTCAGGCAATG; MO2 forward, GAAATAAATATCTCCACTGTCCTG; MO2 reverse, CTATCTGCCTATCTCTCATCTATC; SMAD6 −71 forward, ATTCTCACGACTTAACAATGC; SMAD6 −71 reverse, CTCTCACGACAGTGGTTG. Mouse ChIP primers were the following: Runx1+23 forward, AAGCTGCCCACGTTATCAGT; Runx1+23 reverse, CAGATGGAGGCATCCTGTTT; Runx1 DP forward, TCTGAAAGCCACCAAATCCG; Runx1 DP reverse, CTTCCTGCCCTCCACCTG; Smad6 RBE forward, CCTCTGCTTCGGTGGATTG; Smad6 RBE reverse, GGCAAGTCTCTCCTGAACG; Smad6 RBE+0.5kb forward, GAAGAAACCCGCTATCCC; Smad6 RBE+0.5kb reverse, CTTTAGAGTCAGTCCAAACC; Smad6 RBE−0.5kb forward,GCACACCACCTTCGCTAC; Smad6 RBE−0.5kb reverse, TCAATGAAAGAAATTCCGCTACTC; Smad6−57 forward, GTGGCTTGAGGCTATCTTAC; Smad6−57 reverse, GGGTGTACTTGGGAGTGG; Afp forward, ACAAGTGACCCCTGCTCTGT; and Afp reverse, CCTGTTTAAGGGATGCCTGTT.
Knockdown of gene expression.
Scl knockdown sequences were designed on the Dharmacon website and subcloned into the pLMP expression vector (XhoI/EcoRI). In brief, viral supernatant was harvested from HEK293T cells and passed through a 0.45-μm filter. HPC-7 cells (2 × 106) were infected with 1 ml of viral supernatant. Levels of infection were determined by flow cytometry for GFP 24 h after transduction. Cells were selected in puromycin at 1 μg/ml. Scl depletion probably affects the viability of the stem cell factor-dependent HPC-7 cell line, and no more than 50% knockdown was seen in puromycin selected clones. The following sequences were used (italics denote sequences that are complementary to the target gene): shScl-d3, TGCTGTTGACAGTGAGCGCCACCAGACAAGAAACTAATAGTGAAGCCACAGATGTATTAGTTTCTTGTCTGGTGGTGCCTACTGCCTCGGA; Shluciferase, TGCTGTTGACAGTGAGCGCACGTACGCGGAATACTTCGAATAGTGAAGCCACAGATGTATTCGAAGTATTCCGCGTACGTGTGCCTACTGCCTCGGA.
Transgenic analysis.
Candidate promoter and enhancer sequences were PCR amplified from mouse and human genomic DNA and subcloned using standard procedures into the pGlacZ vector, and details are available upon request. F0 transgenic mouse embryos were generated by the pronuclear injection of gel-extracted lacZ reporter fragments (Cyagen). For histology, embryos were harvested at day 11.5 postcoitus, fixed, stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), embedded in paraffin, sectioned, and counterstained with neutral red. Whole-mount images were acquired using a Leica DFC 420C digital camera attached to a Leica S8APO microscope. Images of sections were acquired with the same camera attached to a Leica DM2500 microscope. Images were acquired using the Leica Application Suite and processed using Adobe Photoshop (Adobe Systems, CA).
Flow cytometry.
The Runx1rd/+ embryos were from a line generated by Wang et al. (37). For Smad6 expression, at least six embryos from each genotype were used to extract RNA at the given time points. Single-cell suspensions of E11.5 FLs (Runx1+/+ and Runx1rd/+) were stained with anti-c-Kit/APC (BD Pharmingen; 553356). Dead cells were excluded by Hoechst 33258 uptake, and c-Kit+ cells were sorted on a MoFlo (Beckman Coulter).
Expression analysis.
Expression analysis was performed by real-time PCR using SYBR green (Applied Biosystems). Dissociation curves were run to ascertain that only single products were amplified in each reaction. Gene expression was quantified relative to that of b2microglobulin. Known amounts of template DNA were used to generate dilution series and standard curves for each set of primers to help quantify levels of expression in test samples.
SYBR green expression primers (mouse) were the following: Runx1 forward, ACTTCCTCTGCTCCGTGCTA; Runx1 reverse, CGCGGTAGCATTTCTCAGTT; Smad6 forward, CCTATTCTCGGCTGTCTCCTC; Smad6 reverse, CTCGGCTTGGTGGCATCC; Fli-1 forward, GGAGTATGACCACATGAATGG; Fli-1 reverse, GACTCTCCGTTCGTTGGTG; Gata2 forward, AAAGGGGCTGAATGTTTCG; Gata2 reverse, GCGTGGGTAGGATGTGTC; Scl forward, CATGTTCACCAACAACAACCG; Scl reverse, GGTGTGAGGACCATCAGAAATCTC; b2microglobulin forward, GAGACTGATACATACGCCTGCAGA; b2microglobulin reverse, TCACATGTCTCGATCCCAGTAGA.
Statistical analysis.
Data were analyzed using GraphPad Prism 5 software (La Jolla, CA). Comparison between two groups was performed using an unpaired t test. A P value of less than 0.05 was considered significant. The following notation is used throughout: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
RESULTS
Runx1 binds the SMAD6 locus in hematopoietic cells.
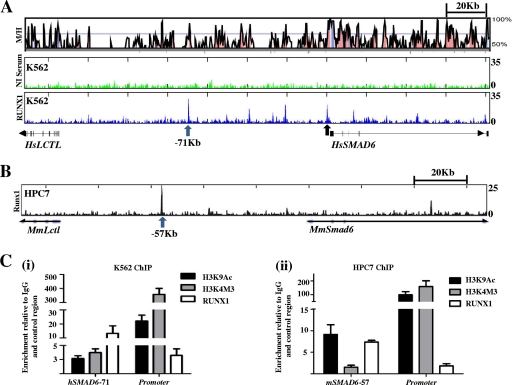
To assess whether Runx1 bound the Smad6 locus in vivo, we performed chromatin immunoprecipitation (ChIP) assays in human K562 erythroleukemic cells and mouse HPC-7 blood stem/progenitor cells. ChIP-seq traces generated in human K562 cells using an anti-Runx1 antibody (Fig. 1A) show peaks of enrichment at the SMAD6 promoter and at multiple sites in and around the SMAD6 locus compared to the traces of the IgG control. Runx1 enrichment is most pronounced at a region 71 kb upstream of the promoter. This peak of enrichment is also upstream of the LTCL gene which, however, encodes an enzyme that is ubiquitously expressed (36) and is unlikely to be under the control of a Runx1-bound tissue-specific enhancer. These traces were compared to the ChIP-seq performed in the HPC-7 hematopoietic stem/progenitor cells (Fig. 1B). This stem cell factor-dependent cell line was used recently as a model to survey the transcriptional landscape of mouse HSCs (40). Interestingly, Runx1 is almost exclusively enriched at a region 57 kb upstream of the mouse Smad6 promoter. This region is homologous to the human SMAD6-71 region, which was bound by RUNX1 in K562 cells. There is no binding of Runx1 at the Smad6 promoter in HPC-7 stem/progenitor cells.
Fig. 1.
(A) RUNX1 binds the SMAD6 promoter and conserved noncoding regions in K562 cells. The upper panel shows a VISTA plot of human/mouse sequence conservation greater than 50% across the SMAD6 locus. Noncoding sequences with greater than 75% conservation are shaded in pink, exons in magenta, and the untranslated regions in cyan. The middle and lower panels show ChIP-seq traces using a nonimmune serum (NI serum) control and a Runx1 antibody in K562 human erythroleukemic cells. Enrichments at the human SMAD6 (HsSMAD6) promoter and a region 71 kb upstream of the promoter are marked with arrows. (B) Runx1 binds a region at kb −57 in mouse HPC-7 stem/progenitor cells. In contrast with K562 cells, Runx1 enrichment in mouse blood stem cells is seen mostly at a region 57 kb upstream of the mSmad6 promoter, which is homologous with the human region at kb −71. (C) There are active chromatin marks and Runx1 enrichment by ChIP-RT-PCR at sites identified by ChIP-seq. (i) Human K562 erythroleukemic cells; (ii) mouse HPC-7 stem/progenitor cells.
To confirm the enrichment of RUNX1 and chromatin accessibility at these regions, ChIP material was analyzed by RT-PCR (Fig. 1C). The acetylation of histone H3K9 marks regions of active transcription (promoters and enhancers), whereas the trimethylation of histone H3K4 is most prominent at transcription start sites (39). The SMAD6 promoter is in an active configuration in both K562 and HPC-7 cells, but as seen with ChIP-seq, it is bound by RUNX1 in the former but not in the latter. The upstream regions are active and bound by RUNX1 in both cell types. Taken together, these data show that RUNX1 binds the SMAD6 locus in human and murine hematopoietic cells. However, the binding of a TF to a particular locus does not in itself imply the regulation of the target gene by the TF. Therefore, the transcriptional activity of the Smad6 promoter and −57 regions were analyzed in detail.
The Smad6 promoter is active in blood progenitors and can be transactivated by Runx1.
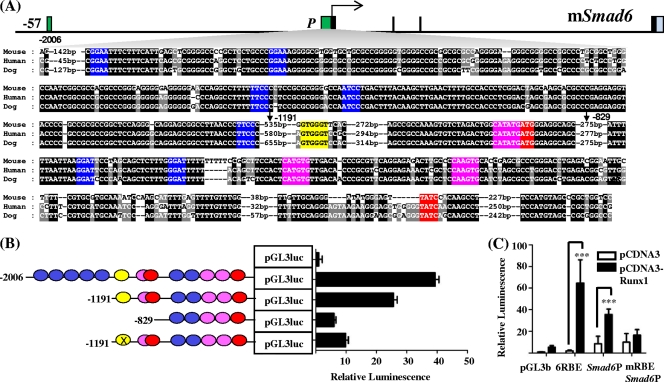
In addition to the previously characterized atypical RBE (GTGGGT) (38), the −2006/+45 Smad6 promoter region has a number of conserved Ets, Gata, and E-box binding sites that tend to be collectively enriched in promoters of genes that are expressed in hematopoietic cells (Fig. 2A). This fragment is known to be active in C2C12 myofibroblasts, and its response to BMP2 is dependent on the atypical RBE (38). A survey of the Smad6 locus for conserved Runx binding sites using CoMoDis (10) failed to identify a single consensus motif (TGYGGT) within 100 kb of the start or end of the gene. To assess the hematopoietic activity of the Smad6 promoter, wild-type and truncated fragments were tested in stable transfection assays in 416B blood progenitors. The full-length 2-kb fragment (−2006/+45) had approximately 40-fold higher activity than that of a promoterless luciferase control plasmid (Fig. 2B), but ∼70% of its basal activity was within a 1.2-kb fragment (−1191/+45). The 0.8-kb fragment (−829/+45) maintains some residual hematopoietic activity but has less than a third of that of the 1.2-kb fragment. The greater part of the basal hematopoietic activity of the promoter therefore resides within the −1191 to −829 region, which contains the conserved RBE and an overlapping Gata/E-box motif. Site-directed mutagenesis of the RBE significantly diminished the hematopoietic activity of the −1191/+45 fragment.
Fig. 2.
Smad6 promoter has conserved Ets, Gata, and E-box motifs and an atypical Runx binding element, is active in hematopoietic cells, and can be transactivated by Runx1. (A) Nucleotide sequence alignment of the −2006/+45 promoter region. Conserved Ets (blue), Gata (red), and E-box (pink) motifs and the Runx (yellow) binding element (RBE) are colored for clarity. Base pair numbering is from the reference transcription start site corresponding to NM008542, and the approximate start sites of the truncated promoter fragments, −1191/+45 and −829/+45, are indicated with arrows. (B) Stable transfection assays. Shown on the left are the reporter constructs of mouse wild-type and mutated Smad6 promoter fragments inserted upstream of the promoterless pGL3basic vector. The conserved Ets, Runx, E-box, and Gata binding sites are represented as circles and are crossed out where mutated. Shown on the right are the results of stable transfection assays in K562 cells corresponding to each construct. The luciferase activities are given as the fold increase above the activity of the basic (pGL3B) vector. Each bar is the means (± standard deviations) of the relative luciferase activity from at least two experiments performed in triplicate. (C) Transactivation assays. A luciferase reporter construct with multiple copies of the RBE subcloned upstream of the SV basal promoter (6RBE-pGL3b) was strongly transactivated by Runx1. A construct carrying the wild-type Smad6 promoter (Smad6P-pGL3b) also was strongly transactivated by Runx1, but this effect was abrogated by the site-directed mutagenesis of the RBE (mRBESmad6P-pGL3b). ***, P < 0.001.
Runx2 has been reported to transactivate the wild-type Smad6 promoter, and this activity is dependent on the atypical RBE (38). To assess the response of this element to Runx1, a Runx1 expression plasmid (pCDNA3-Runx1) was cotransfected with a hexamer of RBE, (6× OSE2-a)-pGL3b, into Cos-7 cells (Fig. 2C). There was a dramatic increase in relative luciferase activity, which also was seen to a lesser degree with the wild-type Smad6 promoter fragment (Smad6P-pGL3b) but not with the mutant RBE-Smad6 promoter fragment. These data indicate that the Smad6 promoter is a transcriptional target of Runx1.
Smad6-57 is active in blood progenitors and can be transactivated by Runx1.
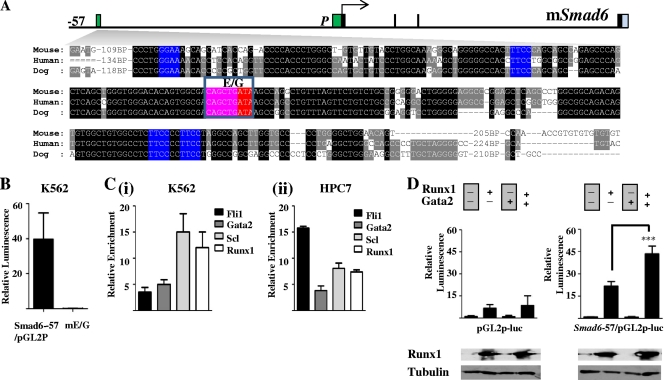
Although the Smad6-57 region is bound by Runx1 in both human and murine hematopoietic cells, it does not contain any recognizable Runx1 consensus motifs. However, there are a number of conserved Ets, Gata, and E-box motifs (Fig. 3A) that often are associated with hematopoietic cis-regulatory modules (13, 27). In a genome-wide survey of hematopoietic TF binding, we recently showed that Runx1 binding overlaps with Scl, Lmo2, Lyl1, Gata2, Fli1, and Erg (40). Furthermore, only a minority of Runx1-bound regions had recognizable Runx1 consensus motifs. Runx1 physically interacts with Scl and Gata2 (40) and likely is targeted to the majority of its binding sites in the absence of direct DNA binding. To test the activity of this region in hematopoietic cells and establish whether its activity is dependent on the Gata and E-box sites, an ∼500-bp fragment from this region was subcloned into the pGL2p reporter vector, and wild-type and mutant constructs were tested in K562 stable transfection assays (Fig. 3B). The wild-type fragment had approximately 40-fold higher activity than the control plasmid, and this activity was abolished by mutating the E-box/Gata site. If Runx1 binding to the Smad6-57 enhancer is mediated by TFs that bind E-box/Gata motifs, we would expect to see the enrichment of candidate factors at this region in hematopoietic cells. We performed RT-PCR on ChIP material obtained from HPC-7 cells with antibodies against Scl, Fli1, and Gata2. We have shown previously that these three factors form a cross-regulating triad at hematopoietic sites in the developing embryo. Indeed, there is enrichment of these TFs at the enhancer in both K562 (Fig. 3Ci) and HPC-7 (Fig. 3Cii) cells. Moreover, Runx1, in conjunction with the core binding factor β (CBFβ), was able to transactivate the mouse Smad6-57 (mSmad6-57) enhancer in vitro (although some activity was noted on the control pGL2p plasmid as well), and this activity was significantly enhanced by cotransfecting Gata2, which had no activity of its own (Fig. 3D). We have shown recently that although Runx1 and Gata2 heterozygotes are not viable and show a failure in fetal liver blood stem/progenitor cell expansion, emphasizing the in vivo cooperativity of these two TFs (40). Taken together, these data show that Runx1 activity at the Smad6-57 enhancer is facilitated by the concomitant activity of Gata2.
Fig. 3.
Smad6-57 region has conserved Ets, Gata, and E-box motifs, is active in hematopoietic cells, and can be transactivated by Runx1 in the absence of a Runx1 binding motif. (A) Nucleotide sequence alignment of the kb −57 mSmad6 (homologous to the kb −71 hSMAD6) region. Conserved Ets (E), Gata (G), and E-box (EB) motifs are colored in blue, red, and pink, respectively. (B) A 500-bp fragment from the kb −57 mSmad6 region (−57) is active in K562 cells. Stable transfection assays in K562 cells with wild-type and E-box/Gata-mutated (mE/G) fragments subcloned into the pGL2p vector. The luciferase activities are given as the fold increase above the activity of the pGL2p vector. Each bar is the means (± standard deviations) of the relative luciferase activity from at least two experiments performed in triplicate. (C) ChIP assays in human and mouse cell lines. (i) The hSMAD6 −71 enhancer is bound by SCL, FLI1, and GATA2 in K562 erythroleukemic cells. (ii) The mSmad6-57 enhancer also is bound by Scl, Fli1, and Gata2 in HPC-7 stem/progenitor cells. (D) The mSmad6-57 enhancer (Smad6-57/pGL2p-luc) can be transactivated by Runx1, but this effect is greatly enhanced by Gata2. The pGL2p-luc vector was used as a control. ***, P < 0.001.
The Smad6-57 enhancer targets embryonic hematopoietic tissues in conjunction with the endogenous promoter.
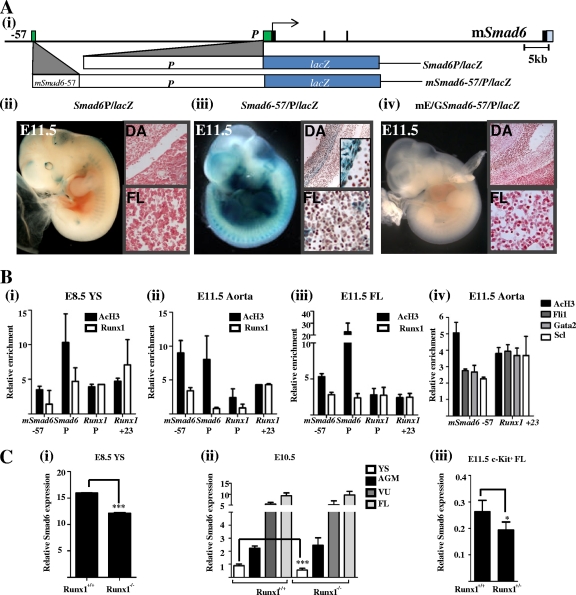
Transgenic assays are the gold standard for assessing the tissue activity and specificity of transcriptional regulatory elements in vivo. In light of this and the cell type-dependent variation in Runx1 binding to the Smad6 promoter, founder transgenic mouse embryos were generated using a construct with the −2006/+45 promoter region cloned upstream of a promoterless lacZ reporter gene (Smad6P/lacZ) (Fig. 4Ai). Reporter expression was analyzed by whole-mount X-Gal staining of E11.5 embryos. Five transgenic embryos were generated and showed faint staining that appeared to be localized to skeletal primordia and brain, which are known to express Smad6 (Fig. 4Aii) (12). However, there was no expression in blood or endothelium, indicating that despite in vitro activity, the Smad6 promoter requires additional regulatory elements to boost in vivo expression in these tissues. In contrast, in vivo transgenics generated using a construct with the mSmad6-57 enhancer subcloned upstream of the −2006/+45 mSmad6 promoter (mSmad6-57/P/lacZ) (Fig. 4Ai) showed staining in the dorsal aorta (DA) and fetal liver (FL) in six out of nine E11.5 F0 transgenic embryos (Fig. 4Aiii). The expression of the lacZ reporter along the ventral aspect of the E11.5 AGM closely mirrors that of endogenous Smad6 and Runx1 expression (21, 28). These data show that the mSmad6-57 enhancer boosts promoter activity in hematopoietic tissues in vivo. This activity was lost in 9/9 transgenic F0 embryos when the E-box/Gata sequence in the Smad6-57 enhancer was replaced with a NotI restriction enzyme digestion site (mE/GSmad6-57/P/lacZ) (Fig. 4Aiv). The human SMAD6-57 enhancer (homologous to mSmad6-47), which targets hematopoietic tissues in conjunction with the simian virus 40 (SV40) minimal promoter (28), failed to enhance the endogenous Smad6 promoter in vivo (data not shown). The functional relationship between these two enhancers is unclear, except that in contrast with mSmad6-57, which shows the robust binding of hematopoietic transcription factors in HPC-7 stem/progenitor cells, the mSmad6-47 region shows little or no enrichment (see Fig. S2 in the supplemental material).
Fig. 4.
(A) Analysis of Smad6 regulatory elements using F0 transgenics. (i) A schematic diagram of the mouse Smad6 locus is shown with the exons and promoter/enhancer regions drawn to scale and represented as black and green rectangles, respectively. The 3′ UTR is shown in cyan. Shown directly below are promoter and promoter/enhancer lacZ reporter constructs used to generate F0 transgenic embryos. (ii) The Smad6 promoter alone does not target reporter expression to blood in vivo. Shown are a representative X-Gal-stained whole-mount embryo and corresponding tissue sections from an E11.5 F0 Smad6P/lacZ embryo showing staining in primordial skeletal tissues and in brain. There is no staining in the DA or FL. (iii) The in vivo activity of the promoter is significantly enhanced by the mSmad6-57 fragment. Shown are a representative X-Gal-stained whole-mount embryo and corresponding tissue sections from an E11.5 F0 Smad6-57/P/lacZ embryo showing staining along the ventral surface of the DA and in FL cells. (iv) A representative X-Gal-stained whole-mount embryo and corresponding tissue sections from an E11.5 F0 mE/GSmad6-57/P/lacZ embryo. The E-box/Gata motif in the Smad6-57 enhancer was replaced with a NotI restriction enzyme digestion site. There was very faint staining in 9/9 transgenic embryos. DA, dorsal aorta; FL, fetal liver. (B) Runx1 binds the Smad6 and Runx1 promoter/enhancer regions in embryonic hematopoietic tissues. (i to iii) Histone H3 acetylation and Runx1 binding at the Smad6 and Runx1 promoter and enhancer regions assessed by ChIP assays in primary tissues. (i) In E8.5 YSs, the Smad6 and Runx1 promoter and enhancer regions show active chromatin marks (AcH3) and Runx1 binding at the Smad6 promoter and at both promoter and enhancer regions of Runx1 but not at the Smad6-57 enhancer. (ii) In E11.5 DAs, the Smad6 and Runx1 promoters show active chromatin marks but little Runx1 binding. However, there is a 3- to 4-fold enrichment of Runx1 at the enhancers of both genes relative to levels for a control region. (iii) In E11.5 FLs, the promoters and enhancers of both Runx1 and Smad6 show the acetylation of H3 and Runx1 binding. AGM, aorta-gonad-mesonephros; DA, dorsal aorta; FL, fetal liver; YS, yolk sac. (iv) The mSmad6-57 and mRunx1+23 enhancers are bound by Fli1, Gata2, and Scl in the E11.5 DAs. (C) Smad6 gene expression is reduced in the absence of Runx1. (i) Smad6 gene expression, in pooled E8.5 YSs dissected from Runx1−/− embryos, was significantly reduced compared to expression in wild-type embryos from the same litter. (ii) Smad6 gene expression in E11.5 Runx1−/− embryos was significantly reduced in YSs but was not apparent in whole-organ RT-PCR of other hematopoietic tissues. (iii) Smad6 gene expression was reduced in c-Kit+ FL blood progenitors FACS sorted from Runx1+/− E11.5 FLs compared to that of cells sorted from wild-type embryos from the same litter. *, P < 0.05; ***, P < 0.001.
The Smad6 promoter and Smad6−57 enhancer are active and bound by Runx1 in vivo.
In light of the tissue-specific activity of the mSmad6-57 enhancer, we performed ChIP assays in E8.5 YSs and E11.5 DAs and FLs to assess in vivo accessibility and Runx1 binding at the Smad6 regulatory elements. The promoters of both Smad6 and Runx1 have active chromatin marks in all primary hematopoietic tissues (Fig. 4Bi to iii). The hematoendothelial enhancers of both Runx1 (Runx1+23) and Smad6 (Smad6-57) also have active chromatin marks. Runx1 is bound to the hematopoietic enhancers or promoters of both Smad6 and Runx1 in all three hematopoietic tissues. Given the cellular heterogeneity in primary tissues, the levels of transcription factor enrichment at functional stem/progenitor enhancers are significantly lower than in clonal stem cell lines, such as HPC-7 (see Fig. S1 in the supplemental material). Bearing this in mind, Runx1 binding to the mSmad6-57 enhancer is most prominent in the E11.5 DA and FL, where the Fli1-Gata2-Scl triad also is active. Indeed, the components of the triad also are bound to the mSmad6-57 enhancer (Fig. 4Biv). These data are consistent with Runx1 binding the hematopoietic regulatory elements of Smad6 in embryonic tissues in combination with Fli1, Gata2, and Scl.
Hematopoietic expression of Smad6 in the developing embryo is regulated by Runx1.
Multipotent blood progenitors originate in the YS from E8.5 embryos, and stem cells originate from the AGM region from E10.5 onwards with subsequent amplification in the FL (11). To examine whether Smad6 expression in hematopoietic tissues is compromised in the absence of Runx1, we dissected YSs from E8.5 Runx1−/− embryos and their wild-type litter mates, quantified Smad6 transcripts by RT-PCR, and noted a significant reduction in Smad6 expression at this developmental time point (Fig. 4Ci). Smad6 expression also was quantified in YS, AGM, vitello-uterine (VU) vessels, and FLs from E11.5 Runx1−/− embryos and their wild-type litter mates; no difference in expression was noted, except in YS (Fig. 4Cii). It is important to note, however, that Smad6 expression is upregulated by the signaling Smad, p-Smad1, in response to Bmp stimulation. There is strong expression of Bmp4 along the ventral surface of the dorsal aorta in the AGM/VU region that could obviate any reduction caused by the absence of Runx1. Furthermore, whole-organ RT-PCR does not account for differences in cell subsets, especially when the cells that are being assayed are a minority in the tissue. To assess whether there is a Runx1 dose-dependent reduction in Smad6 expression in c-Kit+ blood stem/progenitor cells, as opposed to whole FL, single-cell suspensions from FLs harvested from Runx1+/− and wild-type littermates were sorted by flow-cytometry for RT-PCR. There was a significant reduction in Smad6 expression in c-Kit+ FL cells that lacked one copy of Runx1 (Fig. 4Ciii). These data show that although Smad6 expression in the developing embryo is not wholly dependent on Runx1 signaling and is largely unchanged in whole tissues, subtle variations are seen in embryonic blood stem/progenitor cells.
Smad6 and Runx1 levels in hematopoietic cells are maintained in a dynamic equilibrium by a Smad6-Runx1 rheostat.
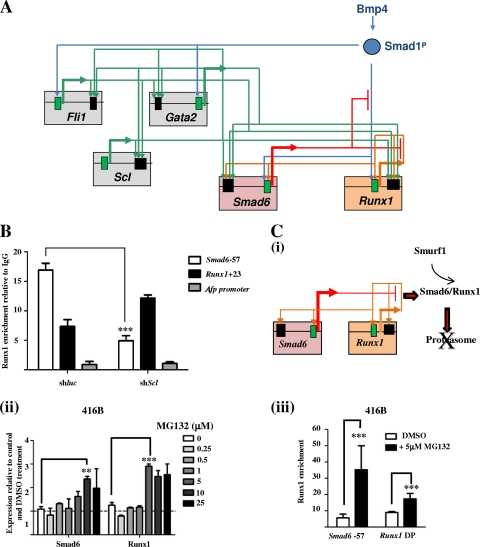
Bmp4 is required for mesoderm specification and the development of blood cells from the mesoderm (18). Smad6 and Runx1 expression in the AGM also is linked to the Bmp signaling pathway (28). Based on our current understanding, it is possible to construct a Smad6-Runx1 transcriptional circuit that integrates both the Bmp4 signaling cascade and the Fli1-Gata2-Scl triad linked via tissue-specific enhancers (Fig. 5A) (29). The activity of the Smad6-57 enhancer, which lacks a Runx binding motif, is dependent on an E-box/Gata motif. The Runx1+23 HSC enhancer, on the other hand, has both a Runx binding and an E-box motif. In HPC-7 cells there is overlapping binding of Runx1 and Scl at both enhancers (see Fig. S2 in the supplemental material, HPC-7 Runx1 and Scl ChIP-seq tracks). If Scl facilitates Runx1 binding to the Smad6-57 enhancer (via its E-box), then one can predict that Runx1 binding falls in Scl-depleted cells. Indeed, in contrast with the control (shluc), Runx1 enrichment at the Smad6-57 enhancer is significantly reduced relative to enrichment at the Runx1+23 enhancer in Scl-depleted HPC-7 cells (Fig. 5B).
Fig. 5.
(A) Schematic diagrams showing Runx1-Smad6 interactions. Smad6 and Runx1 are targets of Bmp4 signaling and are independently regulated by the Fli1-Gata2-Scl triad. Components of the triad stabilize the Smad6-Runx1 rheostat by facilitating Runx1 binding to the Smad6-57 enhancer. (B) Runx1 ChIP enrichments in HPC-7 cells transduced with either an Scl knockdown (shScl) or control (shluc) vector. Runx1 enrichment ratios at the Smad6-57 and Runx1+23 enhancers are reversed following Scl knockdown and are unchanged at the control region (Afp promoter). The ratios of Runx1 binding at the Smad6-57, Runx1+23, and Afp regulatory regions in control (shluc) HPC-7 cells are 18, 8, and 1, respectively. The ratios of Runx1 binding at these regions in shScl HPC-7 cells (∼50% Scl mRNA) are 4.5, 11, and 1, respectively. (C) Perturbation of the Runx1-Smad6 rheostat by proteasome inhibition. (i) A schematic showing Smurf1-mediated targeting of the Smad6-Runx1 complex to the proteasome. (ii) MG132 interrupts the degradation of Runx1 by inhibiting the proteasome, which leads to the increased expression of both Smad6 and Runx1. (iii) The increase in Smad6 and Runx1 gene expression is associated with increased Runx1 binding at both the Smad6 and Runx1 gene promoters. **, P < 0.01; ***, P < 0.001.
If Runx1 and Smad6 exist in a dynamic equilibrium in hematopoietic cells, with Runx1 providing both positive (by acting on its own promoter and enhancer) and negative (by upregulating its degrader, Smad6) inputs to maintain its level, it should be possible to perturb the circuit and evaluate changes in gene expression and measure functional outcomes (Fig. 5Ci). To this end, we incubated 416B blood progenitors with increasing concentrations of MG132, a potent proteasome inhibitor, to determine whether inhibiting Runx1 degradation would read out as an increase in Smad6 and Runx1 expression. As shown in Fig. 5Cii, at a concentration of 5 to 10 μM MG132 there was an increase in Smad6 and Runx1 expression. This coincided with the increased binding of Runx1 at the Smad6-57 enhancer and Runx1 promoter by ChIP (Fig. 5Ciii). Taken together, these data are consistent with proteasome inhibition increasing Runx1 and Smad6 levels by increased Runx1 binding at specific regulatory elements of these genes.
DISCUSSION
Runx1 regulates the emergence of definitive hematopoietic cells in the embryo, and its activity is under tight spatiotemporal control. Smad6, an inhibitor of Bmp4 signaling, binds and modulates Runx1 activity by facilitating its degradation by the proteasome. Here, we show that Runx1 in turn regulates Smad6 transcription in vivo via a novel hematopoietic enhancer. This enhancer, however, lacks Runx1 binding sites, and the activity of the Runx1-Smad6 rheostat is facilitated by the concurrent activity of the Fli1-Gata2-Scl transcriptional triad that operates in the AGM. Interrupting the Runx1-Smad6 rheostat by inhibiting the catalytic activity of the 26S proteasome results in an increase in Runx1 levels and increased binding of Runx1 to Smad6 and Runx1 regulatory elements. Taken together, we describe a mechanism that could account for the fine-tuning of Runx1 concentrations in hematopoietic tissues during embryonic development.
The Runx1-Smad6 rheostat operates within the context of a Bmp4 signaling pathway and a wider HSC transcriptional network (28, 30). Bmp4 is required for the specification of the mesoderm and subsequent blood production from cells derived from the mesoderm. pSmad1 is a mediator of Bmp4 signaling and is upstream of the Fli1-Gata2-Scl transcriptional triad that operates in the AGM (28). pSmad1 and components of the triad drive transcription of both Runx1 and Smad6. The HSC transcriptional network therefore is geared to drive the transcription of Runx1 and its inhibitor, Smad6, independently of each other. The existence of a rheostat links these two otherwise-independent events in cells that have the concomitant expression of other key hematopoietic TFs, such as Fli1, Gata2, and Scl. The Fli1-Gata2-Scl triad functions as a fully connected triad, a network motif that in theory helps maintain the expression of each component TF without an extraneous input following its initial activation (20, 30). Such motifs are ideal for circulating stem cells, such as HSCs, that are generated in the AGM but circulate to other sites, such as the FL, placenta, and BM, retaining their “stemness” without recourse to morphogens such as Bmp4 and Notch, which are thought to activate the triad in the AGM. This also may apply to Runx1 and Smad6 expression in HSCs, in that they become more reliant on the Fli1-Gata2-Scl triad and less so on the Bmp4-pSmad1 pathway, when cells lose their attachment to the DA. It is significant in this regard that the signaling Smads, Smad1 and Smad5, no longer are required for HSC maintenance after these cells have been specified (34). Runx1 levels in circulating HSCs likely are maintained by the activity of Fli1-Gata2-Scl on Runx1 regulatory elements such as the Runx1+23 enhancer and promoter (3). The role of the Runx1-Smad6 rheostat would be to stabilize Runx1 levels within a set range during hematopoiesis.
An intriguing feature of the Runx1-Smad6 rheostat is the lack of a recognized Runx1 binding motif in an enhancer that is robustly bound by Runx1 in hematopoietic cells. This phenomenon is increasingly recognized due to the availability of genome-wide transcription factor binding profiles and bioinformatic tools to analyze target DNA sequences for TF binding motifs. The combinatorial binding of TFs that physically interact with each other is one demonstrated mechanism by which factors bind enhancers that lack motifs for a specific factor. Indeed, only 39% of DNA sequences corresponding to heptad binding (Runx1, Fli1, Gata2, Scl, Lyl1, Lmo2, and Erg) contained a Runx consensus motif (40). The physical interaction between RUNX1 and SCL/TAL1 recently has been shown to be critically required for TAL1 binding to genes that modulate T-cell differentiation (25). In hematopoietic stem/progenitor cells, Runx1 binding to the Smad6 enhancer overlaps with Scl (see Fig. S1 in the supplemental material), which facilitates its binding to the enhancer (Fig. 5B).
A fundamental principle of developmental biology is that the spatial patterning of gene expression is directly determined by a heritable cis-regulatory DNA sequence code (7). These sequence codes more or less function as information-processing units that help determine whether a gene is expressed (or not), and if it is expressed, the level at which it is expressed. At a transcriptional level, these regulatory modules anchor protein-DNA interactions within gene regulatory networks. Biological complexity does not scale with gene number but rather the intricacy of gene regulation, and the information-processing units that regulate gene expression in complex metazoans by and large reside not in gene promoters but in distal regulatory modules that can be recruited in a tissue- and cell-specific manner (29). This is evident in the Runx1-Smad6 rheostat, where the key functional interactions occur not at gene promoters (neither the Smad6 nor the Runx1 promoters are sufficient to target reporter expression to embryonic hematopoietic tissues in transgenic assays) but at distal enhancers. In other words, this control mechanism is wired to operate not at random but in cells where the Runx1 dose is critical for cell fate decisions.
Targeted mechanisms to control leukemogenic transcription factor activity are of considerable scientific and clinical interest. Runx1 is one of the most frequently mutated genes in acute myeloid leukemia, representing the first hit that expands blood progenitors, creating a reservoir for further genetic alterations to occur (15). The knowledge that Runx1 facilitates its own degradation by promoting the Runx1-Smad6-Smurf1-proteasome axis could be harnessed as a therapeutic strategy if Runx1 fusion proteins can be shuttled to the proteasome by Smad6 as efficiently as wild-type Runx1. Nonspecific proteasome inhibitors, on the other hand, are licensed for clinical use to treat hematological malignancies such as multiple myelomas (32). Although it would be difficult to tease out effects on individual proteins, proteasome inhibition could increase Runx1 concentration with potentially deleterious effects on Runx1-dependent malignant processes.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Health and Medical Research Council of Australia, The Australian Research Council, The Leukemia Foundation, The Viertel Foundation, The Ramaciotti Foundation, Cancer Institute of New South Wales, The Medical Research Council, United Kingdom, Leukemia & Lymphoma Research, the Wellcome Trust, and the Israel Science Foundation.
We thank Qing Wang and Di Chen for wild-type and mutated Smad6 promoter luciferase constructs.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Alon U. 2007. An introduction to systems biology: design principles of biological circuits. Chapman & Hall/CRC, Boca Raton, FL [Google Scholar]
- 2. Alon U. 2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8:450–461 [DOI] [PubMed] [Google Scholar]
- 3. Bee T., et al. 2009. The mouse Runx1+23 hematopoietic stem cell enhancer confers hematopoietic specificity to both Runx1 promoters. Blood 113:5121–5124 [DOI] [PubMed] [Google Scholar]
- 4. Ben-Ami O., Pencovich N., Lotem J., Levanon D., Groner Y. 2009. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc. Natl. Acad. Sci. U. S. A. 106:238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cai Z., et al. 2000. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity 13:423–431 [DOI] [PubMed] [Google Scholar]
- 6. Chen M. J., Yokomizo T., Zeigler B. M., Dzierzak E., Speck N. A. 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457:887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davidson E. H. 2006. The regulatory genome: gene regulatory networks in development and evolution. Elsevier/Academic Press, Amsterdam, The Netherlands [Google Scholar]
- 8. Davidson E. H., et al. 2002. A genomic regulatory network for development. Science 295:1669–1678 [DOI] [PubMed] [Google Scholar]
- 9. Deplancke B., et al. 2006. A gene-centered C. elegans protein-DNA interaction network. Cell 125:1193–1205 [DOI] [PubMed] [Google Scholar]
- 10. Donaldson I. J., Gottgens B. 2007. CoMoDis: composite motif discovery in mammalian genomes. Nucleic Acids Res. 35:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dzierzak E., Speck N. A. 2008. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 9:129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Galvin K. M., et al. 2000. A role for smad6 in development and homeostasis of the cardiovascular system. Nat. Genet. 24:171–174 [DOI] [PubMed] [Google Scholar]
- 13. Göttgens B., et al. 2002. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21:3039–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ishida W., et al. 2000. Smad6 is a Smad1/5-induced Smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J. Biol. Chem. 275:6075–6079 [DOI] [PubMed] [Google Scholar]
- 15. Jacob B., et al. 2010. Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood 115:1610–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landry J. R., et al. 2008. Runx genes are direct targets of Scl/Tal1 in the yolk sac and fetal liver. Blood 111:3005–3014 [DOI] [PubMed] [Google Scholar]
- 17. Levanon D., Groner Y. 2004. Structure and regulated expression of mammalian RUNX genes. Oncogene 23:4211–4219 [DOI] [PubMed] [Google Scholar]
- 18. Maeno M., et al. 1996. The role of BMP-4 and GATA-2 in the induction and differentiation of hematopoietic mesoderm in Xenopus laevis. Blood 88:1965–1972 [PubMed] [Google Scholar]
- 19. Marson A., et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134:521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Narula J., Smith A. M., Gottgens B., Igoshin O. A. 2010. Modeling reveals bistability and low-pass filtering in the network module determining blood stem cell fate. PLoS Comput. Biol. 6:e1000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. North T., et al. 1999. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126:2563–2575 [DOI] [PubMed] [Google Scholar]
- 22. North T. E., et al. 2002. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16:661–672 [DOI] [PubMed] [Google Scholar]
- 23. Nottingham W. T., et al. 2007. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 110:4188–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okuda T., van Deursen J., Hiebert S. W., Grosveld G., Downing J. R. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321–330 [DOI] [PubMed] [Google Scholar]
- 25. Palii C. G., et al. 2011. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J. 30:494–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pimanda J. E., et al. 2006. Endoglin expression in the endothelium is regulated by Fli-1, Erg, and Elf-1 acting on the promoter and a −8-kb enhancer. Blood 107:4737–4745 [DOI] [PubMed] [Google Scholar]
- 27. Pimanda J. E., et al. 2008. Endoglin expression in blood and endothelium is differentially regulated by modular assembly of the Ets/Gata hemangioblast code. Blood 112:4512–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pimanda J. E., et al. 2007. The SCL transcriptional network and BMP signaling pathway interact to regulate RUNX1 activity. Proc. Natl. Acad. Sci. U. S. A. 104:840–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pimanda J. E., Gottgens B. 2010. Gene regulatory networks governing haematopoietic stem cell development and identity. International J. Developmental Biology. 54:1201–1211 [DOI] [PubMed] [Google Scholar]
- 30. Pimanda J. E., et al. 2007. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc. Natl. Acad. Sci. U. S. A. 104:17692–17697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pinto do O P., Kolterud A., Carlsson L. 1998. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 17:5744–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rajkumar S. V., Richardson P. G., Hideshima T., Anderson K. C. 2005. Proteasome inhibition as a novel therapeutic target in human cancer. J. Clin. Oncol. 23:630–639 [DOI] [PubMed] [Google Scholar]
- 33. Shen R., et al. 2006. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J. Biol. Chem. 281:3569–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singbrant S., et al. 2010. Canonical BMP signaling is dispensable for hematopoietic stem cell function in both adult and fetal liver hematopoiesis, but essential to preserve colon architecture. Blood 115:4689–4698 [DOI] [PubMed] [Google Scholar]
- 35. Speck N. A., Gilliland D. G. 2002. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer 2:502–513 [DOI] [PubMed] [Google Scholar]
- 36. Su A. I., et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 101:6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Q., et al. 1996. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 87:697–708 [DOI] [PubMed] [Google Scholar]
- 38. Wang Q., et al. 2007. Bone morphogenetic protein 2 activates Smad6 gene transcription through bone-specific transcription factor Runx2. J. Biol. Chem. 282:10742–10748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Z., et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40:897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilson N. K., et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7:532–544 [DOI] [PubMed] [Google Scholar]
- 41. Zhang Y., et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.