Abstract
Activated Syk, an essential tyrosine kinase in B cell signaling, interacts with Vav guanine nucleotide exchange factors and regulates Vav activity through tyrosine phosphorylation. The Vav SH2 domain binds Syk linker B by an unusual recognition of two closely spaced Syk tyrosines: Y342 and Y346. The binding affinity is highest when both Y342 and Y346 are phosphorylated. An investigation in B cells of the dependence of Vav phosphorylation and NFAT activation on phosphorylation of Y342 and Y346 finds that cellular response levels match the relative binding affinities of the Vav1 SH2 domain for singly and doubly phosphorylated linker B peptides. This key result suggests that the uncommon recognition determinant of these two closely spaced tyrosines is a limiting factor in signaling. Interestingly, differences in affinities for binding singly and doubly phosphorylated peptides are reflected in the on rate, not the off rate. Such a control mechanism would be highly effective for regulating binding among competing Syk binding partners. The nuclear magnetic resonance (NMR) structure of Vav1 SH2 in complex with a doubly phosphorylated linker B peptide reveals diverse conformations associated with the unusual SH2 recognition of two phosphotyrosines. NMR relaxation indicates compensatory changes in loop fluctuations upon binding, with implications for nonphosphotyrosine interactions of Vav1 SH2.
INTRODUCTION
Syk is a cytoplasmic protein tyrosine kinase that regulates cellular responses mediated by a variety of membrane receptors with intracellular signaling modules known as immunoreceptor tyrosine-based activation motifs (ITAMs) (23, 52). Examples of ITAM/Syk receptors include the B and T cell receptors for antigen, the immunoglobulin receptors FcεRI, FcγRI, and FcγRIIa, the activating NK cell receptors, and integrins. When these receptors are engaged, a pair of tyrosines within the ITAM become phosphorylated to create a docking site for Syk's N-terminal pair of SH2 domains. Syk binding to the ITAM leads to Syk activation and phosphorylation on several tyrosines to generate sites that participate in protein-protein interactions. Thus, activated Syk at the site of the clustered receptor participates in the formation of a signaling complex. This “signalosome” contains multiple effector proteins that include members of the Vav family of guanine nucleotide exchange factors (GEFs) (13, 14).
Among the three members of the Vav family, Vav1 is restricted largely to hematopoietic cells while Vav2 and Vav3 are more widely distributed (8, 55, 67, 77). Vav proteins have both unique and redundant functions in the immune system, as a loss of Vav1 produces severe defects in T cell development and signaling, while the loss of more than one Vav family member is needed to produce similar defects in B cells (12, 17, 21, 75, 76). Vav proteins enhance many signaling pathways mediated by Syk-associated receptors, including the activation of phospholipase C-γ (PLC-γ) and the mobilization of intracellular calcium, the activation of phosphoinositide 3-kinase (PI3K) and Akt, the activation of the Ras/Erk pathway, the promotion of cytoskeletal rearrangements, and the inside-out activation of integrins. While Vav proteins serve as GEFs for Rac/Rho family GTPases, they also act as scaffolds, a function that promotes the assembly of signaling complexes but does not require GEF activity (66).
Syk associates with Vav family proteins and phosphorylates them on tyrosines that regulate their activity (13, 43, 50, 53, 59, 69, 74, 75). This physical association occurs between the Vav SH2 domain and the Syk linker B region, which separates the tandem SH2 domains from the catalytic domain and contains phosphotyrosines 342 and 346 (based on the numbering system for murine Syk). Syk linker B interacts with multiple binding partners depending on which tyrosines in linker B are phosphorylated and the stoichiometry of this phosphorylation (23). As such, Syk linker B harbors the unusual site of two closely space tyrosines, which are recognized by a single SH2 domain of particular Syk binding partners. These interactions influence the ability of the kinase to couple ITAM-bearing receptors to the many different intracellular signaling pathways that are regulated following receptor engagement (23, 52).
Vav1 SH2 adopts the typical fold of an SH2 domain, including a central β-sheet flanked by two α-helices (Protein Data Bank [PDB] designation: 2CRH). The nonaromatic hydrophobic Ile residue at the βD5 position indicates that Vav1 SH2 is in the new group IIA, defined from the most recent SH2 classification based on phosphopeptide library screening (30). SH2 domains in this subgroup have a loose preference for the motif pTyr-hydrophobic amino acid-X-hydrophobic amino acid (where pTyr is phosphotyrosine and X is any amino acid). The Syk linker B sequences Y342ESP and Y346ADP fit the specificity of Vav1 for either pY342 or pY346 binding of the canonical pocket. The hydrophobic Pro at the pTyr +3 position is a specificity determinant according to peptide library studies (30). Furthermore, the binding site sequences from the proteins known to interact with Vav1 SH2 show that proline is the most preferred amino acid at the pTyr +3 position (7, 10, 24, 32, 33, 38, 46). Glu is predicted from phosphopeptide screening to be the second most favored residue at the pTyr +1 position for Vav1 SH2 (30) and appears most frequently in other binding partners (7, 10, 24, 33, 38). Pro occurs at both pY342 +3 and pY346 +3 positions. The pTyr +1 amino acid is Glu for Y342 but for Y346 is Ala, which is neither selected in the screening nor found in any binding partner although it is not an excluded amino acid. This simple sequence analysis therefore predicts that pY342 has a higher affinity for the canonical pTyr binding site of Vav1 SH2; however, pY346 binding in this site cannot be ruled out.
In this study, we examined the interaction between Vav1 and Syk. Of particular interest is the dependence of signaling through the B cell antigen receptor (BCR) on linker B tyrosines and, on a molecular level, how two closely spaced phosphotyrosines are recognized. Importantly, we found that binding affinity correlates with cellular response levels in signaling. Determination of the solution structure by NMR of the Vav1 SH2 domain in complex with a peptide derived from linker B containing both phosphotyrosines indicated that both pY342 and pY346 are recognized simultaneously by a single Vav1 SH2 domain, with pY346 accommodated within a second, lysine-rich binding pocket. Conformational flexibility of the second pocket in the unligated state of Vav1 SH2 likely facilitates recognition of alternative phosphotyrosine sites. The molecular recognition mechanism of two closely spaced phosphotyrosines and its relationship with cellular regulation are discussed.
MATERIALS AND METHODS
NFAT-luciferase reporter assay.
The DT40 chicken B cell lines (73) were provided by T. Kurosaki (Kansai Medical University) and E. Puré (Wistar Institute). Cells were cultured in RPMI 1640 medium supplemented with 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol, 7.5% heat-inactivated fetal bovine serum (FBS), 50 U/ml penicillin, 50 μg/ml streptomycin, and 1% heat-inactivated chicken serum. Cells were transfected by electroporation with 10 μg of the luciferase reporter plasmid pNFAT-luc (Stratagene) and 20 μg of an expression plasmid coding for a form of Syk-enhanced green fluorescent protein (EGFP) (82) and allowed to recover overnight. For overexpression of Vav1, cells were transfected with 20 μg of a plasmid coding for Vav1-EGFP (Addgene; plasmid 14557) (78). Cells were activated using anti-IgM antibodies or a combination of 0.1 μg/ml phorbol myristate acetate (PMA) and 0.75 μg/ml ionomycin as an internal control and incubated at 37°C for 6 h. Lysates were combined with luciferin substrate, and luciferase activity of the reporter gene was quantified on a luminometer using the luciferase assay system kit (Promega). The values reported indicate the activity resulting from anti-IgM activation divided by that resulting from treatment with PMA plus ionomycin to correct for transfection efficiency. In some experiments, cells were cotransfected with an internal control plasmid, pRL-TK, expressing renilla luciferase, luciferase activity was measured using the dual reporter assay system kit (Promega), and the data represent firefly luciferase units divided by renilla luciferase units. All data represent the averages and standard errors of a minimum of three trials. Protein expression levels were determined by Western blotting using antibodies against Syk (N-19; Santa Cruz) or Vav1 (ZV003; Invitrogen).
Immunoprecipitations and Western blotting.
Cells were lysed in NP-40 lysis buffer (1% NP-40, 100 mM sodium chloride, 5 mM EDTA, 50 mM sodium fluoride, 1 mM sodium orthovanadate, 20 μg/ml leupeptin, and 20 μg/ml aprotinin in 50 mM Tris-HCl, pH 8.0). For immunoprecipitations, cleared lysates were incubated with immobilized GFP-binding protein (GBP; Chromotek) on ice for 15 min. Bound proteins were separated by SDS-PAGE, transferred to membranes, and analyzed by Western blotting using antibodies to Syk, Vav, or phosphotyrosine (4G10; Millipore). Peptide or phosphopeptide pulldown assays were conducted using lysates prepared from human DG75 B cells (ATCC) as described previously (25).
Protein purification and sample preparation.
A pETTEV411 expression vector derived from pET41a (Novagen), provided by Etti Harms, was used to carry the sequence encoding Vav1 SH2 (residues 664 to 767) and was transformed into Echerichia coli BL21(DE3) cells. Glutathione S-transferase (GST)-tagged Vav1 SH2 protein was expressed at 20°C in M9 minimal medium containing 13C/15N isotopes. The fusion protein was initially purified with a glutathione-agarose column. Protein was further digested with AcTEV protease (Invitrogen), and GST was removed by a HisTrap HP affinity column (GE Healthcare).
The high-purity (>98%) pYpY peptide was obtained from EZBiolab. The peptide sequence is DTEVpYESPpYADPE, with N-terminal acetylation and C-terminal amidation modifications. Lyophilized peptide was dissolved in water and dialyzed in the same buffer as that for Vav1 SH2 protein. The purity of the peptide was confirmed by mass spectrometry.
The concentration of Vav1 SH2 protein in 20 mM Tris-HCl (pH 7.0), 100 mM NaCl, 1 mM dithiothreitol (DTT), and 0.02% NaN3 was determined by UV spectroscopy using an extinction coefficient of 11,460 M−1 cm−1 at 280 nm. The unlabeled pYpY peptide was titrated into 13C/15N-labeled Vav1 SH2 protein to form a 1:1 peptide-protein complex. The nuclear magnetic resonance (NMR) sample of 1.1 mM Vav1 SH2 with 10% D2O was placed in a Shigemi tube for NMR measurement.
NMR experiments.
NMR experiments were performed at 298 K either on a Bruker Avance-III-800 equipped with a Z-gradient TXI probe or a Bruker DRX500 equipped with a cryoprobe. NMR data were processed with NMRPipe (15), and spectra were visualized and analyzed with Sparky (T. D. Goddard and D. G. Kneller, University of California, San Francisco).
CBCA(CO)NH, HNCACB, HNCA, and HN(CO)CA were collected for backbone resonance assignments. Initial assignments were made using the program MARS (35) and then manually checked. Aliphatic side chain assignments were made by analyzing C(CO)NH-TOCSY, H(CCO)NH-TOCSY, TOCSY-HSQC, HBHANH, H(C)CH-TOCSY, and (H)CCH-TOCSY. Aromatic assignments were made using HBCBCGCDHD, HBCBCGCDCEHE, and 13C-HSQC (81). Peptide resonances were assigned using two-dimensional (2D) doubled-filtered TOCSY and NOESY experiments (60). Intraprotein distance constraints were obtained from simultaneous 15N/13C-NOESY-HSQC (120-ms mixing time) and aromatic 13C-NOESY-HSQC (120-ms mixing time). Intermolecular distance restraints were measured using a three-dimensional (3D) 13C/15N-filtered 13C-edited NOESY-HSQC (200-ms mixing time) (60).
An alignment medium for residual dipolar coupling (RDC) experiments was prepared using a polyethylene glycol-alcohol mixture of pentaethylene glycol monododecyl ether (C12E5) and hexanol at a molar ratio of 0.96 (64). In-phase and antiphase spectra in 2D 1H-15N-IPAP-HSQC were recorded in an interleaved manner to measure both isotropic and partially aligned samples (62). The backbone 1H-15N RDCs were obtained from the frequency difference between the 15N splittings in the isotropic and partially aligned states.
Structure calculations.
Protein structure was calculated by molecular dynamics simulation using nuclear Overhauser effect (NOE) distance and dihedral angle restraints in torsion angle space. NOE cross peaks in simultaneous 15N/13C-NOESY-HSQC and aromatic 13C-NOESY-HSQC were manually picked, and intraprotein NOEs were assigned automatically using CYANA 2.1 (27, 28). ϕ and ψ dihedral angle restraints were predicted using TALOS based on the chemical shifts of Cα, Cβ, Hα, and HN (11). Iterative assignments and structure calculations of CYANA yielded 2,947 NOE upper distance restraints and a set of Vav1 SH2 structures with the backbone root mean square deviation (RMSD) of 0.78 Å (residues 664 to 765).
Fifty-six NOE resonances in 3D (13C/15N)-filtered 13C-edited NOESY-HSQC were manually assigned and converted to the intermolecular distance restraints between pYpY peptide and the Vav1 SH2 domain using CYANA macro calibration. These restraints were categorized as very weak, weak, medium, or strong for structure calculations. Peptide coordinates were generated for an extended strand, and the peptide was placed ∼12 Å from the Vav1 SH2 ligand binding surface to initiate the structure calculations. The complex structures were calculated using a standard simulated annealing refinement protocol with XPLOR-NIH (68) and further refined using gentle simulated annealing refinement, which included a force field with full van der Waals and electrostatic interactions. The 20 final structures were selected based on the absence of any NOE violation, small total energy values, and correct geometry. The quality of the final structures was assessed using PROCHECK-NMR (42). The structure figures were generated using the program PyMOL.
15N relaxation and model-free analysis.
NMR relaxation experiments were executed at 298 K on a Bruker DRX500. R1 data were recorded with relaxation delays of 65, 145, 245, 366, 523, 755, and 1,120 ms, and R2 data were acquired using relaxation delays of 18, 35, 53, 70, 88, 106, and 141 ms. To estimate the uncertainties, duplicate spectra were measured for the delays of 245 and 523 ms in R1 experiments and the delays of 18 and 70 ms in R2 experiments. 1H-15N steady-state NOE values were determined by collecting spectra with or without an initial 3-s proton saturation pulse (20). The model-free analysis was carried out using Mathematica scripts provided by L. Spyracopoulos (72). The final calculated three-dimensional structures of free Vav1 SH2 and the pYpY-bound complex structure were used to determine axially symmetric diffusion tensor parameters.
Chemical shift perturbations.
The unlabeled pYpY peptide was titrated into the 0.2 mM 15N-labeled Vav1 SH2 protein until the protein was fully saturated, as determined by monitoring changes in the 1H-15N HSQC spectra. The final SH2/pYpY molar ratio was 1:4.7.
Chemical shift perturbations (CSP) are specified by the equation
| (1) |
where ΔδH and ΔδN are the chemical shift differences between the unbound and peptide-bound states in the proton and nitrogen dimensions, respectively.
Surface plasmon resonance.
Surface plasmon resonance (SPR) experiments were performed at 25°C on a BIAcore 3000 instrument. The Vav1 SH2 protein was dialyzed in the running buffer, which contains 20 mM Tris at pH 7.0, 100 mM NaCl, 1 mM DTT, 0.005% surfactant P20, and 0.02% NaN3. For N-biotinylated phosphopeptides (EZBiolab), two residues of 6-aminohexanoic acid were inserted between the biotin moiety and the N terminus of the peptide to increase the accessibility of phosphopeptides immobilized on the surface of sensor chip SA. One flow cell without immobilized peptide was used as a reference to correct for signals from bulk refractive index changes, instrument drift, nonspecific binding, and systematic artifacts (54). Unbound streptavidin on the reaction and reference surfaces was blocked with free biotin.
Proteins were injected at a flow rate of 20 μl/min for a period of 60 s to reach an equilibrium state at each concentration. To confirm that mass transport was not a limiting factor, certain response curves were also measured at faster flow rates up to 75 μl/min and were essentially identical to those at 20 μl/min. The dissociation of Vav1 SH2 was monitored under the same flow rate for 2 min. The response from the injection of running buffer was also measured for double referencing to remove systematic deviations (57). Regeneration between cycles was employed with the solution containing 0.05% SDS in the buffer at a flow rate of 10 μl/min for 1 min.
The program Scrubber 2.0, provided by David Myszka, was used for processing the SPR data and analyzing the binding interaction. The SPR equilibrium response (Req) is a function of injected protein concentration ([P]) described by the equation Req = Rmax/{1 + (KD/[P])}. A dissociation constant (KD) was extracted from curve fitting.
The association rate (ka) and dissociation rate (kd) were obtained by fitting the real-time binding response to the differential equations depicting changes of phosphopeptide-bound complex concentration (57). The dissociation constant KD was calculated as kd/ka.
RESULTS
Effect of Syk Y342 and Y346 phosphorylation on recognition by Vav and signaling. (i) Binding preference of Vav1 and PLC-γ for Syk linker B.
Binding preferences for Syk-interacting proteins were examined in pulldown assays in which proteins present in human DG75 B cell lysates were adsorbed to immobilized peptides matching the sequence of Syk between amino acids 338 and 353 and containing phosphotyrosines at position 342 (pYY), 346 (YpY), both 342 and 346 (pYpY), or neither position (YY). Bound proteins were detected by Western blotting. Vav1 bound to the pYY peptide and to the pYpY peptide, with the stronger interaction being with the doubly phosphorylated peptide (Fig. 1A). Vav1 did not bind appreciably to either the nonphosphorylated YY peptide or the YpY peptide, which contained phosphate only at Y346. This mode of interaction was in contrast to that observed for PLC-γ2, which showed selective binding for the pYpY peptide and minimal binding for either pYY or YpY. As reported previously, the binding characteristics of PLC-γ are conferred by its C-terminal SH2 domain (PLC-γ c-SH2) (25).
Fig. 1.
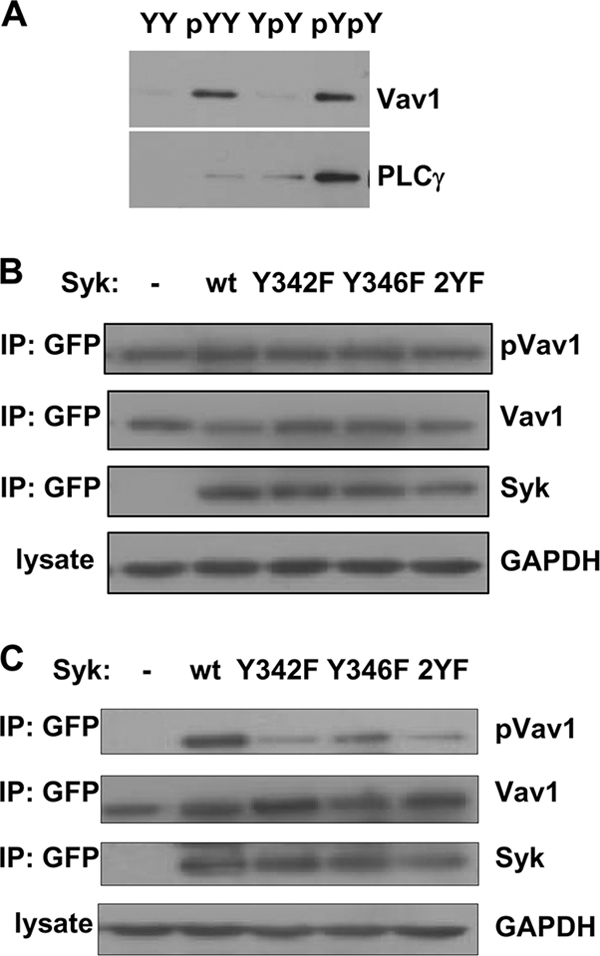
Syk phosphorylation on Y342 and Y346 regulates Vav1 phosphorylation. (A) Peptides corresponding in sequence to Syk residues 338 to 353 and containing no phosphotyrosines (YY), pY342 (pYY), pY346 (YpY), or both pY342 and pY346 (pYpY) were immobilized on a solid support and incubated with detergent lysates generated from DG75 B cells. Proteins bound to each resin were analyzed by Western blotting with antibodies against Vav1 or PLC-γ2. (B) Syk-deficient DT40 cells were transfected with plasmids encoding Vav1-EGFP and either a mutant of Syk-EGFP or EGFP vector control. Cells were stimulated for 5 min with 5 μg anti-IgM. GFP-tagged proteins were immunoprecipitated (IP) with GBP-Sepharose and analyzed by immunoblotting. wt, wild type; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (C) Syk/Lyn double knockout DT40 cells were transfected, stimulated, and analyzed as described for panel B.
(ii) Dependence of Vav1 phosphorylation on Syk linker B tyrosines.
The interaction of Vav with active Syk leads to Vav phosphorylation on tyrosine and the activation of its GEF activity (13, 43, 50, 53, 59, 69, 74, 75). We examined how the different linker B phosphotyrosines influence this interaction by taking advantage of the availability of DT40 B cells engineered to lack endogenous Syk (73). We transfected Syk-deficient DT40 cells with expression vectors coding for Vav1 and for various forms of Syk in which either Y342 or Y346 or both in linker B were replaced by phenylalanines. Both proteins had enhanced green fluorescent protein tags at their C termini. Cells were activated by cross-linking the B cell antigen receptor (BCR) using antibodies against surface IgM. Vav1-EGFP was recovered from cell lysates using immobilized single-chain GFP antibodies and examined for extent of tyrosine phosphorylation by Western blotting with antiphosphotyrosine antibodies. As shown in Fig. 1B, receptor engagement led to the phosphorylation of Vav1 on tyrosine in all cells, including those that lacked Syk. Thus, Syk is not the only kinase in DT40 cells that can contribute to the phosphorylation of Vav1 on tyrosine. Lyn, which is the only Src family kinase that is expressed in wild-type DT40 cells at a detectable level (73), has been described as a Vav1 kinase (22) and is a likely candidate.
To examine the Syk-dependent phosphorylation of Vav1, we turned to DT40 cells that lack both Syk and Lyn. In these cells, exogenously expressed Vav1-EGFP was not phosphorylated on tyrosine in response to anti-IgM (Fig. 1C). However, the expression of Syk-EGFP restored receptor-mediated phosphorylation of Vav1 to the double-knockout cells, allowing us to measure selectively the contribution of Syk to the phosphorylation of Vav1. We expressed the various linker B mutants of Syk-EGFP in the Syk- and Lyn-deficient cells and activated these by cross-linking the BCR. The extent of phosphorylation of Vav1 was highly dependent on the presence of specific linker B tyrosines (Fig. 1C). Phosphorylation was highest in cells expressing the wild-type kinase. The replacement of Y342 by phenylalanine substantially attenuated Vav1 phosphorylation, while the replacement of Y346 reduced phosphorylation by a more modest amount. The greatest reduction in phosphorylation was observed when a form of Syk that lacked both Y342 and Y346 was expressed. Thus, the ability of various forms of Syk to phosphorylate Vav1 in DT40 cells correlated well with the ability of Vav1 in cell lysates to bind to the linker B phosphopeptides.
(iii) Dependence of NFAT activation on Syk linker B tyrosines.
We then asked if the dependence of Vav1 binding and phosphorylation on linker B tyrosines extended to signaling through the BCR. Vav proteins are known to be critical components of the receptor-mediated mobilization of calcium leading to the activation of the transcription factor NFAT (13, 37, 76, 79). Consequently, we measured NFAT activity in Syk- and Lyn-deficient cells transfected to express the various Syk linker B mutants. BCR-stimulated signaling was most robust in cells expressing the wild-type kinase that contained both Y342 and Y346 (Fig. 2A). The inability to phosphorylate Y346 because of replacement by Phe led to a moderate decrease in signaling, while the same replacement of Y342 led to a more substantial deficit. Signaling was dampened most dramatically by replacement of both Y342 and Y346 with phenylalanines. Since chicken Vav3 is the principal endogenous Vav isoform in DT40 cells, we repeated the experiment in cells transfected to overexpress human Vav1-EGFP along with the various Syk mutants. A similar pattern of signaling was observed in cells expressing the exogenous, tagged Vav1. These results indicate that the site or sites on Syk that become phosphorylated play a critical role in the ability of the kinase to couple the BCR both to the phosphorylation of Vav and to the activation of NFAT.
Fig. 2.
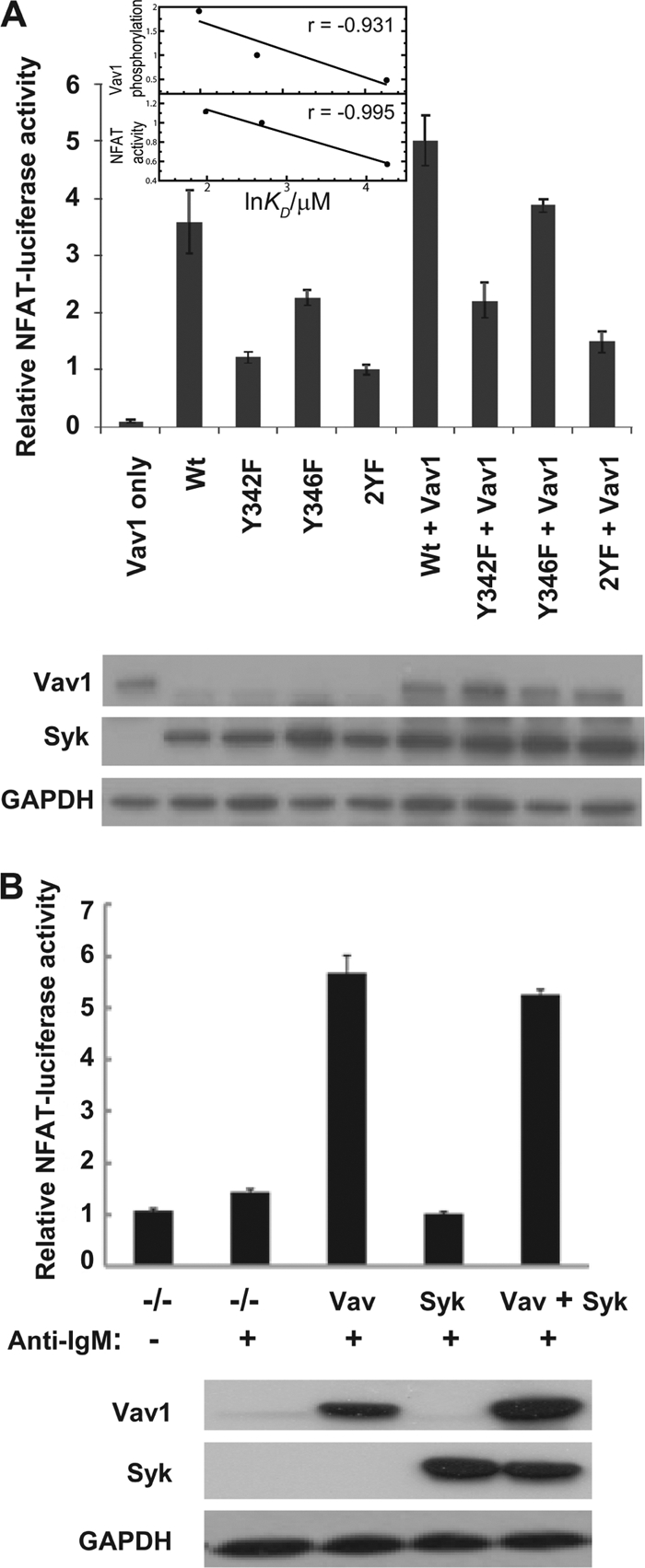
(A) Syk phosphorylation on Y342 and Y346 regulates NFAT activation. Syk/Lyn double knockout DT40 cells were transfected with an NFAT-luciferase reporter plasmid and Vav1-EGFP or Syk-EGFP mutants as labeled. Cells were stimulated for 6 h with 5 μg anti-IgM or with PMA plus ionomycin as an internal control. The plotted luciferase activity measured from luminescence is a ratio of luciferase units obtained following anti-IgM stimulation divided by luciferase units in the presence of PMA plus ionomycin. Protein expression levels were confirmed by Western blotting with antibodies against each protein and are shown in the gels. (Inset) Vav1 SH2 binding affinity for Syk linker B peptides (SPR data from Table 1) correlated with cellular Vav1 phosphorylation (data from Fig. 1) (top) and correlated with cellular NFAT activity (data from this figure) (bottom). Wt, wild type. (B) Dependence of NFAT activation on the expression of Vav. Vav-deficient DT40 B cells were transfected with an NFAT-luciferase expression plasmid and an internal control plasmid, pRL-TK, in the absence (basal; −/−) or presence of plasmids expressing Vav1 (Vav), wild-type Syk-EGFP (Syk), or both (Vav + Syk). Cells were left untreated (basal) or were stimulated with anti-IgM antibodies, and luciferase activity of the reporter genes was measured after 6 h. The expression of Vav1 and Syk-EGFP (Syk) was confirmed by Western blotting with antibodies against each protein (bottom). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected as a loading control.
To confirm a requirement for Vav for BCR signaling to NFAT in DT40 cells, we examined cells engineered to lack endogenous Vav3 (31). BCR engagement with anti-IgM failed to stimulate NFAT activity above that observed in unstimulated cells (Fig. 2B). Signaling was not increased by the expression of exogenous Syk-EGFP but was restored by the expression of Vav1.
(iv) Binding affinities of Vav1 SH2 for Syk linker B.
To better characterize and quantify the contribution of the linker B phosphotyrosines to the Vav1-Syk interaction, we used surface plasmon resonance (SPR) methods to measure the affinity of Vav1 SH2 for binding peptides derived from linker B in alternatively phosphorylated forms: pYpY (DTEVpYESPpYADPE), pYY (DTEVpYESPYADPE), and YpY (DTEVYESPpYADPE) peptides. The dissociation constants for Vav1 SH2 binding to the phosphopeptides were determined from the SPR sensorgrams. Equilibrium constants from nonlinear fitting of the binding curves agreed well with the kinetic constants obtained from the time dependence of the response (Table 1). The sensorgram data and fitted curves, as well as plots for fitting to equilibrium binding curves, are available upon request. The KD values are 7.27 ± 0.04 μM (pYpY), 14.7 ± 0.1 μM (pYY), and 70.9 ± 0.4 μM (YpY) from equilibrium SPR analysis (Table 1). While the dissociation rates are similar for all phosphopeptides, the association rates are the principal cause of differences in the binding affinities. The 2-fold difference between pYpY and pYY interactions suggests that the phosphorylation of Y346 contributes limited binding energy in the pYpY complex. In contrast, YpY has a 1 order of magnitude lower affinity than pYpY. Together these effects on binding affinity indicate that the primary energetic contribution to the interaction is from pY342, as expected for SH2 domain association in general (4).
Table 1.
Kinetic rate constants and binding affinities measured by SPR
| Rate or constant | Valuec for: |
||
|---|---|---|---|
| pYpY | pYY | YpY | |
| ka (103 M−1 s−1)a | 97 (±3) | 47 (±2) | 5.6 (±0.2) |
| kd (s−1)a | 0.71 (±0.02) | 0.68 (±0.02) | 0.4 (±0.01) |
| KD (μM)b | 7.4 (±0.4)/7.27 (±0.04) | 14.6 (±0.7)/14.7 (±0.1) | 71 (±4)/70.9 (±0.4) |
Values were determined by kinetic analysis of the time-dependent SPR response data.
The first value is kd/ka; the second was determined from the SPR response values in the steady state.
Values in parentheses are standard deviations from multiple experiments.
The relative binding affinities of Vav1 SH2 for Syk-derived phosphopeptides match the signaling response profile in B cells. Vav1 SH2 binds with highest affinity to pYpY, and the expression of wild-type Syk results in the highest levels of Vav1 phosphorylation and NFAT activation (Table 1; Fig. 1 and 2). Cells expressing the Y346F variant, analogous to pYY, have an approximate 2-fold reduction in Vav1 phosphorylation and lower NFAT activity. This reduced level of phosphorylated Vav1 matches the 2-fold decrease in the binding affinity of Vav1 SH2 for pYY. Vav1 SH2 has lowest affinity for YpY, the analogue of the Y342F variant, and correspondingly the least amounts of Vav1 phosphorylation and NFAT activation are observed with expression of the Y342F variant. A quantitative analysis of the data yields a correlation coefficient between ln KD and the level of Vav1 phosphorylation or NFAT activation of −0.931 or −0.995, respectively (Fig. 2A, inset). This correlation suggests that modulating the binding affinity of Vav SH2 by alternative forms of Syk 342/346 phosphorylation is significant for controlling the phosphorylation level of Vav1 and that linker B phosphorylation contributes to the regulation of NFAT activity.
Structural basis of the Syk-Vav1 interaction.
We determined the NMR structure of Vav1 SH2 in complex with Syk-derived doubly phosphorylated peptide pYpY (DTEVpYESPpYADPE) to investigate the molecular mechanism for its association with the phosphorylated Syk linker B and structural characteristics in the recognition of dual phosphotyrosines. Comparison of this Vav1 SH2-pYpY complex with the structures of free Vav1 SH2 (PDB designation: 2CRH) and Vav1 SH2 in complex with a singly phosphorylated peptide, pY128 (GEDDGDpYESPNEEEE), derived from SLP-76 (PDB designation: 2ROR) reveals the structural variation involved with binding alternative phosphorylation forms and the conformational changes for recognition of the second phosphotyrosine by Vav1 SH2.
(i) NMR structure calculation.
Chemical shifts of Vav1 SH2 in the pYpY-bound state were assigned for all backbone amide and Cα atoms and aliphatic side chain atoms, except the two residues at the N terminus and seven residues in the BC loop (see Fig. 3B for labeling of SH2 domain elements) for which resonances were not observed. Aromatic resonances were assigned for only 66% of the Hδ/Cδ and Hε/Cε due to the intrinsic insensitivity of the experiments (81). Unobserved resonances for atoms in the BC loop suggest segmental motions and flexibility in this region. Disorder in the BC loop has been reported in other NMR and X-ray structural studies on SH2 complexes (48, 58).
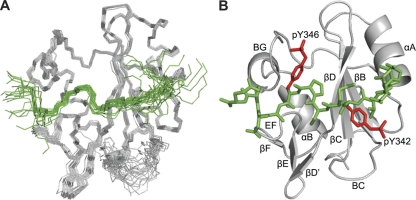
Fig. 3.
Solution structure of Vav1 SH2 in complex with pYpY. The protein is gray, and the peptide is green. (A) Main chain backbone trace for SH2 and pYpY from overlay of the 20 final structures. Structures were aligned by the N, Cα, and C′ coordinates of residues 664 to 697 and 706 to 767 of the Vav1 SH2 domain, excluding the disordered first three residues and BC loop. (B) Ribbon diagram of the energy-minimized average structure. The peptide heavy atoms are shown as vectors, and the side chain heavy atoms for the pTyr residues are highlighted in red. The conventional nomenclature of secondary structural elements and loop regions is used (18). The figure was drawn using the PyMOL molecular graphics system, version 1.3 (Schrödinger, LLC).
3D structures of the Vav1 SH2-pYpY complex were determined on the basis of proton-proton distance restraints obtained from the NMR nuclear Overhauser effects (NOEs). The NOESY assignments and structure determination of the Vav1 SH2 domain were conducted using the program CYANA 2.1 (27, 28). A total of 2,947 NOE distance restraints (1,108 long-range, 560 medium-range, and 1,279 short-range NOEs) were obtained after seven cycles of iterative automated assignment. Additional NOEs between 13C/15N-labeled Vav1 SH2 and unlabeled pYpY were manually assigned and provided 56 intermolecular distance restraints for the final complex structure calculation. Additional constraints for 126 dihedral angles in β-strand or α-helical regions were obtained from the TALOS method (11). Structural statistics for 20 final structures are shown in Table 2.
Table 2.
Structural statistics of 20 final structures
| Parameter | Value |
|---|---|
| NOE distance restraints | |
| Protein NOE | |
| Total | 2,947 |
| Short range (|i − j| ≤ 1) | 1,279 |
| Medium range (1 < |i − j| < 5) | 560 |
| Long range (|i − j| ≥ 5) | 1,108 |
| Intermolecular NOE | 56 |
| Dihedral angle restraints from TALOS | 126 |
| RMSD to mean coordinates (Å) (backbone/heavy atoms) | |
| Complexa | 1.31 ± 0.25/1.70 ± 0.23 |
| SH2 domain residues 664–697 and 706–767 | 0.40 ± 0.09/0.78 ± 0.10 |
| Peptide residues 341–347 | 0.87 ± 0.25/1.21 ± 0.29 |
| Ramachandran plot (%) | |
| Most-favored regions | 71.6 |
| Additionally allowed regions | 20.6 |
| Generously allowed regions | 7.8 |
| Disallowed regions | 0 |
Vav1 SH2 residues 661 to 767 and pYpY residues 338 to 350.
(ii) Structural quality of SH2.
The final structures of the Vav1 SH2-pYpY complex have high precision and satisfy the experimental restraints without NOE violations. The RMSDs for the ordered regions in the Vav1 SH2 domain (residues 664 to 697 and 706 to 767) are 0.40 ± 0.09 Å for the backbone and 0.78 ± 0.10 Å for heavy atoms of the SH2 domain. The RMSDs for the structured region of the pYpY peptide (residues 341 to 347) are 0.87 ± 0.25 Å for the backbone and 1.21 ± 0.29 Å for heavy atoms. As expected, the structure ensemble exhibits disordered conformations in the BC loop and the N and C termini of pYpY because of the lack of structural restraints (Fig. 3A). Ramachandran plots of the backbone dihedral Φ and Ψ angles demonstrate that none of these angles fall in a disallowed region. Residual dipolar couplings (RDCs), which measure the orientation of NH bond vectors relative to a global framework, were not used in the structure calculations and thus provide an independent assessment of the accuracy of the NMR structure. The experimental RDCs were compared to RDCs back-calculated from the energy-minimized average structure using singular value decomposition in the program PALES (83). The observed RDCs agree well with back-calculated RDCs and give a correlation coefficient (R) of 0.82 (data available upon request) (2).
(iii) SH2-pYpY interaction.
The superposition of the 20 final structures and the average structure of Vav1 SH2 in complex with pYpY are shown in Fig. 3A and B, respectively. The structure in most regions of the Vav1 SH2-pYpY complex is well defined, which allows a detailed analysis of the Vav1-Syk interactions. pY342 occupies the canonical phosphotyrosine binding site, with the pYpY peptide lying approximately perpendicular to the central β-sheet of Vav1 SH2 and displaying a two-pronged binding mode for recognition of the two closely spaced phosphotyrosines (Fig. 3B). The two prongs, pY342 and pY346, are separately located in the N- or C-terminal half of Vav1 SH2, respectively, and insert into binding pockets created by surrounding residues. The intermediate residues cross the central β-strand, and the main chain bends to accommodate the interactions in the specificity-determining region in the C-terminal half of the Vav1 SH2 domain. The locations of pY342, pY346, and proline at the pY342 +3 position are well defined by more than 11 intermolecular NOEs per residue. Even though fewer NOEs are observed for E340 (4 NOEs), V341 (4 NOEs), and A347 (7 NOEs), the structure determined for these residues is expected to be reasonably accurate (19). The residues beyond the pY342 +5 position are poorly defined due to the lack of NOE restraints. The residues at the bend, E343 and S344, make little contact with the protein, a result that is supported by the fact that only one intermolecular NOE between S344 and I718 (βD5) is observed for these two residues.
In the canonical pTyr binding site, pY342 interacts with a positively charged region formed by R678 (αA2), R696 (βB5), R698, and K719 (βD6) (Fig. 4A). The guanidinium group of the universally conserved R696 (βB5) directly interacts with the phosphate group of pY342. In the energy-minimized average structure, pY342 interacts with the highly conserved R698 as a result of the molecular mechanics force field given that the resonances, and thus NOEs, for R698 in the BC loop are not observed.
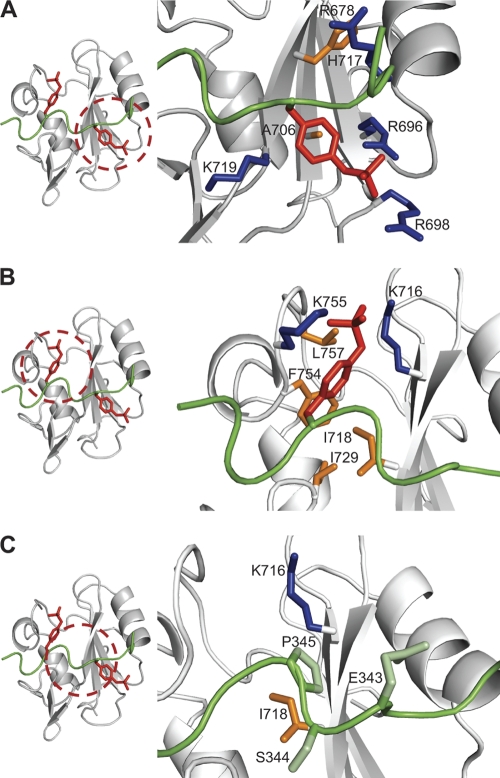
Fig. 4.
Detailed view of the interactions between pYpY and Vav1 SH2. The structures are shown in ribbon representations for SH2 (gray) and pYpY (green). The important arginines and lysines in the protein are blue, and the other highlighted SH2 side chains are orange. Shown are interactions for pYpY side chains pY342 in the primary pTyr binding pocket (A) (red) and pY346 in the secondary pTyr binding pocket (B) (red) and between the two pTyrs, E343, S344, and P345 (C) (pale green). The locations of interaction sites are indicated by red dashed circles.
An unusual aspect of linker B association is recognition of a second phosphotyrosine; pY346 inserts in a binding pocket created by the βD strand, EF loop, and BG loop (Fig. 4B). The negatively charged phosphate group of pY346 interacts with the ε-amino groups of K716 (βD3) and K755 (BG7) on opposite sides of the binding site. As well as the electrostatic complementarity, binding involves recognition of the aromatic ring of pY346 and the cyclic side chain of P345 on the hydrophobic surface. NOE interactions for P345 with the side chains of K716 (βD3) and I718 (βD5) were observed (Fig. 4C). Interactions with the βD strand generally occur with the first residue C-terminal to the pTyr in other SH2-phosphopeptide complexes (6, 56, 58, 61, 63, 80). As noted above for Vav1 SH2, E343 and S344 straddle the central β-sheet with limited contact, while P345 is drawn closer toward the βD strand to allow pY346 binding in the second pocket.
(iv) Chemical shift perturbation and SH2 conformational changes upon binding.
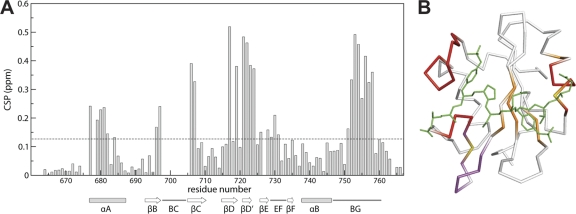
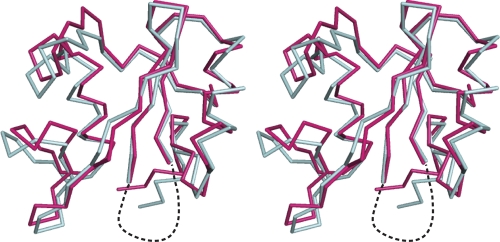
Structural effects on Vav1 SH2 upon binding pYpY are reflected by perturbations in the 15N-HSQC resonance positions that result from either direct contact with pYpY or a remote conformational change of the protein. As shown in Fig. 5A, several residues exhibit significant chemical shift perturbations (CSP; equation 1). The average CSP value is 0.13 ppm, and considerably larger values are observed for residues that interact with pY342 in the primary pTyr binding pocket: residues R696 (βB5), Q697, A706 (βC2), I707, H717 (βD4), and K719 in the central β-sheet and residues E677 and A679 to E682 in the N terminus of the αA helix. It is likely that R678 (αA2) also has a large CSP value; however, the resonance of this residue is not observed in the unbound state due to motional broadening. Relatively larger CSP values also occur for resonances from residues that contact pYpY in the EF loop (I729 [EF1] to E731) and BG loop (K751 [BG3] to D758). Residues in the βD′ and βE strands—M721 (βD′1) to R728 (βE3) excluding Y727 (βE2) (also exchange broadened in the free state)—have no contact with pYpY but have large CSP values, implying a change in the conformation of this small β-sheet upon binding. The change in conformation suggested by large CSP values is confirmed by a comparison of the pYpY complex structure with the unligated structure reported previously (PDB designation: 2CRH). From the overlay of main chain atoms from the unligated structure of Vav1 SH2 and the pYpY complex (Fig. 6), it is evident that this region is altered upon binding. Comparison also finds that the BG and EF loops in contact with pYpY differ in the two structures; the BG loop, including the single-turn helix, is displaced in a manner that expands the specificity pocket, and the EF loop rearrangement is likely a result of a salt bridge between residue K755 and the pYpY residue pY346. A smaller change occurs in the N terminus of αA, where pY342 interacts in the primary pTry pocket. These structural effects from CSP are mapped onto the structure (Fig. 5B). Therefore, the association with the doubly phosphorylated pYpY peptide induces reorientation of the BG and EF loops, as well as the small β-sheet connected to the EF loop, for recognition of the secondary pY346, while binding pY342 in the primary pocket, which typically contributes the major binding free energy, occurs with relatively small conformational change. This conformational behavior fits the general characteristic of high-affinity binding occurring with relatively little change in the protein structure, while specificity is conferred by induced fit that would reduce potential free energy contributions of the corresponding intermolecular interactions.
Fig. 5.
Chemical shift perturbation for Vav1 SH2 upon the binding of pYpY. (A) CSP values are plotted as a function of Vav1 SH2 residue number. The dashed line indicates the average perturbation (0.13 ppm). Secondary structural elements and loop regions are shown at the bottom. (B) Residues with CSP values greater than the average mapped onto the structure of the Vav1 SH2-pYpY complex to indicate residues mainly in direct contact with pYpY (orange), those that change conformation but do not contact pYpY (purple), or both (red). The CSP values for R678 (αA2) and Y727 (βE2) are undetermined because of exchange broadening (yellow).
Fig. 6.
Stereoview of the backbone overlay of Vav1 SH2 in the unligated state (cyan) and in complex with pYpY (fuchsia). The structures were aligned on the basis of the central β-sheet of the protein. The flexible BC loop is shown with dashed lines.
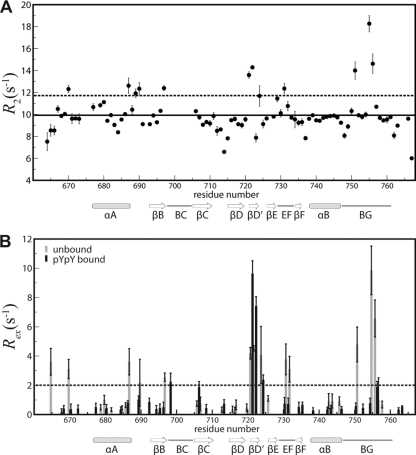
(v) Compensatory changes in dynamics of Vav1 SH2 upon pYpY binding.
Heteronuclear relaxation experiments were performed on unligated Vav1 SH2 and on the pYpY complex to investigate the conformational heterogeneity suggested from the exchange-broadened resonances in the 1H-15N HSQC spectrum. In the case of unligated Vav1 SH2, relatively large values for the transverse relaxation rate, R2, indicative of contributions from an exchange between two or more conformational states, were determined for residues in the βD′ strand (residues M721 and T722) and BG loop (residues K751, K755, and S756) (Fig. 7A). A model-free analysis of the heteronuclear relaxation rates R1 and R2 and NOE finds above-average contributions from chemical exchange, or large Rex values, for these residues (Fig. 7B). We sought to define the actual timescale of the underlying exchange process in the unbound state using relaxation dispersion methods, which measure μs to ms timescales; however no dispersion was observed in a Carr-Purcell-Meiboom-Gill (CPMG) transverse relaxation experiment (data available upon request). We estimate that the timescale of the motions is therefore faster than ∼500 μs and too fast for detection by CPMG experiments (51, 65).
Fig. 7.
Heteronuclear relaxation for unbound and pYpY-bound Vav1 SH2. (A) Transverse relaxation rate, R2, for the residues in the free Vav1 SH2 plotted as a function of residue number. The solid line indicates the average rate (9.97 s−1), and the dashed line is shown at 11.70 s−1 for 1 standard deviation above the mean. (B) Rex values (gray, free Vav1 SH2; black, Vav1 SH2-pYpY complex) from the model-free analysis as a function of Vav1 SH2 residue number. The dashed line is shown at 2 s−1.
Interestingly, the internal motions of Vav1 SH2 are altered upon peptide binding by having both residues with increased mobility and those with decreased mobility, although overall the complex appears to be more rigid given that there are fewer missing resonances due to exchange broadening. Rex values in the pYpY-bound state are significantly diminished for residues in the BG loop (residues K751, K755, and S756) and EF loop (residues E731 and K732), where substantial intermolecular contact is made (Fig. 7B). In contrast, residues in the βD′ strand (residues M721 and T722), which have no contact with the peptide ligand, actually increase in mobility when the peptide binds. As such, the opposing changes in Rex reflect dynamic differences that would have compensatory consequences for binding entropy (26); the greater flexibility of βD′ offsets the entropic penalty associated with rigidifying the BG and EF loops so that the configurational entropy of binding would be overall less unfavorable.
Conformational diversity in SH2 recognition of alternative phosphorylation patterns. (i) Recognition of the second phosphotyrosine, pY346, by PLC-γ and Vav1.
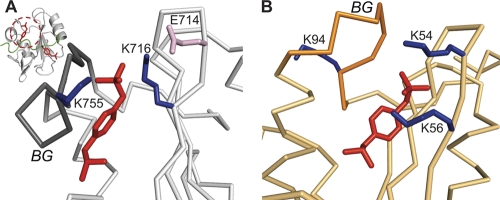
Pulldown experiments using B cell lysates have identified several SH2 domains that bind Syk containing both pY342 and pY346, and all of these SH2 domains have a Lys residue at the βD3 position (23). The NMR structural studies reported here for Vav1 and previously for PLC-γ (25) show that this positively charged residue (K716 or K56, respectively) forms a salt bridge with pY346 in the secondary binding pocket (Fig. 8). In both Vav1 SH2 and PLC-γ c-SH2, an additional salt bridge with pY346 exists but is formed by alternative basic residues: K755 (BG7) in Vav1 SH2 (Fig. 8A) and K54 (βD1) in PLC-γ c-SH2 (Fig. 8B). Sequence alignment of Vav1 SH2 and PLC-γ c-SH2 shows that K755 is aligned with K94 in PLC-γ c-SH2; however, K94 lies in the shorter BG loop of PLC-γ c-SH2, and its orientation away from the pocket impairs interaction with pY346 (Fig. 8B). For the PLC-γ c-SH2 salt bridge, K54 at the βD1 position of PLC-γ c-SH2 aligns with the acidic residue E714 in Vav1 SH2, which does not contact the linker B pYpY peptide. Lys at the βD1 position is found only in PLC-γ c-SH2 among all the known SH2 domains capable of binding to a doubly phosphorylated region of Syk. The distinctness of Lys at the βD1 position for PLC-γ c-SH2 suggests the binding orientation of phosphotyrosine in the secondary pocket shown in Fig. 8B may be specific to PLC-γ c-SH2. Overall, Lys at βD3 is possibly a key recognition determinant for the second phosphotyrosine of Syk linker B, while additional basic residues form alternative interactions with pY346 (Fig. 8).
Fig. 8.
Comparison of pY346 interactions in the secondary pTyr binding pockets in the SH2-pYpY complexes for Vav1 (A) (gray) and PLC-γ (B) (light orange; PDB designation: 2FCI). The location of the secondary pocket is highlighted with a red dashed circle (A). Residue numbering for PLC-γ is from reference 25. The side chains of lysine and glutamate in the SH2 domains are blue and pink, respectively. The backbone and side chain heavy atoms of pY346 are red. The BG loops in both complexes are highlighted in darker colors.
(ii) Conformational diversity in SH2 ligand association.
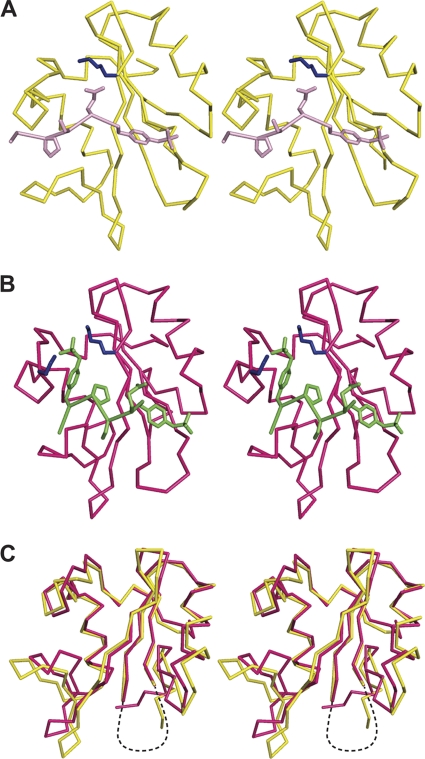
Vav1 SH2 associates with phosphotyrosine by diverse recognition modes. A comparison of the peptide ligand structures from the Vav1 SH2 in complex with pYpY or the peptide pY128, a single-phosphotyrosine site from SLP-76 (Fig. 9), finds that binding a second phosphotyrosine at the pY342 +4 position alters the bound conformation of the specificity residues C-terminal to the primary pTyr. The three residues C-terminal to the pTyr of pY128 (…pY128ESPN…) are identical to those in pYpY (…pY342ESPpY346…), yet the positions of these residues on the surface of Vav1 SH2 differ in the two complexes. In the Vav1 SH2-pY128 complex, the peptide chain from pY128 to pY128 +4 is relatively extended, such that Pro, the pTyr +3 specificity residue for the SH2 class IIA, contacts the BG loop (Fig. 9A) and pY128 +1 interacts with K716 (βD3). This peptide conformation is in contrast to pYpY, whereby binding in the specificity pocket appears to be driven by the interaction of pY346 with K716 and K755, which displaces P345 toward the central β-sheet and bends the main chain straddling the central β-sheet, and E343 becomes solvated (Fig. 9B). Conformational differences associated with the Syk linker B interaction with the central β-sheet of the SH2 domain were also observed in the PLC-γ c-SH2 complex with pYpY; pY346 binding in the specificity pocket occurs with a less extended main chain conformation than in a singly phosphorylated peptide (25).
Fig. 9.
Stereoview of the Vav1 SH2 structures. In the pY128-bound (A) and pYpY-bound (B) complexes, the important side chains of the residues in the peptides (pY128, pink; pYpY, green) and in the proteins (lysine, blue) are highlighted. (C) Backbone overlay of Vav1 SH2 in complex with pY128 (yellow) or pYpY (fuchsia). The structures were aligned on the basis of the central β-sheet of the protein. The flexible BC loop is shown with dashed lines.
The Vav1 SH2 domain structures for the singly phosphorylated pY128 complex and the doubly phosphorylated pYpY complex also differ (Fig. 9C). As noted above, relative to the unligated SH2 structure, binding pYpY displaces the BG and EF loops (Fig. 6). A similar change in the BG loop occurs upon binding pY128 so that the BG loops overlay closely in the structural alignment shown in Fig. 9C. On the other hand, there is less contact with the EF loop by pY128, so a smaller structural change in the EF loop and small β-sheet is found for the pY128 complex. The EF and BG loops are key in determining the specificity of Vav1 SH2 for binding the residues C-terminal to the pTyr. The conformations of these loops vary to adapt to phosphopeptides containing single or double phosphotyrosine without changing the main fold of a SH2 domain.
As described previously (25), the association of Syk linker B peptide pYpY is accompanied by substantial changes in the structure of the PLC-γ c-SH2 domain compared to the complex with singly phosphorylated peptide pY1021 derived from the platelet-derived growth factor (PDGF) receptor (data available upon request) (63). The BG loop of the PLC-γ c-SH2 domain moves outward to a larger degree than observed in Vav1 SH2, and the K54 and K56 side chains on the βD strand are reoriented to interact with the phosphoryl group of pY346 (Fig. 8B) so that the second pY346 has more surface area buried by βC and βD strands. In contrast, the unopened BG loop of Vav1 SH2 provides less binding surface for pY346. The extensive conformational rearrangement for adapting to pYpY in PLC-γ c-SH2 results in a total of 1,189 Å2 buried surface area of the peptide, appreciably greater than 841 Å2 upon the binding to Vav1 SH2. The large changes in PLC-γ c-SH2 induced by pYpY are not found in other SH2-phosphopeptide complexes.
(iii) BG loop conformational mobility.
The conformational heterogeneity in the BG and EF loops of unligated Vav1 SH2 may be functionally relevant and play a role in binding multiple phosphopeptides given that there are conformational rearrangements in the EF and BG loops of Vav1 SH2 complexes. These regions undergo conformational exchange on the submillisecond timescale in the absence of ligands (Fig. 7) and are constrained in the bound state, especially K755 in the BG loop, which directly interacts with pY346 in the Vav1 SH2-pYpY complex. Multiple conformations resembling the bound states for various phosphopeptides may be populated in free Vav1 SH2, which would allow pYpY or pY128 peptides to select the most favored conformation for binding. Besides Syk and SLP-76, Vav1 SH2 has been shown to have the ability to associate with the phosphorylated regions from several other proteins, such as ZAP-70, 3BP2, c-Cbl, CD19, HS1, BLNK, and SHP-1 (7, 10, 24, 32, 33, 38, 39, 46). The phosphopeptide library screening results also indicate binding by diverse sequences, and the importance of the interaction between the EF and BG loops and the pTyr +3 residue (30, 36). Conformational flexibility of the critical regions can facilitate Vav1 SH2 recognition of these different targets, as bound-state conformations are populated to greater extent in the unligated state.
DISCUSSION
Biological activity and affinity correlation.
A primary finding presented here is that the binding affinity of the Vav1 SH2 domain for singly and doubly phosphorylated forms of Syk linker B correlates with the cellular response levels in B cells (Fig. 2A, inset). The relative binding affinities of the isolated Vav1 SH2 domain for pYpY, pYY, and YpY (Table 1) match the relative levels of Vav1 phosphorylation (Fig. 1) and NFAT activation (Fig. 2A) measured in cells expressing different forms of Syk. Notably, the affinity is highest for the uncommon recognition of two closely spaced, phosphorylated tyrosines. Higher affinity corresponds to increased levels of Vav1 phosphorylation and NFAT activity. Coefficients for the correlation between the cellular signaling response for Vav1 phosphorylation or NFAT activation and ln KD are −0.931 and −0.995, respectively, albeit only three values are possible for the different phosphorylated forms of Syk Y342 and Y346. Strong linear relationships between affinity and biological activity have also been observed for Ras/Raf and Sho1p/Pbs2p interactions (3, 47). The direct correlation between affinity and signaling levels identifies protein-protein interactions that are key in the regulation of cellular signaling (3, 47). Accordingly, the data reported here suggest that modulation by Syk linker B phosphorylation of the Syk-Vav SH2 binding affinity is one such primary element of regulation in B cell signaling. Such modulation would affect the competition for Syk interaction among the multiple binding partners of Syk.
PLC-γ is another component of the signaling pathway in B cells leading to NFAT activation, and its association, mediated by its c-SH2, with Syk linker B also depends on the phosphorylation of Y342 and Y346 (25); however, the affinities are substantially higher than those of Vav1 SH2. The dissociation constants, measured by the same SPR methods as those used for Vav1 SH2, are 66 nM for pYpY and 0.49 μM for pYY. These KD values are 1 to 3 orders of magnitude smaller than the values for Vav1 SH2 (Table 1) and also relatively small compared to the range of values, 0.1 to 10 μM, generally observed for phosphotyrosine binding to SH2 domains (41). Thus, the affinities of PLC-γ for Syk linker B are higher than what is typically associated with biological control. Clearly, local concentrations and availability of proteins in cells vary so that it is difficult to predict the degree of saturation from equilibrium binding constants (44). Nevertheless, the direct correlation of signaling outcome with Vav1 SH2 affinity and the magnitudes of KD values suggest that the Syk-Vav1 interaction exerts cellular control, while the higher-affinity Syk–PLC-γ association may not allow a sufficient change in complex concentration in response to phosphorylation for regulation. Moreover, Vav1 is a critical component of the signalosome that mediates the Syk-dependent activation of PLC-γ. In mast cells, for example, the Syk-dependent phosphorylation and activation of PLC-γ in response to the engagement of FcεRI are largely attenuated by the absence of Vav1 (45). In these cells, the phosphorylation of Vav1 also is dependent on the presence of Syk Y342 and is strongly compromised when both Y342 and Y346 are replaced by phenylalanines (71). Thus, both Vav1 and the pair of Syk linker B phosphotyrosines are critical mediators of the receptor-mediated mobilization of calcium and activation of NFAT.
Binding affinity modulated by on rate.
The differences in Vav1 SH2 affinity to bind Syk linker B peptides arise from variations in the kinetic on rates, ka, and not the off rates, kd (Table 1), as might be expected for ligands with small differences. Compared to singly phosphorylated pYY and YpY, the phosphorylation of Y346 in pYpY causes faster association (4.7 × 104 and 5.6 × 103 M−1 s−1 versus 9.7 × 104 M−1 s−1) but has almost no influence on the dissociation rate. Net charge on the peptide is not the only determining factor for ka given that the values differ by 5-fold for pYY and YpY, peptides with equal charge, whereas the values for pYY and the more highly charged pYpY vary by only a factor of 2. The differences in KD for linker B peptide binding to PLC-γ c-SH2 also have their origin in ka and not kd. Moreover, it is also the case that the 1- to 3-order-of-magnitude difference in affinity between PLC-γ c-SH2 and Vav1 SH2 is a result of the association rates, while the dissociation rates differ by 6-fold at most. Whatever the recognition mechanism that generates these kinetic effects might be, it is interesting to consider that controlling association rather than dissociation by tyrosine phosphorylation would be a more effective way to regulate via chemical modification kinetic selection of one binding partner among a pool of potential partners in the cell.
Potential allostery and second binding site.
The observation that binding affects the structure and dynamics of the secondary β-sheet distant from the binding site, which has no contact with pYpY, suggests that this region is a potential site for non-pTyr interactions. Even though there is no contact between the secondary β-sheet and pYpY, comparison of the unligated and pYpY complex structures for Vav1 SH2 and differences in Rex values indicate a change in conformation and increased, rather than decreased, internal motions. The binding of Vav1 SH2 to Syk linker B induces the phosphorylation of Vav1. Vav1 comprises seven domains, and conformational rearrangement in the secondary β-sheet region upon pYpY binding may be related to domain-domain interaction in the activation of Vav1 (5, 40). This region has been reported to be an alternative site for phosphotyrosine-independent binding in other SH2 domains (16, 29). Growing evidence supports the notion that nonclassical binding outside the pTyr and specificity pockets of as SH2 domain can be significant for inter- or intramolecular interactions (1, 9, 34, 49, 70). Future mutagenesis study might provide insight into the possibility of multiple domain interactions between Vav1 and Syk that affect downstream signaling.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants GM039478 (C.B.P.) and CA037372 (R.L.G.), a Purdue University reinvestment grant, and the Purdue Center for Cancer Research (CA23568). C.C. was supported by a Purdue Research Foundation Fellowship.
Footnotes
Published ahead of print on 23 May 2011.
REFERENCES
- 1. Bae J. H., et al. 2009. The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell 138:514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bax A., Grishaev A. 2005. Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr. Opin. Struct. Biol. 15:563–570 [DOI] [PubMed] [Google Scholar]
- 3. Block C., Janknecht R., Herrmann C., Nassar N., Wittinghofer A. 1996. Quantitative structure-activity analysis correlating Ras/Raf interaction in vitro to Raf activation in vivo. Nat. Struct. Biol. 3:244–251 [DOI] [PubMed] [Google Scholar]
- 4. Bradshaw J. M., Waksman G. 2003. Molecular recognition by SH2 domains. Adv. Protein Chem. 61:161–210 [DOI] [PubMed] [Google Scholar]
- 5. Brath U., Akke M. 2009. Differential responses of the backbone and side-chain conformational dynamics in FKBP12 upon binding the transition-state analog FK506: implications for transition-state stabilization and target protein recognition. J. Mol. Biol. 387:233–244 [DOI] [PubMed] [Google Scholar]
- 6. Breeze A. L., et al. 1996. Structure of a specific peptide complex of the carboxy-terminal SH2 domain from the p85α subunit of phosphatidylinositol 3-kinase. EMBO J. 15:3579–3589 [PMC free article] [PubMed] [Google Scholar]
- 7. Brooks S. R., Li X. L., Volanakis E. J., Carter R. H. 2000. Systematic analysis of the role of CD19 cytoplasmic tyrosines in enhancement of activation in Daudi human B cells: clustering of phospholipase C and Vav and of Grb2 and Sos with different CD19 tyrosines. J. Immunol. 164:3123–3131 [DOI] [PubMed] [Google Scholar]
- 8. Bustelo X. R. 2000. Regulatory and signaling properties of the Vav family. Mol. Cell. Biol. 20:1461–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan B., et al. 2003. SAP couples Fyn to SLAM immune receptors. Nat. Cell Biol. 5:155–160 [DOI] [PubMed] [Google Scholar]
- 10. Chiu C. W., Dalton M., Ishiai M., Kurosaki T., Chan A. C. 2002. BLNK: molecular scaffolding through ‘cis’-mediated organization of signaling proteins. EMBO J. 21:6461–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cornilescu G., Delaglio F., Bax A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289–302 [DOI] [PubMed] [Google Scholar]
- 12. Costello P. S., et al. 1999. The Rho-family GTP exchange factor Vav is a critical transducer of T cell receptor signals to the calcium, ERK, and NF-κB pathways. Proc. Natl. Acad. Sci. U. S. A. 96:3035–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deckert M., Tartare-Deckert S., Couture C., Mustelin T., Altman A. 1996. Functional and physical interactions of Syk family kinases with the Vav proto-oncogene product. Immunity 5:591–604 [DOI] [PubMed] [Google Scholar]
- 14. DeFranco A. L. 2001. Vav and the B cell signalosome. Nat. Immunol. 2:482–484 [DOI] [PubMed] [Google Scholar]
- 15. Delaglio F., et al. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 16. Donaldson L. W., Gish G., Pawson T., Kay L. E., Forman-Kay J. D. 2002. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proc. Natl. Acad. Sci. U. S. A. 99:14053–14058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doody G. M., et al. 2001. Signal transduction through Vav-2 participates in humoral immune responses and B cell maturation. Nat. Immunol. 2:542–547 [DOI] [PubMed] [Google Scholar]
- 18. Eck M. J., Shoelson S. E., Harrison S. C. 1993. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature 362:87–91 [DOI] [PubMed] [Google Scholar]
- 19. Eisenmesser E. Z., Zabell A. P. R., Post C. B. 2000. Accuracy of bound peptide structures determined by exchange transferred nuclear Overhauser data: a simulation study. J. Biomol. NMR 17:17–32 [DOI] [PubMed] [Google Scholar]
- 20. Farrow N. A., et al. 1994. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33:5984–6003 [DOI] [PubMed] [Google Scholar]
- 21. Fischer K. D., et al. 1995. Defective T-cell receptor signaling and positive selection of Vav-deficient CD4+ CD8+ thymocytes. Nature 374:474–477 [DOI] [PubMed] [Google Scholar]
- 22. Fujimoto M., Poe J. C., Jansen P. J., Sato S., Tedder T. F. 1999. CD19 amplifies B lymphocyte signal transduction by regulating Src-family protein tyrosine kinase activation. J. Immunol. 162:7088–7094 [PubMed] [Google Scholar]
- 23. Geahlen R. L. 2009. Syk and pTyr'd: signaling through the B cell antigen receptor. Biochim. Biophys. Acta 1793:1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gomez T. S., et al. 2006. HS1 functions as an essential actin-regulatory adaptor protein at the immune synapse. Immunity 24:741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Groesch T. D., Zhou F., Mattila S., Geahlen R. L., Post C. B. 2006. Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by Syk tyrosine kinase. J. Mol. Biol. 356:1222–1236 [DOI] [PubMed] [Google Scholar]
- 26. Grunberg R., Nilges M., Leckner J. 2006. Flexibility and conformational entropy in protein-protein binding. Structure 14:683–693 [DOI] [PubMed] [Google Scholar]
- 27. Guntert P., Mumenthaler C., Wuthrich K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283–298 [DOI] [PubMed] [Google Scholar]
- 28. Herrmann T., Guntert P., Wuthrich K. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319:209–227 [DOI] [PubMed] [Google Scholar]
- 29. Hof P., Pluskey S., Dhe-Paganon S., Eck M. J., Shoelson S. E. 1998. Crystal structure of the tyrosine phosphatase SHP-2. Cell 92:441–450 [DOI] [PubMed] [Google Scholar]
- 30. Huang H. M., et al. 2008. Defining the specificity space of the human Src homology 2 domain. Mol. Cell Proteomics 7:768–784 [DOI] [PubMed] [Google Scholar]
- 31. Inabe K., et al. 2002. Vav3 modulates B cell receptor responses by regulating phosphoinositide 3-kinase activation. J. Exp. Med. 195:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jevremovic D., Billadeau D. D., Schoon R. A., Dick C. J., Leibson P. J. 2001. Regulation of NK cell-mediated cytotoxicity by the adaptor protein 3BP2. J. Immunol. 166:7219–7228 [DOI] [PubMed] [Google Scholar]
- 33. Jordan M. S., et al. 2006. Functional hierarchy of the N-terminal tyrosines of SLP-76. J. Immunol. 176:2430–2438 [DOI] [PubMed] [Google Scholar]
- 34. Joseph R. E., Severin A., Min L., Fulton D. B., Andreotti A. H. 2009. SH2-dependent autophosphorylation within the Tec family kinase Itk. J. Mol. Biol. 391:164–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jung Y. S., Zweckstetter M. 2004. Mars—robust automatic backbone assignment of proteins. J. Biomol. NMR 30:11–23 [DOI] [PubMed] [Google Scholar]
- 36. Kaneko T., et al. 2010. Loops govern SH2 domain specificity by controlling access to binding pockets. Sci. Signal. 3:ra34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Katzav S. 2004. Vav1: an oncogene that regulates specific transcriptional activation of T cells. Blood 103:2443–2451 [DOI] [PubMed] [Google Scholar]
- 38. Katzav S., Sutherland M., Packham G., Yi T. L., Weiss A. 1994. The protein-tyrosine kinase ZAP-70 can associate with the SH2 domain of proto-Vav. J. Biol. Chem. 269:32579–32585 [PubMed] [Google Scholar]
- 39. Kon-Kozlowski M., Pani G., Pawson T., Siminovitch K. A. 1996. The tyrosine phosphatase PTP1C associates with Vav, Grb2, and mSos1 in hematopoietic cells. J. Biol. Chem. 271:3856–3862 [DOI] [PubMed] [Google Scholar]
- 40. Kristensen S. M., Siegal G., Sankar A., Driscoll P. C. 2000. Backbone dynamics of the C-terminal SH2 domain of the p85α subunit of phosphoinositide 3-kinase: effect of phosphotyrosine-peptide binding and characterization of slow conformational exchange processes. J. Mol. Biol. 299:771–788 [DOI] [PubMed] [Google Scholar]
- 41. Ladbury J. E., et al. 1995. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc. Natl. Acad. Sci. U. S. A. 92:3199–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laskowski R. A., Rullmann J. A. C., MacArthur M. W., Kaptein R., Thornton J. M. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477–486 [DOI] [PubMed] [Google Scholar]
- 43. Lopez-Lago M., Lee K., Cruz C., Movilla N., Bustelo X. R. 2000. Tyrosine phosphorylation mediates both activation and downmodulation of the biological activity of Vav. Mol. Cell. Biol. 20:1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Machida K., Mayer B. J. 2005. The SH2 domain: versatile signaling module and pharmaceutical target. Biochim. Biophys. Acta 1747:1–25 [DOI] [PubMed] [Google Scholar]
- 45. Manetz T. S., et al. 2001. Vav1 regulates phospholipase Cγ activation and calcium responses in mast cells. Mol. Cell. Biol. 21:3763–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marengere L. E. M., et al. 1997. Proto-oncoprotein Vav interacts with c-Cbl in activated thymocytes and peripheral T cells. J. Immunol. 159:70–76 [PubMed] [Google Scholar]
- 47. Marles J. A., Dahesh S., Haynes J., Andrews B. J., Davidson A. R. 2004. Protein-protein interaction affinity plays a crucial role in controlling the Sho1p-mediated signal transduction pathway in yeast. Mol. Cell 14:813–823 [DOI] [PubMed] [Google Scholar]
- 48. Mikol V., Baumann G., Keller T. H., Manning U., Zurini M. G. M. 1995. The crystal structures of the SH2 domain of p56lck complexed with 2 phosphonopeptides suggest a gated peptide binding site. J. Mol. Biol. 246:344–355 [DOI] [PubMed] [Google Scholar]
- 49. Min L., Joseph R. E., Fulton D. B., Andreotti A. H. 2009. Itk tyrosine kinase substrate docking is mediated by a nonclassical SH2 domain surface of PLCγ1. Proc. Natl. Acad. Sci. U. S. A. 106:21143–21148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miranti C. K., Leng L., Maschberger P., Brugge J. S., Shattil S. J. 1998. Identification of a novel integrin signaling pathway involving the kinase Syk and the guanine nucleotide exchange factor Vav1. Curr. Biol. 8:1289–1299 [DOI] [PubMed] [Google Scholar]
- 51. Mittermaier A. K., Kay L. E. 2009. Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci. 34:601–611 [DOI] [PubMed] [Google Scholar]
- 52. Mocsai A., Ruland J., Tybulewicz V. L. J. 2010. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat. Rev. Immunol. 10:387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moores S. L., et al. 2000. Vav family proteins couple to diverse cell surface receptors. Mol. Cell. Biol. 20:6364–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morton T. A., Myszka D. G. 1998. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Methods Enzymol. 295:268–294 [DOI] [PubMed] [Google Scholar]
- 55. Movilla N., Bustelo X. R. 1999. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol. Cell. Biol. 19:7870–7885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mulhern T. D., Shaw G. L., Morton C. J., Day A. J., Campbell I. D. 1997. The SH2 domain from the tyrosine kinase Fyn in complex with a phosphotyrosyl peptide reveals insights into domain stability and binding specificity. Structure 5:1313–1323 [DOI] [PubMed] [Google Scholar]
- 57. Myszka D. G. 2000. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol. 323:325–340 [DOI] [PubMed] [Google Scholar]
- 58. Narula S. S., et al. 1995. Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide. Structure 3:1061–1073 [DOI] [PubMed] [Google Scholar]
- 59. Obergfell A., et al. 2002. Coordinate interactions of Csk, Src, and Syk kinases with αIIbβ3 initiate integrin signaling to the cytoskeleton. J. Cell Biol. 157:265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ogura K., Terasawa H., Inagaki F. 1996. An improved double-tuned and isotope-filtered pulse scheme based on a pulsed field gradient and a wide-band inversion shaped pulse. J. Biomol. NMR 8:492–498 [DOI] [PubMed] [Google Scholar]
- 61. Ogura K., et al. 1999. Solution structure of the SH2 domain of Grb2 complexed with the Shc-derived phosphotyrosine-containing peptide. J. Mol. Biol. 289:439–445 [DOI] [PubMed] [Google Scholar]
- 62. Ottiger M., Delaglio F., Bax A. 1998. Measurement of J. and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 131:373–378 [DOI] [PubMed] [Google Scholar]
- 63. Pascal S. M., et al. 1994. Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-γ1 complexed with a high affinity binding peptide. Cell 77:461–472 [DOI] [PubMed] [Google Scholar]
- 64. Ruckert M., Otting G. 2000. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 122:7793–7797 [Google Scholar]
- 65. Saitoh T., et al. 2007. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 26:4777–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Saveliev A., et al. 2009. Function of the nucleotide exchange activity of Vav1 in T cell development and activation. Sci. Signal. 2:ra83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schuebel K. E., et al. 1996. Isolation and characterization of murine vav2, a member of the vav family of proto-oncogenes. Oncogene 13:363–371 [PubMed] [Google Scholar]
- 68. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160:65–73 [DOI] [PubMed] [Google Scholar]
- 69. Schymeinsky J., et al. 2006. The Vav binding site of the non-receptor tyrosine kinase Syk at Tyr 348 is critical for β2 integrin (CD11/CD18)-mediated neutrophil migration. Blood 108:3919–3927 [DOI] [PubMed] [Google Scholar]
- 70. Severin A., Joseph R. E., Boyken S., Fulton D. B., Andreotti A. H. 2009. Proline isomerization preorganizes the Itk SH2 domain for binding to the Itk SH3 Domain. J. Mol. Biol. 387:726–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Simon M., Vanes L., Geahlen R. L., Tybulewicz V. L. J. 2005. Distinct roles for the linker region tyrosines of Syk in FcεRI signaling in primary mast cells. J. Biol. Chem. 280:4510–4517 [DOI] [PubMed] [Google Scholar]
- 72. Spyracopoulos L. 2006. A suite of Mathematica notebooks for the analysis of protein main chain 15N NMR relaxation data. J. Biomol. NMR 36:215–224 [DOI] [PubMed] [Google Scholar]
- 73. Takata M., et al. 1994. Tyrosine kinases Lyn and Syk regulate B cell receptor-coupled Ca2+ mobilization through distinct pathways. EMBO J. 13:1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tartare-Deckert S., et al. 2001. Vav2 activates c-fos serum response element and CD69 expression but negatively regulates nuclear factor of activated T cells and interleukin-2 gene activation in T lymphocytes. J. Biol. Chem. 276:20849–20857 [DOI] [PubMed] [Google Scholar]
- 75. Tedford K., et al. 2001. Compensation between Vav-1 and Vav-2 in B cell development and antigen receptor signaling. Nat. Immunol. 2:548–555 [DOI] [PubMed] [Google Scholar]
- 76. Turner M., et al. 1997. A requirement for the Rho-family GTP exchange factor Vav in positive and negative selection of thymocytes. Immunity 7:451–460 [DOI] [PubMed] [Google Scholar]
- 77. Tybulewicz V. L. J., Henderson R. B. 2009. Rho family GTPases and their regulators in lymphocytes. Nat. Rev. Immunol. 9:630–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wilsbacher J. L., Moores S. L., Brugge J. S. 2006. An active form of Vav1 induces migration of mammary epithelial cells by stimulating secretion of an epidermal growth factor receptor ligand. Cell Commun. Signal. 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wu J., Katzav S., Weiss A. 1995. A functional T-cell receptor signaling pathway is required for p95vav activity. Mol. Cell. Biol. 15:4337–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xu R. X., et al. 1995. Solution structure of the human pp60c-src SH2 domain complexed with a phosphorylated tyrosine pentapeptide. Biochemistry 34:2107–2121 [DOI] [PubMed] [Google Scholar]
- 81. Yamazaki T., Formankay J. D., Kay L. E. 1993. Two-dimensional NMR experiments for correlating 13Cβ and 1Hδ/ε chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J. Am. Chem. Soc. 115:11054–11055 [Google Scholar]
- 82. Zhang X. Y., Shrikhande U., Alicie B. M., Zhou Q., Geahlen R. L. 2009. Role of the protein tyrosine kinase Syk in regulating cell-cell adhesion and motility in breast cancer cells. Mol. Cancer Res. 7:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zweckstetter M., Bax A. 2000. Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J. Am. Chem. Soc. 122:3791–3792 [Google Scholar]