Abstract
The nuclear factor of activated T cells (NFAT) family of transcription factors is expressed in a wide range of cell types and regulates genes involved in cell cycle, differentiation, and apoptosis. NFAT proteins share two well-conserved regions, the regulatory domain and the DNA binding domain. The N- and C-terminal ends are transactivation sites and show less sequence similarity, whereas their molecular functions remain poorly understood. Here, we identified a transcriptional repressor, interferon regulatory factor 2 binding protein 2 (IRF-2BP2), which specifically interacts with the C-terminal domain of NFAT1 among the NFAT family members. IRF-2BP2 was described as a corepressor by inhibiting both enhancer-activated and basal transcription. Gene reporter assays demonstrated that IRF-2BP2 represses the NFAT1-dependent transactivation of NFAT-responsive promoters. The ectopic expression of IRF-2BP2 in CD4 T cells resulted in decreased interleukin-2 (IL-2) and IL-4 production, supporting a repressive function of IRF-2BP2 for NFAT target genes. Furthermore, NFAT1 and IRF-2BP2 colocalized in the nucleus in activated cells, and the mutation of a newly identified nuclear localization signal in the IRF-2BP2 rendered it cytoplasmic, abolishing its repressive effect on NFAT1 activity. Collectively, our data demonstrate that IRF-2BP2 is a negative regulator of the NFAT1 transcription factor and suggest that NFAT1 repression occurs at the transcriptional level.
INTRODUCTION
The regulation of eukaryotic gene expression is a coordinated action of basal transcriptional machinery, chromatin remodeling factors, and transcriptional factors that bind specific elements in promoters and enhancers; these components form a complex network of protein-protein interactions for the proper regulation of mRNA transcription (37). Nuclear factor of activated T cells (NFAT) transcriptional factor, first identified as an inducible nuclear factor that binds the interleukin-2 (IL-2) promoter in activated T cells (57), plays an important role in the control of gene expression in a wide range of cell types and tissues (42, 65). The NFAT family consists of four members that are regulated by calcium and the calcineurin signaling pathway, known as NFAT1 (also called NFATp or NFATc2), NFAT2 (NFATc or NFATc1), NFAT3 (NFATc4), and NFAT4 (NFATx or NFATc3) (42, 52). A fifth member, NFAT5 (TonEBP or OREBP), is regulated by hyperosmotic stress (40, 45).
All NFAT members share a highly conserved DNA binding domain (DBD) that is structurally related to the DBD of the Rel family of transcriptional factors and confers a common DNA-binding specificity to all NFAT proteins (52). The calcium-regulated members, NFAT1 to NFAT4, have a second conserved domain, the NFAT homology region (NHR). This region contains several serines that are phosphorylated when these proteins are in their inactive cytoplasmic forms, and it also contains the docking site for calcineurin and NFAT kinases (42, 52). The high degree of amino acid sequence conservation of the DBD and NHR among the different NFAT family members may result in some functional redundancies, which may be deduced from the observed phenotypes in mice lacking individual NFAT proteins (24, 71, 76). Generally, a more severe functional impairment is observed when more than one NFAT protein is absent (54). However, some divergent phenotypes in individual NFAT-deficient mice can be noted. NFAT1−/− mice develop normally but show a lymphocyte hyperproliferative phenotype, which leads to splenomegaly and a T-cell response that is biased toward a Th2 phenotype, with enhanced IL-4 expression upon T-cell receptor activation (24). These mice also develop the neoplastic transformation of cartilage cells (50). On the other hand, NFAT2 deficiency in mice is lethal as a result of defects in cardiac valve formation (13); nevertheless, in the RAG-deficient complementation system, NFAT2−/− T cells show reduced proliferation and impaired Th2 response (51). Furthermore, in a recent report, Robbs et al. demonstrated distinct roles for NFAT1 and NFAT2 in cell transformation using NIH 3T3 cells, revealing that NFAT1 acts as a tumor suppressor and NFAT2 as an oncogene (55). Taken together, these data suggest that although there is some redundancy in NFAT-related functions, individual NFAT proteins play specific and distinct roles in cell physiology.
An increase in intracellular calcium levels activates the calcium/calmodulin-dependent phosphatase calcineurin, which dephosphorylates the serines in the NHR. This action allows the exposure of a nuclear localization signal to promote the translocation of NFAT into the nucleus. Once in the nucleus, NFAT proteins can bind to their target genes, activating or repressing transcription, either alone or in cooperation with other nuclear partners (42, 65). NFAT proteins have an important role regulating the expression of the cytokines IL-2, IL-4, IL-5, IL-13, tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) (52, 65). Moreover, the NFAT family of transcription factors has a wide role in normal cell physiology, regulating genes related to cell cycle progression (1, 8, 28), cell differentiation (30, 60), angiogenesis (22, 73), and apoptosis (27, 77).
Activator protein 1 (AP-1) family members are the main NFAT partners during T-cell activation. Fos and Jun dimers form complexes with NFAT and DNA on NFAT-AP-1 composite sites that are present in many genes induced during T-cell activation (25, 32, 42). In addition, NFAT proteins interact with other transcriptional factors, such as Maf, ICER, p21SNFT, GATA, EGR (4, 5, 23), the nuclear receptor peroxisome proliferator-activated receptor γ (72), the helix-turn-helix proteins Oct and IRF-4 (3, 53), FOXP3 (70), and MEF-2D (77). Most of these partners interact with the more conserved regions of NFAT, except for MEF-2D, which binds to the NFAT1 C-terminal domain (77).
The amino- and carboxyl-terminal ends of the NFAT1 to NFAT4 proteins exhibit less sequence conservation and are potent transactivation domains (TAD) (41). The reduced sequence conservation observed in these regions may be biologically relevant, because these TADs may be the sites where distinct NFAT family members interact with specific partners. Here, we identified a new NFAT1 partner that interacts with the less-conserved C terminus of NFAT1, called interferon regulatory factor 2 binding protein 2 (IRF-2BP2). This protein binds IRF-2, acting as a corepressor molecule by inhibiting both enhancer-activated and basal transcription, and presents an N-terminal zinc finger and a C-terminal RING domain (10). Recently, it was demonstrated that IRF-2BP2 can influence the p53-mediated transactivation of p21 and Bax promoters (35). Thus, we described that IRF-2BP2 specifically interacts with NFAT1 protein and acts as a repressor of NFAT1-mediated transactivation. To function as a repressor, IRF-2BP2 requires its zinc finger and RING domain, and its effect occurs in the nucleus. In summary, our data identified IRF-2BP2 as a new regulator of NFAT1 transcriptional activity, suggesting an NFAT1-specific mechanism of repression.
MATERIALS AND METHODS
Cell culture.
HEK293T cells and primary CD4 T cells were maintained in Dulbecco's modified medium supplemented with 10% fetal bovine serum (FBS), l-glutamine, penicillin-streptomycin, essential and nonessential amino acids, sodium pyruvate, vitamins, HEPES, and β-mercaptoethanol (all from Invitrogen) at 37°C and 5% CO2 in a humidified environment. Jurkat cells were cultured in RPMI-1640 medium (Gibco) supplemented with 10% FBS, l-glutamine, penicillin-streptomycin, and β-mercaptoethanol under equal conditions.
Plasmid constructions.
The NFAT1 C-terminal end (amino acids 727 to 925) was amplified by PCR and inserted into the EcoRI and BamHI sites of the pTL-1 vector (7) to generate the pTL-CT-NFAT1 vector. The pTL-TAD-C vectors were constructed by site-directed mutagenesis using the Gene Tailor site-directed mutagenesis system (Invitrogen), performed according to the manufacturer's instructions, or by the PCR amplification of the regions of interest and inserted into the pTL-1 vector. The pTL-CT-NFAT1 vector was used as the template, and specific pairs of primers were designed to delete the target regions. To express the C-terminal end of NFAT1 in Escherichia coli, the corresponding cDNA fragment was transferred from the pTL-CT-NFAT1 vector using the enzymes EcoRI and SalI and was inserted into the pProEX-HTa vector (Invitrogen) digested with the same enzymes; the result was pProex-CT-NFAT1. The NFAT1 N-terminal region (amino acids 4 to 405) was amplified by PCR and cloned in the pET-TEV vector (7) using the SacI and HindIII restriction sites of this vector, and the resulting plasmid was pProex-NT-NFAT1. The E. coli strain containing the NFAT1 DBD domain-expressing vector, pNFATpXS(1-297), was a gift from Anjana Rao (Harvard Medical School, Boston, MA) (31). pLIRES-EGFP, pLIRES-EGFP-CA-NFAT1, pLIRES-EGFP-NFAT1 (55, 61), pcDNA5-NFAT1 (8), and pcDNA3-NFAT2 (58) were constructed as described previously. To generate the pcDNA5-NFAT1ΔC vector, NFAT1 (amino acids 1 to 698) was amplified by PCR and inserted into the HindIII and XhoI sites of pcDNA5. To generate the pcDNA5-NFAT3 vector, NFAT3 was purified from pBlueScript-NFAT3 (18) with BamHI and NotI and cloned into the same restriction sites of pcDNA5. To generate pcDNA4-NFAT4, NFAT4 was amplified by PCR from pBlueScript-NFAT4 (17) and cloned into pcDNA4 vector using EcoRI and XhoI restriction sites in fusion with a c-myc tag. Luciferase reporter constructs 3× NFAT-Luc, which contains three copies of the distal NFAT-AP1 site of the IL-2 promoter (21); 6× NFκB, containing six copies of the NF-κB responsive element (49); κ3-Luc, which contains the κ3 element of the TNF-α promoter (63); and the IL-2 and IL-4 proximal promoters (59) were constructed as described previously and obtained from Addgene (Boston).
The coding sequence of IRF-2BP2B (accession number NM_001077397) was amplified by PCR, using cDNA molecules obtained from the Raji cell line as a template; the primers were 5′ CATGGAATTCGGCTCCTCGGACATGGCCG 3′ and 5′CCTACTCGAGCGAGTCTCTCTC TTTTTTCAC 3′. The amplified molecule was inserted in the EcoRI and XhoI sites of pcDNA4/TO/Myc-HisA (Invitrogen) to generate the vector pcDNA4-IRF-2BP2 in fusion with a c-myc tag. The inserts of pcDNA4-IRF-2BP2Δring (amino acids 1 to 485) and pcDNA4-IRF-2BP2Δzinc (amino acids 302 to 571) were obtained through PCR amplification, using pcDNA4-IRF-2BP2 as a template and specific primers to delete the zinc finger and RING domain. The amplified regions were inserted into the EcoRI and XhoI sites of pcDNA4/TO/Myc-HisA (Invitrogen). The vector pGAD-IRF-2BP2 was constructed by transferring the EcoRI-XhoI fragment from pcDNA4-IRF-2BP2 to the EcoRI and SalI sites of pGAD424 (Clontech). pGAD-IRF-2BP2Δring was constructed using specific primers to amplify the region comprising amino acids 1 to 485. This fragment was inserted in the EcoRI and XhoI sites of the pGAD424 vector. The vectors pACT-IRF-2BP2 and pACT-IRF-2BP2Δzinc contain a partial IRF2-BP2 cDNA sequence (amino acids 302 to 571) isolated from the library used in the two-hybrid screen. The IRF-2BP2 C-terminal end containing the RING domain was cloned in the pET-GST-Tev vector (7) through PCR amplification; we used a partial clone isolated in the yeast two-hybrid screen as a template. The resulting vector, pET-GST-IRF-2BP2, contains the C-terminal end of IRF-2BP2 (amino acids 439 to 571) fused to glutathione S-transferase (GST). The pcDNA4-IRF-2BP2MutNLS vector was constructed by site-directed mutagenesis to mutate a putative nuclear localization signal (NLS) in the amino acid positions 339 to 342 using the primers 5′ GCAGTTGCAAGAACAGCAGCGGCAGCGGCGCCCTCTCCAGAACC 3′ and 5′ TGCTGTTCTTGCAACTGCAGTCAGGGC 3′ and pcDNA4-IRF-2BP2 as a template. To construct the pLIRES-EGFP-IRF2BP2 vector, pcDNA4-IRF-2BP2 was digested with PmeI and BamHI, and the digested fragment containing the IRF-2BP2 cDNA was inserted into the BglII and SalI sites (previously blunted) of pLIRES-EGFP.
Yeast two-hybrid assays.
A large-scale screen was performed using the NFAT1 C-terminal end as the bait. For this purpose, the L40 strain (26) containing the pTL1-CT-NFAT1 vector was transformed with an Epstein-Barr virus (EBV)-transformed human B cell cDNA library fused to the activation domain of Gal4p in the pACT vector (Clontech). Positive clones were selected on plates containing YNB medium supplemented with adenine and 3 mM 3-amino-triazole (3-AT), followed by β-galactosidase filter assays (2). Plasmid DNA was extracted from the positive clones, transformed into E. coli XL-1 blue (Stratagene) for amplification, and subjected to DNA sequencing analysis (48). For further interaction analyses, the L40 strain was cotransformed with pTL-CT-NFAT1/pACT-IRF-2BP2; pTL-Nip7/pACT-Nop8 was used as a positive control (78), and the combinations pTL-CT-NFAT1/pGAD424, pTL-Nip7/pACT-IRF-2BP2, and pTL-Nip7/pGAD424 were used as negative controls. The test and control strains were grown in YNB medium containing either histidine or 5 and 15 mM 3-AT. Filter assays to test β-galactosidase expression also were performed.
Protein interaction assays.
The E. coli BL21(DE3) strain (Stratagene) was transformed with vectors pET-GST-IRF-2BP2 and pET-GST-TEV (7). To express the recombinant proteins, the cultures were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of 0.8. E. coli extracts were prepared in phosphate-buffered saline (PBS) containing 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and incubated with glutathione-Sepharose 4B beads (GE Healthcare) for 1 h at 4°C. Subsequently, the beads loaded with the proteins were washed with 6 ml of PBS and incubated for an additional 1 h at 4°C with Jurkat T cell extract (1.5 × 107 cells) or HEK293T cells extract (5 × 106 cells) transfected with pcDNA5-NFAT1, pCDNA5-NFAT1ΔC, pcDNA3-NFAT2, pcDNA5-NFAT3, or pcDNA5-NFAT4, which were prepared in buffer containing 50 mM sodium phosphate (pH 7.5), 5% glycerol, 100 mM NaCl, 0.5 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 30 mM NaPPi, and 50 mM NaF. The beads then were washed with 3 ml of the same buffer, and the eluted proteins, in buffer containing 10 mM reduced glutathione, were analyzed by Western blotting. In a second test, the GST or GST–IRF-2BP2 recombinant proteins bound to glutathione-Sepharose 4B resin, as described above, were incubated for 1 h at 4°C with E. coli extracts in buffer containing 50 mM sodium phosphate (pH 7.5), 5% glycerol, 100 mM NaCl, and 0.5 mM PMSF. The E. coli extracts were prepared from strains transformed with pProex-NT-NFAT1, pProex-CT-NFAT1, and pNFATpXS(1-297), expressing NT-NFAT1, CT-NFAT1, and DBD-NFAT1, respectively. The beads were washed with 4 ml of PBS, and all proteins were eluted in buffer containing 10 mM reduced glutathione. The inputs and eluted proteins were separated on SDS-PAGE and analyzed by Western blotting.
Coimmunoprecipitation assay.
HEK293T (2 × 107) cells transfected either with pLIRES-EGFP-CA-NFAT1, pcDNA4-IRF-2BP2, or both were harvested and incubated with l ml of hypotonic buffer (10 mM Tris, pH 7.5, 10 mM KCl, 0.1 mM EDTA, 1.5 mM MgCl2, 10 mM iodoacetamide) for 30 min on ice. The nuclear fraction was recovered by centrifugation and resuspended in 1 ml of hypertonic buffer (20 mM Tris, pH 8.0, 0.4 M NaCl, 0.1 mM EDTA, 1.5 mM MgCl2, 10 mM iodoacetamide, 25% [vol/vol] glycerol), followed by incubation on ice for 30 min. After centrifugation, the fractions were incubated with anti-c-myc antibody (Invitrogen) and protein A-Sepharose at 4°C overnight. Beads then were washed with 6 ml of wash buffer (20 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% Nonidet P-40) and eluted for Western blot analysis. All buffers contain a mixture of protease and phosphatase inhibitors as described above.
Western blotting.
Total cell lysate proteins from HEK293T, inputs, and eluted proteins from protein interaction assays were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. Total protein from 106 cells (HEK293T) was obtained in cell lysis buffer (40 mM Tris [pH 7.5], 60 mM NaPPi, 10 mM EDTA, and 5% SDS) and then incubated at 100°C for 15 min. The antibodies used were polyclonal antibody anti-67.1 and anti-T2B1, against peptides from NH2- and COOH-terminal regions of NFAT1, respectively; R59, against peptides from the DBD, kindly provided by Anjana Rao (Harvard Medical School, Boston, MA); anti-NFAT2 (Santa Cruz); anti-NFAT3 (Santa Cruz); anti-c-myc monoclonal antibody (Invitrogen); and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody 6C5 (Santa Cruz). Immunodetection was performed with the ECL Western blotting detection kit (GE Healthcare).
Immunofluorescence analysis.
HEK293T cells were transfected by the calcium phosphate method with pcDNA4-IRF-2BP2, pcDNA4-IRF-2BP2MutNLS, pcDNA4-IRF-2BP2Δring, or pcDNA4-IRF-2BP2Δzinc or cotransfected with pLEGFP-NFAT1 and pcDNA4-IRF-2BP2. Forty-eight hours after transfection, cells grown on glass coverslips were left untreated or were treated for 15 min with ionomycin (5 μM), cyclosporine A (CsA; 2 μM), or both. Cells were fixed with 4% paraformaldehyde, and IRF-2BP2 was detected with an anti-tag primary antibody (anti-c-myc; Invitrogen), followed by a rhodamine-conjugated secondary antibody. NFAT1 was fused to green fluorescent protein (GFP). 4′,6′-Diamidino-2-phenylindole (DAPI) staining was used to stain DNA. The cells were examined with an Olympus BX60 fluorescence microscope.
Luciferase reporter assays.
Jurkat cells (3 × 106 cells/600 μl) were electroporated (950 μF and 250 mV) with empty vectors (8 μg each), pcDNA5-NFAT1 (8 μg), pcDNA5-NFAT1ΔC (8 μg), pcDNA3-NFAT2 (8 μg), or pcDNA4-IRF-2BP2 (0.5 to 8 μg), as indicated in the figure legends, and 1 μg of luciferase reporter plasmids containing 3× NFAT-Luc, pGL4.30 (Promega), 6× NFκB-Luc, κ3-Luc (TNF-α), or IL-2 or IL-4 proximal promoter, as indicated. After 24 h, cells were stimulated for 6 h with PMA (10 nM) and ionomycin (1 μM) and lysed with 50 μl of 1× passive lysis buffer (Promega). The extracts were analyzed in a Veritas microplate luminometer (Turner Biosystems) using a dual-luciferase reporter assay system (Promega) as directed by the manufacturer. The firefly luciferase reporter gene was normalized with the renilla vector pRL-TK (0.1 μg). The average values of the tested constructs were normalized to the activity of the empty vectors.
Retroviral transduction of primary CD4 T cells.
To perform retrovirus generation, the Phoenix packaging cell line was transiently transfected with 20 μg of the retroviral vector pLIRES-EGFP or pLIRES-EGFP-IRF-2BP2 and 7.5 μg of pCL-Eco by the calcium phosphate precipitation method for 24 h. The viral supernatants were collected 48 h after transfection and concentrated by centrifugation using an Amicon Ultracel 50 K (Millipore). Primary CD4 T cells were isolated from lymph nodes (inguinal, brachial, axillary, and superficial cervical) of 8- to 12-week-old C57BL/6 mice and purified (>90% purity) by negative selection according to the manufacturer's instructions (Dynal mouse CD4 negative isolation kit; Invitrogen). Cells (1 × 106 cells/well) were stimulated with anti-CD3 (1 μg/ml) and anti-CD28 (1 μg/ml) in a 12-well plate coated with anti-IgG (0.3 mg/ml). After 48 h, the culture medium was replaced with 1 ml of complete medium containing concentrated retrovirus supplemented with polybrene (8 μg/ml). The plates were centrifuged at 930 × g for 1.5 h at room temperature and then incubated at 37°C.
Intracellular cytokine staining.
CD4 T cells (1.5 × 106 cells/ml) were stimulated with 10 nM PMA and 1 μM ionomycin for 6 h. BD Golgi Stop (10 μg/ml; BD Pharmingen) was added 2 h before the staining procedure. Briefly, cells were harvested, fixed with 2% of formaldehyde in PBS, permeabilized with 0.5% of saponin, and stained with anti-IL-2-phycoerythrin and anti-IL-4-allophycocyanin (eBioscience).
RESULTS
NFAT1 interacts with IRF-2BP2.
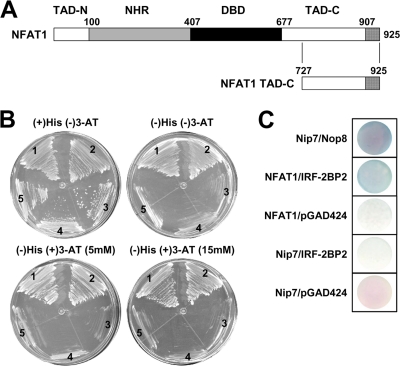
To characterize the molecular mechanisms related to NFAT transcriptional functions, we performed a yeast two-hybrid screening to identify new NFAT1-interacting proteins, using the less-conserved NFAT1 C-terminal end (amino acids 727 to 925) as bait (Fig. 1A). Fifty percent of the sequenced clones isolated from a total of 107 yeast colonies transformed with a human B-cell cDNA library encoded the IRF-2BP2 protein, isoform B. The IRF-2BP2 protein has been identified as an IRF-2-binding protein that acts as a transcriptional corepressor molecule, which contains an N-terminal zinc finger and a C-terminal RING domain (10). To confirm this interaction, the L40 strain was cotransformed with pTL-CT-NFAT1 and pACT-IRF2-BP2. The assay for the HIS3 reporter was performed in medium containing a high 3-AT concentration (up to 15 mM); growth under these conditions is an indication of positive interaction (Fig. 1B). As expected, positive interaction also was observed for the lacZ reporter gene (Fig. 1C). As a positive control, we used the L40 strain cotransformed with the plasmids pTL-Nip7 and pACT-Nop8, which encode interacting proteins (78).
Fig. 1.
IRF-2BP2 interacts with NFAT1 TAD-C in a yeast two-hybrid assay. (A) Schematic representation of the NFAT1 transcriptional factor with its conserved domains: the NFAT homology region (NHR), the DNA binding domain (DBD), and the transactivation domains (TAD-N and TAD-C). The boundary of each region is labeled above the sequence; numbering refers to the amino acid position of the protein. The NFAT1 C-terminal end (amino acids 727 to 925) used in the yeast two-hybrid system also is represented. (B) Yeast two-hybrid system interaction assays on plates containing synthetic minimal medium and 0, 5, or 15 mM 3-AT. Region 1, positive control (pTL-Nip7p + pACT-Nop8p); region 2, test (pTL-CT-NFAT1 + pACT-IRF-2BP2); regions 3, 4, and 5, negative controls (pTL-CT-NFAT + pGAD424, pBTM-Nip7 + pGAD-IRF-2BP2, pBTM-Nip7 + pGAD-424, respectively). (C) β-Galactosidase filter assay. The L40 strain, which contains the HIS3 and lacZ reporter genes, was cotransformed with vectors containing the LexA DBD fusion proteins and Gal4p activation domain fusion proteins. All results are representative of at least three independent experiments.
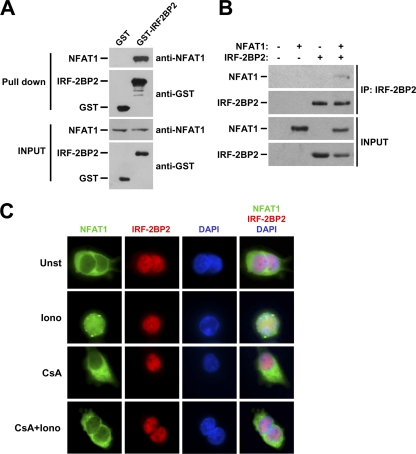
To ensure the interaction between IRF-2BP2 and NFAT1, we performed an in vitro pulldown assay with the purified recombinant IRF-2BP2–RING domain fused to GST. In this test, cell extracts from the Jurkat T-cell line, which expresses NFAT1 endogenously, were incubated with recombinant GST or GST–IRF-2BP2–RING which previously had been immobilized in affinity beads. Our data showed that NFAT1 bound specifically to GST–IRF-2BP2–RING, whereas we did not observe any interaction with GST alone (Fig. 2A). The interaction also was confirmed by coimmunoprecipitating NFAT1 and IRF-2BP2 using nuclear extracts of HEK293T cells cotransfected with pLIRES-EGFP-CA-NFAT1 and pcDNA4-IRF-2BP2 (Fig. 2B).
Fig. 2.
IRF-2BP2 interacts with NFAT1, and these proteins colocalize in the nucleus. (A) In vitro binding assay. Glutathione-Sepharose beads were loaded with GST or GST-IRF-2BP2-RING proteins, washed, and incubated with Jurkat extracts. The proteins then were eluted as described in Materials and Methods. Samples from the extracts and eluted from the resin were fractionated by SDS-PAGE and subjected to immunoblot analysis using anti-GST and anti-NFAT1 antibodies. (B) NFAT1 coimmunoprecipitates (IP) with IRF-2BP2. Nuclear lysates of HEK293T cells cotransfected with pcDNA4-IRF-2BP2 and pLIRES-EGFP-CA-NFAT1 were immunoprecipitated with anti-c-myc antibody. Bound proteins then were analyzed by SDS-PAGE followed by Western blotting using anti-tag antibody (c-myc) for IRF-2BP2 or anti-NFAT1 antibody. (C) Immunofluorescence analysis of IRF-2BP2 and NFAT1. HEK293T cells cotransfected with GFP-NFAT1 and IRF-2BP2, grown on glass coverslips, and treated with ionomycin (Iono) (5 μM), CsA (2 μM), or both for 15 min. Untreated cells were used as a control. NFAT1 was fused to EGFP, and IRF-2BP2 was detected with an anti-tag primary antibody (c-myc) followed by a rhodamine-conjugated antibody. DAPI staining shows the position of the nucleus. All results are representative of at least three independent experiments. Unst, unstimulated.
We then performed an immunofluorescence analysis to determine the subcellular localization of IRF-2BP2 and to investigate whether NFAT1 and IRF-2BP2 colocalize in the cell. HEK293T cells were cotransfected with pLIRES-EGFP-NFAT1 and pcDNA4-IRF-2BP2. After 48 h, transfected cells were left unstimulated or were treated with ionomycin in the presence or absence of CsA (Fig. 2C). NFAT1 was expressed in these cells as a fusion protein with enhanced green fluorescent protein (EGFP), and IRF-2BP2 was fused with the c-myc tag. As shown in Fig. 2C, under all conditions, IRF-2BP2 was localized in the nucleus, whereas NFAT1 was nuclear only when cells were treated with ionomycin. This result indicates that IRF-2BP2 and NFAT1 colocalize only when NFAT1 is in its activated nuclear form.
IRF-2BP2 represses NFAT1-mediated transactivation.
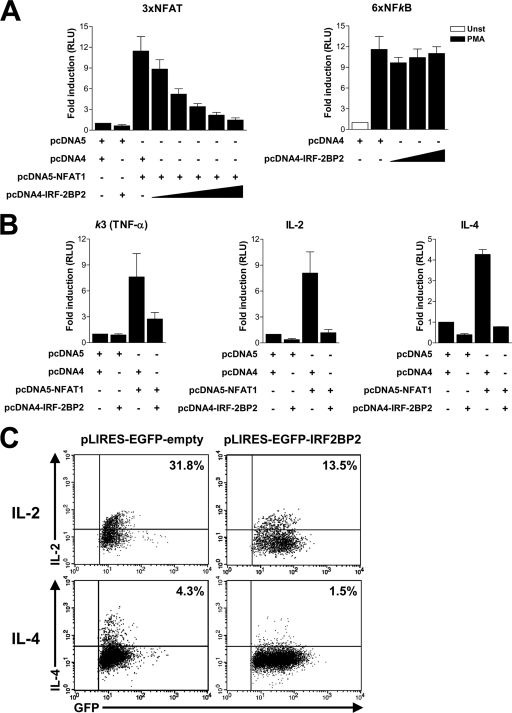
Because IRF-2BP2 has been described as a transcriptional repressor protein (10, 35), we investigated its effect on the transactivation of NFAT1-regulated genes. Jurkat T cells were transfected with NFAT1 and IRF-2BP2 expression constructs along with different luciferase reporter plasmids containing known NFAT element sequences. Transfected cells were stimulated with PMA plus ionomycin to mediate NFAT activation and nuclear translocation. We initially used a luciferase reporter plasmid under the control of three copies of the distal NFAT-AP1 element of the IL-2 promoter (3×NFAT-Luc). As shown in Fig. 3A, NFAT1 was able to drive luciferase expression in activated Jurkat T cells. However, when these cells were cotransfected with NFAT1 and increasing amounts of IRF-2BP2 plasmid, the luciferase signal decreased in a dose-dependent manner (Fig. 3A, left). To evaluate whether IRF-2BP2 affected the transcription mediated by another transcription factor, we analyzed the 6× NF-κB reporter vector, which contains a responsive NF-κB element. Jurkat T cells were transfected with the 6× NF-κB construct and increasing amounts of IRF-2BP2. Twenty-four h after transfection, cells were stimulated for 6 h with PMA and luciferase activity was assessed. As shown in Fig. 3A (right), IRF-2BP2 did not repress NF-κB transcriptional activity.
Fig. 3.
IRF-2BP2 represses NFAT1-mediated transactivation and cytokine expression in primary lymphocytes. (A and B) Jurkat cells were electroporated with empty vectors (8 μg each), the NFAT1 vector (8 μg), the IRF-2BP2 vector (0.5 to 8 μg), and luciferase reporter vectors (1 μg). After 24 h, cells were stimulated for 6 h with PMA (10 nM) plus ionomycin (1 μM) for 3× NFAT vector-transfected cells (left) or PMA (10 nM) alone for 6× NF-κB vector-transfected cells (right). (A) Luciferase assays using reporter plasmids containing the NFAT binding site (3×NFAT-Luc) or NF-κB binding site (6×NFκB). Unst, unstimulated. (B) Luciferase assays using reporter plasmids containing the κ3 element of the TNF-α promoter or IL-2 or IL-4 promoter. For all experiments, the firefly luciferase reporter gene was normalized with a renilla vector (0.1 μg pRL-TK). Results represent the means from three independent experiments ± standard deviations. (C) Primary CD4 T cells from lymph nodes of C57/BL6 mice were stimulated with anti-CD3 and anti-CD28 for 48 h and then transduced with pIRES-EGFP or pILRES-EGFP-IRF-2BP2 vector. Twenty hours after transduction, cells were restimulated with PMA (10 nM) plus ionomycin (1 μM) for 6 h, and the intracellular cytokines IL-2 and IL-4 were stained and analyzed by flow cytometry.
To further characterize the effect of IRF-BP2 on NFAT1-mediated transcription, we have analyzed other reporter vectors of NFAT-regulated promoter genes, including TNF-α, IL-2, and IL-4. The analysis of the NFAT-mediated transactivation of the NFAT element from the κ3 element (TNF-α promoter), which is independent of AP-1 cooperation, showed that the repressive function of IRF-2BP2 also was observed when cells were cotransfected with NFAT1- and IRF-2BP2-expressing constructs (Fig. 3B). In fact, the luciferase signal in cotransfected cells was reduced to 35% of that observed when only NFAT1 was transfected (Fig. 3B). We further evaluated the effect of IRF-2BP2 on NFAT1-mediated transactivation in the context of the IL-2 and IL-4 proximal promoters. Cotransfection with IRF-2BP2- and NFAT1-expressing vectors reduced the NFAT1-dependent activation of IL-2 and IL-4 reporter plasmids to approximately 15 and 20%, respectively, of the signal obtained with NFAT1 alone (Fig. 3B). Taken together, these data suggest that IRF-2BP2 has a repressing effect on the transactivation function of NFAT1 toward its target genes, and that this effect is independent of the ability of NFAT1 to cooperate with the AP-1 transcription factor. Moreover, IRF-2BP2 acts specifically by repressing NFAT-regulated transcription, since the NF-κB reporter vector was not affected by IRF-2BP2.
To reinforce the observation that IRF-2BP2 represses the expression of NFAT target genes, we evaluated the effect of IRF-2BP2 on the endogenous expression of IL-2 and IL-4 in primary CD4 T lymphocytes. CD4 T cells were retrovirally transduced with either pLIRES-EGFP-empty or pLIRES-EGFP-IRF-2BP2 vector. As shown in Fig. 3C, CD4 T cells infected with the pLIRES-EGFP-IRF-2BP2 construct showed a decrease of IL-2 and IL-4 levels compared to those of CD4 T cells infected with the control vector (Fig. 3C). This result corroborates the reporter assays and demonstrates the importance of IRF-2BP2 in repressing the expression of some NFAT target genes.
IRF-2BP2 interacts with NFAT1 C-terminal end, which mediates the IRF-2BP2-repressing effect.
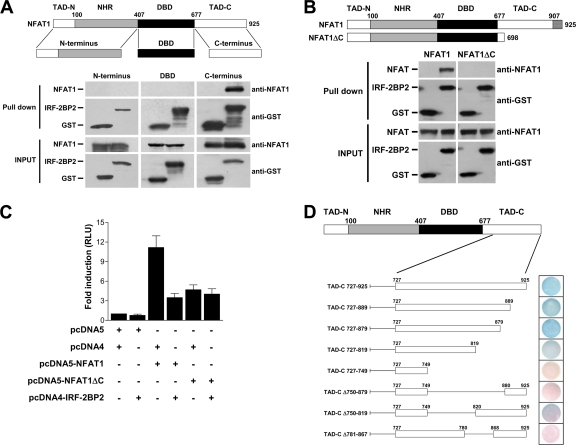
In an effort to map the IRF-2BP2 and NFAT1 interaction, we performed an in vitro binding assay to analyze the interaction of IRF-2BP2 with the NFAT1 domains independently, including the N terminus, DBD, and C terminus (Fig. 4A). As described above, GST and GST–IRF-2BP2–RING proteins were bound to affinity beads and further incubated with E. coli extracts that independently expressed the NFAT domains. As shown in Fig. 4A, we observed that only the C-terminal end of the NFAT1 protein (TAD-C) was able to bind to IRF-2BP2–RING. To confirm the interaction of the NFAT1 TAD-C with IRF-2BP2, we also performed an in vitro binding assay between IRF-2BP2 and a truncated form of NFAT1 deleted at the C-terminal end (NFAT1ΔC). Cell extracts from HEK293T cells transfected with pcDNA5-NFAT1 or pCDNA5-NFAT1ΔC were incubated with recombinant GST or GST–IRF-2BP2–RING, which previously were immobilized in affinity beads. As shown in Fig. 4B, NFAT1ΔC does not bind to IRF-2BP2–RING, as demonstrated by in vitro binding assays.
Fig. 4.
IRF-2BP2 interacts with the C-terminal end of NFAT1, and this region is essential for the repressing phenotype mediated by IRF-2BP2. (A) GST and GST–IRF-2BP2 proteins were immobilized on glutathione-Sepharose beads and further incubated with E. coli extracts expressing each NFAT1 domain independently (N terminus, DBD, or C terminus) as indicated. The beads were washed and then proteins were eluted. Samples from the extracts (inputs) and eluted from the resin (pull down) were fractionated by SDS-PAGE, followed by immunoblot analysis using anti-GST and anti-NFAT1 antibodies. (B) Extracts prepared from HEK293T cells transfected with pcDNA5-NFAT1 or pCDNA5-NFAT1ΔC were incubated with the recombinant proteins GST and GST–IRF-2BP2, which previously were immobilized on glutathione-Sepharose beads. After incubation, the beads were washed, proteins were eluted, and samples from the extracts (inputs) and eluted from the resin (pulldown) were fractionated by SDS-PAGE, followed by immunoblot analysis using anti-GST and anti-NFAT1 antibodies. (C) Jurkat cells were electroporated with empty vectors (8 μg each), NFAT1 vector (8 μg), NFAT1ΔC vector (8 μg), IRF-2BP2 vector (8 μg), and the luciferase reporter vector 3× NFAT (1 μg), as indicated. After 24 h, cells were stimulated for 6 h with PMA (10 nM) plus ionomycin (1 μM). The firefly luciferase reporter gene was normalized with a renilla vector (0.1 μg pRL-TK). Results represent the means from three independent experiments ± standard deviations. (D) The interaction sites of CT-NFAT1 and IRF-2BP2 were mapped by the yeast two-hybrid system in the L40 strain cotransformed with CT-NFAT1 and IRF-2BP2 constructs. A β-galactosidase filter assay was performed to test the lacZ reporter gene. Different deletions of CT-NFAT1 were constructed as shown. The boundary of each region is labeled above the sequence; numbers refer to the amino acid position of the protein. All results are representative of at least three independent experiments.
To confirm the relevance of the NFAT1 C-terminal end for the repressing phenotype exerted by IRF-2BP2 on NFAT1-mediated transcription, we investigated the effect of IRF-2BP2 on NFAT1ΔC by gene reporter assay. Jurkat T cells were transfected with NFAT1 or NFAT1ΔC and IRF-2BP2 expression constructs along with an NFAT reporter plasmid. As shown in Fig. 4C, NFAT1ΔC presents diminished transactivation activity compared to that of full-length NFAT1, confirming that the NFAT1 C-terminal end exhibits transactivation function. Although this truncated form of NFAT1 is less effective at inducing the transactivation of the NFAT reporter plasmid, the NFAT1ΔC-induced luciferase activity was not repressed by IRF-2BP2 (Fig. 4C). These data demonstrated that the NFAT1 TAD-C is the site of IRF-2BP2 interaction, and that this region is extremely relevant for the repressive function of IRF-2BP2.
We then mapped the NFAT1 TAD-C region required for the IRF-2BP2 interaction using the yeast two-hybrid method. Several truncations in the NFAT1 C-terminal end were fused to the LexA DNA binding domain (pTL-TAD-C vectors) and tested for their ability to bind IRF-2BP2 fused to the Gal4p activation domain (Fig. 4D). We observed that the deletion of the C terminus up to amino acid 819 (TAD-C 727-819) still maintained the interaction, although the reporter signal seemed weaker in this construction (Fig. 4D). However, the constructs TAD-C 727-749 and TAD-C Δ750-879 failed to bind to IRF-2BP2 (Fig. 4D). Because the deleted regions were too large, two other constructions were designed to define a smaller interaction site. We observed that the TAD-C Δ781-867 construct failed to interact with IRF-2BP2. Thus, the smallest interaction region mapped in our assay spanned amino acids 781 to 867 (Fig. 4D). The capacity of these truncated NFAT1 constructs to interact with the full-length IRF-2BP2 was confirmed using the HIS3 reporter gene, which corroborated these results (data not shown).
IRF-2BP2 interacts specifically with NFAT1 protein.
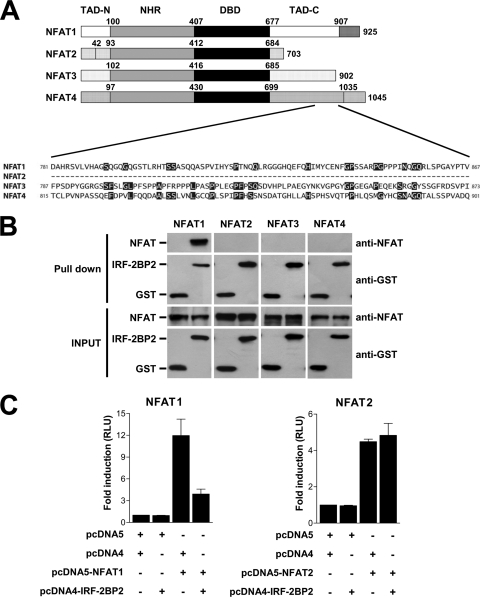
An important characteristic of the C-terminal end of NFAT proteins is its little sequence conservation among NFAT family members. To investigate whether IRF-2BP2 could interact with other NFAT proteins, we first compared the amino acid primary sequence among NFAT1 to NFAT4 proteins comprising the specific region of NFAT1 (amino acids 781 to 867) mapped to interact with IRF-2BP2. As shown in Fig. 5A, we have not seen any amino acid identity between analyzed NFAT family members. This observation suggests that IRF-2BP2 interacts specifically with NFAT1. To address this question, GST and GST–IRF-2BP2–RING proteins were bound to affinity beads and further incubated with extracts of HEK293T cells transfected with pcDNA5-NFAT1, pcDNA3-NFAT2, pcDNA5-NFAT3, or pcDNA4-NFAT4 as described previously. As shown in Fig. 5B, only NFAT1 protein was able to bind to IRF-2BP2–RING. These data demonstrated that IRF-2BP2 interacts specifically with NFAT1, since we have not observed any interaction between IRF-2BP2 and other NFAT proteins, including NFAT2, NFAT3, and NFAT4.
Fig. 5.
IRF-2BP2 interacts specifically with NFAT1. (A) Amino acid sequence alignment comparing the C-terminal end of NFAT1-4 family members. (B) In vitro binding assays. The recombinant proteins GST and GST–IRF-2BP2 expressed in E. coli and immobilized on glutathione-Sepharose beads were incubated with extracts prepared from HEK 293T cells transfected with pcDNA5-NFAT1, pcDNA3-NFAT2, pcDNA5-NFAT3, or pcDNA4-NFAT4. After incubation, the beads were washed, proteins were eluted, and samples from the extracts (inputs) and eluted from the resin (pulldown) were fractionated by SDS-PAGE, followed by immunoblot analysis using anti-GST and anti-NFAT1 antibodies. (C) Jurkat cells were electroporated with empty vectors (8 μg each), NFAT1 vector (8 μg), NFAT2 vector (8 μg), IRF-2BP2 vector (8 μg) as indicated, and the luciferase reporter vector 3× NFAT (1 μg). After 24 h, cells were stimulated for 6 h with PMA (10 nM) plus ionomycin (1 μM). The firefly luciferase reporter gene was normalized with a renilla vector (0.1 μg pRL-TK). Results represent the means from three independent experiments ± standard deviations.
To analyze the specificity of the IRF-2BP2 repression effect on the transactivation mediated by NFAT1, we performed a gene reporter assay using the NFAT reporter plasmid. Jurkat cells were transfected with NFAT1 or NFAT2 and IRF-2BP2 expression constructs with NFAT reporter plasmid. Upon stimulation, IRF-2BP2 repressed the NFAT1-mediated transactivation as previously demonstrated (Fig. 5C). Nevertheless, IRF-2BP2 did not repress the NFAT2-mediated transactivation (Fig. 5C), indicating that the IRF-2BP2 repressor phenotype is NFAT1 specific and dependent on the interaction between these two proteins.
RING domain and the zinc finger are necessary for IRF-2BP2-mediated repression.
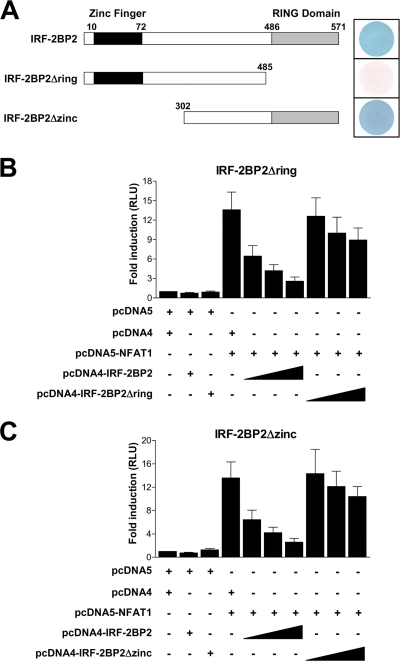
To better characterize the repressive effect of IRF-2BP2 on NFAT1-mediated transcription and to study the importance of the conserved domains of the IRF-2BP2 protein for its function, we investigated the role of the RING domain and the zinc finger in the IRF-2BP2–NFAT1 interaction. We used the yeast two-hybrid system to map the IRF-2BP2 regions that are required for interaction with NFAT1. The L40 strain containing the LexA-NFAT1 vector was transformed with full-length IRF-2BP2, IRF-2BP2Δring (amino acids 1 to 485), or IRF-2BP2Δzinc (amino acids 302 to 571) fused to the Gal4p activation domain (Fig. 6A). Gal4–IRF-2BP2 and Gal4–IRF-2BP2Δzinc were able to interact with NFAT1, as shown by the lacZ reporter gene (Fig. 6A). However, when the IRF-2BP2 RING domain was deleted (IRF-2BP2Δring), the interaction with the NFAT1 C terminus was abolished (Fig. 6A). The assay for the HIS3 reporter gene produced the same results (data not shown).
Fig. 6.
RING domain and zinc finger are necessary for the repressive function of IRF-2BP2. (A) Mapping the IRF-2BP2 interaction sites with NFAT1 via the yeast two-hybrid system (β-galactosidase reporter assay). The L40 strain was cotransformed with the following CT-NFAT1 and IRF-2BP2 constructs: full-length IRF-2BP2, IRF-2BP2Δring, or IRF-2BP2Δzinc. A schematic diagram of the IRF-2BP2 constructs is shown on the left. The boundary of each region is labeled above the sequence; numbers refer to the amino acid position of the protein. Luciferase reporter assays testing the truncated proteins IRF-2BP2Δring (B) and IRF-2BP2Δzinc (C) are shown. Jurkat cells were electroporated with 3× NFAT-Luc (1 μg), empty vectors (8 μg), the NFAT1 vector (8 μg), and increasing amounts (1 to 8 μg) of IRF-2BP2 constructs (full length, Δring, or Δzinc). After 24 h, cells were stimulated for 6 h with PMA (10 nM) plus ionomycin (1 μM). For all experiments, the firefly luciferase reporter gene was normalized with a renilla vector (0.1 μg pRL-TK). Results represent the means from three independent experiments ± standard deviations.
To confirm the relevance of the RING domain and address the role of the zinc finger in the repressive function of IRF-2BP2, we performed a luciferase reporter assay using 3× NFAT-Luc as a reporter plasmid and the IRF-2BP2-deleted RING domain (IRF-2BP2Δring) or zinc finger (IRF-2BP2Δzinc) proteins in Jurkat T cells. The cells were cotransfected with the NFAT1 construct and increasing amounts of vectors expressing full-length IRF-2BP2, IRF-2BP2Δring, or IRF-2BP2Δzinc. Accordingly, the reduction in NFAT1-mediated transactivation no longer was observed after transfection with IRF-2BP2Δring (Fig. 6B), since the RING domain is the interacting region between IRF-2BP2 and NFAT1 (Fig. 6A). Interestingly, when the zinc finger was deleted in IRF-2BP2 (IRF-2BP2Δzinc), the repression of NFAT1 activity was abolished as well (Fig. 6C), although the deletion of the IRF-2BP2 zinc finger did not interfere with the IRF-2BP2–NFAT1 interaction (Fig. 6A). Since IRF-2BP2 has been described as a nuclear protein (10), it was important to find out whether the deletion of the RING domain or the zinc finger would alter the nuclear localization of the truncated proteins. The analysis of HEK293T cells transfected with IRF-2BP2Δring or IRF-2BP2Δzinc constructs demonstrated that these truncated proteins were still in the nucleus (data not shown). Taken together, these data demonstrate that the IRF-2BP2 RING domain is essential for the interaction with NFAT1 TAD-C. Therefore, both the IRF-2BP2 RING domain and the zinc finger are necessary for the repressive function IRF-2BP2.
Interaction with IRF-2BP2 does not lead to NFAT1 degradation.
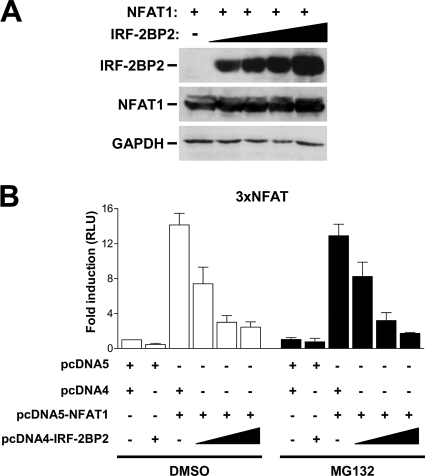
Several reports have demonstrated that RING domains generally are found in ubiquitin E3 ligases. A recent report identified the IRF-2BP1 protein, which is related to IRF-2BP2 and displays a highly conserved RING domain, as a JDP2 ubiquitin ligase (34). Consequently, we decided to investigate whether IRF-2BP2 influences NFAT1 protein stability. To address this question, we transfected HEK293T cells with NFAT1 and increasing amounts of IRF-2BP2 and then evaluated the NFAT1 protein levels in cotransfected cells. Flow-cytometric analysis of EGFP levels was performed to ensure equal transfection efficiency (data not shown). As shown in Fig. 7A, we could not detect any changes in the levels of NFAT1 protein in the presence of IRF-2BP2. To further analyze whether the repressive function of IRF-2BP2 on NFAT1-mediated transactivation involves NFAT1 protein degradation via the ubiquitin-proteasome pathway, we also performed luciferase reporter assays using Jurkat T cells left untreated or treated with the proteasome inhibitor MG132. Cells were cotransfected with an NFAT1 construct and increasing amounts of IRF-2BP2 plasmid. After 24 h, cells were left untreated or were treated with MG132 and then stimulated with PMA plus ionomycin for 6 h. As shown in Fig. 7B, we did not detect any differences in the reporter signal using 3× NFAT-Luc in the presence or absence of the proteasome inhibitor. The proteasome inhibitor was able to prevent NF-κB activation in a similar assay, which demonstrates that we used an effective concentration of MG132 (data not shown). These results indicate that the effect of IRF-2BP2 on the inhibition of NFAT1-mediated transactivation is not related to NFAT1 protein stability, which suggests that IRF-2BP2 repression occurs at the transcriptional level.
Fig. 7.
NFAT1 protein stability is not affected by IRF-2BP2. (A) HEK293T cells were transfected with pLIRES-EGFP-CA-NFAT1 (2 μg) and increasing amounts of pcDNA4-IRF-2BP2 (0.25 to 2 μg). After 48 h, the indicated proteins were detected by Western blotting with specific antibodies. Results are representative of three independent experiments. (B) Jurkat cells were electroporated with the luciferase reporter vector 3× NFAT-Luc (1 μg), empty vectors (8 μg), the pcDNA5-NFAT1 vector (8 μg), and increasing amounts of the pcDNA4-IRF-2BP2 vector (1 to 8 μg). After 24 h, cells were left untreated or were treated with MG132 (20 μM) and stimulated for 6 h with PMA (10 nM) plus ionomycin (1 μM). The firefly luciferase reporter gene was normalized with a renilla vector (0.1 μg pRL-TK). Results represent means from three independent experiments ± standard deviations.
IRF-2BP2 is a nuclear protein that possesses an NLS.
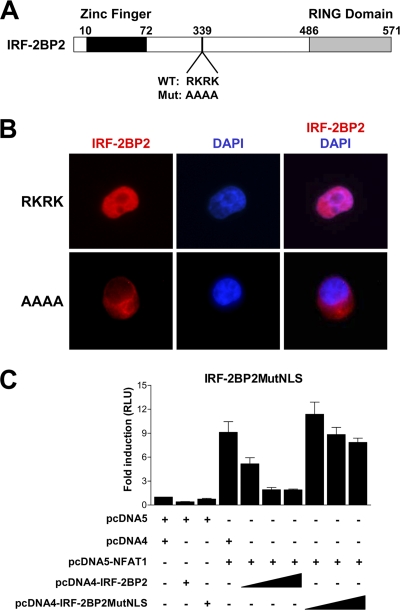
The description of IRF-2BP2 as a nuclear protein (10) is in accordance with our subcellular localization results with HEK293T cells transfected with full-length IRF-2BP2 (Fig. 2C). We investigated whether IRF-2BP2 possesses an NLS that could keep this protein in the nucleus. There is a relatively basic region in IRF-2BP2 which has the sequence RKRK (residues 339 to 342) at its core that could serve as an NLS (Fig. 8A). The ectopic expression of IRF-2BP2 in HEK293T cells shows a correctly regulated intracellular localization, where the protein is restricted to the nucleus (Fig. 8B, upper). The mutation of amino acids RKRK to AAAA in the putative NLS of IRF-2BP2 prevents its nuclear localization. The expression of this mutated protein was restricted to the cytoplasm (Fig. 8B, lower). These findings are consistent with a role for the RKRK residues of IRF-2BP2 in the control of its intracellular localization.
Fig. 8.
IRF-2BP2 repression phenotype depends on its nuclear localization. (A) Schematic representation of IRF-2BP2 and its nuclear localization signal (NLS). The RKRK amino acids from the wild-type protein (WT) were changed to alanine to generate a mutated protein (Mut: AAAA). The boundary of each region is labeled above the sequence; numbers refer to the amino acid position of the protein. (B) Immunofluorescence analysis of HEK293T cells transfected with vectors encoding wild-type IRF-2BP2 (RKRK) or the AAAA-mutated protein. Proteins were labeled with a conjugated rhodamine antibody. DAPI staining shows the position of the nucleus. Results are representative of three independent experiments. (C) Luciferase reporter assay comparing IRF-2BP2 (wild type) and IRF-2BP2MutNLS (AAAA mutation) using the reporter 3× NFAT-Luc. Jurkat cells were electroporated with 3× NFAT-Luc (1 μg), empty vectors (8 μg), the NFAT1 vector (8 μg), and increasing amounts (1 to 8 μg) of IRF-2BP2 constructs. After 24 h, cells were stimulated for 6 h with PMA (10 nM) plus ionomycin (1 μM). The firefly luciferase reporter gene was normalized with a renilla vector (0.1 μg pRL-TK). Results represent means from three independent experiments ± standard deviations.
Further, we evaluated whether the nuclear localization of IRF-2BP2 is important for its activity. We used the 3× NFAT-Luc reporter plasmid to determine whether AAAA-mutated IRF-2BP2 (IRF-2BP2MutNLS) could repress NFAT1-mediated transactivation. Jurkat T cells were transfected with the NFAT1 construct and increasing amounts of wild-type IRF-2BP2 or IRF-2BP2MutNLS vectors, as indicated in Fig. 8C. Twenty-four h after transfection, cells were stimulated for 6 h with PMA plus ionomycin, and luciferase activity was assessed. As shown in Fig. 8C, the mutated IRF-2BP2 (IRF-2BP2MutNLS) that remained in the cytoplasm did not repress NFAT1 transcriptional activity, which indicates that the repressor phenotype observed previously only takes place when the two proteins are colocalized in the nucleus. Furthermore, this result supports the idea that the repression of NFAT1 activity by IRF-2BP2 occurs at the transcriptional level.
DISCUSSION
In this study, we performed a yeast two-hybrid screen using the less-conserved NFAT1 C-terminal end as bait and found a new NFAT1 partner, the transcriptional repressor IRF-2BP2 (Fig. 1). This protein has an N-terminal C4 zinc finger and a C-terminal C3HC4 RING domain and was first identified as an IRF-2-interacting molecule that functions as a corepressor in inhibiting both basal and enhancer-activated transcription (10). The NFAT1–IRF-2BP2 interaction was confirmed by pulldown and coimmunoprecipitation assays (Fig. 2). To understand the phenotype of this interaction, we tested the effect of IRF-2BP2 in NFAT1-mediated transactivation. We observed that IRF-2BP2 represses the NFAT1-mediated transactivation in luciferase assays using NFAT binding sites (3×NFAT and κ3 element) and NFAT1-dependent promoters (IL-2 and IL-4) as reporter plasmids (Fig. 3). It is important to note that IRF-2BP2 does not repress the NF-κB-mediated transactivation (Fig. 3A). Consistently with this, primary CD4 T lymphocytes transduced with IRF-2BP2 vector showed impaired endogenous production of IL-2 and IL-4 cytokines (Fig. 3C), reinforcing the fundamental role of IRF-2BP2 as a repressor of NFAT-regulated genes. The IRF-2BP2 repressor properties also were described more recently by Koeppel et al., who observed that IRF-2BP2 represses the p53-dependent transcription of p21 and Bax genes (35). Interestingly, they did not find direct interaction between p53 and IRF-2BP2, which suggests that the repressor phenotype observed is mediated by endogenous IRF-2 (35). However, it is possible that IRF-2BP2 repression observed in the p21 gene promoter also is mediated by the NFAT1 protein, because this transcription factor regulates p21 in primary mouse keratinocytes (56).
We observed that IRF-2BP2 interacts specifically with the C-terminal end of NFAT1, since the DBD and N terminus do not interact with IRF-2BP2 (Fig. 4A). Another important finding is that the truncated protein at amino acid 698 was not able to interact with IRF-2BP2 (Fig. 4B), and the IRF-2BP2 repressor phenotype observed in NFAT1-mediated transactivation was abolished when the NFAT1 C-terminal end was deleted (Fig. 4C). Furthermore, the NFAT1 and IRF-2BP2 interaction was mapped to a region comprising 86 amino acids at the C-terminal end, spanning amino acids 781 to 867 (Fig. 4D). These data indicated that the repression mediated by IRF-2BP2 is dependent of the interaction between IRF-2BP2 and the NFAT1 C-terminal end. In fact, the analysis of amino acids 781 to 867 demonstrated no sequence identity among NFAT family members (Fig. 5A). Thus, we investigated whether IRF-2BP2 interaction was NFAT1 specific. Our data strongly suggest that IRF-2BP2 is an NFAT1-specific interacting partner, since no interaction with other NFAT proteins was observed (Fig. 5B). Taken together, these data contribute to elucidating a specific regulatory mechanism in the NFAT family, which indicates a new regulatory pathway for NFAT1-mediated gene regulation and may explain the distinct functions of NFAT observed in different studies (13, 24, 50, 51, 55).
Although we observed a complete inhibition of NFAT1 activity in the presence of IRF-2BP2, the signaling events that control IRF-2BP2 regulation are not fully understood, thus other factors may contribute to NFAT regulation under different conditions. Accordingly, accumulating evidence demonstrated that NFAT activity is negatively regulated by coordinated posttranslational mechanisms, which differentially regulate the NFAT proteins in response to specific cellular signals. Inside the nucleus NFAT is inactivated by rephosphorylation by different kinases, including GSK3, DYRK, and CK1 (42, 43, 65), resulting in NFAT export to the cytoplasm. In addition to phosphorylation, several other mechanisms that control NFAT activation have been described, including a noncoding RNA called NRON (noncoding repressor of NFAT) (68), cleavage by caspase-3 (69), ubiquitylation (74, 75), and sumoylation (47, 62). Here, our data demonstrated that IRF-2BP2 is a central negative regulator of NFAT1 protein, which seems to suppress NFAT1 activity at the transcriptional level. However, further studies will be necessary to address the physiological contribution of IRF-2BP2 to the NFAT1 regulatory system and its potential interactions with other regulators.
IRF-2BP2 possesses a RING domain at its C-terminal end. Our data suggest that NFAT1 recruits IRF-2BP2 to the target gene via its C terminus and IRF-2BP2 RING domain, since when its RING domain was deleted, the repressor phenotype of IRF-2BP2 was abolished in luciferase reporter assays (Fig. 6). RING domains of many proteins mediate ubiquitin ligase activity (14). Because IRF-2BP1, an IRF-2BP2-related protein, was identified as a JDP2 ubiquitin ligase (34) and NFAT1 is regulated through ubiquitination in an Akt-dependent manner (75), we tested whether IRF-2BP2 could influence the stability of the NFAT1 protein. We did not observe any reduction in NFAT1 levels in the presence of IRF-2BP2 (Fig. 7A), and we did not see any difference in the luciferase reporter signal in cells left untreated or treated with a proteasome inhibitor (Fig. 7B). These data indicate that the decreased signal observed in the luciferase assays was not related to ubiquitin-dependent NFAT1 degradation. However, our data did not rule out whether IRF-2BP2 targets other protein substrates for ubiquitination or whether it ubiquitinates NFAT1 for nonproteolytic purposes, such as intracellular trafficking or altering the three-dimensional structure of NFAT1.
The mechanism by which IRF-2BP2 represses its target genes is not yet clear. It is possible that IRF-2BP2 interferes with the following functions: the binding of transcriptional activators to DNA, the assembly of basal transcriptional machinery, and the recruitment of histone deacetylases or chromatin-remodeling complexes, which would alter the chromatin structure at the promoters of target genes. Our data showed that when the IRF-2BP2 N terminus was deleted (IRF-2BP2Δzinc), the repressor phenotype mediated by IRF-2BP2 was abolished, although the interaction with NFAT1 was maintained (Fig. 6). The deleted region contains the C4 zinc finger, which suggests that the zinc finger domain is necessary for IRF-2BP2 to function as a repressor.
The C4 zinc finger at the N-terminal end of IRF-2BP2 is an interesting domain that should be considered in further analyses. Zinc finger proteins can bind to DNA and regulate target genes both positively and negatively (36). In addition to interaction with nucleic acids, protein-protein interactions mediated by zinc fingers have been described extensively (44, 46). The MDM2 C4 zinc finger plays an important role in mediating binding to ribosomal proteins and in the degradation of p53 (38). It is possible that the IRF-2BP2 zinc finger binds to DNA and helps to stabilize the interaction with the target promoter, because the affinity of NFAT for DNA is enhanced by interaction with other transcriptional factors or cofactors (52). Another possibility is that the zinc finger interacts with other proteins, such as other transcriptional factors that may bind to the promoter adjacent to NFAT1 elements, histone deacetylases, chromatin-remodeling complexes, or the basal transcriptional machinery. NFAT1 interacts with p300/CBP (16), which has an intrinsic histone acetylase activity and interacts with the basal transcription machinery (9). Thus, IRF-2BP2 may prevent NFAT1 from recruiting p300/CBP to promoters.
IRF-2BP2 has been described as a nuclear protein (10), and NFAT1 translocates to the nucleus when it is dephosphorylated by calcineurin after the activation of the calcium-signaling pathway (39). Our data confirmed the nuclear localization of IRF-2BP2, and we observed that IRF-2BP2 and NFAT1 colocalize in the cell only when NFAT1 is also in the nucleus (Fig. 2C). We also identified an NLS in the IRF-2BP2 protein at amino acid positions 339 to 342 (the RKRK sequence) (Fig. 8A). The mutation of the RKRK sequence to AAAA retained IRF-2BP2 in the cytoplasm (Fig. 8B), which demonstrates that the RKRK sequence acts as a functional NLS. Furthermore, the repressor phenotype mediated by IRF-2BP2 was abolished in the mutated IRF-2BP2 (IRF-2BP2MutNLS) (Fig. 8C); this finding suggests that IRF-2BP2 functions as a repressor in the nucleus. Interestingly, the analysis of the secondary structure of IRF-2BP2 demonstrated that the central region of this protein (amino acids 70 to 490), which includes the NLS, is predicted to be intrinsically disordered. Unstructured regions may adopt rigid structures upon binding to specific ligands (15). However, even if the central region of IRF-2BP2 could fold under special conditions, the NLS appears to always be exposed, because IRF-2BP2 was constitutively nuclear under all conditions tested.
Besides NFAT1, IRF-2 is the only IRF-2BP2 partner described thus far. IRF-2 was described as a factor that bound to the IFN-β promoter and antagonized the effect of the transcriptional activator IRF-1 (19). IRF-2 also has been described as an oncogene. The overexpression of IRF-2 in NIH 3T3 cells resulted in the oncogenic transformation of these cells (20). Furthermore, it was shown that IRF-2 upregulates genes involved in cell cycle regulation, such as the histone H4 gene (64) and cyclin D1 (67). Moreover, specific short interfering RNA (siRNA) to downregulate IRF-2 in TF cells resulted in an augmented IRF1/IRF2 ratio, thereby allowing IRF-1 and p21Waf1 tumor suppressor effects (11). The IRF-2BP2 knockdown data from U2OS cells seem to support this model (35). In fact, a discrete upregulation of p21 expression and increased apoptosis rates after chemotherapeutic treatments in the IRF-2BP2 knockdown cells have been demonstrated (35).
Finally, the physiological role of NFAT1–IRF-2BP2 interaction is not completely understood. In addition to their widely known effects on cytokine gene expression, members of the NFAT family of transcription factors have been shown to regulate genes related to cell cycle progression, cell differentiation, and apoptosis, which indicates a broader role for these proteins in normal cell physiology (29, 42, 65). NFAT1 has been implicated in the control of cell proliferation as a repressor of cell cycle progression (6). Furthermore, NFAT1 was shown to be able to negatively regulate two cell cycle genes, CDK4 and cyclin A2 (1, 8). Data generated by the in vivo gene disruption of the NFAT1 showed an enhanced Th2 development in both in vivo and in vitro models of Th differentiation, as demonstrated by increased levels of IL-4 production (24, 33, 66). These results implicate NFAT1 as a negative controller of genes that regulate cell cycle progression and cell differentiation, thereby supporting the idea that NFAT1 acts as a central regulator of the immune system homeostasis. NFAT1 also has been described as a positive regulator of cell death by apoptosis and also as a tumor suppressor gene (50, 55). Moreover, it has been shown that the overexpression of human NFAT1 leads to an increase in activation-induced cell death (AICD) in primary CD4+ T cells (12). We can hypothesize that in view of the fact that IRF-2BP2 acts by repressing gene transcription mediated by NFAT1, IRF-2BP2 impairs the gene suppressor effect of NFAT1; this action, along with NFAT1-induced AICD, may determine the fate of cells. However, the function of IRF-2BP2 still is unclear, and more studies are necessary to better understand the molecular regulation of the cellular transcriptional machinery by this corepressor and its role in cell physiology.
ACKNOWLEDGMENTS
We are especially grateful to Miriam B. F. Werneck and Bruno K. Robbs for comments on the work and manuscript. We also are grateful to A. Rao for kindly providing the NFAT1 reagents and Nilson Zanchin for the plasmids used in the two-hybrid system.
This work was supported by grants to J.P.B.V. from the ICGEB (CRP/BRA09-01), CNPq (473837/2008-0), and FAPERJ (111.486/2008 and 110.617/2009). F.R.G.C. was supported by a Brazilian Ministry of Health fellowship, and R.R.O. and G.P.M. were supported by a FAPERJ fellowship.
Footnotes
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Baksh S., et al. 2002. NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol. Cell 10:1071–1081 [DOI] [PubMed] [Google Scholar]
- 2. Bartel P. L., Fields S. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254:241–263 [DOI] [PubMed] [Google Scholar]
- 3. Bert A. G., Burrows J., Hawwari A., Vadas M. A., Cockerill P. N. 2000. Reconstitution of T cell-specific transcription directed by composite NFAT/Oct elements. J. Immunol. 165:5646–5655 [DOI] [PubMed] [Google Scholar]
- 4. Bodor J., Bodorova J., Gress R. E. 2000. Suppression of T cell function: a potential role for transcriptional repressor ICER. J. Leukoc. Biol. 67:774–779 [DOI] [PubMed] [Google Scholar]
- 5. Bower K. E., Zeller R. W., Wachsman W., Martinez T., McGuire K. L. 2002. Correlation of transcriptional repression by p21(SNFT) with changes in DNA.NF-AT complex interactions. J. Biol. Chem. 277:34967–34977 [DOI] [PubMed] [Google Scholar]
- 6. Caetano M. S., et al. 2002. NFATC2 transcription factor regulates cell cycle progression during lymphocyte activation: evidence of its involvement in the control of cyclin gene expression. FASEB J. 16:1940–1942 [DOI] [PubMed] [Google Scholar]
- 7. Carneiro F. R., et al. 2006. Spectroscopic characterization of the tumor antigen NY-REN-21 and identification of heterodimer formation with SCAND1. Biochem. Biophys. Res. Commun. 343:260–268 [DOI] [PubMed] [Google Scholar]
- 8. Carvalho L. D., et al. 2007. The NFAT1 transcription factor is a repressor of cyclin A2 gene expression. Cell Cycle 6:1789–1795 [DOI] [PubMed] [Google Scholar]
- 9. Chan H. M., La Thangue N. B. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363–2373 [DOI] [PubMed] [Google Scholar]
- 10. Childs K. S., Goodbourn S. 2003. Identification of novel co-repressor molecules for interferon regulatory factor-2. Nucleic Acids Res. 31:3016–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choo A., et al. 2008. siRNA targeting the IRF2 transcription factor inhibits leukaemic cell growth. Int. J. Oncol. 33:175–183 [PubMed] [Google Scholar]
- 12. Chuvpilo S., et al. 2002. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity 16:881–895 [DOI] [PubMed] [Google Scholar]
- 13. de la Pompa J. L., et al. 1998. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 392:182–186 [DOI] [PubMed] [Google Scholar]
- 14. Deshaies R. J., Joazeiro C. A. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434 [DOI] [PubMed] [Google Scholar]
- 15. Dunker A. K., Cortese M. S., Romero P., Iakoucheva L. M., Uversky V. N. 2005. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 272:5129–5148 [DOI] [PubMed] [Google Scholar]
- 16. García-Rodríguez C., Rao A. 1998. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 187:2031–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graef I. A., Chen F., Chen L., Kuo A., Crabtree G. R. 2001. Signals transduced by Ca(2+)/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 105:863–875 [DOI] [PubMed] [Google Scholar]
- 18. Graef I. A., et al. 1999. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature 401:703–708 [DOI] [PubMed] [Google Scholar]
- 19. Harada H., et al. 1989. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58:729–739 [DOI] [PubMed] [Google Scholar]
- 20. Harada H., et al. 1993. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science 259:971–974 [DOI] [PubMed] [Google Scholar]
- 21. Hedin K. E., et al. 1997. Delta-opioid receptors expressed by Jurkat T cells enhance IL-2 secretion by increasing AP-1 complexes and activity of the NF-AT/AP-1-binding promoter element. J. Immunol. 159:5431–5440 [PubMed] [Google Scholar]
- 22. Hernández G. L., et al. 2001. Selective inhibition of vascular endothelial growth factor-mediated angiogenesis by cyclosporin A: roles of the nuclear factor of activated T cells and cyclooxygenase 2. J. Exp. Med. 193:607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ho I. C., Hodge M. R., Rooney J. W., Glimcher L. H. 1996. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell 85:973–983 [DOI] [PubMed] [Google Scholar]
- 24. Hodge M. R., et al. 1996. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity 4:397–405 [DOI] [PubMed] [Google Scholar]
- 25. Hogan P. G., Chen L., Nardone J., Rao A. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205–2232 [DOI] [PubMed] [Google Scholar]
- 26. Hollenberg S. M., Sternglanz R., Cheng P. F., Weintraub H. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holtz-Heppelmann C. J., Algeciras A., Badley A. D., Paya C. V. 1998. Transcriptional regulation of the human FasL promoter-enhancer region. J. Biol. Chem. 273:4416–4423 [DOI] [PubMed] [Google Scholar]
- 28. Horsley V., Aliprantis A. O., Polak L., Glimcher L. H., Fuchs E. 2008. NFATc1 balances quiescence and proliferation of skin stem cells. Cell 132:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horsley V., Pavlath G. K. 2002. NFAT: ubiquitous regulator of cell differentiation and adaptation. J. Cell Biol. 156:771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ishida N., et al. 2002. Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J. Biol. Chem. 277:41147–41156 [DOI] [PubMed] [Google Scholar]
- 31. Jain J., Burgeon E., Badalian T. M., Hogan P. G., Rao A. 1995. A similar DNA-binding motif in NFAT family proteins and the Rel homology region. J. Biol. Chem. 270:4138–4145 [PubMed] [Google Scholar]
- 32. Jain J., McCaffrey P. G., Valge-Archer V. E., Rao A. 1992. Nuclear factor of activated T cells contains Fos and Jun. Nature 356:801–804 [DOI] [PubMed] [Google Scholar]
- 33. Kiani A., Viola J. P., Lichtman A. H., Rao A. 1997. Down-regulation of IL-4 gene transcription and control of Th2 cell differentiation by a mechanism involving NFAT1. Immunity 7:849–860 [DOI] [PubMed] [Google Scholar]
- 34. Kimura M. 2008. IRF2-binding protein-1 is a JDP2 ubiquitin ligase and an inhibitor of ATF2-dependent transcription. FEBS Lett. 582:2833–2837 [DOI] [PubMed] [Google Scholar]
- 35. Koeppel M., et al. 2009. The novel p53 target gene IRF2BP2 participates in cell survival during the p53 stress response. Nucleic Acids Res. 37:322–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laity J. H., Lee B. M., Wright P. E. 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11:39–46 [DOI] [PubMed] [Google Scholar]
- 37. Levine M., Tjian R. 2003. Transcription regulation and animal diversity. Nature 424:147–151 [DOI] [PubMed] [Google Scholar]
- 38. Lindström M. S., Deisenroth C., Zhang Y. 2007. Putting a finger on growth surveillance: insight into MDM2 zinc finger-ribosomal protein interactions. Cell Cycle 6:434–437 [DOI] [PubMed] [Google Scholar]
- 39. Loh C., et al. 1996. Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J. Biol. Chem. 271:10884–10891 [DOI] [PubMed] [Google Scholar]
- 40. Lopez-Rodríguez C., Aramburu J., Rakeman A. S., Rao A. 1999. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc. Natl. Acad. Sci. U. S. A. 96:7214–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo C., Burgeon E., Rao A. 1996. Mechanisms of transactivation by nuclear factor of activated T cells-1. J. Exp. Med. 184:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macian F. 2005. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 5:472–484 [DOI] [PubMed] [Google Scholar]
- 43. Mancini M., Toker A. 2009. NFAT proteins: emerging roles in cancer progression. Nat. Rev. Cancer 9:810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCarty A. S., Kleiger G., Eisenberg D., Smale S. T. 2003. Selective dimerization of a C2H2 zinc finger subfamily. Mol. Cell 11:459–470 [DOI] [PubMed] [Google Scholar]
- 45. Miyakawa H., Woo S. K., Dahl S. C., Handler J. S., Kwon H. M. 1999. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc. Natl. Acad. Sci. U. S. A. 96:2538–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morgan B., et al. 1997. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 16:2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nayak A., et al. 2009. Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J. Biol. Chem. 284:10935–10946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Otto T. D., et al. 2008. ChromaPipe: a pipeline for analysis, quality control and management for a DNA sequencing facility. Genet. Mol. Res. 7:861–871 [DOI] [PubMed] [Google Scholar]
- 49. Pahl H. L., Baeuerle P. A. 1995. A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-kappa B. EMBO J. 14:2580–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ranger A. M., et al. 2000. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J. Exp. Med. 191:9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ranger A. M., et al. 1998. Delayed lymphoid repopulation with defects in IL-4-driven responses produced by inactivation of NF-ATc. Immunity 8:125–134 [DOI] [PubMed] [Google Scholar]
- 52. Rao A., Luo C., Hogan P. G. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707–747 [DOI] [PubMed] [Google Scholar]
- 53. Rengarajan J., et al. 2002. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195:1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rengarajan J., Tang B., Glimcher L. H. 2002. NFATc2 and NFATc3 regulate T(H)2 differentiation and modulate TCR-responsiveness of naive T(H)cells. Nat. Immunol. 3:48–54 [DOI] [PubMed] [Google Scholar]
- 55. Robbs B. K., Cruz A. L., Werneck M. B., Mognol G. P., Viola J. P. 2008. Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol. Cell. Biol. 28:7168–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Santini M. P., Talora C., Seki T., Bolgan L., Dotto G. P. 2001. Cross talk among calcineurin, Sp1/Sp3, and NFAT in control of p21(WAF1/CIP1) expression in keratinocyte differentiation. Proc. Natl. Acad. Sci. U. S. A. 98:9575–9580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shaw J. P., et al. 1988. Identification of a putative regulator of early T cell activation genes. Science 241:202–205 [PubMed] [Google Scholar]
- 58. Sherman M. A., Powell D. R., Weiss D. L., Brown M. A. 1999. NF-ATc isoforms are differentially expressed and regulated in murine T and mast cells. J. Immunol. 162:2820–2828 [PubMed] [Google Scholar]
- 59. Szabo S. J., Gold J. S., Murphy T. L., Murphy K. M. 1993. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: roles for NF-Y and NF-ATc. Mol. Cell. Biol. 13:4793–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Takayanagi H., et al. 2002. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3:889–901 [DOI] [PubMed] [Google Scholar]
- 61. Teixeira L. K., et al. 2005. IFN-gamma production by CD8+ T cells depends on NFAT1 transcription factor and regulates Th differentiation. J. Immunol. 175:5931–5939 [DOI] [PubMed] [Google Scholar]
- 62. Terui Y., Saad N., Jia S., McKeon F., Yuan J. 2004. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J. Biol. Chem. 279:28257–28265 [DOI] [PubMed] [Google Scholar]
- 63. Tsai E. Y., Jain J., Pesavento P. A., Rao A., Goldfeld A. E. 1996. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol. Cell. Biol. 16:459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vaughan P. S., et al. 1995. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature 377:362–365 [DOI] [PubMed] [Google Scholar]
- 65. Viola J. P., Carvalho L. D., Fonseca B. P., Teixeira L. K. 2005. NFAT transcription factors: from cell cycle to tumor development. Braz. J. Med. Biol. Res. 38:335–344 [DOI] [PubMed] [Google Scholar]
- 66. Viola J. P., Kiani A., Bozza P. T., Rao A. 1998. Regulation of allergic inflammation and eosinophil recruitment in mice lacking the transcription factor NFAT1: role of interleukin-4 (IL-4) and IL-5. Blood 91:2223–2230 [PubMed] [Google Scholar]
- 67. Wang Y., et al. 2007. Involvement of IFN regulatory factor (IRF)-1 and IRF-2 in the formation and progression of human esophageal cancers. Cancer Res. 67:2535–2543 [DOI] [PubMed] [Google Scholar]
- 68. Willingham A. T., et al. 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309:1570–1573 [DOI] [PubMed] [Google Scholar]
- 69. Wu W., et al. 2006. Proteolytic regulation of nuclear factor of activated T (NFAT) c2 cells and NFAT activity by caspase-3. J. Biol. Chem. 281:10682–10690 [DOI] [PubMed] [Google Scholar]
- 70. Wu Y., et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126:375–387 [DOI] [PubMed] [Google Scholar]
- 71. Xanthoudakis S., et al. 1996. An enhanced immune response in mice lacking the transcription factor NFAT1. Science 272:892–895 [DOI] [PubMed] [Google Scholar]
- 72. Yang X. Y., et al. 2000. Activation of human T lymphocytes is inhibited by peroxisome proliferator-activated receptor gamma (PPARgamma) agonists. PPARgamma co-association with transcription factor NFAT. J. Biol. Chem. 275:4541–4544 [DOI] [PubMed] [Google Scholar]
- 73. Yiu G. K., Toker A. 2006. NFAT induces breast cancer cell invasion by promoting the induction of cyclooxygenase-2. J. Biol. Chem. 281:12210–12217 [DOI] [PubMed] [Google Scholar]
- 74. Yoeli-Lerner M., Chin Y. R., Hansen C. K., Toker A. 2009. Akt/protein kinase b and glycogen synthase kinase-3beta signaling pathway regulates cell migration through the NFAT1 transcription factor. Mol. Cancer Res. 7:425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yoeli-Lerner M., et al. 2005. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol. Cell 20:539–550 [DOI] [PubMed] [Google Scholar]
- 76. Yoshida H., et al. 1998. The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production. Immunity 8:115–124 [DOI] [PubMed] [Google Scholar]
- 77. Youn H. D., Chatila T. A., Liu J. O. 2000. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J. 19:4323–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zanchin N. I., Goldfarb D. S. 1999. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol. Cell. Biol. 19:1518–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]