Abstract
How early- and late-firing origins are selected on eukaryotic chromosomes is largely unknown. Here, we show that Mrc1, a conserved factor required for stabilization of stalled replication forks, selectively binds to the early-firing origins in a manner independent of Cdc45 and Hsk1 kinase in the fission yeast Schizosaccharomyces pombe. In mrc1Δ cells (and in swi1Δ cells to some extent), efficiency of firing is stimulated, and its timing is advanced selectively at those origins that are normally bound by Mrc1. In contrast, the late or inefficient origins which are not bound by Mrc1 are not activated in mrc1Δ cells. The enhanced firing and precocious Cdc45 loading at Mrc1-bound early-firing origins are not observed in a checkpoint mutant of mrc1, suggesting that non-checkpoint function is involved in maintaining the normal program of early-firing origins. We propose that prefiring binding of Mrc1 is an important marker of early-firing origins which are precociously activated by the absence of this protein.
INTRODUCTION
Initiation of eukaryotic DNA replication proceeds in two steps: assembly of prereplicative complexes (pre-RCs) on the chromosomes during early G1 and activation of selected pre-RCs during S phase (22, 28). Although the components of pre-RCs and the assembly process as well as regulation of their formation are well understood, the processes of firing still remain largely elusive. Especially, the mechanisms of selection of the origins to be fired and regulation of the timing of firing are two critical issues of eukaryotic DNA replication which are now under intensive study. Studies in the budding yeast Saccharomyces cerevisiae indicate the presence of the origins that are fired efficiently in early to mid-S phase and of others that are fired late in S phase (38). Suppression of late-origin firing is regulated by the Mec1-Rad53/Rad3-Cds1 checkpoint pathway in response to replication stress (25, 39, 42) and by chromatin structures through histone acetylation (2, 12, 49). These are conserved in other species as well. However, it is not clear whether there is a critical determinant for early replication on eukaryotic chromosomes during the normal course of S phase. It was also reported in the fission yeast Schizosaccharomyces pombe that early recruitment of origin recognition complex (ORC) and pre-RC components correlates with early firing in S phase (52).
Studies in the fission yeast or mammalian cells indicate a great deal of flexibility in origin selection, and it was even proposed that origins are selected from the preformed pre-RCs in a stochastic manner (6, 36). Indeed, single-cell analyses in yeast cells showed that the site selection for firing occurs rather randomly in each cell cycle of a single cell (36). A reverse correlation between the fork progression rate and firing frequency was reported, and this could also be explained by stochastic activation of pre-RCs (5, 26). On the other hand, genome-wide determination of origins that are fired in the presence of hydroxyurea (HU) clearly indicates the presence of specific pre-RCs that are not restrained by checkpoint pathways, and most of these origins fire in early to mid-S phase (9, 14, 15). In mammalian cells, a genome-wide program of replication timing has been determined in several cell lines, and the presence of early- and late-replication domains on the chromosomes was clearly demonstrated (20). The presence of early- and late-replicating domains was suggested even in budding yeast (30). Furthermore, these replication domains can undergo drastic reorganization during the course of differentiation (17). These results indicate the presence of domains on the chromosome where replication occurs more frequently during early S phase and others where firing occurs much less frequently.
Mrc1, Swi1, and Swi3 are conserved replication fork factors which function to stabilize stalled forks and are also required for transmission of checkpoint signals (1, 21, 35, 44). By using a chromatin immunoprecipitation-microarray analysis (ChIP-chip) method, we show that Mrc1 is loaded selectively onto origins that are actively fired in the early S phase in a manner independent of Hsk1 kinase and Cdc45. In mrc1Δ or swi1Δ cells, firing is enhanced and advanced in timing at those origins bound with Mrc1 but not at unoccupied origins. This stimulation is not observed in cds1Δ cells or a checkpoint-defective mutant of mrc1. Precocious binding of Cdc45 was observed in mrc1Δ cells at these stimulated origins. These results indicate that Mrc1 may somehow selectively recognize early-firing origins, which are hyper-activated in the mutants lacking fork-stabilizing factors. Our results show a novel checkpoint-independent function of Mrc1 which may define early-firing origins among the potential origins scattered on the chromosome.
MATERIALS AND METHODS
Strains and general methods.
All the strains used in this study are listed in Table S1 in the supplemental material. All epitope-tagged strains were made by integration of a 3FLAG- or 13Myc-tagged gene fragment into the original chromosome loci. All the tags were at the C terminus. The herpes simplex thymidine kinase (TK) gene from pAUR-GDP-TK (21) and the human equilibrative nucleoside transporter 1 (ENT1) gene whose codon usage was optimized for yeast (24) were expressed by placing them between the Adh1 promoters and terminator amplified from S. pombe genomic DNA. These genes and the aur1r gene amplified from pAUR224 (Takara Bio, Inc.) were cloned into pBluescript SK(−). The plasmid was digested with SmiI and transformed and integrated into the aur1 locus.
Yeast cultures and FACS analysis.
Cells were cultured in YES medium containing 0.5% yeast extract, 2% glucose, and 0.1 mg/ml each of adenine, uracil, and leucine (Sigma). An nda3-KM311 mutant was incubated at 20°C for 5 h (or 8 h for the hsk1-89 background), unless indicated otherwise in the figure legends, to induce arrest in M phase and then released into growth by shifting to 30°C, a restriction temperature for hsk1-89 cells. A cdc25-22 mutant was incubated at 36.5°C for 3 h to cause G2 phase arrest and then released into growth at 25°C. For fluorescence-activated cell sorting (FACS) analyses, cells were fixed with 70% ethanol. After treatment with RNase A at 37°C for 2 h, DNA was stained with 4 μg/ml propidium iodide.
Two-dimensional gel electrophoresis analysis of replication intermediates.
Genomic DNA was prepared as previously described (3). Briefly, 1.2 × 108 cells, fixed with 1% (wt/vol) NaN3, were suspended in SP1 buffer at pH 5.6 (1.2 M sorbitol, 50 mM citrate phosphate, and 40 mM EDTA) with 0.7 mg/ml lyticase. After incubation at 37°C for 1 h, the spheroplasts were suspended in 0.5% low-melting-point agarose and poured into CHEF Plug molds (Bio-Rad) to make agarose plugs. The plugs were treated with DB buffer (1% lauryl sarcosine, 0.25 mg/ml proteinase K, and 25 mM EDTA) and an appropriate restriction enzyme (40 U of EcoT22I, BglII, or EcoRI per plug) followed by incubation at 37°C for 1 h. The plugs were then melted by incubation at 70°C for 10 min with gentle mixing. The restriction enzyme (40 U of enzyme/plug) was added again, and the solid samples were incubated at 37°C for 1 h. Subsequently, the solid samples were treated with 4 U of β-agarase I (NEB) and 40 μg/ml RNase A (Sigma) at 37°C for 1 h. DNA was precipitated by addition of 0.7 volume of isopropanol and 0.1 volume of 3 M sodium acetate (NaOAc). The DNA samples were analyzed on 0.32% agarose in the first dimension and on 0.9% agarose in the second dimension (both in Tris-borate-EDTA [TBE] buffer at 4°C). The DNA was transferred to N+ membrane (Amersham) and cross-linked with UV. The DNA probes were prepared by using a Megaprime DNA Labeling System (Amersham). The specific signals were detected by imaging plates (Fuji BAS2000). Individual sequences of the primers used for probes are as follows: ars2004 HaeIII, 5′-AAAGTGCGTGCATGGCTTTAGG-3′ (sense) and 5′-TGAGAGAGTACAGTCAAGCGTAGAG-3′ (antisense); ars1 BglII, 5′-GGTTGCCCTGCAGGAAATTG-3′ (sense) and 5′-TTCTTTGTGGGGCTAGCCTGTA-3′ (antisense); ori1–200 EcoRI, 5′-TGGGCAAAACGCTTGAAA-3′ (sense) and 5′-GCTGATTCCTTCACTAACTCT-3′ (antisense); non-ori1–223 EcoRI, 5′-GAAATCACCACCAGGAACA-3′ (sense) and5′-GAATTGGATGAAGAAGAGGAG-3′ (antisense); AT2080 HindIII, 5′-CCGCCATTTGATGTAATCCT-3′ (sense) and 5′-TTGGTGTTTGTGGTCTGCT-3′ (antisense).
Chromatin immunoprecipitation assay and real-time PCR.
A ChIP assay was performed as previously described with some modifications (21, 54). We disrupted collected cells (2.5 × 108 cells) by a Multibead Shocker (Yasui-kikai Co., Osaka), and DNA was sheared to 600 bp by sonication. Immunoprecipitation was performed by using monoclonal anti-c-Myc antibody (Nacalai Tesque) or monoclonal anti-FLAG M2 antibody (Sigma). After incubation of lysate with antibody and protein G beads (Dynal) for 3 h, beads were washed three times with the lysis buffer (54). Coimmunoprecipitated DNA was purified by a MiniElute Reaction Cleanup Kit (Qiagen). The following primers were used for amplifying the DNA: ars2004, 5′-CTTTTGGGTAGTTTTCGGATCC-3′ (sense) and 5′-ATGAGTACTTGTCACGAATTC-3′ (antisense); non-ori1 5′-TCGAAGATCCTACCGCTTTC-3′ (sense) and 5′-GATTCACATAACCCGCTAGC-3′ (antisense); non-ori2, 5′-ATGTATAGCTGGAACGCCTG-3′ (sense) and 5′-TTCCTCAAATCACCCCACGT-3′ (antisense); ars1, 5′-AGACAGAATGGGATACAAG-3′ (sense) and 5′-GGCAGTTACATTGTTGGA-3′ (antisense). Real-time PCR was performed by using SYBR Premix Ex Taq (Takara) and LightCycler 480 (Roche Diagnostics). The extent of enrichment of origin DNA was determined by calculating the ratio of ars2004 to non-ori1 (30 kb away from ars2004) or that of ars1 to non-ori1, and measurements were determined by real-time PCR at each time point. Bromodeoxyuridine (BrdU) incorporation and immunoprecipitation (IP) analyses were performed as described previously (21).
ChIP-chip.
Chromatin-immunoprecipitated DNA was amplified, labeled, and hybridized with a GeneChip S. pombe tiling array (Affymetrix), as described previously (14). ChIP signals were validated by using previously described criteria (21, 51). A detection P value of ≤0.001 was used in this experiment. A summary of BrdU incorporation and Mcm4 and Mrc1 binding in the wild-type cells is presented in Table S2 in the supplemental material. The entire raw data for some of the ChIP-chip analyses are presented in Fig. S5 to S10 in the supplemental material.
Microarry data accession number.
The raw and processed microarray data are accessible through GEO series accession number GSE28182 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28182).
RESULTS
Mrc1 associates with early-firing origins.
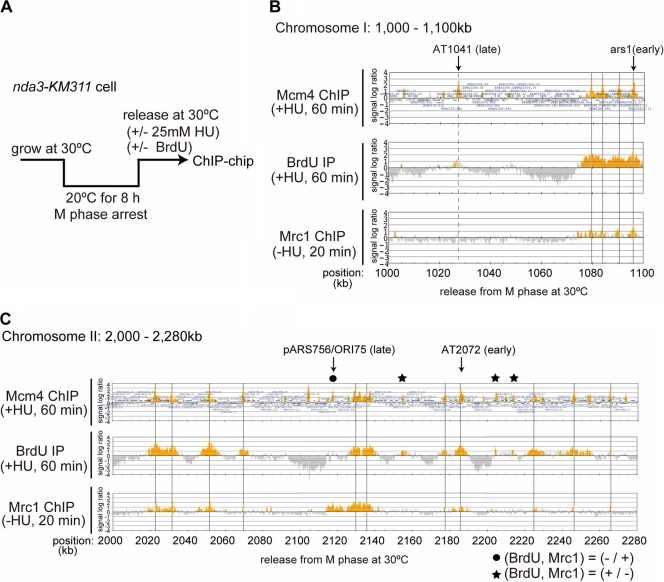
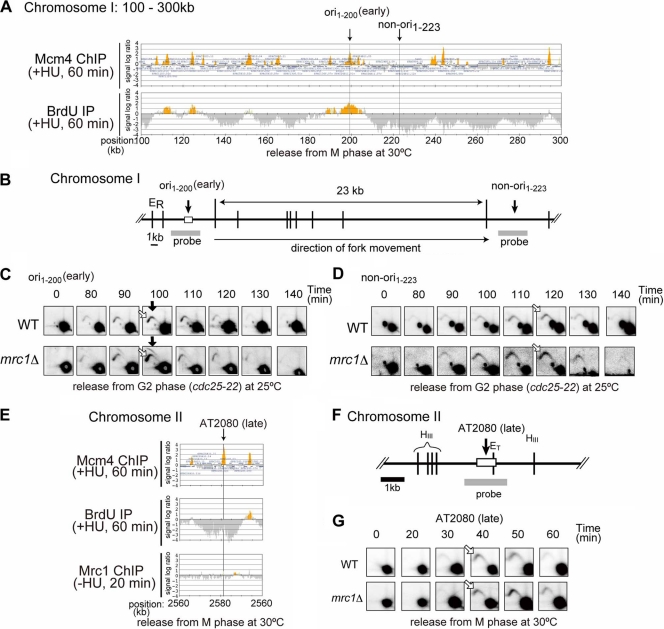
We employed ChIP (chromatin immunoprecipitation)-chip assays using a tiling array to determine Mrc1 localization on fission yeast chromosomes. First, locations of pre-RC and the early-firing origins were determined by immunoprecipitation of Mcm4 and BrdU-incorporating DNA, respectively, in cells released from nda3-KM311-induced metaphase arrest (16) into HU-containing medium for 1 h. Under this condition, cells arrested at early S phase can be collected (Fig. 1A; see also Fig. 4A). We detected 982 potential origins (MCM4 binding sites), and among them 561 fired in the presence of HU and were defined as early-firing origins (see Table S2 in the supplemental material). As pre-RC assembly sites (potential replication origins) were clustered in subtelomere and centromere regions, we did not count origins in these regions. Excluding these regions, most of the previously reported pre-RC locations that were detected in the presence of HU (354 out of the 431 origins reported) (14) were identified in our studies. Although ars1 and ars2004, well-characterized early-firing origins, fired under this condition (Fig. 1B; see Fig. S1A in the supplemental material), AT1041, to which Mcm4 bound, did not fire, as reported previously (Fig. 1B) (14). This is consistent with the notion that ars1 and ars2004 are early-firing origins and that AT1041 is an inefficient or late-firing origin (Fig. 1B, Mcm4 ChIP and BrdU IP). Mrc1 was immunoprecipitated by using cells collected at 20 min after release in the absence of HU to obtain a cell population which is predominantly in late G1 to early S phase. The result shows that Mrc1 binds to ars1 and ars2004 but not to AT1041 (Fig. 1B; see also Fig. S1A, Mrc1 ChIP-HU, in the supplemental material). Figure 1C presents the result of another genomic segment, showing again the colocalization of Mrc1 at the majority of the early-firing origins. Genome-wide examination shows that among 982 Mcm4 binding sites, 342 were bound with Mrc1. The majority (90.0%, or 308/342) of the Mrc1-bound sites were at early-firing origins (origins firing in the presence of HU) (Fig. 1B and C, gray lines), and only a subset (10.0%, or 34/342) of the Mrc1-bound sites were at silent origins (Fig. 1C, filled circles; see also Table S2 in the supplemental material). pARS756/ORI75 was previously classified as an early-firing origin (14), but we did not detect initiation signals either in ChIP-chip or in two-dimensional gel analyses (data not shown). We speculate that firing may be suppressed at this origin due to origin interference from the adjacent origins. Mrc1 did not bind to some early-firing origins (39.0%, or 219/561) which fired in the presence of HU (Fig. 1C; see also Table S2). However, these origins were generally very weak, incorporating only a low level of BrdU (Fig. 1C, stars). We have attempted to detect firing at such weak origins (ori1-563 and ori1-1255) during a normal cell cycle by neutral/neutral two-dimensional gel electrophoresis analysis in the absence of HU, but we detected no signals (ori1-563) or detected only a very weak initiation signal during late S phase (ori1-1255), as if the origins are late-firing origins in the normal cell cycle (see Fig. S2 in the supplemental material).
Fig. 1.
Mrc1 localizes at early-firing replication origins. (A) Scheme of the experiment. nda3-KM311 cells grown at 30°C were arrested in M phase by incubation at 20°C for 8 h and then released into cell cycle. (B and C) Mrc1 preferentially binds to an early-firing origin in G1/S phase. After incubation at 30°C for 1 h in the presence of 25 mM HU and 200 μM BrdU (Mcm4 ChIP and BrdU IP, respectively) or for 20 min in the absence of HU (Mrc1 ChIP −HU), cells were collected, and immunoprecipitation with anti-FLAG antibody (for Mcm4 and Mrc1) or with anti-BrdU antibody (for newly synthesized DNA) was conducted. Precipitated DNA was purified and analyzed by using a GeneChip S. pombe tiling array (Affymetrix). Cells used were the following: Mcm4 and BrdU, aur1r-Adh-TK-Adh1-scENT1 mcm4-3flag nda3-KM311; Mrc1 +/−HU, mrc1-5flag nda3-KM311. ChIP-chip data of selected genome loci (B, 1,000 kb to 1,100 kb of chromosome I; C, 2,000 kb to 2,280 kb of chromosome II) are indicated. Vertical gray lines indicate the positions of active origins which colocalize with Mrc1 binding sites including some of the previously characterized origins (ars1 and AT2072 [early origins]). The gray dotted line in panel B indicates AT1041 (late or dormant origin which does not fire in the presence of HU) to which Mrc1 is not bound. A filled circle (pARS756/ORI75) indicates a silent (or late) origin to which Mrc1 is bound, and stars indicate weak early-firing origins which are not bound by Mrc1.
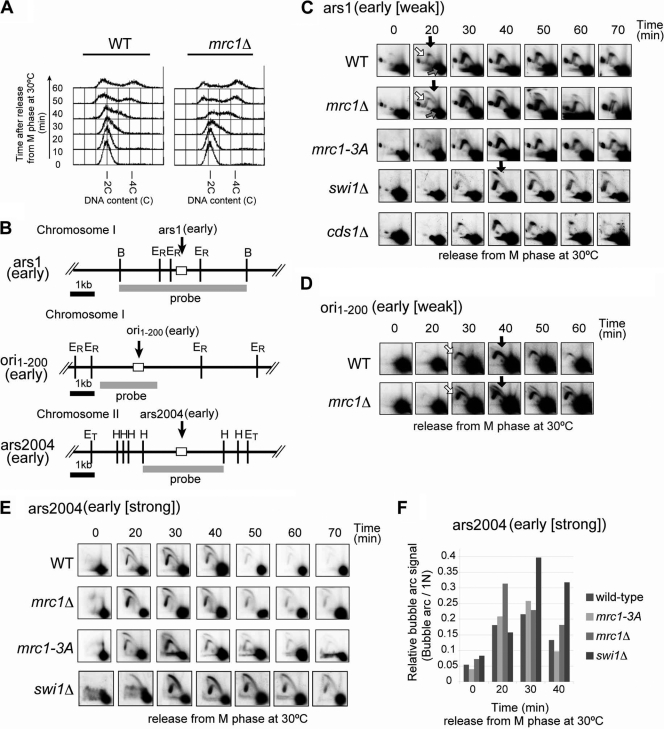
Fig. 4.
Timing of initiation is advanced, and initiation is stimulated at early-firing origins in mrc1Δ cells but not in a checkpoint-deficient mutant of mrc1 or cds1Δ cells. Wild-type, mrc1Δ, mrc1-3A (S604A T645A T653A), swi1Δ, and cds1Δ cells were arrested at M phase and released into cell cycle at 30°C. Cells were harvested at indicated time points. (A) FACS analyses of DNA contents in wild-type and mrc1Δ cells. (B) Schematic representations of the segment containing three early-firing origins, ars1 (upper), ori1–200 (middle), and ars2004 (lower) on chromosome I, I, and II, respectively. Gray bars indicate probes used for two-dimensional gel electrophoresis analyses. Relevant restriction sites are indicated. (ET, EcoT22I; H, HaeIII; B, BglII; ER, EcoRI). Open boxes indicate the segments of ars1, ori1–200, and ars2004. (C, D, and E) Replication intermediates in various strains analyzed by neutral/neutral two-dimensional gel electrophoresis at ars1 (C), ori1–200 (D), and ars2004 (E). (F) Relative bubble arc signal was calculated by dividing the intensity of the bubble arc by that of the 1N spot at ars2004. Genomic DNA isolated was digested with BglII (ars1), EcoRI (ori1–200), or EcoT22I (ars2004). Filled arrows, bubble arc representing initiation; open arrows, Y arc representing forks emanating from the adjacent segments; gray arrows, 1N DNA spot.
Timing of origin loading of Mrc1.
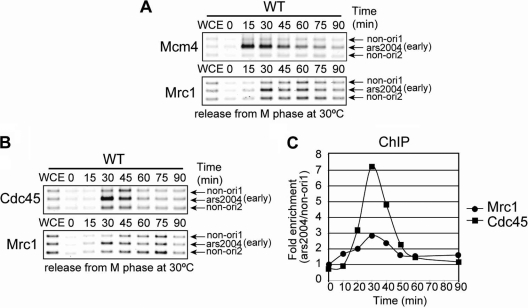
Preferential binding of Mrc1 to the early-firing origins in the early S phase may simply represent association of Mrc1 with an activated replication fork as a result of firing of origins. Therefore, we examined the timing of Mrc1 binding to early-firing origins during the course of the G1 to S transition. We used cells in which Mcm4 and Mrc1 were simultaneously tagged with Flag and Myc, respectively, and compared the timing of their loading onto ars2004 in synchronized culture by ChIP analyses, as described earlier (54). Cells were harvested at various time points after release from M phase, and immunoprecipitated DNA was amplified by using primer sets for ars2004 as well as for non-ori1 and non-ori2 present at 30 kb and 18 kb, respectively, away from ars2004 (33). Mcm4 associated with ars2004 at 15 min after release and dissociated from it in the following 60 to 90 min. In contrast, the peak of Mrc1 loading onto ars2004 was at 30 min, and it moved away from the origin (Fig. 2 A). This indicates that Mrc1 is loaded onto origins following MCM complex loading. Next, we compared the timing of Mrc1 loading with that of Cdc45 by using an Mrc1-13Myc and Cdc45-3Flag double-tagged strain. The peak of association of both Cdc45 and Mrc1 with ars2004 was at 30 min (Fig. 2B and C). However, close examination indicated that Mrc1 started to be associated with ars2004 slightly earlier than Cdc45 (Fig. 2C).
Fig. 2.
Mrc1 associates with a replication origin slightly earlier than Cdc45. Cells, released into cell cycle at 30°C from M phase arrest after incubation at 20°C for 5 h, were harvested at indicated times, and ChIP assays were conducted. Coimmunoprecipitated DNA was PCR amplified with the primer sets for ars2004, non-ori1, and non-ori2. PCR products were analyzed on 2.3% agarose gel electrophoresis (A and B) or by real-time PCR (C). In panel C, signal enrichment was quantified by normalizing the amount of precipitated ars2004 by that of precipitated non-ori1 DNA. mcm4-3flag mrc1-13myc (A) and cdc45-3flag mrc1-13myc (B and C) cells were used. The lower level of enrichment of Mrc1 may be partly due to instability of Mrc1 protein (41). WT, wild type.
Prefiring binding of Mrc1 to potential early-firing origins.
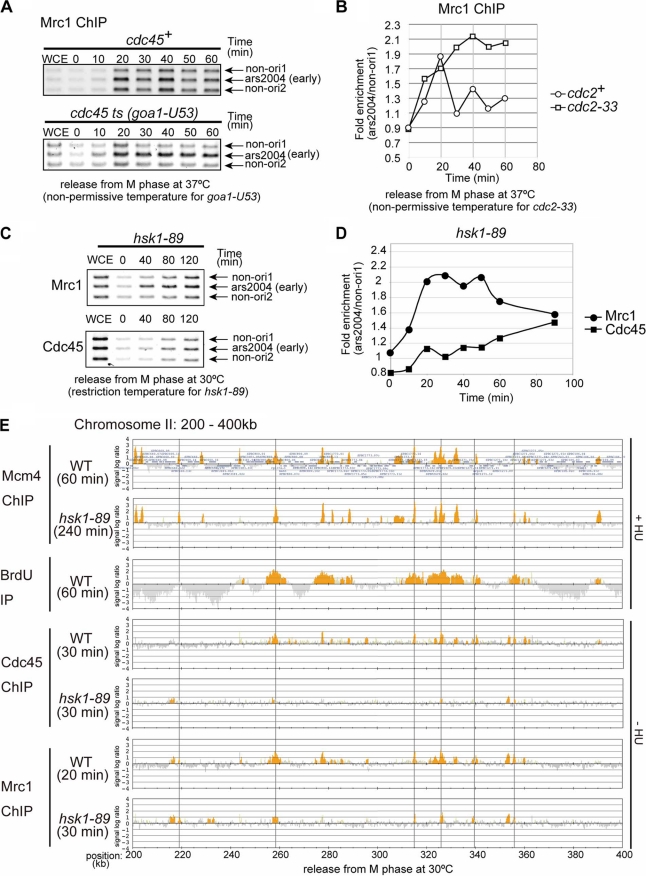
We then examined whether loading of Mrc1 onto pre-RC depends on Cdc45 by using a goa1-U53 mutant, an initiation-type mutant of Cdc45 (48). Mrc1 was loaded onto ars2004 in the goa1-U53 mutant at a nonpermissive temperature as well but appeared to accumulate and stay at the origin, presumably due to the block of replication fork progression from the origin in this mutant (Fig. 3A). We next examined cdc2-33 since it is well known that Cdc45 loading requires the Cdk function (27). Mrc1 is loaded onto ars2004 in cdc2-33 at a nonpermissive temperature and stays bound to the origin (Fig. 3B). These results indicate that Mrc1 is loaded onto origins in a Cdc2- and Cdc45-independent manner.
Fig. 3.
Mrc1 binds to origins in a manner independent of Hsk1, CDK, and Cdc45. (A) Mrc1 association with ars2004 was examined in goa1-U53 cells, a temperature-sensitive (ts) mutant of cdc45. Cells, arrested in M phase as described in the legend of Fig. 2, were released at 37°C, the nonpermissive temperature for goa1-U53 cells. DNA coimmunoprecipitated with Mrc1 was amplified and separated on 2.3% agarose gel. (B) mrc1-13myc cells in the cdc2+ or cdc2-33 (ts) strain background arrested at M phase were released into cell cycle at 37°C, the nonpermissive temperature for cdc2-33 cells. Binding of Mrc1 to ars2004 was quantified with real-time PCR using ars2004 and non-ori1 primer sets. Open circles, cdc2+; open squares, cdc2-33 (ts). The timing and enrichment of Mrc1 binding to ars2004 are not the same as those shown in Fig. 2C since the cells were released at 37°C. (C) mrc1-13myc cdc45-3flag nda3-KM311 cells in the hsk1-89 (ts) strain background were arrested at M phase by incubation at 20°C for 8 h and released into cell cycle at 30°C, the restriction temperature for hsk1-89 (ts) cells. Mrc1 and Cdc45 were immunoprecipitated with anti-Myc and anti-FLAG antibody, respectively. Coimmunoprecipitated DNA was PCR amplified with the primer sets for ars2004, non-ori1, and non-ori2. PCR products were analyzed by 2.3% agarose gel electrophoresis. (D) DNA collected by ChIP assay in panel C was quantified with real-time PCR as described in panel B. Filled circles, Mrc1 IP; filled squares, Cdc45 IP. (E) ChIP-chip analyses of BrdU incorporation and binding of MCM4, Mrc1, and Cdc45 in wild-type and hsk1-89 (ts) cells. Procedures for immunoprecipitation of Mcm4, BrdU, Mrc1, and Cdc45 in wild-type cells were basically as described in the legend of Fig. 1A. Mcm4 was immunoprecipitated using the cells collected at 60 min or 240 min after release in the presence of 25 mM HU (Mcm4 ChIP WT or Mcm4 ChIP hsk1-89 cells, respectively). BrdU was immunoprecipitated using the cells collected at 60 min after release in the presence of 25 mM HU in the wild type. Cdc45 was immunoprecipitated using the cells collected at 30 min after release in the absence of HU (in both WT and hsk1-89 cells). Mrc1 was immunoprecipitated using the cells collected at 20 min or 30 min after release in the absence of HU (Mrc1 ChIP WT or Mrc1 ChIP hsk1-89, respectively). The area of 200 to 400 kb on chromosome II is shown (E). Gray lines indicate the early-firing origins to which Mrc1 is bound in both wild-type and hsk1-89 cells. In panels B and D, signal enrichment was quantified by normalizing the amount of precipitated ars2004 by that of precipitated non-ori1 DNA.
In fission yeast, Hsk1, the Cdc7 homologue, is required for chromatin loading of Sld3, which precedes the loading of Cdc45 (54). Therefore, we next examined the requirement of Hsk1 for loading of Mrc1. ChIP analyses of synchronized cells showed that Cdc45 bound to ars2004 at early S phase in the wild-type background (Fig. 2B and C), whereas the binding was severely abrogated in hsk1-89 cells (Fig. 3C and D), as expected from requirement of Hsk1 for Cdc45 loading. In contrast, Mrc1 was loaded at ars2004 even under the hsk1-89 background with a timing similar to that in the wild type (Fig. 3C and D). Mrc1 stayed at the origin up to 60 min after release and started to dissociate after that. This may be correlated to the abortive initiation of DNA replication in hsk1-89 cells at a nonpermissive temperature, which starts to be detected at 80 min after release (results of two-dimensional gel [data not shown]). Mrc1 loading onto ars2004 in hsk1-89 cells (2.1-fold enrichment) was slightly lower than that in the wild type (3-fold enrichment) (compare Fig. 2C and 3D). This may be due to poor cell cycle synchronization in the former or to a higher level of Mrc1 protein in hsk1-89 cells, which can potentially cause adverse effects on signal enrichment in a ChIP assay (41).
In order to further confirm that Mrc1 is bound to early-firing origins in hsk1-89 cells, we conducted ChIP-chip assays to compare genome-wide distribution of Mrc1 binding sites between the wild-type and hsk1-89 cells (Fig. 3E; see Fig. S1 in the supplemental material). We first analyzed Mcm4 binding in the two strains. Almost identical binding patterns were observed, confirming that Hsk1 kinase is not required for pre-RC assembly. On the other hand, strong Cdc45 binding at early-firing origins observed in the wild-type cells was almost completely wiped out in hsk1-89 cells under the same condition, consistent with the results of the quantitative PCR (Fig. 3E; see also Fig. S1 in the supplemental material). This result confirms the strict requirement of Hsk1 function for loading of Cdc45 onto pre-RC.
We then analyzed Mrc1 binding in hsk1-89 cells by ChIP-chip assays. Basically, the binding pattern was similar between the hsk1-89 and wild-type strains. In the arm region proximal to the telomere segment where the origin activity is relatively weak, the locations and extents of Mrc1 binding are very similar between the wild-type and hsk1-89 strains (Fig. 3E, gray lines).
In the arm segment near the centromere, where initiation activity is generally strong, the locations of Mrc1 binding in hsk1-89 cells also largely overlap with those in the wild-type cells, whereas the Mrc1 binding signals are generally stronger and spread over longer DNA segments in the wild type (see Fig. S1 in the supplemental material). This is probably due to the progression of DNA replication and migration of replication forks to which Mrc1 is associated. In hsk1-89 cells, the replication fork would not be established, and therefore the pre-RC and associated proteins would be stuck at the origins. Mrc1, although loaded onto origins in hsk1-89 cells, cannot participate in the fork and thus eventually dissociates from the complex, resulting in reduced binding signals. The above results clearly show that Mrc1 is loaded onto pre-RC in a manner independent of Hsk1 kinase, namely, before the firing of origins. There are some Mrc1 binding sites that are observed only in the wild-type and not in hsk1-89 cells or some that are observed only in hsk1-89 cells. There are also some Mrc1 binding sites detected in both the wild-type and hsk1-89 cells which do not overlap with the pre-RC sites (Fig. 3E; see Fig. S1 in the supplemental material). The significance of these binding events is currently unknown.
Accelerated bubble formation in mrc1Δ cells but not in a checkpoint-defective mutant of mrc1.
The above results indicate that Mrc1 somehow selectively binds to early-firing origins prior to firing. In order to disclose the potential role of Mrc1 in the initiation, we analyzed replication intermediates at ars1 and ars2004 (11, 34) in synchronized cell populations of the wild-type and mrc1Δ cells by two-dimensional gel electrophoresis. FACS analyses indicated that DNA replication initiated at 20 to 30 min in wild-type cells after release from M phase (Fig. 4A). S phase progression is slightly accelerated at the early stage of S phase in mrc1Δ cells compared to progression in wild-type cells. Next, we analyzed replication intermediates in the same cell populations at ars1 and ars2004 by two-dimensional gel electrophoresis. A strong Y arc (representing passive replication) and very weak bubble arc (representing initiation) were detected at ars1 in the wild-type cells, consistent with rather inefficient firing at this origin (15) and indicating that ars1 is largely replicated passively from neighboring origins (Fig. 4C). However, in mrc1Δ cells, a strong bubble arc appeared at 20 min and mostly disappeared by 60 min, indicating that firing efficiency and its timing are significantly facilitated (Fig. 4C). Stimulation of initiation was observed in mrc1Δ cells also at ori1–200, another early firing origin, whose replication efficiency in the absence of HU, as estimated by two-dimensional gel electrophoresis, is similar to that of ars1 (Fig. 4D). At ars2004, a strong bubble arc was detected at 20 min and disappeared by 50 min in the wild type, consistent with efficient firing at this origin (34). A slightly stronger bubble arc signal was observed also at ars2004 in mrc1Δ cells, suggesting that initiation at early-firing origins is generally stimulated in the absence of Mrc1 (Fig. 4E and F). However, the stimulation was weak at ars2004, presumably because the firing at this origin is already nearly maximally efficient.
In order to determine whether this effect is dependent on the checkpoint function of Mrc1, we examined an mrc1-3A mutant (S604A T645A T653A), which is specifically defective in checkpoint function (see Fig. S3 in the supplemental material) (53, 55). The mutation did not facilitate initiation at ars1 or ars2004 (Fig. 4C and E). The stimulation was not observed in cds1Δ cells either (Fig. 4C). These results indicate that the absence of Mrc1 somehow leads to more efficient initiation at early-firing origins in a checkpoint-independent manner.
Firing at early origins is enhanced also in swi1Δ cells.
Swi1 forms a complex with Swi3, generating a fork protection complex (32). Swi1-Swi3 are evolutionally conserved (Tof1-Csm3 in budding yeast and Tim1-Tipin in metazoan), and these proteins physically interact with Mrc1 (4, 13, 41, 45) and are known to be required for stabilization of stalled replication fork and induction of replication checkpoint (7, 31). Bubble formation at ars1 was stimulated also in swi1Δ cells (Fig. 4C). The Swi1-Swi3 complex or its homologues are known to facilitate the loading of Mrc1 or Claspin onto chromatin (8, 41). Indeed, binding of Mrc1 to ars2004 is reduced, and its timing was also delayed in swi1Δ cells (see Fig. S4 in the supplemental material), suggesting that reduced loading of Mrc1 at origins in swi1Δ cells may be responsible for stimulation of firing. We noted that the timing of the appearance of the bubble was delayed in swi1Δ cells compared to mrc1Δ cells at both ars1 and ars2004 (Fig. 4C and E). At ars2004, a stronger bubble was observed at 30 min, and the bubble was detected even at 70 min after release, reflecting the slow movement of replication fork and/or asynchronous initiation in swi1Δ cells (Fig. 4C and E). Slow progression of the replication fork was reported also for tof1Δ cells in budding yeast (46). Thus, it is also possible that slowed fork progression leads to more firing events in swi1Δ cells.
Fork progression rate is not significantly affected in mrc1Δ cells.
In budding yeast and human cells, Mrc1 and its homolog of Claspin are required for normal replication fork progression (37, 43, 46). In budding yeast, S-phase progression and fork rate are decreased in the absence of Mrc1, and this function is independent of the replication checkpoint function of Mrc1. To determine whether Mrc1 affects the fork progression rate in fission yeast, we performed two-dimensional gel analysis at ori1–200 (equivalent to 1018, an AT-rich island reported before [40]), which is fired in the presence of HU (Fig. 5A). The closest origin to the right is 125 kb away from ori1–200, and a weak origin is present at 10 kb to its left. We designed a probe at ori1–200 and a region 23 kb to the right of ori1–200 (non-ori1–223) that is replicated passively by a replication fork emanating from ori1–200 or further left (Fig. 5A and B). Two-dimensional gel analysis was performed with the cells synchronously growing at 25°C from G2 arrest under the cdc25-22 background. This condition would slow down the fork movement and permit more accurate measurement of the fork rate. At ori1–200, a Y arc but not a bubble arc was detected in the wild-type cells, and the Y-arc signal peaked at 100 min (Fig. 5C). Only a very weak bubble arc signal was detected at 100 min after long exposure (data not shown). A stronger bubble arc signal was detected at the ori1–200 in mrc1Δ cells, as was shown in Fig. 4D (release from M phase arrest), and signal intensity changed with timing similar to that of the wild type. Y-arc signal on ori1–200 largely disappeared by 130 min in both wild-type and mrc1Δ cells and started to appear on non-ori1–223 at 90 min and peaked at 120 min in both cells (Fig. 5D), indicating that the replication fork progression rate is not significantly affected by mrc1Δ in fission yeast, precluding the possibility that a slowed fork rate indirectly induced origin firing.
Fig. 5.
Effect of mrc1Δ mutant on replication fork rate and firing of a checkpoint-restrained late-replicating origin. (A) BrdU incorporation profile of the 100- to 300-kb region on chromosome I in the wild-type cells determined as described in the legend to Fig. 1. The position of ori1–200, an early-firing origin, is shown. (B) Schematic representations of the segment containing ori1–200 and non-ori1–223. (C and D) cdc25-22 (WT) and cdc25-22 mrc1Δ (mrc1Δ) cells were arrested at G2 phase by incubation at 36.5°C for 3 h. Cells were then released into synchronous cell cycle at 25°C and were collected at the indicated times. Genomic DNA was digested with EcoRI, and replication intermediates were analyzed by neutral/neutral two-dimensional gel electrophoresis at ori1–200 (C) and at a region 23 kb away from ori1–200 (non-ori1–223) (D). Filled arrow, bubble arc representing initiation; open arrows, Y arc. (E) ChIP-chip analyses of MCM4 binding, BrdU incorporation, and Mrc1 binding at AT2080, which was previously identified as a late/dormant and checkpoint-restrained origin (14). The data confirm this and further show that Mrc1 does not bind to this origin in the presence of HU. (F) Schematic representation of the segment containing AT2080. HIII, HindIII. (G) Two-dimensional gel electrophoresis analysis of replication intermediates at AT2080 from wild-type and mrc1Δ cells was performed by using the cells released from M phase. Genomic DNA was digested with HindIII. For panels B and F, see also the legend to Fig. 4B for details.
Enhanced firing in the absence of Mrc1 is observed only on those origins which are bound by Mrc1.
Genome-wide examination of DNA replication in the presence of HU revealed the presence of late or inefficient origins which are not fired under this condition but could be fired in the absence of a replication checkpoint (rad3Δ or cds1Δ or mrc1Δ cells) (S. Matsumoto et al., submitted for publication) (9, 14, 15). Mrc1 is not bound to the majority of these potential origins at early S phase (Fig. 1C and 5E). Initiation at one of these origins, AT2080, was examined by two-dimensional gel electrophoresis analyses. Either in the wild-type or mrc1Δ mutant background, a bubble arc was not detected even at 60 min after release, and only a weak Y arc was detected at 40 to 50 min, representing the incoming replication fork from the adjacent segment (Fig. 5G). Similar results were obtained at ARS727, another inefficient origin to which Mrc1 is not bound (data not shown). Taken together, the above results indicate that enhanced firing is observed on early-firing origins bound by Mrc1 but not on the checkpoint-restrained origins which are not bound by Mrc1.
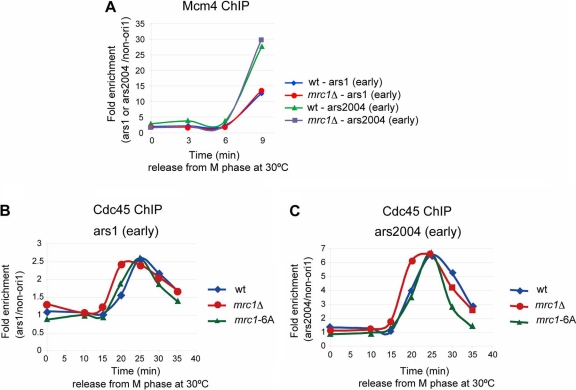
The mrc1Δ mutant facilitates loading of Cdc45 onto early-firing replication origins.
The above results indicate that the absence of Mrc1 somehow promotes initiation at a subset of potential origins on the chromosome. Bubbles in two-dimensional gel electrophoresis indicate the presence of unwound duplex for a certain length, indicating that replication forks have been generated in the DNA fragments examined. Therefore, we examined the timing and efficiency of the loading of Mcm4 and Cdc45 onto origins since this event is essential for generation of pre-RC and a replication fork. It was also reported that the timing of pre-RC assembly correlates with the timing of origin firing (52). Cells were collected at various times after release from M phase arrest, and Flag-tagged Mcm4 and Cdc45 were immunoprecipitated with anti-Flag antibody. Mcm4 was bound to ars1 and ars2004 by 9 min after the release, and Mcm4 binding patterns were identical between wild-type and mrc1Δ cells (Fig. 6A), suggesting that the timing and efficiency of pre-RC formation are not affected by the mrc1Δ mutation. In contrast, the peak of Cdc45 loading onto ars1 was at 25 min after release in the wild-type cells, whereas that in mrc1Δ cells was at 20 min (Fig. 6B). Timing of Cdc45 loading was similarly advanced in mrc1Δ cells at ars2004 (Fig. 6C). Binding efficiency of Cdc45 was similar between wild-type and mrc1Δ cells. The precocious Cdc45 loading was not observed in an mrc1-6A (S599A S604A S614A T634A S637A T645A) mutant, which is deficient in DNA replication checkpoint (55), consistent with the absence of enhanced initiation in a similar checkpoint mutant, mrc1-3A (Fig. 4C and E). The data indicate that Mrc1 regulates the timing of Cdc45 loading at early-firing replication origins in a checkpoint-independent manner.
Fig. 6.
Loading of Cdc45 onto early-firing origins is facilitated in mrc1Δ cells but not in a checkpoint mutant of mrc1. Cells were released from M phase as described in the legend to Fig. 2. Mcm4-3Flag (A) and Cdc45-3Flag (B and C) were immunoprecipitated with anti-FLAG antibody at various time points in wild-type and mrc1Δ cells. Coimmunoprecipitated DNA at ars1 and ars2004 was quantified by real-time PCR. Signal enrichment was quantified by normalizing the amount of precipitated origin DNA (ars1 or ars2004) by that of precipitated non-ori1 DNA. We conducted the same experiment at least three times and obtained basically identical results. Representative data are presented. Cells used were as follows: for panel A, mcm4-3Flag nda3-KM311 and mcm4-3Flag mrc1Δ nda3-KM311 cells; for panels B and C, cdc45-3Flag nda3-KM311, cdc45-3flag nda3-KM311 mrc1Δ, and cdc45-3flag nda3-KM311 mrc1-6A cells.
DISCUSSION
Mrc1 binds to early-firing origins prior to initiation: Mrc1 as a potential marker for early-firing origins.
We have determined genome-wide localization of Mrc1 at the early S phase (Fig. 1; see also Fig. S1 in the supplemental material). The data indicate that Mrc1 binds preferentially to early-firing origins, which initiate DNA replication in the presence of HU. This binding is independent of Hsk1, Cdk, and Cdc45 (Fig. 3) and thus occurs prior to activation of pre-RC. This indicates that Mrc1 may somehow specifically recognize and become associated with pre-RC which will be fired at early S phase. Although 39.0% of the origins firing in the presence of HU are not bound with Mrc1, incorporation of BrdU is very low at these origins, suggesting that these origins may not be efficiently utilized during the normal course of initiation. Indeed, initiation signals in two-dimensional gel electrophoresis analysis were not detected at these origins during the early S phase of the normal cell cycle (see Fig. S2 in the supplemental material). These classes of origins appear to be more enriched in the telomere-proximal arm segment (Fig. 1C, indicated by stars), where replication initiation activity is generally low.
What feature of the prospective early-firing origins would be recognized by Mrc1? It could be specific chromatin structure (e.g., special modification of histones) or chromatin organization within the nuclei. Alternatively, it could be other factors associated with pre-RC or modification of pre-RC (such as phosphorylation or acetylation) that might occur during the prereplication stage or a combination of above. The specific features associated with early-firing origins may be more enhanced with earlier recruitment of the ORC (52).
We previously reported that Hsk1 interacts with Mrc1 (41). It is tempting to speculate that Hsk1 is recruited to pre-RC through interaction with Mrc1, thus facilitating the initiation at early-firing origins. Mrc1 is vigorously phosphorylated by Hsk1 in vitro (19) and also undergoes Hsk1-dependent phosphorylation in HU-treated cells (29) and in normally growing cells at the G1-S boundary (our unpublished data). It was previously reported that Cdc7 interacts with Claspin in both human cells and Xenopus egg extracts, and Claspin is phosphorylated in a manner dependent on Cdc7 in human cells (10, 23).
Regulation of origin firing and its timing by Mrc1.
The precocious appearance of strong bubble signals at selected origins in two-dimensional gel electrophoresis in the mrc1Δ strain (Fig. 4) suggests that firing is negatively regulated by Mrc1. What would be the mechanism for this regulation?
Wu and Nurse reported that the timing of pre-RC assembly in G1 correlates with that of origin firing in the following S phase (52). Thus, Mrc1 could negatively regulate the timing of pre-RC assembly. However, the timing and efficiency of pre-RC formation do not appear to be facilitated at early-firing origins in mrc1Δ cells (Fig. 6A and our unpublished results of a ChIP-chip assay of Mcm4 in mrc1Δ cells), precluding the possibility that Mrc1 affects the origin firing by altering the pre-RC assembly step.
The second possibility is that a slow fork rate in mrc1Δ or swi1Δ cells increases the chance of origin firing, which could occur in a stochastic manner, at those origins normally fired with low frequency (such as ars1). In fact, a slow fork rate has been reported in budding yeast mrc1Δ and tof1Δ cells (18). However, our results show that the replication fork rate is not significantly affected by loss of mrc1 in fission yeast (Fig. 5). Fork rate appears to be significantly slowed down in swi1Δ cells (Fig. 4C and E), but the stimulation of initiation is less efficient in swi1Δ than in mrc1Δ cells (Fig. 4C). These considerations make this possibility less likely as well.
Another possibility is that Mrc1 directly inhibits the firing at selected origins by somehow temporally inhibiting Cdc45 loading. Mrc1 protein, recruited at provisional early-firing origins, may limit the time window for Cdc45 loading by transiently inhibiting the function of pre-RC. The subsequent recruitment of Hsk1 kinase and phosphorylation events by Hsk1 may antagonize this inhibition and permit more synchronous activation of early origins. The absence of Mrc1 may result in more active pre-RC, which can be precociously activated for Cdc45 loading. This speculation is supported by the observation that bubble formation is stimulated in swi1Δ cells, which is at least partially defective in timely loading of Mrc1 onto chromatin (see Fig. S4 in the supplemental material) (41).
Summary and perspectives.
Another intriguing finding is that the hyper-firing in the absence of Mrc1 appears to occur selectively at origins bound by Mrc1 but not at other unoccupied origins (Fig. 5G). Mrc1-bound origins may be embedded in the chromatin context, which is permissive for firing of pre-RC, and may tend to fire early in S phase in the absence of Mrc1. In contrast, Mrc1-free origins may be in a chromatin context less permissive for firing and would not be affected by Mrc1 but would require additional chromatin remodeling for firing.
In summary, we show here that Mrc1 recognizes and is associated with the early-firing and active potential origins, which are likely to be embedded in a permissive chromatin context, and we propose that this specific recognition will play a crucial role in defining the early-firing origins versus less active, late-firing origins. We also speculate that Mrc1 may play a role in recruiting Cdc7/Hsk1 kinase for efficient firing. Further experiments are needed to determine whether the binding of Mrc1 to origins directly inhibits firing (i.e., the origin loading of Cdc45) or whether altered higher-order structures of the replication complex (in the presence and absence of Mrc1) indirectly affect the efficiency of initiation. It should also be noted that regulation of early-firing origins by Mrc1 is mediated by its checkpoint-independent function (Fig. 4 and 6). Checkpoint-independent functions of Mrc1 were previously reported to be involved in efficient S phase progression, in replication through the hard-to-replicate segment, and in telomere maintenance (43, 46, 47, 50). It remains to be determined whether the function of Mrc1 in origin regulation is related to these functions.
Supplementary Material
ACKNOWLEDGMENTS
We sincerely thank Katsuhiko Shirahige for his generosity in permitting us to use his facility for the ChIP-chip analyses and for his helpful advice. We thank Paul Russell for valuable discussion and various materials, Kanji Furuya and Hisao Masukata for strains, advice and the protocol on two-dimensional gel electrophoresis, and Katsunori Tanaka and Eishi Noguchi for yeast strains. We also thank members of our laboratory for helpful discussion.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by Takeda Science Foundation, and by Astellas Foundation for Research on Metabolic Disorders (to H.M.) and by research fellowships of the Japan Society for Promotion of Science for young scientist (to M.H.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 25 April 2011.
REFERENCES
- 1. Alcasabas A. A., et al. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965 [DOI] [PubMed] [Google Scholar]
- 2. Aparicio J. G., Viggiani C. J., Gibson D. G., Aparicio O. M. 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arcangioli B. 1998. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 17: 4503–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bando M., et al. 2009. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J. Biol. Chem. 284: 34355–34365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Courbet S., et al. 2008. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 [DOI] [PubMed] [Google Scholar]
- 6. Czajkowsky D. M., Liu J., Hamlin J. L., Shao Z. 2008. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J. Mol. Biol. 375: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dalgaard J. Z., Klar A. J. 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102: 745–751 [DOI] [PubMed] [Google Scholar]
- 8. Errico A., Costanzo V., Hunt T. 2007. Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc. Natl. Acad. Sci. U. S. A. 104: 14929–14934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feng W., et al. 2006. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 8: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gold D. A., Dunphy W. G. 2010. Drf1-dependent kinase interacts with Claspin through a conserved protein motif. J. Biol. Chem. 285: 12638–12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gomez M., Antequera F. 1999. Organization of DNA replication origins in the fission yeast genome. EMBO J. 18: 5683–5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goren A., Tabib A., Hecht M., Cedar H. 2008. DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev. 22: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gotter A. L. 2003. Tipin, a novel timeless-interacting protein, is developmentally co-expressed with timeless and disrupts its self-association. J. Mol. Biol. 331: 167–176 [DOI] [PubMed] [Google Scholar]
- 14. Hayashi M., et al. 2007. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J. 26: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heichinger C., Penkett C. J., Bahler J., Nurse P. 2006. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hiraoka Y., Toda T., Yanagida M. 1984. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell 39: 349–358 [DOI] [PubMed] [Google Scholar]
- 17. Hiratani I., et al. 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hodgson B., Calzada A., Labib K. 2007. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol. Biol. Cell 18: 3894–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kakusho N., Taniyama C., Masai H. 2008. Identification of stimulators and inhibitors of Cdc7 kinase in vitro. J. Biol. Chem. 283: 19211–19218 [DOI] [PubMed] [Google Scholar]
- 20. Karnani N., Taylor C., Malhotra A., Dutta A. 2007. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 17: 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Katou Y., et al. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- 22. Kelly T. J., Brown G. W. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69: 829–880 [DOI] [PubMed] [Google Scholar]
- 23. Kim J. M., et al. 2008. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene 27: 3475–3482 [DOI] [PubMed] [Google Scholar]
- 24. Kitamura E., Blow J. J., Tanaka T. U. 2006. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell 125: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar S., Huberman J. A. 2004. On the slowing of S phase in response to DNA damage in fission yeast. J. Biol. Chem. 279: 43574–43580 [DOI] [PubMed] [Google Scholar]
- 26. Lebofsky R., Heilig R., Sonnleitner M., Weissenbach J., Bensimon A. 2006. DNA replication origin interference increases the spacing between initiation events in human cells. Mol. Biol. Cell 17: 5337–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacNeill S. A., Creanor J., Nurse P. 1991. Isolation, characterisation and molecular cloning of new mutant alleles of the fission yeast p34cdc2+ protein kinase gene: identification of temperature-sensitive G2-arresting alleles. Mol. Gen. Genet. 229: 109–118 [DOI] [PubMed] [Google Scholar]
- 28. Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M. 2010. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79: 89–130 [DOI] [PubMed] [Google Scholar]
- 29. Matsumoto S., et al. 2010. Hsk1 kinase and Cdc45 regulate replication stress-induced checkpoint responses in fission yeast. Cell Cycle 9: 4627–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCune H. J., et al. 2008. The temporal program of chromosome replication: genomewide replication in clb5Δ Saccharomyces cerevisiae. Genetics 180: 1833–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noguchi E., Noguchi C., Du L. L., Russell P. 2003. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol. 23: 7861–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Noguchi E., Noguchi C., McDonald W. H., Yates J. R., Russell P. 2004. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell. Biol. 24: 8342–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogawa Y., Takahashi T., Masukata H. 1999. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol. 19: 7228–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okuno Y., Okazaki T., Masukata H. 1997. Identification of a predominant replication origin in fission yeast. Nucleic Acids Res. 25: 530–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osborn A. J., Elledge S. J. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel P. K., Arcangioli B., Baker S. P., Bensimon A., Rhind N. 2006. DNA replication origins fire stochastically in fission yeast. Mol. Biol. Cell 17: 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petermann E., Helleday T., Caldecott K. W. 2008. Claspin promotes normal replication fork rates in human cells. Mol. Biol. Cell 19: 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raghuraman M. K., et al. 2001. Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- 39. Santocanale C., Diffley J. F. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- 40. Segurado M., de Luis A., Antequera F. 2003. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 4: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shimmoto M., et al. 2009. Interactions between Swi1-Swi3, Mrc1 and S phase kinase, Hsk1 may regulate cellular responses to stalled replication forks in fission yeast. Genes Cells 14: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shirahige K., et al. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- 43. Szyjka S. J., Viggiani C. J., Aparicio O. M. 2005. Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol. Cell 19: 691–697 [DOI] [PubMed] [Google Scholar]
- 44. Tanaka K., Russell P. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3: 966–972 [DOI] [PubMed] [Google Scholar]
- 45. Tanaka T., et al. 2010. Fission yeast Swi1-Swi3 complex facilitates DNA binding of Mrc1. J. Biol. Chem. 285: 39609–39622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tourriere H., Versini G., Cordon-Preciado V., Alabert C., Pasero P. 2005. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol. Cell 19: 699–706 [DOI] [PubMed] [Google Scholar]
- 47. Tsolou A., Lydall D. 2007. Mrc1 protects uncapped budding yeast telomeres from exonuclease EXO1. DNA Repair (Amst.) 6: 1607–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Uchiyama M., Arai K., Masai H. 2001. sna41goa1, a novel mutation causing G1/S arrest in fission yeast, is defective in a CDC45 homolog and interacts genetically with polα. Mol. Genet. Genomics 265: 1039–1049 [DOI] [PubMed] [Google Scholar]
- 49. Vogelauer M., Rubbi L., Lucas I., Brewer B. J., Grunstein M. 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10: 1223–1233 [DOI] [PubMed] [Google Scholar]
- 50. Voineagu I., Surka C. F., Shishkin A. A., Krasilnikova M. M., Mirkin S. M. 2009. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat. Struct. Mol. Biol. 16: 226–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wendt K. S., et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- 52. Wu P. Y., Nurse P. 2009. Establishing the program of origin firing during S phase in fission yeast. Cell 136: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu Y. J., Davenport M., Kelly T. J. 2006. Two-stage mechanism for activation of the DNA replication checkpoint kinase Cds1 in fission yeast. Genes Dev. 20: 990–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yabuuchi H., et al. 2006. Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J. 25: 4663–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao H., Tanaka K., Nogochi E., Nogochi C., Russell P. 2003. Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast. Mol. Cell. Biol. 23: 8395–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.