Abstract
The treatment of cells with histone deacetylase inhibitors (HDACi) was reported to reveal the acetylation of STAT1 at lysine 410 and lysine 413 (O. H. Krämer et al., Genes Dev. 20:473–485, 2006). STAT1 acetylation was proposed to regulate apoptosis by facilitating binding to NF-κB and to control immune responses by suppressing STAT1 tyrosine phosphorylation, suggesting that STAT1 acetylation is a central mechanism by which histone deacetylase inhibitors ameliorate inflammatory diseases (O. H. Krämer et al., Genes Dev. 23:223–235, 2009). Here, we show that the inhibition of deacetylases had no bearing on STAT1 acetylation and did not diminish STAT1 tyrosine phosphorylation. The glutamine mutation of the alleged acetylation sites, claimed to mimic acetylated STAT1, similarly did not diminish the tyrosine phosphorylation of STAT1 but precluded its DNA binding and nuclear import. The defective transcription activity of this mutant therefore cannot be attributed to STAT1 acetylation but rather to the inactivation of the STAT1 DNA binding domain and its nuclear import signal. Experiments with respective cDNAs provided by the authors of the studies mentioned above confirmed the results reported here, further questioning the validity of the previous data. We conclude that the effects and potential clinical benefits associated with histone deacetylase inhibition cannot be explained by promoting the acetylation of STAT1 at lysines 410 and 413.
INTRODUCTION
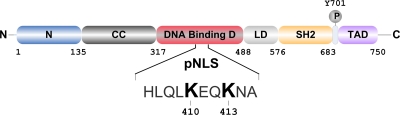
The inactivation of histone deacetylases by chemicals termed histone deacetylase inhibitors (HDACi), such as trichostatin A (TSA) and valproic acid (VPA), has found important clinical applications ranging from psychiatry to cancer and inflammation (1, 13). Best characterized are their effects on the acetylation of histones resulting in altered chromatin accessibility and, hence, gene transcription (7). Additionally, there is a growing number of nonhistone proteins whose acetylation has been reported to be increased by HDACi (13), including the transcription factor STAT1 (22, 23), which is indispensable for the actions of interferons (IFNs) (8). The IFNs have key roles in antiviral and antigrowth responses and in modulating immunity, essentially all of which entail the activation of STAT1 by the phosphorylation of Tyr701 (8, 39). STAT1 activation has important functional consequences. It transforms STAT1 into a sequence-specific DNA binding protein and transcription activator (24). Contrary to unphosphorylated STAT1, which is a nucleocytoplasmic shuttling protein, phosphorylated STAT1 can only enter the nucleus (42). In further contrast to unphosphorylated STAT1, the nuclear import of the activated molecule is dependent on metabolic energy and importin transport factors (42). Importin recruitment requires a nuclear localization signal (NLS) situated in the DNA binding domain (Fig. 1), which was named the dimer-specific NLS (dsNLS) (29). However, as it is now known that STAT1 is dimeric before activation, we propose renaming it NLS of phosphorylated STAT1 (pNLS). Since activated STAT1 needs to be dephosphorylated before nucleocytoplasmic shuttling can resume, activation is associated with the transient trapping of STAT1 in the nucleus, which is readily apparent as nuclear accumulation in interferon-stimulated cells (30).
Fig. 1.
Functional domains (demarcated by residue number) and nuclear localization signal sequence (pNLS) of STAT1 (29). N, N domain; CC, coiled-coil domain; LD, linker domain; SH2, src homology 2 domain; TAD, transactivation domain.
Interferon signaling was demonstrated to require deacetylase activity, but initial studies did not establish a direct link to STAT1 acetylation or phosphorylation (5, 33). Nonetheless, micromolar concentrations of the deacetylase inhibitor TSA were shown to decrease the phosphorylation of STAT1; however, this effect was found to be indirect, as the STAT1-activating JAK tyrosine kinases were inhibited by HDACi (19, 20). Nanomolar concentrations of TSA, in contrast, were shown to reduce the expression of IFN-regulated genes without affecting STAT1 phosphorylation (5, 33).
Krämer et al., however, linked the inhibition of IFN-regulated gene expression directly to the acetylation of STAT1 (21–23, 38). They reported that the treatment of cells with IFN-α or low HDACi concentrations, namely, 30 nM TSA or 1.5 mM VPA, revealed the acetylation of STAT1 at Lys410 and Lys413 (22, 23). Moreover, reference 22 reported that the IFN-α- and IFN-γ-induced tyrosine phosphorylation of STAT1 was suppressed at the same low HDACi concentrations, which was explained by the recruitment of phosphatase TC45 (41) by acetylated STAT1 (22). Consequently, STAT1 remained unphosphorylated in HDACi-treated cells. It accordingly failed to accumulate in the nucleus of IFN-stimulated cells, and it did not bind to the STAT1 recognition site on DNA (termed the GAS element), such that IFN-induced gene expression was lost. However, the inhibitory effects of HDACi on STAT1 phosphorylation were reported to be reversed when cellular tyrosine phosphatases were inactivated by vanadate treatment (22). As both HDACi and vanadate are not STAT1-specific reagents, the authors produced a STAT1 mutant that mimicked the acetylated state, i.e., KK410/413QQ (25), to link HDACi-associated effects directly to the acetylation of STAT1 (22, 23). In perfect agreement with the results obtained with HDACi and wild-type STAT1, the acetylation-mimicking mutant was reported to remain unphosphorylated in IFN-treated cells, it did not accumulate in the nucleus or bind DNA, and hence it too was incapable of activating IFN target genes (22). Importantly, according to the authors, the glutamine mutations per se did not interfere with any of these STAT1 activities, as they reported that the treatment of cells with vanadate or small interfering RNA to inactivate tyrosine phosphatases completely rescued the phenotype of the acetylation-mimicking mutant KK410/413QQ. Remarkably, when tyrosine phosphatase activity was suppressed, the IFN stimulation of cells was shown to trigger Tyr701 phosphorylation of the mutant STAT1, which now accumulated in the nucleus and bound to GAS sites on DNA, culminating in the ability to activate IFN-responsive genes. Thus, based on this consistent data set the authors posited that the phosphorylation/dephosphorylation cycle of STAT1 and, hence, interferon signaling was controlled by the acetylation of residues 410 and 413 (22).
However, residues 410 and 413 are crucial for the NLS of activated STAT1 (29), a fact that received no attention in these studies. In agreement with general NLS requirements, lysines in these positions facilitate the association of activated STAT1 with importin-α, which is required for subsequent nuclear import (26, 29, 32). Thus, we wondered how the acetylation of NLS residues and the replacement of the lysines in positions 410 and 413 with glutamines to mimic acetylation affected STAT1 nucleocytoplasmic mobility and IFN signaling. Table 1 summarizes published results concerning the effects of Lys410/413 mutations on the tyrosine phosphorylation of STAT1, as well as DNA binding and the nuclear import of the activated molecule. As our results from the outset did not conform to the model proposed by Krämer et al., the key experiments published in references 22 and 23 were repeated to evaluate the biological significance of STAT1 acetylation. This exercise confirmed that the model presented by Krämer et al. in references 22 and 23 is incorrect. We show here that neither IFN nor HDACi triggered STAT1 acetylation. We show furthermore that the treatment of cells with HDACi did not interfere with the IFN-induced tyrosine phosphorylation of STAT1. In addition, the alleged acetylation-mimicking glutamine mutant of STAT1, i.e., KK410/413QQ, is demonstrated to be defective per se both in nuclear import and DNA binding. In further stark contrast to the previously published results (22, 23), phosphatase inhibition could not rescue the nuclear import and DNA binding deficiencies of acetylation-mimicking STAT1. Finally, we have repeated the previously published experiments with the original cDNAs encoding wild-type and acetylation-mimicking STAT1 that were provided by the authors of the published studies (22, 23). The experiments with these reagents did not reproduce the published results, whereas the contradictory findings reported here were fully confirmed. In sum, the claims that STAT1 acetylation controls immune responses by suppressing STAT1 tyrosine phosphorylation (22) and regulates apoptosis by facilitating STAT1 binding to NFκB (23) were not upheld by the results presented here.
Table 1.
Functional characterization of wild-type STAT1 and STAT1 variants mutated in positions 410 and 413
| STAT1 construct | Functional statusa |
Reference(s) | ||
|---|---|---|---|---|
| pTyr701 | DNA binding | pTyr701 STAT1 nuclear import | ||
| WT | + | + | + | |
| RR | + | + | + | 22 |
| − | + | + | 22 | |
| EE | + | − | − | 29 |
| AA | + | − | − | 26, 28 |
+, Functionality; −, loss of functionality.
MATERIALS AND METHODS
Cell culture, cDNAs, and reagents.
Human HeLa S3, U3A, and 293T cells were grown at 37°C in a humidified 5% CO2 atmosphere in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS; Sigma) and antibiotics. Mammalian expression plasmids for wild-type (pSTAT1-GFP) and mutant KK410/413EE (EE) STAT1 fused to green fluorescent protein (GFP) have been described already (29); STAT1 was C-terminally Flag tagged. The site-directed mutagenesis of pSTAT1-GFP (QuikChange; Stratagene) generated mutants KK410/413QQ (QQ) and KK410/413RR (RR). Mutations were verified by DNA sequencing. Transient transfections were carried out using Lipofectamine Plus (Invitrogen) as described previously (3). Unless stated otherwise, the treatment of cells was with human IFN-α (103 U/ml; Calbiochem), human IFN-γ (20 ng/ml; Calbiochem), 1.5 mM VPA (Sigma), 30 nM TSA (Sigma), and 1 mM pervanadate. Mammalian expression constructs published by Krämer et al. (22) encoding the wild type and QQ mutant of STAT1 were requested from the authors in February 2010, and dried cDNA was obtained on 14 October 2010. DNA was extracted with Tris-EDTA (TE) and was used to transform DH5α bacteria. Single bacterial colonies were amplified, and plasmid DNA was purified using the Qiagen Maxiprep protocol. The resulting cDNAs were of comparable quality, as judged by readings of the optical density at 260 and 280 nm, which were 1.92 for STAT1-QQ and 1.87 for STAT1-WT, respectively. Clones of both plasmids were partially sequenced in house using the Big Dye sequencing protocol (Applied Biosystems), followed by capillary electrophoresis using a commercial source (Gene Services Nottingham).
Quantitative Western blotting and immunoprecipitation.
Cell extractions were performed in whole-cell extraction buffer (50 mM Tris-HCl, pH 7.4, 280 mM NaCl, 5 mM EDTA, 2 mM EGTA, 10% [vol/vol] glycerol, 50 mM NaF, 10 mM glycerol phosphate, 1 mM sodium pervanadate [VO43−], 0.2% [vol/vol] NP-40, 3 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF], and Complete [Roche] protease inhibitors) for 30 min on ice. Soluble components were separated by centrifugation at 14,000 × g for 15 min at 4°C and used for immunoprecipitation (30) and Western blotting. Western blotting entailed the labeling of primary antibodies with IRdye800-conjugated secondary immunoglobulin (Licor Biosciences). Control readings were obtained with diluted cell extracts to confirm that signal intensities were within the linear range. Where the presence of multiple proteins/protein modifications was examined, this was done consecutively for each antibody using the same blot. Bound antibodies were stripped off nitrocellulose membranes by incubation for 30 min twice in stripping buffer (25 mM glycine, 2% SDS, pH 2) at 65°C. Primary antibodies were purchased from Cell Signaling (anti-pTyr701 STAT1, order no. 9171 and anti-acetylated lysine, order no. 9441 [formerly sold by NEB]), Santa Cruz (anti-STAT1, C24, E23, anti-simian virus 40 [SV40] T antigen [Pab 108], order no. sc-148), Abcam (anti-acetyl-histone 3 and H3[lys9]), and Sigma (anti-FLAG and anti-β-actin).
Immunofluorescence microscopy.
Cells were fixed with methanol at −20°C, washed in phosphate-buffered saline (PBS), and blocked with 20% (vol/vol) FCS–PBS. Samples were incubated overnight with either anti-STAT1 antibody (C24; dilution, 1:6,000) or anti-pTyr701 STAT1 antibody diluted 1:250 in 20% (vol/vol) FCS–PBS. After three washes with PBS, the cells were incubated with appropriate species-specific affinity-purified Cy3-conjugated immunoglobulins (Jackson Research) in 20% (vol/vol) FCS–PBS. A Leica DMLB microscope was used as described previously (3).
EMSA.
For electrophoretic mobility shift assay (EMSA), U3A cells (1.8 × 105 to 2 × 105) were transiently transfected. Thirty-six h later, the cells were left untreated or were treated as described in the figure legends. After two washes with ice-cold PBS, the cells were incubated on ice for 5 min in 70 μl cytoplasmic extraction buffer (10 mM KCl, 1 mM EDTA, 20 mM HEPES-NaOH, pH 7.4, 0.1 mM sodium pervanadate, 1.5 mM MgCl2, 10% [vol/vol] glycerol, 1 mM DTT, Complete protease inhibitors, 0.2% [vol/vol] NP-40, and 0.1 mM PMSF), followed by centrifugation for 20 s at 4°C and 14,000 × g. The supernatant (cytoplasmic extract) was isolated and kept on ice. The pellet was resuspended in 70 μl nuclear extraction buffer (420 mM KCl, 1 mM EDTA, 20 mM HEPES-NaOH, pH 7.4, 0.1 mM pervanadate, 1.5 mM MgCl2, 20% [vol/vol] glycerol, 1 mM DTT, Complete protease inhibitors, and 0.1 mM PMSF), and incubated for 30 min on ice. After centrifugation for 15 min at 4°C and 14,000 × g, the nuclear extract was isolated and combined with the corresponding cytoplasmic extract. A Bradford assay (Bio-Rad) was used to confirm equal protein concentrations for the different extracts. Five micrograms of extracted protein was incubated in a 14-μl reaction mix {containing 1 mg/ml bovine serum albumin (BSA), 1 mM DTT, 2 μg poly(dI-dC), 20 mM HEPES-NaOH, pH 7.9, 40 mM KCl, 1 mM MgCl2, 0.4 mM EDTA, 0.1 mM EGTA, 4% (vol/vol) Ficoll, and 1 nM double-stranded [γ-32P]CTP-labeled DNA probe} for 20 min at room temperature. The DNA probe (5′-TGATTTCCCCGAAATGACGGC-3′ with 5′-TGAG-3′ overhangs at the 5′ end of both strands) harbored a single GAS site (underlined) from the IRF-1 gene promoter (35). For the immune detection of DNA-bound STAT1 (supershift), anti-STAT1 antibody (C24) was added to the reaction mix (1 μl). DNA-protein complexes were resolved at 4°C and 400 V on 4.7% nondenaturing polyacrylamide gels using Tris-borate-EDTA buffer (140 mM Tris-HCl, pH 8.0, 22 mM boric acid, and 2 mM EDTA). After a running time of 90 min, gels were dried; radioactivity was detected and quantified by phosphorimaging (FLA-3000; Fujifilm).
RESULTS AND DISCUSSION
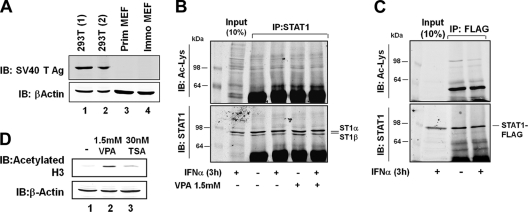
Our analyses of the effects of lysine acetylation on the function of STAT1 started with an attempt to confirm STAT1 acetylation. The previously published experiments were done with several different cell lines; however, crucial experiments were performed with 293T cells, which express SV40 large T antigen (22, 23). Because SV40 large T antigen binds to cellular factors, including the transcriptional coactivators p300 and CBP (2), and CBP is responsible for the acetylation of STAT1 according to the published results (22, 23), we first confirmed by Western blotting that our 293T cells, but not primary and spontaneously immortalized mouse embryonic fibroblasts (MEF), expressed SV40 large T antigen. Figure 2A shows that this was indeed the case. We then stimulated 293T cells with IFN-α for 3 h as described by Krämer et al. (see Fig. 1A of reference 22), followed by the immunoprecipitation of the endogenous STAT1 from whole-cell extracts and the probing of the immunoprecipitates with antiacetyllysine-specific antibody. Additionally, 293T cells were treated with 1.5 mM VPA for 24 h, either alone or in conjunction with subsequent IFN-α for 3 h, as this was described to boost the acetylation of STAT1 (see Fig. 6A of reference 23). However, despite repeated attempts, in our hands neither approach gave a signal for acetylated STAT1 (Fig. 2B). Human fibrosarcoma 2fTGH cells are another cell line that was reported to harbor acetylated STAT1 upon stimulation with IFN-α (see Fig. 6F of reference 23). We thus used 2fTGH-derived U3A cells (31), which lack STAT1 expression, and reconstituted them with Flag-tagged STAT1. However, immunoprecipitated Flag-tagged STAT1 did not show reactivity with acetyllysine antibody irrespective of IFN-α treatment (Fig. 2C). To verify that the HDACi were active at the concentrations used, we performed control reactions with acetylated histone 3-specific antibody, which demonstrated increased histone 3 acetylation in 293T cells upon treatment with TSA or VPA (Fig. 2D). As STAT1 acetylation has been demonstrated exclusively by immune reactivity but was not validated to date by another method, e.g., mass spectrometry, we could not decide whether this outcome reflected the absence of acetylated STAT1 or was due to unreliable immunodetection. To our knowledge, STAT1 acetylation has been observed in two other instances, but the data quality was not definitive (16, 40), and according to those studies there was no impairment of STAT1 phosphorylation associated with acetylation. A third study on STAT1 acetylation reported a lack of immunodetectable STAT1 lysine acetylation in B-cell lines irrespective of treatment with IFN-α or TSA (27). Our failure to detect acetylated STAT1 is unlikely to be explained by antibody differences, as preparations of the same commercially available acetyllysine antibody were used in the present and the earlier studies (22, 23). Another factor relevant for successful detection is the abundance of acetylated STAT1 in cells, which to our knowledge has not been quantified. However, Krämer et al. report that the target of acetylation is phosphorylated STAT1 (22, 23), which makes up 30 to 40% of STAT1 in interferon-stimulated cells (43). According to reference 22, acetylation is required to recruit phosphatase TC45 to the activated STAT1. If we assume that at least one monomer of each phosphodimer needs to be acetylated for efficient phosphatase recruitment, it is safe to presume that in IFN-stimulated cells about 10 to 20% of STAT1 molecules are acetylated at steady state. Contrary to labile posttranslational modifications, like the SUMO conjugation of STAT1, which is difficult to preserve (12), lysine acetylation is efficiently stabilized by deacetylase inhibitors according to the published experiments (22, 23). Moreover, treatment with deacetylase inhibitors was reported to completely suppress the IFN-induced phosphorylation of STAT1 (see Fig. 1E in reference 22), suggesting that the quantitative acetylation of STAT1 is readily achieved under these conditions. Thus, it is reasonable to assume that a large fraction, probably in excess of 30 to 40% of the cellular STAT1 content, should have been acetylated in our experiments. Therefore, a very small modified and, hence, difficult-to-detect fraction of acetylated STAT1 is an unlikely explanation for the lack of antiacetyllysine immunoreactivity.
Fig. 2.
STAT1 lacks immunoreactivity with antiacetyllysine antibody irrespective of treatment with interferon and HDACi. (A) Whole-cell extracts from 293T cells used in this study (lane 1), control 293T cells obtained from LGC Standards (ATCC CRL-1573) (lane 2), primary MEFs (lane 3), and a spontaneously immortalized MEF cell line (lane 4). Shown are Western blotting (IB) results with anti-SV40 T antigen antibody and the reprobing with anti-β-actin antibody. (B) 293T cells were incubated with 1.5 mM VPA (+) or left untreated for 24 h, followed by the addition of 1,000 U/ml IFN-α for 3 h where indicated (+). STAT1 was immunoprecipitated (IP) from whole-cell extracts, followed by Western blotting. Shown are the consecutive probings with antiacetyllysine antibody (upper) and anti-STAT1 antibody (lower). (C) U3A cells were transfected with vector encoding C-terminally Flag-tagged STAT1. Twenty-four hours later, cells were stimulated with 1,000 U/ml IFN-α for 3 h (+), followed by whole-cell extraction and immunoprecipitation using anti-Flag antibody. Shown are Western blotting results for the precipitates probed consecutively with antiacetyllysine antibody and anti-STAT1 antibody. (D) 293T cells were left untreated or were treated with VPA or TSA for 24 h as indicated. Shown are Western blotting results for whole-cell extracts probed with anti-acetyl-Lys9 histone 3 antibody and subsequently reprobed with anti-β-actin antibody.
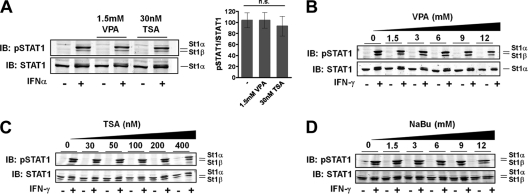
We therefore focused on the reported consequences of STAT1 acetylation, namely, the inhibition of IFN-α- and IFN-γ-induced STAT1 Tyr701 phosphorylation. It was reported that a 24-h pretreatment of 293T cells with HDACi VPA (1.5 mM) or TSA (30 nM) precludes the subsequent 20-min IFN-α-induced tyrosine phosphorylation of STAT1 (see Fig. 1E in reference 22). To attempt to reproduce this experiment, 293T cells were left untreated or were treated with HDACi VPA (1.5 mM) or TSA (30 nM) for 24 h, followed by stimulation with IFN-α for 20 min as described above. IFN treatment alone caused the expected Tyr701 phosphorylation of STAT1 (Fig. 3A). However, in stark contrast to the published results, pretreatment with HDACi TSA (30 mM) or VPA (1.5 mM) left the IFN-α-induced tyrosine phosphorylation of STAT1 undiminished (Fig. 3A). Three quantitative Western blotting experiments generated no statistically significant differences regarding IFN-α-induced Tyr701 phosphorylation between untreated and HDACi-treated cells (Fig. 3A). We additionally studied the effects on STAT1 phosphorylation of increasing concentrations of VPA (Fig. 3B), TSA (Fig. 3C), or another HDACi, sodium butyrate (NaBu) (Fig. 3D). Even at more than 10-fold higher than published HDACi concentrations, STAT1 was strongly phosphorylated in response to IFN-γ, not at all resembling the complete loss of IFN-α- or γ-induced Tyr701 phosphorylation reported in Fig. 1E and data not shown of reference 22. We thus inferred that low HDACi concentrations did not alter the activation of STAT1. This conclusion agrees with numerous previous studies, which reported the unchanged or even prolonged tyrosine phosphorylation of STAT1 at low HDACi concentrations similar to those used here (5, 33, 36).
Fig. 3.
STAT1 activation continues in the presence of histone deacetylase inhibitors. (A) 293T cells were left untreated or were treated with VPA or TSA. After 24 h the cells were stimulated for 20 min with 1,000 U/ml IFN-α where indicated (+), followed by whole-cell extraction. Shown are representative Western blotting (IB) results of consecutive probing with anti-pTyr701 STAT1 antibody and anti-STAT1 antibody. The positions of STAT1α (St1α) and its C-terminally truncated splice variant STAT1β (St1β) are indicated. Note that the anti-STAT1 antibody C24 used here does not recognize STAT1β. The bar diagram on the right combines the data from three independent experiments. Shown are the means and standard deviations for the specific tyrosine phosphorylation of STAT1 in IFN-stimulated cells, as determined by quantitative Western blotting. The mean value for untreated cells was set to 100. For statistical analysis, a one-way analysis of variance test was performed. n.s., not significant. (B) HeLa cells were incubated with increasing concentrations of VPA as indicated. Twenty-four hours later, cells were stimulated with 20 ng/ml IFN-γ for 1 h where indicated (+). Tyr701 phosphorylation of STAT1 was assessed by Western blotting as described for panel A. (C) Cells were treated as described for panel B, but 293T cells were used and treated with TSA. Note that anti-STAT1 antibody E23 was used here, which detects both STAT1 splice variants. (D) Cells were treated as described for panel C, except that Na-butyrate, another HDACi, was used.
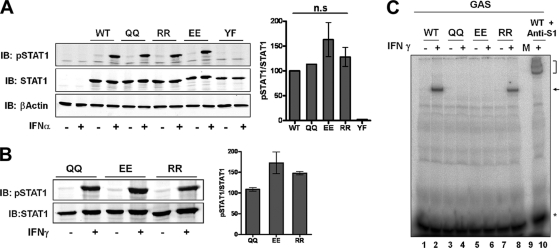
We next studied the IFN-induced activation of the acetylation-mimicking mutant of STAT1, KK410/413QQ (termed QQ). This mutant was reported to remain unphosphorylated in U3A cells after 1 h of IFN-α treatment, thus justifying its characterization as an acetylation-mimicking STAT1 variant (see Fig. 1G in reference 22). We repeated this experiment in U3A cells as described previously (22). However, in contrast to the published results, we found robust phosphorylation of this STAT1 mutant (Fig. 4A). We also analyzed the Tyr701 phosphorylation of additional STAT1 variants, where the lysine residues at positions 410 and 413 were changed to arginine (RR) or glutamate (EE). Three independent quantitative immunoblotting experiments were done, and the specific tyrosine phosphorylation of the STAT1 variants was determined. This experiment demonstrated that there were no statistically significant phosphorylation differences between wild-type STAT1 and any of these mutants (Fig. 4A). Specifically, the tyrosine phosphorylation of STAT1 KK410/413QQ was not weaker than that of the wild type. As a negative phosphorylation control, we included a Tyr701-to-phenylalanine mutant in this analysis (37). Strong tyrosine phosphorylation of the alleged acetylation-mimicking mutant was seen in response to IFN-γ (Fig. 4B), again in clear contradiction to results reported previously (22). We thus concluded that the alleged acetylation-mimicking mutant does not differ from wild-type STAT1 regarding tyrosine phosphorylation.
Fig. 4.
Side chain chemistry of residues 410 and 413 is irrelevant for STAT1 phosphorylation, but positive charges in these positions are required for sequence-specific DNA binding. (A, B) U3A cells were transiently reconstituted with the STAT1 wild type (WT), mutant KK410/413QQ (QQ), mutant KK410/413EE (EE), mutant KK410/413RR (RR), and mutant Tyr701Phe (YF). Forty-eight hours after transfection, cells were treated with 1,000 U/ml IFN-α (A) or 20 ng/ml IFN-γ (B) for 1 h where indicated (+), followed by whole-cell extraction. Shown are representative Western blotting (IB) results obtained by consecutive probing with anti-pTyr701 STAT1 antibody and anti-STAT1 antibody (C24). The bar diagrams combine the data from three independent experiments each. Shown are the means and standard deviations for the specific tyrosine phosphorylation of STAT1 in IFN-stimulated cells, as determined by quantitative Western blotting. The mean value for wild-type cells (A) or QQ mutant cells (B) was set to 100. For statistical analysis, one-way analysis of variance tests were performed. n.s., not significant. (C) U3A cells were transfected with the indicated STAT1 constructs described for panel A. Thirty-six h later, consecutive cytoplasmic and nuclear cell extractions were done as described in Materials and Methods. Shown are EMSA results using [γ-32P]CTP-labeled DNA probe containing the IRF-1 GAS binding site. The binding reaction mixture analyzed in lane 10 contained anti-STAT1 antibody. Free probe (*), STAT1-DNA complex (arrow), and the antibody-bound STAT1-DNA complex (bracket) are indicated.
We then examined the DNA binding of the acetylation-mimicking STAT1 mutant KK410/413QQ, which was shown to bind to GAS sites (see Fig. 5F in reference 22). However, to reveal the mutant's DNA binding activity, previously cells needed to be treated with a phosphatase inhibitor, e.g., vanadate (22). According to the authors, this treatment was required to overcome the excessive dephosphorylation caused by the phosphatase recruitment of the pseudoacetylated STAT1. The QQ mutant therefore bound to DNA in the presence of a tyrosine phosphatase inhibitor (see Fig. 5F in reference 22) but not in its absence (see Fig. 5F, S1H, and S2D in reference 22). According to the published data, these results establish that acetylation-mimicking STAT1 KK410/413QQ, if phosphorylated, is capable of binding to GAS-containing DNA. The dependence of DNA binding and transcriptional activity on phosphatase inhibition (as shown in Fig. 5E in reference 22) is of pivotal importance for the proposed phosphorylation-acetylation switch model. For one thing, it confirms the acetylation-mimicking activity of the QQ mutant. More crucially, it represents the cornerstone of the proposed model on which the authors rest their central conclusion that acetylation inhibits STAT1 signaling by facilitating the recruitment of phosphatase activity to the activated transcription factor (Table 1).
Before we report the results of experiments evaluating the validity of this set of published data, we first need to comment on details of the methodology that was used to demonstrate STAT1 DNA binding in the published study (22). To evaluate the sequence-specific DNA binding of a protein, the sequence of the DNA must be known. “GAS consensus oligonucleotides” and “GAS sequences” are mentioned numerous times in reference 22, yet the only precise information about the actual GAS sequence that was used can be found in the legend to Fig. S1H, where it is stated, “A pulldown assay with a biotinylated GAS oligonucleotide (ABCD-assay) was used to detect STAT1-DNA binding. The oligonucleotide represents two palindromic TCC half-sites, contacted by the DNA binding domains of an activated STAT1 dimer.” This statement does not fully clarify the issue, since GAS sites are nonameric (TTCNNNGAA) and thus are not represented by two palindromic TCC half sites (9). Nonetheless, this and other statements suggest that the authors wanted to study the DNA binding of dimers of activated STAT1. However, instead of treating cells with IFN-γ, which is the most potent and specific STAT1-activating stimulus, IFN-α was used to assess DNA binding. IFN-α stimulation gives rise predominantly to STAT1/STAT2 heterodimers that bind together with another protein, IRF-9, to an unrelated DNA site (9, 15), such that relatively few STAT1 dimers capable of GAS binding remain. These incomplete descriptions and the very difficult-to-interpret study design precluded the exact repetition of the experiments as published. We decided therefore to follow established procedures that generate unambiguous results to examine the DNA binding of dimers of wild-type and mutant STAT1, respectively. This included the use of electrophoretic mobility shift assay (EMSA) rather than precipitation methods like the ABCD assay, as the latter uses denaturing conditions to resolve DNA binding complexes, thus destroying information on the size and composition of the DNA-associated protein complexes. This limitation makes precipitation assays less useful for studying sequence-specific DNA binding proteins. As the probe, we used radiolabeled DNA containing a single high-affinity STAT1 binding site, the GAS element of the IRF-1 promoter (35). Also, cells were treated with IFN-γ to generate large quantities of activated STAT1 dimers, which facilitates analyses of GAS-specific DNA binding activity.
Using whole-cell extracts from U3A cells transiently reconstituted with STAT1 variant proteins as described previously (22), we observed a single protein-DNA complex for wild-type STAT1 (Fig. 4C, lanes 1 and 2) that was reactive with STAT1 antibody (lane 10), indicating that the DNA binding complex contained STAT1 as expected. We then tested the DNA binding of the acetylation-mimicking mutant KK410/413QQ. Although abundant activated STAT1 was present in the extracts (Fig. 4B), there was no binding of STAT1 QQ to GAS-containing DNA (lanes 3 and 4). The identical negative outcome was seen with mutant KK410/413EE (lanes 5 and 6), which we had shown previously to dimerize yet to be defective in DNA binding (29), while the arginine mutant KK410/413RR retained binding to the GAS site (lanes 7 and 8). These results demonstrated that positive charges in positions 410 and 413 are indispensable for the binding of STAT1 to its recognition site on DNA.
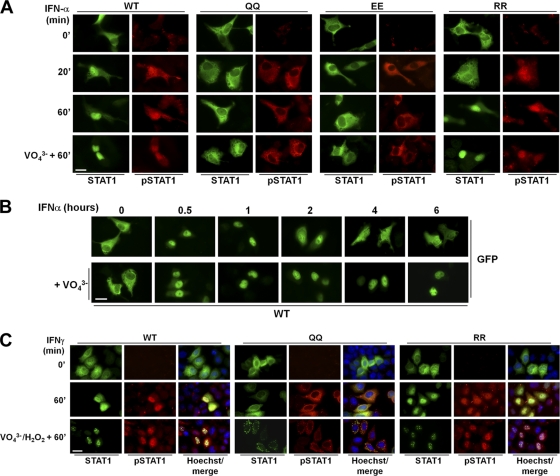
Lysines 410 and 413 are part of the nuclear import signal for activated STAT1 (29). However, little is known about how the acetylation of lysine residues or their charge-neutralizing mutation affects NLS activity. It was reported for STAT1 that the charge-neutralizing mutation of lysines 410 and 413 to glutamine does not per se impair the nuclear import of the phosphorylated protein (see Fig. 5D in reference 22). We therefore transiently reconstituted U3A cells with wild-type or mutant STAT1 and used immunofluorescence microscopy to observe the activation and intracellular distribution of STAT1 in response to IFN-α stimulation. Wild-type STAT1 showed the well-known IFN-α-induced activation and concomitant nuclear accumulation (42) (Fig. 5A). The acetylation-mimicking QQ mutant showed similarly strong IFN-α-induced activation, substantiating our Western blotting results but again contradicting the published experiments (see Fig. 1G in reference 22). Importantly, however, the phosphorylated mutant did not accumulate in the nucleus but rather stayed in the cytoplasm (Fig. 5A), in further contradiction of the published results (see Fig. 5C and D in reference 22). The glutamate mutation of residues 410 and 413 also precluded nuclear accumulation, and the activated protein stayed in the cytoplasm, whereas the arginine mutation of these positions did not impair STAT1 nuclear accumulation (Fig. 5A). We additionally tested whether treatment with vanadate could overcome the nuclear import defect of the glutamine mutants of STAT1, as shown in Fig. 5D of the earlier study (22). However, this was not the case, as the phosphorylated glutamine as well as the glutamate mutants remained in the cytoplasm (Fig. 5A). Control experiments verified the tyrosine phosphatase-inhibiting activity of the vanadate preparation used here. Vanadate measurably prolonged the IFN-induced nuclear accumulation of wild-type STAT1 (Fig. 5B), which is explained by the inability of tyrosine-phosphorylated STAT1 to return to the cytoplasm (30). When this experiment was performed in HeLa cells using IFN-γ stimulation, the same outcome was observed, namely, strong phosphorylation yet defective nuclear import of the acetylation-mimicking STAT1 QQ mutant, while the phosphorylated arginine mutant retained nuclear import (Fig. 5C). Interestingly, in the presence of H2O2, which was used in this experiment in conjunction with pervanadate to inhibit tyrosine phosphatase activity, phosphorylated STAT1 formed paracrystals. This phenomenon is explained by the concomitant H2O2-dependent inactivation of STAT1 SUMO conjugation (12). Of note, the QQ mutant formed paracrystals in the cytoplasm, providing corroborating evidence for the efficient phosphorylation and loss of nuclear import. We thus concluded that positive charges in positions 410 and 413 are required both for sequence-specific DNA binding and the nuclear import of activated STAT1. These results agree with data independently obtained for STAT1 (28), Max transcription factor (14), and the Rothmund-Thomson-syndrome gene product RECQL4 (11), whose nuclear import is precluded by charge-neutralizing replacements with glutamine or alanine of lysine residues required for NLS functioning.
Fig. 5.
Acetylation-mimicking STAT1 mutant is phosphorylated but not imported in the nucleus. (A) U3A cells were transiently reconstituted with GFP-tagged wild-type STAT1 (WT), mutant KK410/413QQ (QQ), mutant KK410/413EE (EE), or mutant KK410/413QRR (RR). After 48 h the cells were treated with 1,000 U/ml IFN-α for the indicated times. Where indicated (VO43−), cells were pretreated for 45 min with 1 mM pervanadate before the addition of IFN-α for another 60 min. Shown are immunofluorescence images depicting the localization of STAT1 (determined by GFP epifluorescence) and the localization of phosphorylated STAT1 (pSTAT1), determined by labeling with anti-pTyr701-STAT1 antibody. (B) U3A cells were transfected with GFP-tagged wild-type STAT1. Forty-eight hours later, cells were treated for 0 to 6 h with 1,000 U/ml IFN-α alone or together with 1 mM pervanadate (+VO43−). Shown are GFP epifluorescence micrographs that depict the distribution of STAT1. (C) HeLa cells were transiently transfected with GFP-tagged wild-type STAT1 (WT) or mutant QQ or RR as indicated. After 36 h the cells were left untreated or were treated for 60 min with IFN-γ (20 ng/ml). Where indicated (VO43−/H2O2), cells were pretreated for 45 min with 0.8 mM pervanadate and 0.2 mM H2O2 prior to the addition of IFN-γ. Shown are immunofluorescence images depicting the localization of STAT1 (determined by GFP epifluorescence) and the localization of phosphorylated STAT1 (pSTAT1), determined by labeling with anti-pTyr701-STAT1 antibody. Nuclei were labeled with Hoechst dye. Note the appearance of STAT1 paracrystals in the presence of H2O2. Bars, 10 μm.
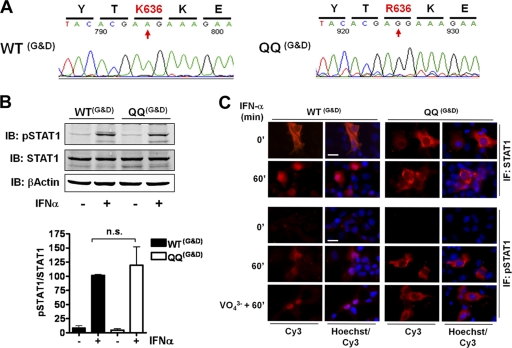
Given the complete lack of agreement with the reported results, we requested the original mammalian expression vectors encoding wild-type STAT1 and the acetylation-mimicking mutant that were used in the published studies (22, 23). DNA sequencing of ∼100 bp upstream and ∼800 bp downstream of the STAT1 coding sequence revealed no differences between the two constructs (data not shown), and they matched the sequence of pcDNA3.1 TOPO vector (Invitrogen), thus confirming the published information (23). The wild-type STAT1-encoding sequence was identical to the GenBank entry NM_007315 for human STAT1. The cDNA of STAT1 acetylation-mimicking mutant QQ showed the expected (single-base) changes of codons 410 and 413 resulting in mutations to glutamine (see Fig. S1 in the supplemental material). We found three additional single-base changes affecting codons 636, 740, and 742, one of which resulted in the mutation of the STAT1 protein at Lys636→Arg (Fig. 6A; also see Fig. S1 in the supplemental material). Residue 636 is situated in the SH2 domain and is part of a conserved antiparallel β-sheet that constitutes the core of this domain (6), but subsequent experiments showed that the protein mutated in position 636 was expressed in mammalian cells, indicating that this amino acid change did not alter protein structure. U3A cells then were transfected with these constructs to determine tyrosine phosphorylation and the nuclear translocation of the published proteins. As shown in Fig. 6B, quantitative Western blotting demonstrated that wild-type STAT1 and the acetylation mutant QQ both were equally well tyrosine phosphorylated, with no statistically significant differences between them. Similarly, immunofluorescence microscopy confirmed both the undiminished tyrosine phosphorylation of acetylation-mimicking STAT1 KK410/413QQ in response to IFN-α and its inability to accumulate in the nucleus (Fig. 6C). Thus, irrespective of the origin of the reagents used, the same results were obtained.
Fig. 6.
Published results (22, 23) are not reproducible using published reagents. (A) DNA sequencing of the published constructs reveals an undisclosed amino acid exchange in position 636. (B) U3A cells were transiently transfected with the published constructs encoding wild-type STAT1 (WTG&D) or mutant KK410/413QQ (QQG&D). After 36 h, the cells were left untreated or were stimulated with 1,000-U/ml IFN-α for 1 h, followed by whole-cell extraction. Shown are representative Western blotting (IB) results obtained by consecutive probing with anti-pTyr701-STAT1 antibody, anti-STAT1 antibody (C24), and anti-β-actin antibody. The bar diagram combines the data from three independent experiments. Shown are the means and standard deviations for the specific tyrosine phosphorylation of STAT1 before and after IFN stimulation, as determined by quantitative Western blotting. The mean value for IFN-stimulated wild-type cells was set to 100. For statistical analysis, a t test was applied. n.s., not significant. (C) U3A cells were transfected as described for panel B. After 36 h, cells were left untreated or were treated with 1,000-U/ml IFN-α for 60 min. Where indicated (VO43−), IFN was added after a 45-min pretreatment with 1 mM pervanadate. After fixation and permeabilization in ice-cold methanol, cells were labeled with anti-STAT1 antibody (C24, upper) or anti-pTyr701-STAT1 antibody (lower), followed by Cy3-labeled secondary antibody. Shown are immunofluorescence micrographs depicting the localization of total STAT1 (upper) and the localization of Tyr701-phosphorylated STAT1 (lower). Nuclei were stained with Hoechst dye (Hoechst). Bar, 10 μm.
In summary, the experiments reported here generated no evidence that the stimulation of cells with interferons or treatment with HDACi result in the acetylation of STAT1. However, we cannot formally rule out this possibility, nor do we exclude that STAT1 might be acetylated in circumstances other than those investigated. That said, the results presented here conclusively exclude the possibility that STAT1 signaling is regulated by a phosphorylation-acetylation switch as proposed by Krämer et al. (22). The proposed switch mechanism is based on the experimental demonstration (22) that the treatment of cells with HDACi prevents the IFN-induced phosphorylation of STAT1, and that the resulting inhibition of STAT1 signaling can be overcome by the inactivation of cellular phosphatases. We disprove this claim by showing that the treatment of cells with HDACi results in the undiminished phosphorylation of STAT1. Validation of the model proposed in reference 22 furthermore hinges on the use of a single reagent, namely, the supposed acetylation-mimicking glutamine 410/413 mutant of STAT1. It was reported that glutamine mutations of residues 410 and 413 prevented IFN-induced phosphorylation of STAT1, and that the resulting inhibition of STAT1 signaling caused by these mutations could be overcome by the inactivation of cellular phosphatases (22). We show here that the IFN-induced phosphorylation of the glutamine mutant of STAT1 is not diminished, which disproves the alleged resistance of this mutant to IFN stimulation and hence invalidates its characterization as an acetylation-mimicking STAT1 variant. In addition, we show that the nuclear import and sequence-specific DNA binding of the glutamine mutant both are inactivated, which prevents this protein from functioning as a signal transducer and activator of transcription, i.e., STAT1. The invalidated so-called acetylation-mimicking glutamine mutant is pivotal for Krämer et al. to demonstrate that acetylated STAT1 controls both IFN signaling and cell death. At present, the claims put forth in references 22 and 23 were found to be without experimental justification, as are attempts to link the protective effects of histone deacetylase inhibitors against cancer and inflammation to the Lys410/Lys413 acetylation of STAT1 (4, 10, 17, 18, 34, 44). As reported here, despite of our best attempts to reproduce exactly the experiments of references 22 and 23, we found no experimental confirmation of the results presented there. We are unable to provide a scientifically acceptable explanation for the complete dichotomy of our findings from those reported by Krämer et al.
Supplementary Material
ACKNOWLEDGMENTS
We note that the publisher of references 22 and 23 was first notified of the results reported here in November 2009.
This work was supported by Deutsche Forschungsgemeinschaft (KF0192) grants VI 218/4 and VI 218/2 (to U.V.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 16 May 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Adcock I. M. 2007. HDAC inhibitors as anti-inflammatory agents. Br. J. Pharmacol. 150:829–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ali S. H., DeCaprio J. A. 2001. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 11:15–23 [DOI] [PubMed] [Google Scholar]
- 3. Begitt A., Meyer T., van Rossum M., Vinkemeier U. 2000. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. U. S. A. 97:10418–10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buchwald M., Krämer O. H., Heinzel T. 2009. HDACi-targets beyond chromatin. Cancer Lett. 280:160–167 [DOI] [PubMed] [Google Scholar]
- 5. Chang H. M., et al. 2004. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc. Natl. Acad. Sci. U. S. A. 101:9578–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X., et al. 1998. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93:827–839 [DOI] [PubMed] [Google Scholar]
- 7. Cress W. D., Seto E. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1–16 [DOI] [PubMed] [Google Scholar]
- 8. Darnell J. E., Jr 2007. Interferon research: impact on understanding transcriptional control. Curr. Top. Microbiol. Immunol. 316:155–163 [DOI] [PubMed] [Google Scholar]
- 9. Decker T., Kovarik P., Meinke A. 1997. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 17:121–134 [DOI] [PubMed] [Google Scholar]
- 10. Dickinson M., Johnstone R. W., Prince H. M. 2010. Histone deacetylase inhibitors: potential targets responsible for their anti-cancer effect. Investig. New Drugs 28:S3–S20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dietschy T., et al. 2009. p300-mediated acetylation of the Rothmund-Thomson-syndrome gene product RECQL4 regulates its subcellular localization. J. Cell Sci. 122:1258–1267 [DOI] [PubMed] [Google Scholar]
- 12. Droescher M., Begitt A., Marg A., Zacharias M., Vinkemeier U. 2011. Cytokine-induced paracrystals prolong the activity of Signal Transducers and Activators of Transcription (STAT) and provide a model for the regulation of protein-solubility by Small Ubiquitin-like Modifier (SUMO). J. Biol. Chem. 286:18731–18746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drummond D. C., et al. 2005. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu. Rev. Pharmacol. Toxicol. 45:495–528 [DOI] [PubMed] [Google Scholar]
- 14. Faiola F., et al. 2007. Max is acetylated by p300 at several nuclear localization residues. Biochem. J. 403:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fu X. Y., Schindler C., Improta T., Aebersold R., Darnell J. E., Jr 1992. The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc. Natl. Acad. Sci. U. S. A. 89:7840–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo L., et al. 2007. Stat1 acetylation inhibits inducible nitric oxide synthase expression in interferon-gamma-treated RAW264.7 murine macrophages. Surgery 142:156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu X., Ivashkiv L. B. 2009. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity 31:539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khodarev N., et al. 2010. Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer. Oncogene 29:920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klampfer L., Huang J., Sasazuki T., Shirasawa S., Augenlicht L. 2003. Inhibition of interferon gamma signaling by the short chain fatty acid butyrate. Mol. Cancer Res. 1:855–862 [PubMed] [Google Scholar]
- 20. Klampfer L., Huang J., Swaby L. A., Augenlicht L. 2004. Requirement of histone deacetylase activity for signaling by STAT1. J. Biol. Chem. 279:30358–30368 [DOI] [PubMed] [Google Scholar]
- 21. Krämer O. H., Heinzel T. 2010. Phosphorylation-acetylation switch in the regulation of STAT1 signaling. Mol. Cell Endocrinol. 315:40–48 [DOI] [PubMed] [Google Scholar]
- 22. Krämer O. H., et al. 2009. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 23:223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krämer O. H., et al. 2006. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 20:473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levy D. E., Darnell J. E., Jr 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3:651–662 [DOI] [PubMed] [Google Scholar]
- 25. Li M., Luo J., Brooks C. L., Gu W. 2002. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277:50607–50611 [DOI] [PubMed] [Google Scholar]
- 26. McBride K., Banninger G., McDonald C., Reich N. 2002. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 21:1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McLaren J., Rowe M., Brennan P. 2007. Epstein-Barr virus induces a distinct form of DNA-bound STAT1 compared with that found in interferon-stimulated B lymphocytes. J. Gen. Virol. 88:1876–1886 [DOI] [PubMed] [Google Scholar]
- 28. Melén K., Kinnunen L., Julkunen I. 2001. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J. Biol. Chem. 276:16447–16455 [DOI] [PubMed] [Google Scholar]
- 29. Meyer T., Begitt A., Lödige I., van Rossum M., Vinkemeier U. 2002. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 21:344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyer T., Marg A., Lemke P., Wiesner B., Vinkemeier U. 2003. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev. 17:1992–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Müller M., et al. 1993. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 12:4221–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nardozzi J., Wenta N., Yasuhara N., Vinkemeier U., Cingolani G. 2010. Molecular basis for the recognition of phosphorylated STAT1 by importin alpha5. J. Mol. Biol. 402:83–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nusinzon I., Horvath C. M. 2003. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl. Acad. Sci. U. S. A. 100:14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pang M., et al. 2009. Inhibition of histone deacetylase activity attenuates renal fibroblast activation and interstitial fibrosis in obstructive nephropathy. Am. J. Physiol. Renal Physiol. 297:F996–F1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pine R., Canova A., Schindler C. 1994. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN alpha and IFNgamma, and is likely to autoregulate the p91 gene. EMBO J. 13:158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roger T., et al. 2011. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood 117:1205–1217 [DOI] [PubMed] [Google Scholar]
- 37. Shuai K., Stark G. R., Kerr I. M., Darnell J. E., Jr 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 261:1744–1746 [DOI] [PubMed] [Google Scholar]
- 38. Spange S., Wagner T., Heinzel T., Krämer O. H. 2009. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 41:185–198 [DOI] [PubMed] [Google Scholar]
- 39. Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264 [DOI] [PubMed] [Google Scholar]
- 40. Tang X., et al. 2007. Acetylation-dependent signal transduction for type I interferon receptor. Cell 131:93–105 [DOI] [PubMed] [Google Scholar]
- 41. ten Hoeve J., et al. 2002. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 22:5662–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vinkemeier U. 2004. Getting the message across, STAT! Design principles of a molecular signaling circuit. J. Cell Biol. 167:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wenta N., Strauss H., Meyer S., Vinkemeier U. 2008. Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc. Natl. Acad. Sci. U. S. A. 105:9238–9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yao Y. L., Yang W. M. 2011. Beyond histone and deacetylase: an overview of cytoplasmic histone deacetylases and their nonhistone substrates. J. Biomed. Biotechnol. 2011:146493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.