Abstract
The Mediator complex is required for the regulated transcription of nearly all RNA polymerase II-dependent genes. Here we demonstrate a new role for Mediator which appears to be separate from its function as a transcriptional coactivator. Mediator associates directly with heterochromatin at telomeres and influences the exact boundary between active and inactive chromatin. Loss of the Mediator Med5 subunit or mutations in Med7 cause a depletion of the complex from regions located near subtelomeric X elements, which leads to a change in the balance between the Sir2 and Sas2 proteins. These changes in turn result in increased levels of H4K16 acetylation near telomeres and in desilencing of subtelomeric genes. Increases in H4K16 acetylation have been observed at telomeres in aging cells. In agreement with this observation, we found that the loss of MED5 leads to shortening of the Saccharomyces cerevisiae (budding yeast) replicative life span.
INTRODUCTION
In Saccharomyces cerevisiae (budding yeast), telomeric DNA consists of imperfect tandem repeats of the consensus sequence (TG1–3)n, with a combined length of about 300 nucleotides (21). Next to the telomere is the subtelomeric region, which often contains two types of repeats, the Y′ and X elements. The Y′ elements are between 4 and 12 kb long and are located next to the telomeres at many chromosome ends (26). The sizes of X elements vary, but they always contain a “core X” repeat region that is found at nearly all telomeres. Depending on how the Y′ and X elements are distributed, S. cerevisiae chromosome ends can be divided into X and X-Y′ types (22, 23).
Genes situated close to telomeres undergo reversible silencing, a phenomenon that has been termed the telomere position effect (TPE) (28). This effect was first observed when a reporter gene was inserted next to a telomeric TG1–3 tract of an artificial telomere. TPE can also be observed at native yeast telomeres, but the phenomenon appears to be a bit more complex at these locations, since TPE varies between telomeres and in different strain backgrounds (29, 40, 41). The molecular basis of TPE is believed to be the Rap1, Ku, and Sir protein-mediated spreading of heterochromatin-like structures from the telomeric DNA inwards, which represses genes located in the subtelomeric region (42). According to this model, the Rap1/Ku/Sir structures are formed at telomeres and propagate toward the subtelomeres via interactions between the Sir proteins and histone tails. Sir2 is an active histone deacetylase that removes the acetyl group on lysine 16 of histone H4 (H4K16), which allows Sir3 and Sir4 to bind the nonacetylated histone tails (11, 35).
As mentioned, TPE varies between individual chromosome ends, and the exact repeat structure of the subtelomeric region may in fact influence the spread of heterochromatin. Y′ elements counteract the spread of Sir proteins, and the Y′ regions display high levels of H4K16 acetylation. Furthermore, Y′ elements are highly enriched in nucleosomes and are transcriptionally active. In contrast, even X elements situated at some distance from the telomeric ends appear to be repressed transcriptionally. The X elements lack a defined nucleosomal structure, are bound by Sir proteins, and have very low levels of H4K16 acetylation (45). At some telomeres, an actively transcribed Y′ element may even separate the telomeric repeat region from a repressed X element bound by Sir proteins.
Deacetylation of H4K16 by Sir2 stimulates the spread of the Rap1/Ku/Sir structures, whereas Sas2, an H4K16-acetylating enzyme, antagonizes this process (36). The opposing effects of Sir2 and Sas2 generate a gradient of H4K16 acetylation, which in turn marks the boundary between active and silenced chromatin near telomeres. How the balance between Sir2 and Sas2 is regulated is not understood in detail, but a recent report implicated the SAGA subunit Ada2 as a possible regulator of this process. Ada2 was shown to bind telomeric chromatin and the silencing protein Sir2 in vivo, and loss of ADA2 increased the levels of Sir2 and Sir3 in subtelomeric regions, concomitant with decreased H4K16 acetylation (13).
The SIR2 gene has also been identified as a major determinant of the replicative life span in budding yeast (18). Inactivation of SIR2 reduces the yeast life span, whereas an increase in SIR2 dosage extends the life span. The Sir2 protein represses homologous recombination within ribosomal DNA repeats, which reduces the formation of extrachromosomal ribosomal DNA circles and directly reduces the pace of aging in yeast (33). Longevity in yeast might also be regulated by H4K16 acetylation and the chromatin state at telomeres, since there is an age-associated decrease in Sir2 protein occupancy at telomeres in replicatively old yeast cells (7). This change leads to an increase in H4K16 acetylation and causes compromised transcriptional silencing in the subtelomeric region. Interestingly, Sas2 appears to antagonize the effects of Sir2 on chromatin and life span, suggesting that the exact boundary between active and inactive chromatin may directly influence the replicative life span in budding yeast (7).
Mediator is an evolutionarily conserved coregulator complex required for transcription of almost all RNA polymerase II (Pol II)-dependent genes (5). One function of this multiprotein complex is to serve as a functional bridge between gene-specific regulatory proteins bound to upstream elements and the general transcription machinery bound at the promoter. Several pieces of evidence also link Mediator to the maintenance of telomeric heterochromatin. Deletion of genes encoding Mediator components (MED1, MED15, MED18, and MED20) leads to shortening of telomere repeat lengths (2). In addition, deletion of MED15 suppresses silencing defects in a rap1 mutant strain and restores telomere repeat length to near wild-type (wt) levels (37).
We previously observed that purified Mediator interacts directly with reconstituted mononucleosomes (20). Here we demonstrate that Mediator associates with heterochromatin at telomeres and regulates the balance between Sir2 and Sas2. This appears to be a completely new role for the Mediator complex that is distinct from its role as a transcriptional coregulator. Loss of the Med5 component of Mediator causes decreased Mediator occupancy and a defect in the heterochromatin block, which in turn leads to impaired telomeric silencing. The observed effects appear to be functionally significant, since the change of the boundary between active and inactive chromatin at telomeres in the med5Δ strain is associated with a shortening of the replicative life span.
MATERIALS AND METHODS
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (46). Briefly, we cultured yeast cells to an optical density at 600 nm (OD600) of 0.6 to 0.8, followed by cross-linking by incubation with 1% formaldehyde for 10 min at room temperature. The reactions were quenched with 125 mM glycine, and the cells were spun down for 5 min at 3,000 rpm and 4°C. Pellets were washed twice in cold phosphate-buffered saline (PBS) and resuspended in 400 μl cold lysis buffer with protease inhibitors (Roche). The fixed cells were lysed with 400 μl glass beads (Sigma-Aldrich), using a FastPrep-24 machine (MP Biomedicals). The extracts were sonicated to obtain chromatin fragments of about 250 bp (Bioruptor UCD-200; Diagenode). Antibodies used for chromatin immunoprecipitation were against histone H4 (ab2423; Abcam), the C-terminal domain of RNA polymerase II (4H8; Abcam), acetylated H4K16 (ab61240; Abcam), Med1 (a gift from Stefan Björklund, Umeå University, Sweden), Sir2 (ab4626; Abcam), and Sas2 (a gift from Jerry L. Workman, Stowers Institute for Medical Research). After overnight incubation with antibody, 30 μl of protein A Sepharose slurry (GE Healthcare) was added to the reaction mixtures, followed by incubation for 1 h. After being washed and eluted, samples were treated with proteinase K and the cross-link was reversed by overnight incubation at 65°C. DNA was purified by phenol-chloroform extraction, ethanol precipitation, and incubation with RNase A. The purified DNA was used for real-time PCR analysis (Bio-Rad). The names and sequences of the primers used are available at on request. Quantifications were performed using real-time PCR software (Bio-Rad) and Excel (Microsoft); ratios of IP/input are depicted in the figures after subtracting ratios obtained with a nonantibody control.
Global gene expression profiles and data analysis.
Expression profiling data were obtained from a previously published study (39). Moving average analysis was carried out with Microsoft Excel and Access (Microsoft) as previously described (43). For each coding gene, the distance to the nearest chromosome end was calculated based on the translation start site (ATG).
Yeast strains.
Saccharomyces cerevisiae strains used in this study are listed at http://mitochondria.home.dyndns.org/medkem/SuppL.
Electrophoretic mobility shift assays.
The Mediator complex was purified from yeast as previously described (25). Flag-tagged Sir3 protein was overexpressed and purified from yeast as described previously (27). To test binding of Mediator to 32P-labeled mononucleosomes, we performed electrophoretic mobility shift assays as previously described (9, 20, 44), with the following modifications. Binding reaction mixtures (12 μl) comprised a buffer containing 10 mM Tris-Cl (pH 8.0), 5% glycerol, 50 mM NaCl, 0.1 mg/ml bovine serum albumin (BSA), 1 mM dithiothreitol (DTT), and 0.16 nM nucleosomes, with or without Sir3 or Mediator complex at the final concentrations indicated in the figure legends.
Life span analysis.
To determine the replicative life span, wild-type (BY4741) and med5Δ cells were grown to mid-log phase (OD600 ≈ 0.5) at 30°C and plated on yeast extract-peptone-dextrose (YPD) medium. Experiments were performed with 64 virgin cells per plate. Plates were incubated at 30°C during working hours and kept at 4°C overnight. Daughter cells generated by each individual mother were removed and counted by a micromanipulator (Singer Instruments), and the life span was calculated as described previously (15). Statistical assessment of life span differences was determined using the Wilcoxon rank sum test.
RESULTS
Mediator mutant strains affect telomeric silencing.
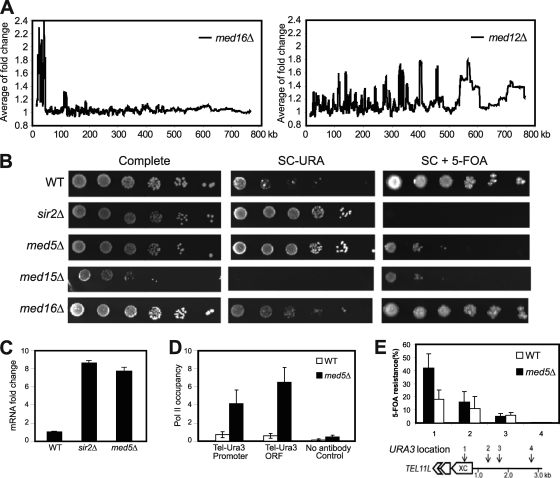
Components of the Cdk8 module of Mediator have been shown to repress transcription of many genes, and genome-wide analysis demonstrated that this effect was independent of gene localization relative to the telomere (Fig. 1A and data not shown). In contrast, deletion of the MED16 gene caused a specific increase in the transcription of genes located in subtelomeric regions (Fig. 1A). We previously reported that Mediator binds near the telomere region (34), and the effect of the med16Δ mutation that we now observed could therefore indicate that Mediator directly regulates transcription activity in the subtelomeric region. To investigate this possibility, we used a URA3 marker inserted next to the (TG1–3)n repeat region of telomere 7L (tel7L) (34). In wt cells, this URA3 marker is subjected to telomeric silencing, but deletion of genes regulating the telomeric structure, e.g., SIR2, has previously been shown to activate expression of this telomeric marker gene. We used this approach to investigate effects in the med16Δ strain. We also investigated effects of MED5 and MED15 deletions. The Med5 protein forms a distinct Mediator submodule together with the Med16 protein. The Med15 protein is also associated with Med16 in the tail module of the Mediator complex but forms its own subcomplex with two other Mediator components, Med2 and Med3. The med15Δ strain displayed a slow-growing phenotype, but we could not observe significant effects on URA3 gene expression in this strain (Fig. 1B). In contrast, loss of MED16 caused a silencing defect, demonstrating that the Med16 protein may be required for telomeric silencing. Surprisingly, the med16Δ strain was also able to survive in the presence of 5-fluoroorotic acid (5-FOA), a substrate that should kill cells expressing URA3. This observation suggested that loss of MED16 might cause a metastable situation, with fluctuation between an active and a repressed state for the URA3 gene, similar to the incomplete suppression of the his4-912δ gene construct by deletion of MED16 (14). Heterochromatin silencing occurs not only at telomeres but also at the mating type loci. In S. cerevisiae, a cells respond to the α cell mating pheromone (the α factor) by growing a projection known as a shmoo. To investigate if the med16Δ cells generated a metastable chromatin phenotype at mating type loci, we investigated shmoo formation after α factor treatment of MATa cells (8). Among wild-type cells, 79% (n = 320) responded to α factor to activate shmoo formation, whereas only 46% of med16Δ cells (n = 518) displayed shmoo formation. Therefore, med16Δ cells display a metastable phenotype at both telomeres and mating type loci. Even though we found this effect very interesting, it made further analysis of subtelomeric silencing in the med16Δ strain difficult.
Fig. 1.
Deletion of Mediator components disrupts transcription silencing at telomeres. (A) Moving average analysis of gene expression changes in med16Δ and med12Δ mutant strains. The distance from a gene to the telomere was calculated based on the position of the translation start site. The window size is 150. (B) Fivefold dilutions of the indicated deletion strains were spotted on complete medium, on synthetic complete medium lacking uracil (SC−URA), and on synthetic complete medium containing 5-FOA (SC + 5-FOA). All strains (UCC3505, CGC209, CGC210, CGC211, and CGC212) contained the URA3 reporter gene inserted at tel7L. (C) Real-time PCR analysis shows fold changes of tel7L URA3 gene transcription in strains UCC3505, CGC210, and CGC209. (D) A ChIP assay using the 4H8 antibody followed by real-time PCR analysis shows Pol II occupancy at URA3 promoter and coding regions of the indicated strains. Background values were calculated using a no-antibody control. Error bars indicate standard deviations based on experiments from at least three independent cultures. (E) The X core elements of telomeres show silencing defects in med5Δ cells. The positions (1 to 4) of URA3 gene insertions near tel11L are indicated. Silencing assays were performed with wild-type and med5Δ cells bearing the URA3 gene inserted at positions 1 to 4. The extent of silencing is expressed as the fraction of cells resistant to 5-FOA (n = 4). Error bars shows standard deviations. (Schematic diagram adapted from reference 7 with permission of the publisher.)
The med5Δ strain also displayed a subtelomeric silencing defect, but in contrast to the case for the med16Δ strain, the phenotype appeared to be stable, since the strain was unable to grow on 5-FOA (Fig. 1B). Real-time quantitative PCR confirmed this observation and revealed upregulation of URA3 transcription in med5Δ cells similar to that seen in the sir2Δ strain (Fig. 1C). Deletion of MED5 affects very few genes, and it was therefore difficult to observe statistically significant changes between different chromatin regions. We could conclude, however, that the frequency of genes affected in the med5Δ strain was slightly higher near telomeres than at other regions, further supporting a role for the Med5 protein in subtelomeric silencing (data not shown). Telomeres have a distinct chromosome conformation which can block binding of Pol II and other transcription factors. We performed a ChIP assay to monitor Pol II occupancy at the promoter and coding regions of the telomeric URA3 gene. In agreement with our observation of increased gene transcription, we found increased Pol II occupancy at both the promoter and coding regions of the gene (Fig. 1D). The tel7L-URA3 strain lacks the PPR1 gene, which encodes an activator of URA3 gene transcription. The absence of the Ppr1 protein may increase the distance over which telomeric silencing of URA3 is observed and therefore create somewhat artificial effects. We therefore also investigated silencing in strains with the PPR1 gene intact in which the URA3 marker had been inserted at different locations of the native yeast telomere 11L (29). In wt cells, URA3 transcription was strongly repressed in the subtelomeric X element, located close to the telomeric repeat region, whereas URA3 located further downstream of the telomere end displayed gradually weaker repression (Fig. 1E). In the med5Δ cells, we observed derepression at the X element as monitored by decreased 5-FOA resistance, demonstrating a role for Med5 in repression of transcription at native telomeres, particularly at X elements.
Reduced Mediator complex occupancy in Med5 mutant.
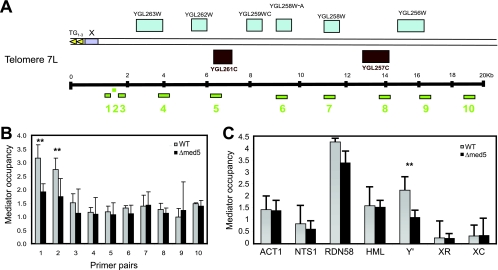
Previously published genome-wide studies have demonstrated the presence of Mediator in subtelomeric regions (1, 10). Given the observed effects of the med5Δ mutation on telomeric silencing, we decided to test whether deletion of MED5 leads to reduced Mediator complex occupancy at this location. To monitor Mediator occupancy, we performed ChIP analysis with an antibody against the Mediator component Med1 (Fig. 2). For our analysis, we used 10 primer pairs detecting genomic regions situated at various distances (0.9 to 19 kb) from the TG1–3 repeats of tel7L (Fig. 2A). We observed a distinct peak of Mediator occupancy near the X element (primer pairs 1 and 2), whereas Mediator occupancy was lower at more distant positions (Fig. 2B). Mediator occupancy at the peak position near the telomeric end (primer pairs 1 and 2) was significantly reduced in the med5Δ mutant strain, whereas other sites were unaffected.
Fig. 2.
Mediator complex occupancy near telomeres. (A) Schematic diagram of primer pairs detecting positions at various distances (0.9 kb to 20 kb) from tel7L. (B) Mediator occupancy at tel7L. ChIP analyses were performed with material from wt (BY4741) and med5Δ mutant (CGC117) strains. Mediator occupancy was monitored using an antibody against the Med1 protein, followed by real-time PCR quantification. Error bars indicate standard deviations for at least three independent cultures. (C) Mediator occupancy at different genomic loci, including euchromatin (ACT1), ribosomal DNA (NTS1 and RDN58), a mating type locus (HML-α1), and subtelomeric regions (the X and Y′ elements of telomere 5R). X elements include X core (XC) positions and X repeat (XR) positions. The analysis was performed as described for panel B. **, P < 0.05.
The subtelomeric region of tel7L contains an X element (positions −781 to −35) situated directly adjacent to the TG1–3 repeats, but very similar X elements are also present at other chromosome ends. Therefore, we could not construct a primer pair that would specifically amplify the X element at tel7L. We could, however, use primer pairs against unique sequences within the X and Y′ elements in the subtelomeric region of tel5R (7). For comparison, we used primers detecting other chromosome regions, i.e., the ACT1 promoter (active euchromatin), NTS1 and RDN58 (transcribed ribosomal DNA), and HML-α1 (mating type locus). We noted that Mediator occupancy was distinctly different between X and Y′ elements in wild-type cells, with the levels of Mediator occupancy being about 10- to 15-fold higher at the Y′ elements than at X elements (Fig. 2C). In fact, we observed nearly no Mediator binding to the tel5R X element, suggesting that Mediator is more or less absent from this region. Interestingly, loss of Med5 led to a reduction of Mediator at Y′ elements but did not affect the other genomic locations investigated. Our data for Mediator occupancy at telomeres C5R and C7L therefore suggested that Mediator is located in genomic regions bordering chromosome ends and X elements, e.g., in the Y′-element region, but that the complex is absent from the X element itself.
Loss of Med5 changes Sir2 and Sas2 protein occupancy at X elements.
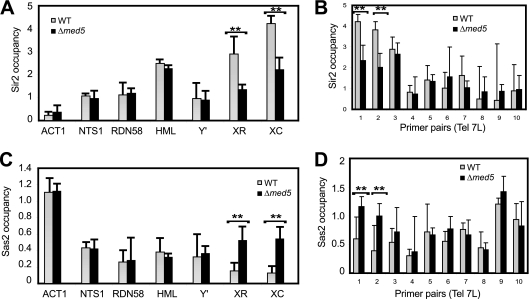
Both TG1–3 repeat regions and X elements lack a defined nucleosome structure and instead are bound by Sir proteins. The opposing effects of Sir2 and Sas2 generate a gradient of H4K16 acetylation, which in turn marks the boundary between active and silenced chromatin near telomeres. Given its genomic location and its requirement for subtelomeric silencing, we wondered if Mediator could directly contribute to the formation of this border. Indeed, deletion of MED5 caused a significant reduction of Sir2 levels at the X element in both telomeres 5R and 7L (Fig. 3A and B). The observed changes were not due to an overall effect on Sir2 levels, since a Western blot analysis of whole-cell extracts demonstrated that the global amounts of the Sir2 protein remained unchanged in the Med5 mutant strain (Fig. 4A).
Fig. 3.
Sir2 and Sas2 protein occupancy changes in med5Δ mutant cells. (A and B) Sir2 occupancy was determined by ChIP assays with wt (BY4741) and med5Δ (CGC117) mutant cells, using the primers shown in Fig. 2. (C and D) Sas2 occupancy was determined as described for panels A and B. Data shown are averages for at least three independent experiments. Error bars represent standard deviations. **, P < 0.05.
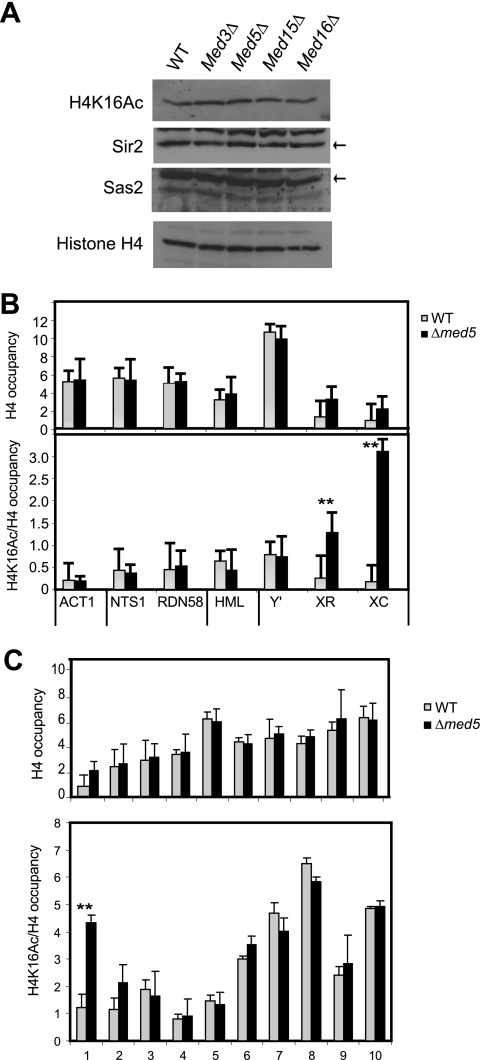
Fig. 4.
H4K16 acetylation levels change in med5Δ mutant cells. (A) Western analyses revealed that the overall levels of Sir2, Sas2, H4K16 acetylation, and H4 remained unchanged in a med5Δ strain (CGC117) compared to a wt strain (BY4741). The Sir2 and Sas2 proteins are indicated with black arrows. (B) ChIP analysis of histone H4 levels and H4K16 acetylation in med5Δ and wt cells. The genomic locations are indicated in Fig. 2. (C) ChIP analysis of histone H4 and H4K16 acetylation levels near telomere 7L. H4K16 acetylation and histone H4 occupancy were normalized to the input. H4K16 acetylation was also normalized to the histone H4 level. Error bars indicate standard deviations calculated for at least three independent cultures. **, P < 0.05.
Given the effects on Sir2 occupancy, we also monitored binding of Sas2, the dominant H4K16 acetyltransferase, which has been shown to antagonize Sir2 in telomeric regions (16, 36). In this experiment, we observed a significant increase in the level of Sas2 occupancy in med5Δ mutant cells at X elements (Fig. 3C and D), whereas Sas2 occupancy at other regions remained unchanged. Our results therefore demonstrated that the Mediator complex could affect the balance between Sir2 and Sas2 in subtelomeric regions.
Loss of Med5 leads to increased H4K16 acetylation levels in the subtelomeric region.
To investigate the functional consequences of the observed changes in Sir2 and Sas2 occupancy, we investigated if deletion of MED5 could influence levels of H4K16 acetylation and the genomic distribution of this modification. Total levels of H4K16 acetylation were monitored by immunoblotting with whole-cell extracts and remained unchanged in the Mediator mutant strains investigated (Fig. 4A). We also monitored the levels of the Sas2 proteins, which remained constant in the wt and mutant cell extracts, similar to the case for Sir2.
We next investigated if loss of MED5 could induce changes in histone occupancy and H4K16 acetylation at different genomic locations. In wt cells, histone H4 occupancy is very low at X elements, and we observed a slight but consistent increase of H4 at these locations in the med5Δ mutant strain (Fig. 4B). No other gross alterations in histone H4 occupancy were observed. We next monitored the levels of H4K16 acetylation normalized to total histone H4 occupancy and noticed a dramatic increase in H4K16 acetylation levels at the X core element of tel5R. We also noted a significant increase at the neighboring X repeat element (Fig. 4B). No other genomic regions were affected.
We next examined the effects of the MED5 gene deletion on the C7L chromosome end that lacks a conserved Y′ element but contains an X element situated next to the telomeric repeat region. In wt cells, we found that histone H4 occupancy and H4K16 acetylation levels were at their lowest close to the X element and telomere end (Fig. 4C). In the med5Δ strain, we observed a strong increase of H4K16 acetylation near the telomere end by use of primer pairs 1 and 2, whereas the other locations were unaffected. Since primer pairs 1 and 2 are located near the boundary of the X element in tel7L, this observation supported the idea that loss of Med5 leads to increased levels of H4K16 acetylation near X elements. Similar to our observations at the tel5R X element, we observed a slight increase in H4 occupancy near the tel7L X element in the med5Δ mutant strain.
Our data therefore demonstrate that Mediator mutations lead to decreased Sir2 occupancy, which is linked to increased Sas2 and H4K16 acetylation levels at X elements. Mediator appears to cause this effect by binding to genomic regions adjacent to X elements but not to the X element itself (Fig. 2C). We next investigated if other Mediator mutations could influence the exact boundary between active and inactive chromatin. We found that a temperature-sensitive mutation in the gene encoding Med7 caused a dramatic change in H4K16 acetylation and in the distribution of Sas2 and Sir proteins (data not shown). We analyzed the tel7L region by using 10 different primer pairs as described above. In the Med7 mutant strain (med7-163), the levels of H4K16 acetylation and Sas2 were increased about 3-fold at positions close to the telomere, whereas Sir3 occupancy was decreased >2-fold at the same positions upon a shift from the permissive to the nonpermissive temperature. The changes observed in the med7-163 strain were therefore similar to but even more dramatic than those observed in the med5Δ mutant strain.
Sir3 and Mediator compete for binding to nucleosomes.
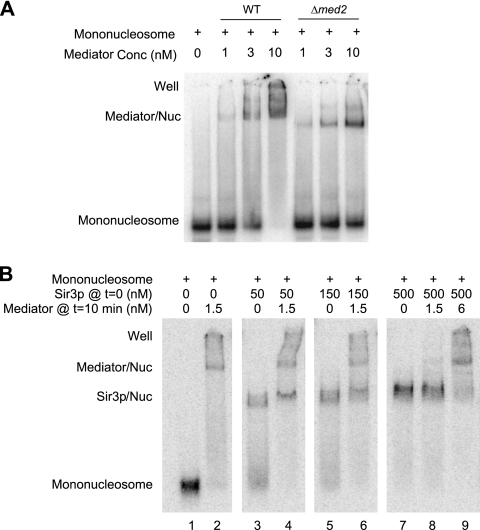
As demonstrated here, both Mediator and Sir proteins bind to subtelomeric regions. Our ChIP data suggested mutually exclusive targeting of Mediator and Sir proteins and that Mediator is not a stoichiometric component of heterochromatin akin to the Sir proteins. We therefore wanted to investigate if Mediator and Sir3 could co-occupy the same nucleosomes in vitro. To investigate this phenomenon in vitro, we followed previously published protocols and purified Mediator to near homogeneity (see Fig. 2C in reference 3). We preincubated purified Mediator with reconstituted nucleosomes and analyzed complex formation in gel retardation experiments (Fig. 5). We noticed the formation of a Mediator-nucleosome complex (Fig. 5A), as previously reported (20). To further verify the identity of this complex, we also used a Mediator complex, purified from Δmed2 cells, that lacks the Med2, Med3, and Med15 subunits (24). We incubated the smaller Δmed2 Mediator with mononucleosomes and compared the results with those obtained for the wild-type Mediator complex. Indeed, the Δmed2 mutant Mediator bound mononucleosomes with an affinity comparable to that observed with wild-type Mediator (Fig. 5A), but the shifted band was lower due to the decreased molecular weight of the mutant Mediator.
Fig. 5.
The Mediator complex and the Sir3 protein compete for binding to mononucleosomes. Electrophoretic mobility shift assays investigated the binding of wild-type and mutant Mediator complexes to 32P-labeled mononucleosomes. Each panel shows the result of an individual experiment where all samples were run in the same gel. Mediator/Nuc, Mediator-mononucleosome cocomplex; Sir3p/Nuc, Sir3-mononucleosome cocomplex; well, material that was unable to enter the gel. (A) Wild-type and Δmed2 Mediator complexes bind to mononucleosomes with comparable affinities. Different concentrations of Mediator were incubated with an invariant concentration of mononucleosomes (0.16 nM), and the reaction mixture was loaded onto the gel. (B) Various concentrations of Sir3 (as shown in the figure) were incubated with 32P-labeled reconstituted mononucleosomes (0.16 nM) for 10 min, after which Mediator (at the concentrations specified in the figure) was added to the binding reaction mixtures, followed by incubation for another 10 min.
Next, we investigated if Mediator could compete with Sir3 for binding to mononucleosomes. We first created Sir3-nucleosome complexes with different concentrations of Sir3. We noted the formation of a distinct complex between Sir3 and mononucleosomes (Fig. 5B, lanes 3, 5, and 7). We added Mediator to free mononucleosomes or to mononucleosomes that had been preincubated for 10 min with the indicated concentrations of Sir3. We noted that Mediator could compete with Sir3 for binding to mononucleosomes in a manner that depended on the concentrations of Sir3 and Mediator (Fig. 5B, lanes 4, 6, 8, and 9). Interestingly, it appeared that Mediator bound to nucleosomes more strongly than did Sir3, since much lower concentrations of Mediator (than of Sir3) were necessary to shift mononucleosomes and to outcompete Sir3 for mononucleosome binding (Fig. 5B).
Deletion of MED5 shortens replicative life span.
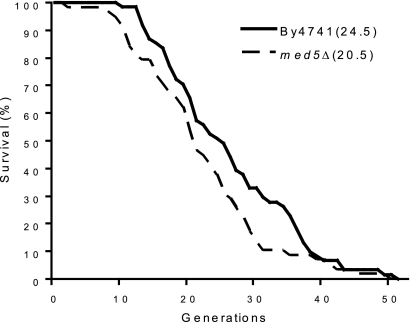
H4K16 acetylation has been implicated in the regulation of cellular life span in S. cerevisiae (7). In old yeast cells, the total protein level of Sir2 was decreased, which in turn led to an increase in H4K16 acetylation and to compromised subtelomeric transcriptional silencing. Since we had observed an effect of Mediator on the balance between Sir2 and Sas2, we decided to examine the effect of a MED5 deletion on replicative life span. In agreement with the idea that Med5 may be a key factor in the regulation of subtelomeric H4K16 acetylation patterns, we observed an approximately 20% decrease of the replicative life span in the med5Δ mutant compared to that for wt cells (Fig. 6).
Fig. 6.
The med5Δ strain displays a shorter replicative life span than that of the wild-type strain (for BY4741, median replicative life span = 24.5 generations; for med5Δ strain, median replicative life span = 20.5 generations; P = 0.029). Life spans were determined by counting the total number of daughters generated by each mother cell. Experiments were performed with 64 virgin cells per plate.
DISCUSSION
In this work, we demonstrate that Mediator associates with telomeric regions and influences the exact boundary between active and inactive chromatin. We suggest that Mediator influences the delicate balance that exists between Sir2 and Sas2. The opposing effects of these enzymes are believed to generate a gradient of H4K16 acetylation, which in turn marks the boundary between active and silenced chromatin near telomeres. Our data suggest that Mediator may interact directly with the subtelomeric region and physically regulate this balance. Mutations that lead to a loss of Mediator from this region cause a decrease of subtelomeric silencing and a shift in the H4K16 acetylation gradient.
Our findings are distinctly different from those presented in a recent report that identified the SAGA subunit Ada2 as a possible regulator of the Sir2-Sas2 balance at telomeres (13). Ada2 was shown to bind telomeric chromatin and the silencing protein Sir2 in vivo, and loss of ADA2 caused a spread of Sir2 and Sir3 into subtelomeric regions and decreased histone H4K16 acetylation. Interestingly, a series of publications demonstrated a close link between the SAGA complex and Mediator (17, 31). The two protein complexes appear to be required for stepwise activation of transcription at many promoters. In addition, recruitment of Mediator and the SAGA complexes by Gcn4 has been shown to be interdependent (30, 38). Furthermore, the SAGA complex makes physical contacts with Mediator, and mutations/deletions of many genes encoding SAGA components (including ADA2) synthetically interact with mutations/deletions of Mediator-encoding genes (6, 19).
In our studies, we observed a dramatic increase of H4K16 acetylation at the conserved X core element and also noticed a slight increase of histone H4 levels. The function of this element is not well understood, but in wt cells it is depleted of nucleosomes and instead bound by typical telomeric proteins, e.g., Sir2 and Rap1 (45). Based on our findings, we suggest that Mediator helps to set up the boundary between X elements bound by Sir proteins and surrounding regions bound by Sas2. In support of this notion, we found that deletion of MED5 or temperature-sensitive mutations in MED7 could disturb the balance between Sir2 and Sas2, consequently allowing nucleosomes acetylated at H4K16 to “leak” into nearby X elements.
How Mediator is recruited to X element border regions is not yet clear, but one factor could be the modification status of the histones, since Mediator can interact with histone H4 N-terminal tail peptides and H4K16 acetylation has a strong negative effect on this interaction (X. Zhu et al., submitted for publication). Even if Mediator and Sir3 can both bind to deacetylated H4 N-terminal tails and nucleosomes in vitro, they cannot do so simultaneously. Instead, our experiments suggest that they are mutually exclusive, supporting the idea that Mediator may function as a border element. But Mediator blocks not only Sir3 binding but also spreading of Sas2, which may explain why the loss of Mediator does not lead to increased Sir3 occupancy but instead to a spreading of Sas2 and H4K16 acetylation into X elements. In support of this idea, overexpression of the Sas2 protein causes lower occupancy of the Sir2 protein and higher H4K16 acetylation levels in telomeric regions (32). Interestingly, the increase of H4K16 acetylation seen in aging cells is also especially pronounced within X elements (7).
Med5 is structurally located at the interface of the tail and middle modules of Mediator, and a med5Δ mutant strain displays increased respiration and mitochondrial activity (4). This result could potentially be coupled to our observation of a shortening life span in med5Δ deletion cells, since impaired respiratory chain function has been linked to premature aging in many organisms. However, we determined the life span of another Mediator tail component mutant strain, the med16Δ strain, that also displayed a similar increase in respiratory activity. We failed to observe changes in replicative life span in the med16Δ strain, arguing against changed mitochondrial activity being the reason for aging in the med5Δ mutant cells (data not shown). The Med5 protein has also been identified as an active histone acetyltransferase, but its substrate specificity remains unknown (12). We do, however, find it unlikely that Med5 directly acetylates H4K16, since the observed increase in H4K16 acetylation levels was associated with a decrease of Mediator in the subtelomeric region. In addition, we cannot rule out the possibility that Med5 acts in concert with other components of the Mediator complex. We previously reported that Mediator interacts directly with Med16 and forms a specific Mediator subcomplex in the tail module of the Mediator complex. Previous reports have demonstrated that loss of MED16 is associated with global alterations in chromatin accessibility (1, 46), and as demonstrated here, deletion of MED16 also affects subtelomeric silencing. It is therefore possible that Med5 and Med16 together form a Mediator submodule that affects chromatin structure at specific genomic locations. Med16 (in contrast to Med5) is conserved in evolution, and Mediator may therefore affect chromatin structure in higher eukaryotes as well.
A large number of studies have identified the Mediator complex as the component of the transcription preinitiation complex required to stimulate Pol II-dependent transcription. Genome-wide occupancy studies have slightly modified this view, however, and demonstrated distinctly different patterns for Mediator and Pol II (1, 46). Mediator is present at many regions throughout the subtelomeric regions, whereas Pol II is depleted from these locations. Furthermore, Mediator is also enriched at other heterochromatin regions, such as the centromere (10). Our observations suggest that Mediator could contribute to the formation of a border between active and inactive chromatin regions. In support of this notion, loss of Med5 leads to a pronounced change in histone occupancy and H4K16 acetylation levels in X elements but does not change Mediator occupancy within the X elements. Instead, loss of Med5 affects Mediator occupancy at adjacent regions bordering the X elements. It is tempting to speculate that Mediator may have a similar function at some gene regulatory regions and that a role for Mediator in defining the border between active and inactive chromatin may be a recurring theme at many genomic locations.
ACKNOWLEDGMENTS
We thank Jerry Workman for providing antibodies against Sas2 and Stefan Björklund for antibodies against Med1. We thank Danesh Moazed for the Sir3-Flag yeast expression plasmid. We also thank Edward J. Louis for providing a yeast strain bearing the tel11L URA3 marker.
This work was supported by grants to C.M.G. from the Swedish Research Council, the Swedish Cancer Society, the European Research Council, and the IngaBritt and Arne Lundberg Research Foundation. This work was also supported by NIH grant GM62483 (to L.C.M.).
Footnotes
Published ahead of print on 11 April 2011.
REFERENCES
- 1. Andrau J. C., et al. 2006. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22: 179–192 [DOI] [PubMed] [Google Scholar]
- 2. Askree S. H., et al. 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. U. S. A. 101: 8658–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baidoobonso S. M., Guidi B. W., Myers L. C. 2007. Med19(Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae mediator complex. J. Biol. Chem. 282: 5551–5559 [DOI] [PubMed] [Google Scholar]
- 4. Beve J., et al. 2005. The structural and functional role of Med5 in the yeast Mediator tail module. J. Biol. Chem. 280: 41366–41372 [DOI] [PubMed] [Google Scholar]
- 5. Bjorklund S., Gustafsson C. M. 2004. The mediator complex. Adv. Protein Chem. 67: 43–65 [DOI] [PubMed] [Google Scholar]
- 6. Collins S. R., et al. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- 7. Dang W., et al. 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Day A., Schneider C., Schneider B. L. 2004. Yeast cell synchronization. Methods Mol. Biol. 241: 55–76 [DOI] [PubMed] [Google Scholar]
- 9. Dyer P. N., et al. 2004. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375: 23–44 [DOI] [PubMed] [Google Scholar]
- 10. Esnault C., et al. 2008. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell 31: 337–346 [DOI] [PubMed] [Google Scholar]
- 11. Grunstein M. 1997. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 9: 383–387 [DOI] [PubMed] [Google Scholar]
- 12. Gustafsson C. M., et al. 1998. Identification of new mediator subunits in the RNA polymerase II holoenzyme from Saccharomyces cerevisiae. J. Biol. Chem. 273: 30851–30854 [DOI] [PubMed] [Google Scholar]
- 13. Jacobson S., Pillus L. 2009. The SAGA subunit Ada2 functions in transcriptional silencing. Mol. Cell. Biol. 29: 6033–6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang Y. W., Stillman D. J. 1996. Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev. 10: 604–619 [DOI] [PubMed] [Google Scholar]
- 15. Kennedy B. K., Austriaco N. R., Jr., Guarente L. 1994. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell Biol. 127: 1985–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kimura A., Umehara T., Horikoshi M. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32: 370–377 [DOI] [PubMed] [Google Scholar]
- 17. Larschan E., Winston F. 2005. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 25: 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin S. J., Defossez P. A., Guarente L. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289: 2126–2128 [DOI] [PubMed] [Google Scholar]
- 19. Lin Y. Y., et al. 2008. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 22: 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lorch Y., Beve J., Gustafsson C. M., Myers L. C., Kornberg R. D. 2000. Mediator-nucleosome interaction. Mol. Cell 6: 197–201 [DOI] [PubMed] [Google Scholar]
- 21. Louis E. J. 1995. The chromosome ends of Saccharomyces cerevisiae. Yeast 11: 1553–1573 [DOI] [PubMed] [Google Scholar]
- 22. Louis E. J. 1994. Corrected sequence for the right telomere of Saccharomyces cerevisiae chromosome III. Yeast 10: 271–274 [DOI] [PubMed] [Google Scholar]
- 23. Louis E. J., Naumova E. S., Lee A., Naumov G., Haber J. E. 1994. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics 136: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Myers L. C., Gustafsson C. M., Hayashibara K. C., Brown P. O., Kornberg R. D. 1999. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl. Acad. Sci. U. S. A. 96: 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Myers L. C., Leuther K., Bushnell D. A., Gustafsson C. M., Kornberg R. D. 1997. Yeast RNA polymerase II transcription reconstituted with purified proteins. Methods 12: 212–216 [DOI] [PubMed] [Google Scholar]
- 26. Neidle S., Parkinson G. N. 2003. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 13: 275–283 [DOI] [PubMed] [Google Scholar]
- 27. Onishi M., Liou G. G., Buchberger J. R., Walz T., Moazed D. 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 28: 1015–1028 [DOI] [PubMed] [Google Scholar]
- 28. Ottaviani A., Gilson E., Magdinier F. 2008. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie 90: 93–107 [DOI] [PubMed] [Google Scholar]
- 29. Pryde F. E., Louis E. J. 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18: 2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qiu H., et al. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roberts S. M., Winston F. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shia W. J., Li B., Workman J. L. 2006. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 20: 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sinclair D. A., Lin S. J., Guarente L. 2006. Life-span extension in yeast. Science 312: 195–197 [DOI] [PubMed] [Google Scholar]
- 34. Singer M. S., Gottschling D. E. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- 35. Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11: 83–93 [DOI] [PubMed] [Google Scholar]
- 36. Suka N., Luo K., Grunstein M. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32: 378–383 [DOI] [PubMed] [Google Scholar]
- 37. Sussel L., Vannier D., Shore D. 1995. Suppressors of defective silencing in yeast: effects on transcriptional repression at the HMR locus, cell growth and telomere structure. Genetics 141: 873–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Topalidou I., Thireos G. 2003. Gcn4 occupancy of open reading frame regions results in the recruitment of chromatin-modifying complexes but not the mediator complex. EMBO Rep. 4: 872–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van de Peppel J., et al. 2005. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19: 511–522 [DOI] [PubMed] [Google Scholar]
- 40. Vega-Palas M. A., Martin-Figueroa E., Florencio F. J. 2000. Telomeric silencing of a natural subtelomeric gene. Mol. Gen. Genet. 263: 287–291 [DOI] [PubMed] [Google Scholar]
- 41. Vega-Palas M. A., Venditti S., Di Mauro E. 1997. Telomeric transcriptional silencing in a natural context. Nat. Genet. 15: 232–233 [DOI] [PubMed] [Google Scholar]
- 42. Venditti S., Vega-Palas M. A., Di Mauro E. 1999. Heterochromatin organization of a natural yeast telomere. Recruitment of Sir3p through interaction with histone H4 N terminus is required for the establishment of repressive structures. J. Biol. Chem. 274: 1928–1933 [DOI] [PubMed] [Google Scholar]
- 43. Wiren M., et al. 2005. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 24: 2906–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wittmeyer J., Saha A., Cairns B. 2004. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 377: 322–343 [DOI] [PubMed] [Google Scholar]
- 45. Zhu X., Gustafsson C. M. 2009. Distinct differences in chromatin structure at subtelomeric X and Y′ elements in budding yeast. PLoS One 4: e6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu X., et al. 2006. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol. Cell 22: 169–178 [DOI] [PubMed] [Google Scholar]