Abstract
Gamma interferon (IFN-γ) is an inflammatory cytokine that has complex effects on myogenesis. Here, we show that the IFN-γ-induced inhibition of myogenesis is mediated by the major histocompatibility complex (MHC) class II transactivator, CIITA, which binds to myogenin and inhibits its activity. In IFN-γ-treated myoblasts, the inhibition of muscle-specific genes includes the expression of myogenin itself, while in myotubes, myogenin expression is unaffected. Thus, CIITA appears to act by both repressing the expression and inhibiting the activity of myogenin at different stages of myogenesis. Stimulation by IFN-γ in skeletal muscle cells induces CIITA expression as well as MHC class II gene expression. The IFN-γ-mediated repression is reversible, with myogenesis proceeding normally upon removal of IFN-γ. Through overexpression studies, we confirm that the expression of CIITA, independent of IFN-γ, is sufficient to inhibit myogenesis. Through knockdown studies, we also demonstrate that CIITA is necessary for the IFN-γ-mediated inhibition of myogenesis. Finally, we show that CIITA, which lacks DNA binding activity, is recruited to muscle-specific promoters coincident with reductions in RNA polymerase II recruitment. Thus, this work reveals how IFN-γ modulates myogenesis and demonstrates a key role for CIITA in this process.
INTRODUCTION
Gamma interferon (IFN-γ) is an inflammatory cytokine that was first identified as an antiviral factor. IFN-γ is a pleiotropic cytokine that regulates different immune responses and influences many physiological processes. Many studies have also shown that IFN-γ influences skeletal muscle homeostasis and repair. Transient administration of exogenous IFN-γ has been shown to improve healing of skeletal muscle and limit fibrosis (14). Endogenous IFN-γ is required for efficient muscle regeneration, as mice lacking IFN-γ show impaired muscle regeneration following cardiotoxin-induced damage (6). Expression of IFN-γ is robust in proliferating C2C12 cells, but expression is diminished in differentiated C2C12 cells (6). Exogenous IFN-γ influences the proliferation and differentiation of cultured myoblasts and appears to have a direct role on gene expression (25, 26, 28, 48).
Myoblasts have been shown to express immunological properties such as the complement component of both the classical and alternative pathways and major histocompatibility complex (MHC) genes. Exogenous IFN-γ treatment has been shown to increase the expression of MHC class II genes, complement C components, intracellular adhesion molecule (Icam1), chemokine (C-C motif) ligand 5 (Ccl5; RANTES), chemokine (C-C motif) ligand 2 (Ccl2), and chemokine (C-X-C motif) ligand 10 (Cxcl10; Ip10) (15, 25, 28, 48). It is not currently known how IFN-γ mediates these transcriptional effects in myoblasts.
The positive role for IFN-γ established in muscle healing and repair suggests that this cytokine plays an important role in muscle biology. However, IFN-γ signaling is likely to be tightly regulated, as negative effects of IFN-γ have been observed as well. When IFN-γ is overexpressed at the neuromuscular junction in transgenic mice, the mice demonstrate an age-dependent necrotizing myopathy (51). When cultured myoblasts were stimulated with exogenous IFN-γ, the proliferation of myoblasts and the fusion into myotubes were inhibited (21, 57). In these studies, decreases in creatine kinase, actin, and myosin expression were observed with IFN-γ stimulation. These effects could be observed at relatively low concentrations of IFN-γ; however, even at extremely high doses, IFN-γ was not toxic to myoblasts.
IFN-γ signals through the JAK-STAT pathway. When IFN-γ binds to its receptor, the receptor-associated protein tyrosine kinases Janus kinase I (JAK1) and JAK2 are activated (37). This leads to the phosphorylation of STAT1, which then dimerizes, translocates to the nucleus, and activates its target promoters, including the pIV promoter of Ciita (31). The JAK1-STAT1 pathway has been shown to play important roles in myogenesis (55). JAK1 and STAT1 are required for myoblast proliferation and also have a potent antidifferentiation effect. Intriguingly, the antidifferentiation effect is specific to STAT1 and is not mediated by STAT2, -3, -5A, or -5B (55).
The class II transactivator, CIITA, is required for both constitutive and IFN-γ-inducible expression of MHC class II genes. CIITA lacks DNA binding activity but is recruited to proximal promoters by interactions with sequence-specific DNA binding factors (9, 29, 50, 63). CIITA has been shown to interact with a variety of transcription factors and coactivators, including the histone acetyltransferase, the CREB binding protein (CBP), and the Swi/Snf complex (13, 23, 32, 53). CIITA itself houses acetyltransferase activity that is required for CIITA-mediated transactivation (46). CIITA is encoded by one gene that contains four separate promoters that generate four isoforms (33). CIITA expression is stimulated by IFN-γ, primarily through two of the four promoters, promoters III and IV (41, 42).
CIITA is also critical for IFN-γ-induced repression. IFN-γ suppresses a large family of genes that includes genes required for cell proliferation and cell differentiation, such as those for cyclin D1, c-myc, and n-myc; certain cytokine genes expressed by the TH2 subpopulation, such as IL-4 and IL-10; and genes coding for matrix proteins, such as collagen (Col1a1) and proteoglycan (Col1a1). IL-4, IL-10, and the cathepsin E gene (Ctse) have all been shown to be targets for IFN-γ-mediated CIITA repression. CIITA is a potent repressor of the Col1a1 promoter, and conversely, CBP, a histone acetyltransferase, is an activator of the Col1a1 promoter. Overexpression of CBP in the presence of CIITA allowed reactivation of a Col1a1 reporter, indicating that CIITA represses the Col1a1 promoter by sequestering CBP (64). The CIITA-mediated repression of matrix metalloproteinase 9 (MMP-9) is also mediated by the sequestration of CBP by CIITA (36). Another group has shown that CIITA negatively regulates Ctse by inhibiting a related histone acetyltransferase, p300, required for CtseE promoter activity (61).
CIITA is constitutively expressed in B cells but is expressed in response to IFN-γ in several cell types, including astrocytes, fibroblasts, and aortic smooth muscle cells (4, 10, 60). Surprisingly, when CIITA protein expression was examined in a system-wide approach by tissue immunohistochemistry, CIITA was detected in human skeletal muscle tissue as well (1).
Skeletal muscle differentiation is controlled by four highly related basic helix-loop-helix proteins referred to as the myogenic regulatory factors (MRFs). The MRFs have distinct but overlapping patterns of gene expression during muscle development (20). Gene knockouts of each factor in the mouse have revealed that each MRF has a unique role in skeletal muscle differentiation. Myf5, Myf6 (also known as MRF4), and MyoD are not required for viability, although each mutant has a distinct phenotype (reviewed in reference 39). In the combined absence of Myf5, Myf6, and MyoD, myoblasts are not specified and no skeletal muscle forms, resulting in death. Myogenin is the only MRF singly required for viability (19, 35). The Myog-null mice have myoblasts but very few muscle fibers. This suggests that myogenin is not required for the specification of skeletal muscle but is required for the later stages of myofiber fusion. In normal animals, Myog is downregulated shortly after birth and can be upregulated in response to muscle damage (17) or during aging (22, 34).
Here, we show that IFN-γ inhibits myogenesis through a direct inhibition of myogenin. The inhibition of myogenin is mediated by CIITA, whose expression is induced by IFN-γ signaling in myoblasts. CIITA inhibits myogenesis by two mechanisms. CIITA both represses Myog in myoblasts induced to differentiate and inhibits the activity of myogenin in myotubes. The inhibition of myogenin expression and activity leads to a downregulation of muscle-specific genes required for differentiation, thus halting differentiation. This effect is entirely reversible, with myogenesis proceeding normally once IFN-γ is removed. Thus, IFN-γ signaling allows a temporary halt to terminal differentiation by directly controlling the expression and activity of myogenin.
MATERIALS AND METHODS
Cell culture.
Proliferating C2C12 myoblasts (ATCC) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (HyClone) in a humidified CO2 incubator at 37°C according to standard protocols (44). To induce differentiation into myotubes, cells were grown to 70% confluence and the medium was switched to DMEM supplemented with 2% horse serum (HyClone). C2C12 cells were grown in differentiation medium for the number of days indicated in each experiment. 10T1/2 cells (ATCC) were grown in DMEM supplemented with 10% fetal bovine serum (HyClone).
Isolation of primary myoblasts.
Myoblasts were isolated according to standard protocols (45). The muscle from neonatal mouse limbs was removed and minced. The minced tissue was digested with collegenase/dispase and filtered to remove large pieces of tissue. The cells were resuspended in F-10-based primary myoblast medium and plated onto a collagen-coated culture dish and allowed to grow. Enriched populations of myoblasts were recovered by removing the cells without trypsin and preplating to further reduce fibroblast contamination. These steps were repeated until fibroblasts were no longer observed in the culture.
GST affinity pulldown.
The sequence encoding myogenin was PCR amplified from embryonic limb cDNA and cloned into pGEX6 (GE Healthcare Life Sciences). Glutathione S-transferase (GST) fusion proteins were expressed by transfecting BL21 cells with the GST fusion constructs under the control of the lac promoter. Cells were grown to an optical density at 600 nm (OD600) of 0.7, and recombinant protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 2 h. The cells were harvested and lysed, and the fusion proteins were bound to glutathione Sepharose 4B (GE Healthcare). The bound proteins were washed and eluted upon addition of reduced glutathione. The purified GST and GST-myogenin proteins were rebound to the affinity resin and incubated with 45 mg of nuclear extract isolated from differentiated C2C12 cells. Interacting proteins were eluted with increasing amounts of salt, and eluted fractions were run on SDS-PAGE gels and silver stained. Bands that appeared specifically in the GST-myogenin fractions were excised, trypsin digested, and analyzed by mass spectrometry (Peter Yao, University of Illinois at Urbana Champaign). Gel slices from the corresponding region of the GST-only samples were excised as well.
Coimmunoprecipitations.
HEK293 cells were transiently transfected with the plasmids expressing the MRFs and CIITA. EMSV-myogenin (provided by Diane Edmondson, The University of Texas Medical School at Houston) and pEMCIIs (provided by Andrew Lassar, Harvard Medical School) were used for expressing myogenin and MyoD, respectively. EMSV-Myf5 (Addgene plasmid 14711) and EMSV-Mrf4 (Addgene plasmid 14713) were provided by Michael Rudnicki (49) and used for expressing Myf5 and Myf6 (MRF4). The myc-CIITA plasmid (provided by Jeremy Boss, Emory University) was used for expressing CIITA with a Myc epitope on the N terminus. Following the transfection, whole-cell extracts were made in radioimmunoprecipitation assay (RIPA) buffer. Extract (300 μg) was used for each immunoprecipitation with 1 μg of antibody. The antibodies used included anti-CIITA (H-300 [Santa Cruz Biotechnology] and 7-1H [Sigma]), anti-Myf5 (C-20 [Santa Cruz Biotechnology]), anti-MyoD (5.8A [Santa Cruz Biotechnology]), anti-MyoG (F5D [Developmental Studies Hybridoma Bank]), and anti-Myf6 (C-19 [Santa Cruz Biotechnology]) antibodies. Following an overnight incubation, antibody-antigen complexes were collected with protein A-agarose beads (Invitrogen). The beads were washed with RIPA buffer and resuspended in protein loading dye. Immunoprecipitated samples with appropriate controls were loaded onto SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes for Western blot analysis. For each immunoprecipitation, the blot was probed with both the reciprocal factor, to test for the coimmunoprecipitation, and the antibody used for the immunoprecipitation, to confirm that the IP was successful. All immunoprecipitations were performed at least twice to confirm the results.
Western blot analysis.
Cell extracts were made by lysing phosphate-buffered saline (PBS)-washed cell pellets in RIPA buffer supplemented with protease inhibitors (Complete protease inhibitor; Roche Diagnostics). Following incubation on ice, clear lysates were obtained by centrifugation. Protein concentrations were determined by Bradford's assay (Bio-Rad). For each sample, 30 μg of protein was loaded on each gel. Proteins were transferred onto a PVDF membrane using a tank blotter (Bio-Rad). The membranes were then blocked using 5% milk and 1× TBST (Tris-buffered saline plus Tween 20) and incubated with primary antibody overnight at 4°C. Membranes were then washed with 1× TBST and incubated with the corresponding secondary antibody. Membranes were again washed with 1× TBST, incubated with chemiluminescent substrate according to the manufacturer's protocol (SuperSignal; Pierce), and visualized by autoradiography. The antibodies used included anti-CIITA (7-1H; Sigma), anti-myc (9E10; Roche), anti-Myf5 (C-20; Santa Cruz Biotechnology), anti-MyoD (5.8A; Santa Cruz Biotechnology), anti-MyoG (F5D; Developmental Studies Hybridoma Bank), and anti-Myf6 (C-19; Santa Cruz Biotechnology) antibodies.
Cell transfections and luciferase assays.
10T1/2 cells were transfected with calcium phosphate according to standard protocols. The plasmids EMSV-myogenin (gift of D. Edmondson, University of Texas [UT] Medical School at Houston) and pEMCIIs (provided by Andrew Lassar, Harvard Medical School) were used for expressing myogenin and MyoD, respectively. The myc-CIITA plasmid (provided by Jeremy Boss, Emory University) was used for expressing CIITA with a Myc epitope on the N terminus. For quantitative reverse transcription-PCR experiments, cells were seeded at a density of 5 × 104 cells per well in 6-well plates and transfected with 2 μg of plasmid DNA. Cells were maintained in growth medium for 1 day posttransfection. When the cells reached confluence, low-serum medium (differentiation medium) was placed on the cells for 24 h prior to harvesting RNA. Luciferase activity was determined using the dual-luciferase reporter assay system (Promega). NIH 3T3 cells were seeded at a density of 5 × 103 cells per well in 96-well plates and transfected with 0.2 μg of DNA. Transfections were normalized to Renilla luciferase. Transfections were performed in triplicates, and all data sets were repeated at least twice.
IFN-γ stimulation.
Cells were treated with murine IFN-γ (Peprotech). Except where noted, cells were stimulated with 50 units/ml IFN-γ (5 ng/ml). Cells were harvested for RNA or protein at defined time points after the IFN-γ stimulation. IFN-γ was added to the medium and replenished every time the medium was changed. For differentiating cells, the medium was changed every other day. At least three independent stimulations were assayed for each data point.
Immunohistochemistry.
Cells were grown on coverslips, fixed with paraformaldehyde, incubated with goat serum and 1.0% NP-40 for 1 h, and washed with PBS. Primary antibodies against myosin heavy chain (1:100, MF20, supernatant; Developmental Studies Hybridoma Bank) were incubated overnight at 4°C, washed with PBS, and detected by Alexa Fluor 488 goat anti-rabbit antibody (1:500; Invitrogen). Cell nuclei were then stained by incubating with DAPI (4′,6-diamidino-2-phenylindole; 1 uM; Invitrogen) for 5 min.
Quantitative reverse transcriptase PCR.
RNA was isolated from cells by Trizol extractions (Invitrogen). Following treatment with DNase (Promega), 2 μg of total RNA was reversed transcribed with MultiScribe MuLV reverse transcriptase (Applied Biosystems). The cDNA equivalent to 40 ng was used for quantitative PCR (qPCR) amplification (Applied Biosystems) with SYBR green PCR master mix (Applied Biosystems). Samples in which no reverse transcriptase was added (no RT) were included for each RNA sample. The relative levels of expression of genes were normalized according to that of the gene for hypoxanthine phosphoribosyltransferase (HPRT). qPCR data were calculated using the comparative threshold cycle (CT) method (Applied Biosystems). Standard deviations from the mean of the ΔCT values were calculated from three independent RNA samples. Primers to the coding region of Ciita (forward [F], 5′ TTCACCATTGAGCCATTTAAAGC 3′; reverse [R], 5′ CTGGGTCTGCACGAGACGAT 3′), H2Ea (F, 5′AAGTCATGGGCTATCAAAGAGGA 3′; R, 5′ CTCATCGCCGTCAAAGTCAAA 3′), Acta1 (F, 5′ GGCACCCAGGGCCAGAGTCA3′; R, 5′ TCATCCCCGGCAAAGCCAGC 3′), Mylpf (F, 5′ GGCTGCCGGGGCAGGACTAT 3′; R, 5′ CGGCCCATGGCTGCAAAGGT 3′), Lmod2 (F, 5′ ACCTTATCCCGATTTGCTGAAG; R, 5′ ACCTTGAGCATGTCTGCAAT 3′), Tnni2 (F, 5′ GCCGCCGAGAATCTGAGA 3′; R, 5′ GACATGGAGCCTGGGATGTG 3′), p21 (F, 5′ CCTGGTGATGTCCGACCTG 3′; R, 5′ CCATGAGCGCATCGCAATC 3′), MyoD (F, 5′ GCCGGTGTGCATTCCAA 3′; R, 5′ CACTCCGGAACCCCAACAG 3′), Myf5 (F, 5′ AGCTTGCAAGAGGAAGTCCACTA 3′; R, 5′ CTACGCTCGCGCATGGT 3′), Myog (F, 5′ GACCTGATGGAGCTGTATGAG 3′; R, 5′ CTGAAGGTGGACAGGAAGG 3′), Myf6 (F, 5′ CCCTGAAGCGTCGGACTGT; R, 5′ ATGGCACTCCGCAGAATCTC), and Hprt (F, 5′ TGACACTGGCAAAACAATGCA 3′; R, 5′ GGTCCTTTTCACCAGCAAGCT 3′) were used. Where possible, intron-spanning primers were used. All quantitative PCR was performed in triplicates, and three independent RNA samples were assayed for each time point.
Stable cell lines.
Stable C2C12 cell lines overexpressing exogenous CIITA were made by transfecting C2C12 cells with linearized myc-CIITA plasmid or the empty vector and linearized pcDNA3.1 and by selecting for Geneticin (400 μg/ml)-resistant colonies. Individual clones were isolated and propagated. Stable C2C12 lines expressing both exogenous CIITA and myogenin were constructed by transfecting the CIITA overexpression line with linearized EMSV-myogenin (gift of D. Edmondson, UT Medical School at Houston) and selecting for both puromycin (2 μg/ml)- and Geneticin (400 μg/ml)-resistant colonies. Individual clones were isolated and propagated.
Small hairpin RNA (shRNA) knockdown.
CIITA knockdown lines were constructed with shRNA constructs designed by the RNAi Consortium in the pLOK.1 plasmid (Open Biosystems). Five constructs targeting murine CIITA and one scrambled control were linearized, transfected into C2C12 cells, and selected with puromycin (2 μg/ml). Pooled clones were selected and propagated.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) assays were performed and quantified as described previously (59, 62) with the following modifications: 1 × 107 cells were used for each immunoprecipitation, and protein A-agarose beads (Invitrogen) were used to immunoprecipitate the antibody-antigen complexes. The following antibodies were used: anti-CIITA (7-1H; Sigma), anti-MyoD (5.8A; Santa Cruz Biotechnology), anti-myogenin (F5D; Developmental Studies Hybridoma Bank), and anti-myc (9E10; Roche) antibodies. Rabbit IgG (Santa Cruz Biotechnology) was used as a nonspecific control. Primers spanning the promoters of Tnni2 (F, 5′ GCCAAAGGAGCAAGAGTTAAAAAT 3′; R, 5′ AGGAGAAAGTGTTCCCAAAATGTC 3′), H2Ea (F, 5′ CTCGGATACTAAATAGGACCTGG 3′; R, 5′ TTCAGAAGCGATCGCAGAC 3′), and IgH (F, 5′ GCCGATCAGAACCAGAACACCTGC 3′; R, 5′ TGGTGGGGCTGGACAGAGTGTTTC 3′) were used to detect promoter enrichment. The real-time PCR was performed in triplicates. Values of ΔΔCT were calculated using the following formula based on the comparative CT method: ΔCT, template (antibody) − ΔCT, template (IgG) = ΔΔCT. Fold enrichments were determined using the following formula: 2 − ΔΔCT (experimental)/2 − ΔΔCT (reference, IgH). Standard error of the mean was calculated from replicate ΔΔCT values. The IgH locus was used to normalize the fold enrichments for the individual promoters. All ChIP assay results are representative of at least three individual experiments.
Statistics.
Data are presented as means ± standard errors (SE). Statistical comparisons were performed using paired two-tailed Student's t tests, with a probability value of <0.05 taken to indicate significance.
RESULTS
CIITA interacts with myogenin.
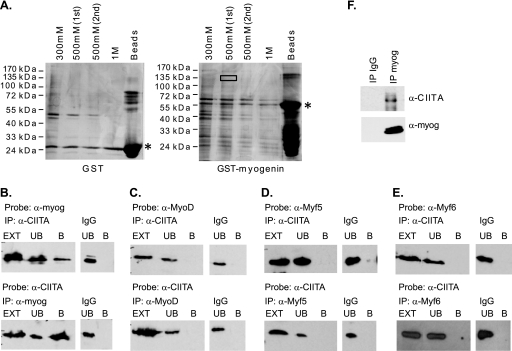
We identified the MHC class II transactivator, CIITA, as an interaction partner of myogenin through an affinity binding approach with GST-myogenin and nuclear extracts from differentiated C2C12 cells. Following elution from the column, proteins were resolved on SDS-PAGE gels and silver stained (Fig. 1A). Bands corresponding to proteins detected in elution fractions from the GST myogenin column and not detected in elution fractions from the GST column were excised and analyzed by mass spectroscopy. CIITA was identified as one of the potential interacting partners of myogenin from this analysis. The region of the gel that was excised for the identification of CIITA is boxed in Fig. 1A. The band was at approximately 135 kDa, consistent with the observed molecular mass of CIITA, 130 kDa. To confirm that the experimental approach could identify known interaction partners of myogenin, the eluted fractions were probed with antibodies against E proteins to detect the presence of known myogenin-interacting proteins. We probed for both E12/47 and HEB and detected both of these E proteins in the elution fractions (data not shown).
Fig. 1.
CIITA interacts with myogenin. (A) Isolation of myogenin-interacting proteins by a GST affinity approach. Nuclear differentiated C2C12 extract (45 mg) was incubated with the indicated constructs. Following incubation, the glutathione beads were washed and interacting proteins eluted with increasing amounts of salt. SDS-PAGE gels were silver stained. The mobility of the GST fusion proteins is marked with a black asterisk. The box indicates the region that was excised and trypsin digested for protein identification by liquid chromatography/mass spectrometry (LC/MS). (B) CIITA interacts with myogenin. HEK cells were transiently transfected with myc-CIITA and EMSV-myogenin and cell extracts immunoprecipitated with antibodies against CIITA or myogenin. (C) CIITA does not interact with MyoD. HEK cells were transiently transfected with myc-CIITA and EMSV-MyoD and cell extracts immunoprecipitated with antibodies against CIITA or MyoD. (D) CIITA does not interact with Myf5. HEK cells were transiently transfected with myc-CIITA and EMSV-Myf5 and cell extracts immunoprecipitated with antibodies against CIITA or Myf5. (E) HEK cells were transiently transfected with myc-CIITA and EMSV-Myf6, and cell extracts were immunoprecipitated with antibodies against CIITA or Myf6. (F) Endogenous interaction of CIITA with myogenin in myotubes. Extracts from C2C12 cells differentiated for 2 days were immunoprecipitated with antibodies against myogenin (F5D), and the blot was probed with antibodies against CIITA (7-1H). The blot was then stripped and reprobed with antibodies against myogenin. For each panel, the antibodies used for the immunoprecipitation and the probe are labeled above the lanes. The immunoprecipitated sample is labeled B (bound fraction) and the supernatant, or unbound fraction, is labeled UB. The lysate used for the immunoprecipitation is labeled EXT (extract). Reciprocal blots are shown. The blots were also probed with the antibody used for the immunoprecipitation to confirm that the target protein was immunoprecipitated in each case (data not shown).
We next sought to confirm the interaction between myogenin and CIITA with coimmunoprecipitation studies. Experiments with CIITA are often performed with exogenous CIITA expression due to the very low levels of endogenous CIITA (54). HEK293 cells were used for these experiments, as they allow for very high transfection efficiencies and levels of expressed proteins. These studies confirmed the binding of CIITA and myogenin and also demonstrated that the interaction could be detected reciprocally (Fig. 1B). Given the high homology of the MRF family, we next sought to determine if the interaction was specific to myogenin or common to the MRFs. We performed similar experiments for MyoD, Myf5, and Myf6 and found that MyoD, Myf5, and Myf6 do not interact with CIITA (Fig. 1C, D, and E). The interaction with CIITA is specific to myogenin. We then sought to confirm the interaction of CIITA and myogenin in differentiated C2C12 cells, as the interaction with CIITA was initially identified in a differentiated cell extract. The endogenous interaction of myogenin and CIITA was confirmed in extracts from differentiated C2C12 cells (Fig. 1F).
CIITA inhibits the activity of myogenin.
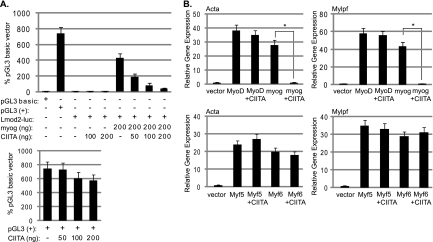
To determine how the interaction with CIITA affects the activity of myogenin, we tested for alterations in myogenin's activity in the presence of CIITA. We first tested the effect of CIITA on a muscle-specific luciferase construct. The construct chosen contained a minimal promoter element of the leiomodin 2 gene (Lmod2), a gene we have previously characterized as highly dependent on myogenin in vivo (7). As we have observed previously (7), transfection of myogenin activates this construct in NIH 3T3 cells (Fig. 2A). Cotransfection with CIITA acts as a potent inhibitor of myogenin-dependent transactivation. To confirm that the inhibition mediated by CIITA was specific to the myogenin-dependent reporter, we also tested the effect of CIITA on the pGL3(+) vector, which drives luciferase with the constitutive cytomegalovirus (CMV) promoter. We found that the transfection of CIITA had no significant effect on the pGL3(+) vector (Fig. 2A). Thus, the effect observed appears to be specific to the myogenin-driven activation of the muscle-specific reporter. We also assayed for the effects of CIITA on muscle-specific genes in an endogenous context. Transfection of the MRFs into the 10T1/2 cell line, a fibroblast cell line considered poised to enter the myogenic fate, activates muscle-specific genes (8). 10T1/2 cells were transfected with MyoD or myogenin in combination with CIITA, and the gene expression changes were determined for two muscle-specific genes that have been previously shown to respond to MyoD and myogenin in this system, those for actin (Acta1) and myosin light chain (Mylpf). Both MyoD and myogenin were tested to determine if the effect of CIITA was specific to myogenin, as would be predicted from the interaction studies. We found that CIITA acts as a potent inhibitor of myogenin-dependent gene activation, without affecting MyoD (Fig. 2B.). Similar experiments were repeated with Myf5 and Myf6, and again no CIITA-dependent inhibition of activity was observed (Fig. 2B).
Fig. 2.
CIITA inhibits myogenin. (A) CIITA inhibits a myogenin-dependent muscle-specific reporter construct. NIH 3T3 cells were transfected with the empty luciferase vector (pGL3 basic), a positive control [pGL3(+)], the muscle-specific reporter (Lmod2-luc), an expression construct for myogenin (EMSV-myogenin), and an expression construct for CIITA (myc-CIITA) as indicated. The data are expressed as percentages of the pGL3 basic vector expression levels. All plasmids were used at a concentration of 200 ng except where indicated. (B) CIITA inhibits endogenous gene activation by myogenin and does not inhibit MyoD, Myf5, or Myf6. 10T1/2 fibroblast cells were transiently transfected with empty vector (vector), MyoD, myogenin, Myf5, or Myf6 alone or with equivalent amounts of myc-CIITA. Gene expression was assayed with real-time PCR. Primers detecting skeletal alpha actin (Acta1) and myosin light chain, phosphorylatable, fast skeletal (Mylpf) were used for the analysis as indicated. For both panels A and B, error bars represent standard deviations of the mean. Asterisks indicate statistical significance. For Acta, the P value was 0.0033 for myogenin+CIITA compared to the sample with myogenin alone. For Mylpf, the P value was 0.0029.
CIITA is induced by IFN-γ in myoblasts.
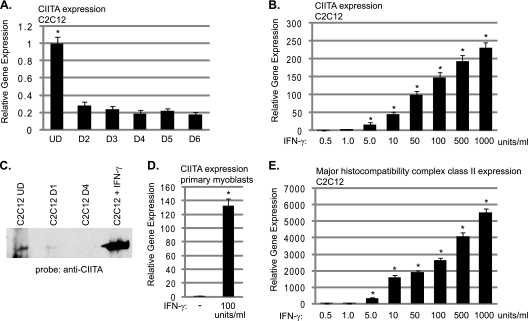
Prior to our work, the expression of CIITA in skeletal muscle was not known beyond its identification in the system-wide immunohistochemistry study mentioned above. To confirm the expression of Ciita in skeletal muscle cells, we assayed for RNA expression in proliferating and differentiated C2C12 cells. We found that Ciita expression is detectable in proliferating C2C12 cells and the level is modestly downregulated as cells begin to differentiate (Fig. 3A). We then stimulated proliferating C2C12 cells with IFN-γ and examined changes in the expression level of Ciita. As it has been shown that tumor necrosis factor alpha (TNF-α) is promyogenic at low concentrations (0.05 ng/ml) but antimyogenic at higher concentrations (5), we tested a wide range of IFN-γ concentrations. We found that the expression of Ciita was greatly stimulated at the RNA level by the addition of IFN-γ (Fig. 3B). We next analyzed protein expression of CIITA by Western blot analysis and found that the results mirrored our gene expression data. CIITA was weakly expressed in proliferating C2C12 cells; this level decreased after 1 day of differentiation, and CIITA was not detectable by Western blot analysis after 4 days of differentiation (Fig. 3C). Following IFN-γ stimulation, robust expression of CIITA was also observed at the protein level (Fig. 3C). To confirm our results, primary myoblasts were also isolated from neonatal animals, and the IFN-γ stimulation was repeated. We found that IFN-γ also robustly stimulates Ciita in primary myoblasts (Fig. 3D). We next examined the expression of a classical CIITA target gene, the major histocompatibility complex class II gene H2Ea. We observed that the expression of H2Ea was also highly stimulated by IFN-γ in C2C12 cells (Fig. 3E). The inductions of Ciita and H2Ea gene expression were detectable with 5 units of IFN-γ (0.5 ng/ml) and increased over the dosage curve.
Fig. 3.
CIITA is expressed in skeletal muscle and is stimulated by IFN-γ. (A) CIITA is expressed in C2C12 cells. Real-time PCR was used to assess Ciita transcript levels. Data are presented as fold changes with respect to the level observed in proliferating C2C12 cells (UD). C2C12 cells differentiated for 2 days are represented by D2, and the other lanes are labeled accordingly. Asterisks indicate statistical significance. The P value for the change between UD and D2 samples was 0.004. (B) IFN-γ stimulates Ciita expression. Seventy-percent-confluent C2C12 myoblasts were stimulated with the indicated amount of IFN-γ, harvested 24 h after stimulation, and assayed for Ciita expression by real-time PCR. All P values for stimulated samples compared to unstimulated controls were <0.01. (C) CIITA protein expression is stimulated by IFN-γ. Lanes are as indicated, and extract from proliferating C2C12 cells stimulated with 100 units IFN-γ for 24 h was labeled C2C12+IFN-γ. Western blot analysis with anti-CIITA antibodies (7-1H) is shown. (D) IFN-γ stimulates Ciita in primary myoblasts. Primary myoblasts were stimulated as described for panel B and assayed for expression of Ciita. The P value of the stimulated sample compared to the untreated control was 0.002. (E) IFN-γ stimulates MHC class II gene expression in C2C12 myoblasts. Cells were stimulated as described for panel B, and gene expression was analyzed for the MHC class II gene H2Ea. All P values for stimulated samples compared to unstimulated controls were <0.01. For each panel, primers to Hprt were used to normalize the samples. Data are shown as fold stimulations, with each sample relative to the untreated sample. Error bars represent standard deviations from the means for the replicate PCR values.
We also examined changes in several of the genes previously shown to be activated by IFN-γ, including Ccl2, Ccl5, and IP-10 (48). While we did observe activation of these targets, the fold changes were 2.1, 2.3, and 2.6, respectively (data not shown). While significant, these fold changes are much smaller than the effects we observe for Ciita and H2Ea.
IFN-γ inhibits myogenesis by repressing muscle-specific genes.
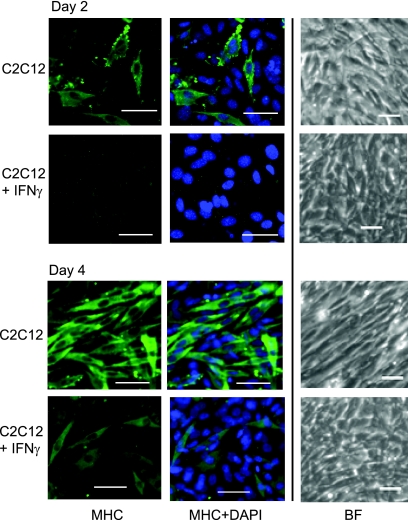
Next, we reasoned that IFN-γ should act as a repressor of myogenin-dependent gene expression and inhibit myogenesis. To test this hypothesis, C2C12 cells were treated with IFN-γ and examined for an inhibition of myogenesis and muscle-specific gene expression changes. Following IFN-γ stimulation, we observed that the cells appeared to halt differentiation prior to myoblast fusion. The addition of IFN-γ inhibited fusion and the expression of myosin heavy chain (Fig. 4). IFN-γ-stimulated cells appear normal prior to fusion, but they do not progress past this point. Cells were stimulated with IFN-γ and differentiated for up to 10 days with no further change in the morphology observed after 2 days of differentiation.
Fig. 4.
IFN-γ inhibits myogenesis. IFN-γ inhibits differentiation. Seventy-percent-confluent C2C12 myoblasts (MB) were stimulated with IFN-γ and allowed to differentiate in the presence of IFN-γ for 2 days (top) or 4 days (bottom). Cells were stained with antibodies against myosin heavy chain (MHC) and DAPI. Fluorescent images are at ×20 magnification. The scale shown is 50 μm. Bright-field (BF) images of a different field are shown at ×10 magnification.
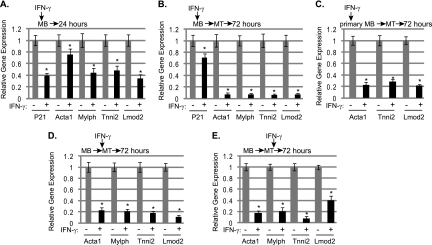
Next, we examined muscle-specific gene expression changes in C2C12 cells treated with IFN-γ. We chose to examine the genes assayed in the 10T1/2 system, those for actin (Acta1) and myosin light chain (Mylpf), but also analyzed changes at the genes for troponin 1 type 2 (Tnni2) and leiomodin 2 (Lmod2), as we have shown that these genes are highly dependent on myogenin in vivo (7). We anticipated observing gene expression changes in differentiating cells, as myogenin is expressed at this stage and each of these genes is highly upregulated in a differentiating cell. We also examined expression of the cdk inhibitor p21, which is upregulated immediately upon differentiation and regulated by MyoD (18, 40). Unlike the genes mentioned above, the expression of p21 proceeds or is coincident with the expression of myogenin. First, C2C12 cells were treated with IFN-γ while proliferating and harvested after 24 h. We observed small reductions in muscle-specific gene expression when IFN-γ was added before cells had begun to differentiate (Fig. 5A). p21 expression was also inhibited (Fig. 5A). Next, C2C12 cells were treated with IFN-γ while proliferating, and the stimulation was maintained as the cells differentiated for three additional days. We found that muscle-specific genes were highly downregulated in C2C12 myotubes differentiated in the presence of IFN-γ (Fig. 5B). These data were consistent with the morphological effects observed in Fig. 4. Unlike the differentiation-specific genes, p21 levels were not highly downregulated under these conditions (Fig. 5B). Thus, it appears that IFN-γ delays the induction of p21 but does not completely block p21 expression in a differentiating cell. The downregulation of muscle-specific genes was confirmed in primary myoblasts. IFN-γ was added to proliferating primary myoblasts, and the stimulation was maintained for 3 days of differentiation. We found that IFN-γ inhibited myogenesis by inhibiting muscle-specific gene expression in primary myoblasts as well (Fig. 5C). We next asked the question of whether IFN-γ could modulate myogenesis once differentiation has initiated. Two experimental conditions were tested. We first stimulated cells with IFN-γ as the cells began to differentiate. In this case, we observed that muscle gene expression was again severely downregulated (Fig. 5D). Next, cells were stimulated with IFN-γ 1 day after differentiation medium was added and allowed to differentiate for two additional days. This was the only instance where IFN-γ was added after myogenin was upregulated. We observed that IFN-γ can inhibit muscle gene expression even after differentiation has initiated (Fig. 5E). We note that the most dramatic and uniform suppression of muscle gene expression occurs when cells are differentiated in the presence of IFN-γ. In each case, we also determined if Ciita and H2Ea were stimulated by IFN-γ in each experimental situation and found that, indeed, both Ciita and H2Ea were activated upon IFN-γ stimulation, regardless of when the cells were treated with IFN-γ (data not shown).
Fig. 5.
IFN-γ inhibits myogenesis differentiation and muscle-specific gene expression. (A) Modest decreases in muscle-specific gene expression are observed when proliferating myoblasts are treated with IFN-γ and assayed after 24 h. Asterisks indicate statistical significance. The P values were <0.01 for all samples except Acta1, for which it was 0.036. (B) IFN-γ severely inhibits differentiation-specific gene expression, but not p21 expression, when cells are differentiated in the presence of IFN-γ. C2C12 cells were stimulated as described above and differentiated for 3 days (myotubes [MT]) in the presence of IFN-γ. The P values were <0.01 for all samples. (C) IFN-γ inhibits muscle-specific gene expression in primary myoblasts. Primary myoblasts were stimulated as described for panel B. The P values were <0.01 for all samples. (D) IFN-γ can inhibit muscle-specific gene expression when added as differentiation initiates. IFN-γ was added to confluent myoblasts as differentiation medium was added. Cells were allowed to differentiate for 3 days before being assayed. The P values were <0.01 for all samples. (E) IFN-γ can inhibit muscle-specific gene expression when added to myotubes. IFN-γ was added after the cells had differentiated for 1 day. The cells were allowed to differentiate for two additional days in the presence of IFN-γ. For panels A to E, real-time PCR gene expression analysis was performed for the indicated genes. Data are shown as fold stimulations, with each sample relative to the untreated sample, which was set as 1. Error bars represent standard deviations from the means for the replicate PCR values.
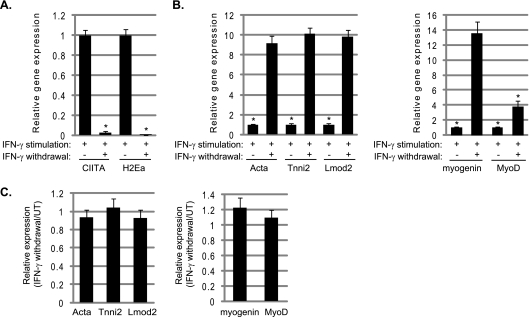
We also examined the effect of IFN-γ on the expression of the MRF family. When C2C12 cells were differentiated in the presence of IFN-γ, a robust downregulation of Myog with a smaller effect on MyoD was observed (Fig. 6A). Myf5 and Myf6 expression levels were not downregulated. The inhibition of Myog expression was observed at the protein level as well (Fig. 6B). We also repeated the same experiment in primary myoblasts and again observed the robust downregulation of Myog, with a more modest effect on MyoD (Fig. 6C). Surprisingly, when MRF levels were examined in C2C12 cells that were differentiated for 1 day prior to IFN-γ treatment, no changes in the levels of Myog or MyoD were observed (Fig. 6D). This result was confirmed in primary myotubes as well (Fig. 6E). However, as shown in Fig. 5E, muscle-specific genes like Acta and Tnni2 were still downregulated under these conditions.
Fig. 6.
IFN-γ inhibits the expression of Myog and MyoD in myoblasts. (A) IFN-γ inhibits the expression of Myog and MyoD in C2C12 cells. Seventy-percent confluent C2C12 cells (myoblasts [MB]) were stimulated with IFN-γ and differentiated for 3 days (myotubes [MT]). Real-time PCR gene expression analysis was performed for the indicated genes. Asterisks indicate statistical significance between the untreated and treated samples. The P values were 0.003 for Myog and 0.04 for MyoD. (B) IFN-γ downregulates myogenin at the protein level as well. Seventy-percent-confluent myoblasts were stimulated with IFN-γ and allowed to differentiate for 2 days in the presence of IFN-γ. The Western blot was probed with anti-myogenin antibodies. (C) IFN-γ inhibits Myog and MyoD expression in primary myoblasts. Primary myoblasts were stimulated as described for panel A, and real-time PCR was performed with primers against Myog and MyoD. The P values were 0.0058 for Myog and 0.021 for MyoD. (D) IFN-γ does not inhibit Myog or MyoD in C2C12 myotubes. C2C12 cells were differentiated for 1 day prior to IFN-γ stimulation, and real-time PCR was performed with primers against Myog and MyoD. (E) IFN-γ does not inhibit Myog expression in primary myotubes. Primary myoblasts were treated as described for panel D and assayed for Myog expression. For panels A, C, D, and E, primers to Hprt were used to normalize the samples. Data are shown as fold stimulations, with each sample relative to the untreated sample, which was set as 1. Error bars represent standard deviations from the means for the replicate PCR values.
The inhibition of myogenesis mediated by IFN-γ is reversible.
Following extended incubations with IFN-γ, we then withdrew IFN-γ and observed that the cells appeared to resume differentiation. To confirm this finding, gene expression profiles were compared in C2C12 cells stimulated with IFN-γ for 4 days to similarly treated cells where IFN-γ was then withdrawn after 2 days and the cells were allowed to recover in medium lacking IFN-γ for two additional days. We observed that the IFN-γ-dependent effects are fully reversible. The expression levels of Ciita and H2Ea were quickly downregulated (Fig. 7A) and muscle-specific genes were upregulated, including the expression of Myog and MyoD (Fig. 7B). The muscle-specific gene expression levels in samples following the withdrawal of IFN-γ were also compared to the expression levels that would normally be observed in cells differentiated for 4 days. We found that the expression levels in the IFN-γ-treated cells were fully restored to untreated expression levels (Fig. 7C).
Fig. 7.
The effects of IFN-γ are fully reversible. (A) Ciita and MHC class II expression is downregulated following the withdrawal of IFN-γ. C2C12 cells were differentiated in the presence of IFN-γ for 2 days, and then IFN-γ was withdrawn for 2 days. Real-time PCR was performed on cDNA samples with primers against Ciita and H2Ea. Asterisks indicate statistical significance. The P value was <0.001 for both Ciita and H2Ea. (B) Muscle-specific gene expression is restored when IFN-γ is withdrawn. The same samples as described above were analyzed with primers against Acta, Tnni2, Lmod2, Myog, and MyoD as indicated. The P values were 0.003 for Acta, 0.001 for Tnni2, 0.002 for Lmod2, 0.005 for Myog, and 0.01 for MyoD. For panels A and B, the value from stimulated cells with no IFN-γ withdrawal was set to one. (C) Removal of IFN-γ restores gene expression levels to unstimulated levels. The IFN-γ-withdrawn samples were compared to samples that were differentiated for 4 days with no IFN-γ treatment. The bar represents the comparison between gene expression observed in the withdrawn sample versus the untreated sample.
CIITA inhibits muscle-specific gene expression.
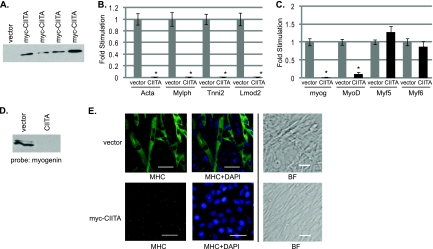
To confirm that CIITA was the mediator of the effects observed with IFN-γ, we stably transfected C2C12 cells with either a plasmid that contains CIITA under the control of the CMV promoter or the empty vector (Fig. 8A). Multiple cell lines were recovered, and three independent clones for both the cells transfected with the CIITA construct and the vector control were assayed. All lines showed equivalent effects for all data shown. We found that the cells that overexpress CIITA mimic the effects observed in IFN-γ-stimulated cells. The expression of muscle-specific genes is dramatically reduced (Fig. 8B). A downregulation of both Myog and MyoD is also observed, while Myf5 and Myf6 are relatively unchanged (Fig. 8C). The downregulation of myogenin is observed at both the RNA and protein levels (Fig. 8C and D). As anticipated from the gene expression results, the cells appear to be blocked in myotube formation and myosin heavy chain expression (Fig. 8E).
Fig. 8.
CIITA overexpression inhibits differentiation and muscle-specific genes, including Myog and MyoD. (A) Stable cell lines overexpress CIITA. Shown is a Western blot of multiple isolates probed with anti-myc antibodies. (B) CIITA inhibits muscle-specific gene expression. Stable cell lines overexpressing CIITA were differentiated for 4 days and harvested for RNA. Real-time PCR was performed on cDNA samples with primers against muscle-specific genes as indicated. Asterisks indicate statistical significance. The P value for gene expression in cells expressing CIITA versus empty vector was <0.001 at every gene tested. (C) CIITA inhibits Myog and MyoD expression. Real-time PCR was performed on the same cDNA samples with primers against the MRFs as indicated. For both panels B and C, data are shown as fold stimulations, with each sample relative to the vector sample, which was set as 1. Error bars represent standard deviations from the means for the replicate PCR values. The P value was 0.0002 for Myog and 0.0006 for MyoD. (D) The downregulation of myogenin is observed at the protein level as well. The Western blot was probed with antibodies against myogenin (F5D, DSHB). (E) Stable cell lines overexpressing CIITA do not appear to differentiate. Cells were differentiated for 4 days and stained with antibodies against myosin heavy chain (MHC) and DAPI. Fluorescent images are ×20 magnification. The indicated scale represents 50 μm. Bright field images of a different field are ×10 magnification. The marker is 50 μm.
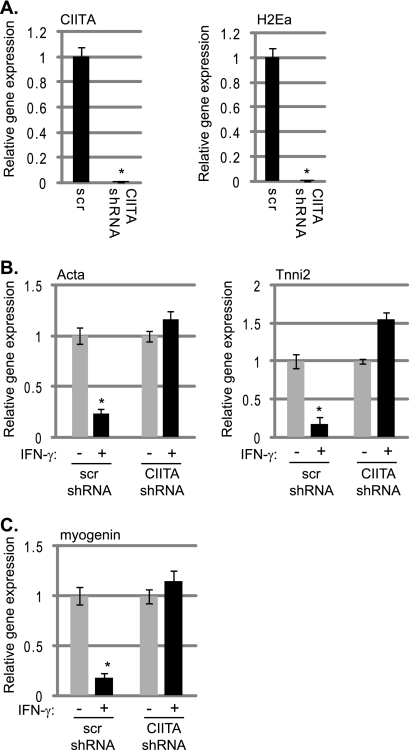
To confirm that CIITA was required for the IFN-γ effects in myoblasts, we knocked down Ciita in C2C12 cells using shRNA constructs. C2C12 cells were transfected with a plasmid-based shRNA construct targeting Ciita. Compared to cells transfected with a scrambled shRNA construct, cells transfected with the Ciita-targeting construct showed the anticipated reduction in Ciita expression (Fig. 9A). We also observed a corresponding reduction in MHC class II gene expression, as assayed by H2Ea expression (Fig. 9A). Cells expressing the scrambled control and the Ciita shRNA construct were stimulated with IFN-γ and assayed for changes in the expression of muscle genes, including MyoD and Myog. We found that the IFN-γ-stimulated Ciita knockdown cell lines did not show reductions in the expression levels of muscle-specific genes (Fig. 9B). We also looked for changes in Myog expression and found that the expression of Myog was unaffected by IFN-γ in the absence of CIITA (Fig. 9C). A second Ciita shRNA construct was also tested, and the results were identical to data presented (data not shown). These data confirm that CIITA is required to mediate the antidifferentiation effects of IFN-γ in muscle cells.
Fig. 9.
CIITA is required for the IFN-γ-mediated inhibition of muscle gene expression. (A) An shRNA construct effectively downregulates Ciita expression. Stable cell lines expressing an shRNA-targeting construct and a scrambled (scr) control were assayed for Ciita and H2Ea expression following IFN-γ stimulation for 3 days. Asterisks indicate statistical significance. The P values were 0.003 for Ciita and 0.001 for H2Ea. (B) Muscle-specific gene expression is unaffected by IFN-γ in the absence of CIITA. IFN-γ-stimulated and nonstimulated cells were differentiated for 3 days, and gene expressions were compared. Results for primers against Acta and Tnni2 are shown. The P value for the scr control was 0.007 for Acta and 0.0036 for Tnni2. (C) The inhibition of Myog expression by IFN-γ requires CIITA. Cells were treated as described for panel B, and the results for primers against Myog are shown. The P value for the scr control was 0.0028. For panels A to C, real-time PCR on cDNA derived from isolated RNA was used to monitor gene expression. Primers against Hprt were used to normalize the data.
CIITA binds to the promoter of muscle-specific genes.
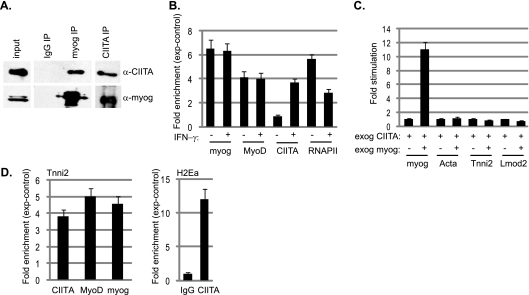
As we found that both myogenin and CIITA are robustly expressed in C2C12 myotubes following IFN-γ treatment, we confirmed the coimmunoprecipitation of the two proteins in these cells (Fig. 10A). To approach how CIITA inhibits myogenin-dependent transcription, we performed a ChIP analysis on C2C12 cells that were differentiated for 2 days and on C2C12 cells differentiated for 2 days where IFN-γ was added after the first day of differentiation. The presence of myogenin, MyoD, CIITA, and RNAPII was assayed on the Tnni2 promoter. As we have previously observed (27), myogenin, MyoD, and RNAPII were detected on the Tnni2 promoter after 2 days of differentiation. In the cells that were stimulated with IFN-γ following 1 day of differentiation and allowed to differentiate one additional day, we found that the recruitment of myogenin and MyoD was unaffected (Fig. 10B). However, in these cells, we also detected CIITA at the Tnni2 promoter, which is transcriptionally downregulated in these cells (Fig. 10B and 8B). We also observed that RNAPII levels decreased compared to the levels in untreated cells. Similar results were obtained on additional muscle-specific promoters (data not shown). Next, we asked if exogenous myogenin expression could overcome the effects of exogenous CIITA. Exogenous myogenin was expressed in the C2C12 cell line expressing exogenous CIITA, and we found that muscle gene expression was not restored (Fig. 10C). ChIP analysis on this cell line revealed that myogenin, MyoD, and CIITA cooccupy muscle-specific promoters in this cell line (shown as Tnni2 in Fig. 10D). As a positive control, we also assayed for the presence of CIITA on the MHC class II promoter for H2Ea. CIITA was also detected on the H2Ea promoter in C2C12 cells (Fig. 10D), and myogenin and MyoD were not detected on the H2Ea promoter (data not shown). Thus, these data argue that CIITA does not block the DNA binding of myogenin but that the interaction with myogenin serves to recruit CIITA to muscle-specific genes. CIITA lacks DNA binding activity and requires the interaction with DNA-bound transcription factors to mediate its activity.
Fig. 10.
CIITA interacts with myogenin in vivo and inhibits the activity of myogenin by binding to target genes. (A) Coimmunoprecipitation of CIITA and myogenin from C2C12 myotubes treated with IFN-γ. Extract was immunoprecipitated with antibodies against CIITA and myogenin and probed with the reciprocal antibodies. Blots were then stripped and reprobed with the antibody used for the IP. (B) CIITA binds to muscle-specific promoters upon IFN-γ stimulation and inhibits RNAPII association but does not affect the binding of myogenin or MyoD. ChIP assays were performed on myotubes treated with IFN-γ or a vehicle control with antibodies against myogenin, MyoD, CIITA, RNAPII, and IgG and analyzed for the Tnni2 promoter. (C) Exogenous myogenin expression cannot overcome the inhibition mediated by CIITA. Gene expression analysis for the indicated genes in the cell line expressing exogenous CIITA and myogenin. Asterisks indicate statistical significance. For Myog, the P value was 0.003. (D) CIITA and myogenin are detected at promoters when both proteins are exogenously expressed. ChIP assays were performed on a cell line stably transfected with exogenous expression constructs for both CIITA and myogenin with antibodies against CIITA, myogenin, MyoD, and IgG and analyzed for the Tnni2 promoter. Primers against the H2Ea promoter were also used to confirm the binding of CIITA to this promoter. The fold enrichment values were calculated relative to the nonspecific control antibody values (IgG). Relative enrichments at the IgH locus were used to normalize the data. For panels B and D, the fold enrichment values were calculated relative to the nonspecific control antibody values (IgG). Relative enrichments at the IgH locus were used to normalize the data.
DISCUSSION
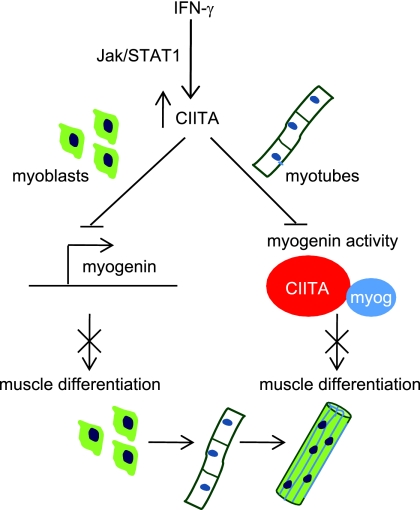
The complex effects of IFN-γ on muscle have remained poorly understood for many years. We show here that IFN-γ acts as a reversible inhibitor of myogenesis by inhibiting the expression and activity of myogenin, the regulator of skeletal muscle differentiation (Fig. 11). In this work, we also revealed a major undiscovered component of the IFN-γ response in skeletal muscle. This component is the well-studied MHC class II transactivator, CIITA. While extensively characterized in cells of the immune system, CIITA is also known to be expressed in several other cell types, including aortic smooth muscle (4). Here, we show that CIITA is expressed in skeletal muscle and also serves an important biological function in muscle. CIITA mediates the activation of the MHC class II genes in muscle, explaining the surprising presence of these molecules in skeletal muscle, and represses myogenic differentiation. The repression of myogenic differentiation occurs at least in part through the interaction of CIITA with myogenin, which represses the activation of muscle-specific genes required for differentiation. This repression includes the expression of Myog and MyoD at specific time points. When IFN-γ or CIITA is introduced before differentiation initiates, myogenin expression is almost completely abolished. Myogenin is only weakly detectable by RNA analysis and is undetectable by Western blot analysis. MyoD is the known activator of Myog expression (2, 11, 12, 56), but we have shown that CIITA does not bind or inhibit MyoD. Myogenin is known to contribute to its own expression (2, 11, 12, 56), so the repression could also occur through the autoregulation of myogenin. It is also possible that CIITA sequesters some other factor that may be required for the activation of Myog. A candidate for this activity might be CBP, which is required for myogenic differentiation and is sequestered by CIITA (43, 64). Equally surprising is the partial repression of MyoD. While it is not unexpected that myogenin would contribute to the expression of MyoD, the expression of MyoD in Myog-null animals is not significantly altered (7, 19, 58). How CIITA represses Myog and MyoD is not currently understood, but we hypothesize that the recruitment of CIITA through the interaction with myogenin causes a repression at promoters that other transcriptional activators cannot overcome. Indeed, our chromatin immunoprecipitation experiments support this hypothesis, as these experiments show that CIITA, myogenin, and MyoD are bound to the troponin promoter (Tnni2) under conditions where Tnni2 expression is repressed. This experiment reveals that MyoD cannot activate transcription of the Tnni2 promoter when CIITA is present.
Fig. 11.
Model for IFN-γ-mediated inhibition of myogenesis. IFN-γ signals through the JAK/STAT pathway, which leads to the phosphorylation of STAT1 (pSTAT1). pSTAT1 translocates to the nucleus and activates CIITA. When cells are stimulated with IFN-γ before myogenin is expressed, CIITA represses the expression of myogenin, which blocks further muscle differentiation. When cells that have already begun to differentiate are exposed to IFN-γ, CIITA binds to myogenin and inhibits its activity, thus blocking further muscle differentiation.
However, we also show that when myotubes, which have already established Myog expression, are treated with IFN-γ, no change in Myog or MyoD expression is observed. Muscle gene expression is still affected, although the effects vary at certain promoters. We find that the troponin gene (Tnni2) is strongly affected, while the leiomodin 2 (Lmod2) gene is less affected. This is an intriguing result, as we have shown that the RNA profiles and transcription factor occupancies of these genes differ over a time course of differentiation (27). As cells begin to differentiate, Lmod2 activates and quickly reaches expression levels that are close to its maximal level (day 3). Tnni2 begins to activate as the cells differentiate, but Tnni2 reaches its maximal expression level much later in the differentiation process (day 6).
Our data suggest to us that CIITA may be able to repress a promoter only if the promoter is not already activated to a high level. Thus, if CIITA is brought to a myogenin-responsive promoter before the promoter is highly active, CIITA can repress the promoter. However, if CIITA is recruited to a myogenin-bound promoter after the promoter is already highly active, CIITA is unable to efficiently block transcription. Consistent with this hypothesis, we find that a gene activated late in differentiation (Tnni2) is more affected by IFN-γ treatment of myotubes than a gene activated earlier in differentiation (Lmod2).
Our overexpression data suggest that CIITA is the mediator of many of the effects of IFN-γ on muscle cells, as the overexpression of CIITA phenocopies the effects observed for IFN-γ stimulation. Knockdown experiments confirm that CIITA is necessary for the antidifferentiation effects observed in IFN-γ-treated cells. Taken together, the overexpression and knockdown experiments demonstrate that CIITA is both necessary and sufficient for the antidifferentiation effects of IFN-γ. Given what we have learned about the role of CIITA in skeletal muscle, it is surprising that we first identified CIITA as an interaction partner of myogenin in differentiated C2C12 cells. However, very low levels of CIITA are detected in differentiated C2C12 cells, and we hypothesize that these levels are not sufficient to block myogenin.
IFN-γ is known to have both positive and negative effects on myogenesis. While IFN-γ is required for efficient muscle repair, constitutive expression causes necrotizing myopathies (51). We believe the data presented here support both of these roles. Based on our findings, we hypothesize that IFN-γ, which is stimulated immediately upon muscle damage, sends an antidifferentiation signal to muscle, which results in an inhibition of myogenin, the MRF required for terminal differentiation. This allows time for satellite cell activation and proliferation before the commitment to terminal differentiation. Once IFN-γ levels fall, the inhibition is reversed and myogenin expression and activity are restored, allowing the final stages of muscle differentiation.
IFN-γ is one of the many proinflammatory cytokines that are delivered to areas of injury by the inflammatory infiltrate. Understanding how the inflammatory infiltrate influences muscle regeneration is essential for designing therapeutic strategies to promote the regeneration of diseased or injured muscle. Another component of the inflammatory infiltrate is tumor necrosis factor alpha (TNF-α), which also regulates muscle regeneration (16, 24). However, studies have shown that IFN-γ and TNF-α have distinct effects on muscle and do not appear to share common signaling pathways (52). Unlike what we have observed for IFN-γ, TNF-α is promyogenic at physiological concentrations (5). TNF-α regulates muscle regulation by activating the promyogenic p38 signaling (5), which promotes the recruitment of polycomb repressive complex 2 (PRC2) to the Pax7 promoter (38). Thus, TNF-α signaling modulates the expression of Pax7, which is a regulator of embryonic muscle progenitors and adult satellite cells (3, 47). Here, we show that IFN-γ controls the activity and expression of myogenin, the regulator of terminal differentiation.
The ability of IFN-γ to harness the activity of myogenin is of particular therapeutic interest given the recent finding that myogenin controls neurogenic atrophy through the regulation of components of the ubiquitin machinery that promote muscle proteolysis and atrophy (30). These findings suggest an unexpected role for myogenin, the regulator of terminal differentiation, in promoting muscle atrophy following denervation. Our results suggest that IFN-γ may have clinical value in the numerous neuropathic disorders, such as amyotrophic lateral sclerosis and Guillain Barré syndrome, that disrupt the nerve supply to muscle and cause a debilitating loss of muscle mass and eventual paralysis.
Like TNF-α, the role of IFN-γ in modulating myogenesis is of particular interest, as myoblasts not only respond to this cytokine during injury but also express IFN-γ (6). The control of myogenin expression and activity is clearly an important component of the response to IFN-γ, but IFN-γ may also halt myogenesis through additional mechanisms. This work also establishes a key role for CIITA in inhibiting differentiation, but the mechanism of action for CIITA on muscle-specific genes remains to be determined. Determining how CIITA represses muscle-specific gene expression will be an important future direction for these studies.
ACKNOWLEDGMENTS
We thank Peter Yao (University of Illinois—Urbana Champaign) for the mass spectroscopy analysis and identification of CIITA. We also thank Jeremy Boss (Emory University) and Nancy Choi (Emory University) for the CIITA expression plasmids and technical advice.
This work was funded by the National Institutes of Health, NIAMS division (grant RAR 060017A). Work in the laboratory of J.K.D. is also supported by grant number 159609 from the American Cancer Society, Illinois Division. This work was additionally supported by a grant from the Excellence in Academic Medicine program, Southern Illinois University School of Medicine.
Footnotes
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Berglund L., et al. 2008. A genecentric human protein atlas for expression profiles based on antibodies. Mol. Cell. Proteomics 7:2019–2027 [DOI] [PubMed] [Google Scholar]
- 2. Braun T., et al. 1989. Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J. 8:3617–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buckingham M., Relaix F. 2007. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu. Rev. Cell Dev. Biol. 23:645–673 [DOI] [PubMed] [Google Scholar]
- 4. Buttice G., Miller J., Wang L., Smith B. D. 2006. Interferon-gamma induces major histocompatibility class II transactivator (CIITA), which mediates collagen repression and major histocompatibility class II activation by human aortic smooth muscle cells. Circ. Res. 98:472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen S. E., Jin B., Li Y. P. 2007. TNF-alpha regulates myogenesis and muscle regeneration by activating p38 MAPK. Am. J. Physiol. Cell Physiol. 292:C1660–C1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng M., Nguyen M. H., Fantuzzi G., Koh T. J. 2008. Endogenous interferon-gamma is required for efficient skeletal muscle regeneration. Am. J. Physiol. Cell Physiol. 294:C1183–C1191 [DOI] [PubMed] [Google Scholar]
- 7. Davie J. K., et al. 2007. Target gene selectivity of the myogenic basic helix-loop-helix transcription factor myogenin in embryonic muscle. Dev. Biol. 311:650–664 [DOI] [PubMed] [Google Scholar]
- 8. Davis R. L., Weintraub H., Lassar A. B. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51:987–1000 [DOI] [PubMed] [Google Scholar]
- 9. DeSandro A. M., Nagarajan U. M., Boss J. M. 2000. Associations and interactions between bare lymphocyte syndrome factors. Mol. Cell. Biol. 20:6587–6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong Y., Rohn W. M., Benveniste E. N. 1999. IFN-gamma regulation of the type IV class II transactivator promoter in astrocytes. J. Immunol. 162:4731–4739 [PubMed] [Google Scholar]
- 11. Edmondson D. G., Brennan T. J., Olson E. N. 1991. Mitogenic repression of myogenin autoregulation. J. Biol. Chem. 266:21343–21346 [PubMed] [Google Scholar]
- 12. Edmondson D. G., Cheng T. C., Cserjesi P., Chakraborty T., Olson E. N. 1992. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol. Cell. Biol. 12:3665–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fontes J. D., Kanazawa S., Jean D., Peterlin B. M. 1999. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol. Cell. Biol. 19:941–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Foster W., Li Y., Usas A., Somogyi G., Huard J. 2003. Gamma interferon as an antifibrosis agent in skeletal muscle. J. Orthop. Res. 21:798–804 [DOI] [PubMed] [Google Scholar]
- 15. Gasque P., Morgan B. P., Legoedec J., Chan P., Fontaine M. 1996. Human skeletal myoblasts spontaneously activate allogeneic complement but are resistant to killing. J. Immunol. 156:3402–3411 [PubMed] [Google Scholar]
- 16. Gopinath S. D., Rando T. A. 2008. Stem cell review series: aging of the skeletal muscle stem cell niche. Aging Cell 7:590–598 [DOI] [PubMed] [Google Scholar]
- 17. Grounds M. D., Garrett K. L., Lai M. C., Wright W. E., Beilharz M. W. 1992. Identification of skeletal muscle precursor cells in vivo by use of MyoD1 and myogenin probes. Cell Tissue Res. 267:99–104 [DOI] [PubMed] [Google Scholar]
- 18. Halevy O., et al. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267:1018–1021 [DOI] [PubMed] [Google Scholar]
- 19. Hasty P., et al. 1993. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364:501–506 [DOI] [PubMed] [Google Scholar]
- 20. Kablar B., Rudnicki M. A. 2000. Skeletal muscle development in the mouse embryo. Histol. Histopathol. 15:649–656 [DOI] [PubMed] [Google Scholar]
- 21. Kalovidouris A. E., Plotkin Z., Graesser D. 1993. Interferon-gamma inhibits proliferation, differentiation, and creatine kinase activity of cultured human muscle cells. II. A possible role in myositis. J. Rheumatol. 20:1718–1723 [PubMed] [Google Scholar]
- 22. Kostrominova T. Y., Macpherson P. C., Carlson B. M., Goldman D. 2000. Regulation of myogenin protein expression in denervated muscles from young and old rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 279:R179–R188 [DOI] [PubMed] [Google Scholar]
- 23. Kretsovali A., et al. 1998. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol. Cell. Biol. 18:6777–6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuang S., Rudnicki M. A. 2008. The emerging biology of satellite cells and their therapeutic potential. Trends Mol. Med. 14:82–91 [DOI] [PubMed] [Google Scholar]
- 25. Legoedec J., Gasque P., Jeanne J. F., Scotte M., Fontaine M. 1997. Complement classical pathway expression by human skeletal myoblasts in vitro. Mol. Immunol. 34:735–741 [DOI] [PubMed] [Google Scholar]
- 26. Llovera M., et al. 1998. Different cytokines modulate ubiquitin gene expression in rat skeletal muscle. Cancer Lett. 133:83–87 [DOI] [PubMed] [Google Scholar]
- 27. Londhe P., Davie J. K. 2011. Sequential association of myogenic regulatory factors and E proteins at muscle specific genes. Skeletal Muscle 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mantegazza R., et al. 1991. Modulation of MHC class II antigen expression in human myoblasts after treatment with IFN-gamma. Neurology 41:1128–1132 [DOI] [PubMed] [Google Scholar]
- 29. Masternak K., et al. 2000. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 14:1156–1166 [PMC free article] [PubMed] [Google Scholar]
- 30. Moresi V., et al. 2010. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 143:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morris A. C., Beresford G. W., Mooney M. R., Boss J. M. 2002. Kinetics of a gamma interferon response: expression and assembly of CIITA promoter IV and inhibition by methylation. Mol. Cell. Biol. 22:4781–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mudhasani R., Fontes J. D. 2002. The class II transactivator requires brahma-related gene 1 to activate transcription of major histocompatibility complex class II genes. Mol. Cell. Biol. 22:5019–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muhlethaler-Mottet A., Otten L. A., Steimle V., Mach B. 1997. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 16:2851–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Musaro A., et al. 1995. Enhanced expression of myogenic regulatory genes in aging skeletal muscle. Exp. Cell Res. 221:241–248 [DOI] [PubMed] [Google Scholar]
- 35. Nabeshima Y., et al. 1993. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364:532–535 [DOI] [PubMed] [Google Scholar]
- 36. Nozell S., Ma Z., Wilson C., Shah R., Benveniste E. N. 2004. Class II major histocompatibility complex transactivator (CIITA) inhibits matrix metalloproteinase-9 gene expression. J. Biol. Chem. 279:38577–38589 [DOI] [PubMed] [Google Scholar]
- 37. O'Shea J. J., Gadina M., Schreiber R. D. 2002. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109:S121–S131 [DOI] [PubMed] [Google Scholar]
- 38. Palacios D., et al. 2010. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 7:455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parker M. H., Seale P., Rudnicki M. A. 2003. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 4:497–507 [DOI] [PubMed] [Google Scholar]
- 40. Parker S. B., et al. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267:1024–1027 [DOI] [PubMed] [Google Scholar]
- 41. Piskurich J. F., Linhoff M. W., Wang Y., Ting J. P. 1999. Two distinct gamma interferon-inducible promoters of the major histocompatibility complex class II transactivator gene are differentially regulated by STAT1, interferon regulatory factor 1, and transforming growth factor beta. Mol. Cell. Biol. 19:431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Piskurich J. F., Wang Y., Linhoff M. W., White L. C., Ting J. P. 1998. Identification of distinct regions of 5′ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of the class II transactivator gene. J. Immunol. 160:233–240 [PubMed] [Google Scholar]
- 43. Polesskaya A., et al. 2001. CBP/p300 and muscle differentiation: no HAT, no muscle. EMBO J. 20:6816–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pollard J. W., Walker J. M. 1997. Basic cell culture protocols, second edition Humana Press, Totawa, NJ [Google Scholar]
- 45. Rando T. A., Blau H. M. 1994. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125:1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Raval A., et al. 2001. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol. Cell 7:105–115 [DOI] [PubMed] [Google Scholar]
- 47. Relaix F., Rocancourt D., Mansouri A., Buckingham M. 2005. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435:948–953 [DOI] [PubMed] [Google Scholar]
- 48. Reyes-Reyna S. M., Krolick K. A. 2000. Chemokine production by rat myocytes exposed to interferon-gamma. Clin. Immunol. 94:105–113 [DOI] [PubMed] [Google Scholar]
- 49. Sabourin L. A., Rudnicki M. A. 2000. The molecular regulation of myogenesis. Clin. Genet. 57:16–25 [DOI] [PubMed] [Google Scholar]
- 50. Scholl T., Mahanta S. K., Strominger J. L. 1997. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc. Natl. Acad. Sci. U. S. A. 94:6330–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shelton G. D., et al. 1999. Necrotizing myopathy induced by overexpression of interferon-gamma in transgenic mice. Muscle Nerve 22:156–165 [DOI] [PubMed] [Google Scholar]
- 52. Smith M. A., Moylan J. S., Smith J. D., Li W., Reid M. B. 2007. IFN-gamma does not mimic the catabolic effects of TNF-alpha. Am. J. Physiol. Cell Physiol. 293:C1947–C1952 [DOI] [PubMed] [Google Scholar]
- 53. Spilianakis C., Papamatheakis J., Kretsovali A. 2000. Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol. Cell. Biol. 20:8489–8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Steimle V., Otten L. A., Zufferey M., Mach B. 1993. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 75:135–146 [PubMed] [Google Scholar]
- 55. Sun L., et al. 2007. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J. Cell Biol. 179:129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thayer M. J., et al. 1989. Positive autoregulation of the myogenic determination gene MyoD1. Cell 58:241–248 [DOI] [PubMed] [Google Scholar]
- 57. Tomita Y., Hasegawa S. 1984. Multiple effects of interferon on myogenesis in chicken myoblast cultures. Biochim. Biophys. Acta 804:370–376 [DOI] [PubMed] [Google Scholar]
- 58. Venuti J. M., Morris J. H., Vivian J. L., Olson E. N., Klein W. H. 1995. Myogenin is required for late but not early aspects of myogenesis during mouse development. J. Cell Biol. 128:563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weinmann A. S., Farnham P. J. 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 26:37–47 [DOI] [PubMed] [Google Scholar]
- 60. Xu Y., Wang L., Buttice G., Sengupta P. K., Smith B. D. 2004. Major histocompatibility class II transactivator (CIITA) mediates repression of collagen (COL1A2) transcription by gamma interferon (IFN-gamma). J. Biol. Chem. 279:41319–41332 [DOI] [PubMed] [Google Scholar]
- 61. Yee C. S., et al. 2004. Cathepsin E: a novel target for regulation by class II transactivator. J. Immunol. 172:5528–5534 [DOI] [PubMed] [Google Scholar]
- 62. Zhang S., Londhe P., Zhang M., Davie J. K. 2011. Transcriptional analysis of the titin cap gene. Mol. Genet. Genomics 285:261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhu X. S., et al. 2000. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol. Cell. Biol. 20:6051–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu X. S., Ting J. P. 2001. A 36-amino-acid region of CIITA is an effective inhibitor of CBP: novel mechanism of gamma interferon-mediated suppression of collagen alpha(2)(I) and other promoters. Mol. Cell. Biol. 21:7078–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]