Abstract
The rate of ribosome biogenesis, which is downregulated in terminally differentiated cells and upregulated in most cancers, regulates the growth rate and is linked to the cell's proliferative potential. The U3 box C/D small nucleolar RNP (snoRNP) is an integral component of the small subunit (SSU) processome and is essential for 18S rRNA processing. We show that U3 snoRNP assembly, and therefore U3 snoRNA accumulation, is regulated through the U3-specific protein hU3-55K. Furthermore, we report that the levels of several SSU processome components, including the U3 snoRNA but not other box C/D snoRNAs, are specifically downregulated during human lung (CaCo-2) and colon (CaLu-3) epithelial cell differentiation. c-Myc is reported to play an integral role in regulating ribosome production by controlling the expression of many ribosome biogenesis factors. Our data, however, indicate that this regulation is not dependent on c-Myc since the level of this protein does not change during epithelial cell differentiation. In addition, depletion of c-Myc had only a mild affect on the levels of SSU processome proteins. CaCo-2 cells are colon adenocarcinoma epithelial cells that are believed to revert to their precancerous state during differentiation. This suggests a significant increase in the levels of specific SSU processome components during tumorogenesis.
INTRODUCTION
The rate of ribosome biogenesis, which is downregulated in terminally differentiated cells and upregulated in most cancers, controls the cellular protein synthesis capacity. This, therefore, regulates the growth rate, which is in turn linked to the proliferative potential of the cell (17, 31). In the nucleolus, rRNA production is regulated both transcriptionally and at the level of pre-rRNA processing (24). Altering ribosome turnover rates and accelerating pre-rRNA processing also regulate ribosome levels (5, 8). Overexpression of the proto-oncogene c-Myc has been shown to increase the levels of mRNAs encoding several pre-rRNA processing factors. It has also been shown that c-Myc directly influences pre-rRNA processing (7, 27, 33).
Ribosome biogenesis requires more than 200 trans-acting factors, which include exonucleases, endonucleases, chaperones, helicases, annealing factors, small nucleolar RNPs (snoRNPs), and the 80 ribosomal proteins (11, 15). There are over 100 different box C/D snoRNPs in human cells, and most function in the 2′-O methylation of rRNA. Box C/D snoRNPs contain four common core proteins, 15.5K (Snu13p), NOP56, NOP58, and fibrillarin (Nop1p), which bind the C/D (C′/D motif in the U3 snoRNA) motif in the small nucleolar RNA (snoRNA) (15). snoRNP biogenesis is mediated by a large pre-snoRNP complex that contains factors involved in assembly, nucleocytoplasmic transport, and snoRNA maturation (3, 4, 25, 44). The formation of the core box C/D snoRNP is thought to be sufficient for the accumulation and nucleolar localization of these complexes. A few snoRNPs, including U3, U8, and U14, are essential for pre-rRNA processing. In addition to the four core proteins, the U3 snoRNP contains a second molecule of 15.5K, which together with the U3-specific protein hU3-55K (Rrp9p) binds to the U3 snoRNA B/C motif (13, 42). Several biogenesis factors involved in core box C/D snoRNP assembly also play a role in box B/C RNP formation (3). In snoRNPs that guide methylation, the core proteins 15.5K, NOP56, and NOP58 are proposed to be organized asymmetrically (6, 40). 15.5K, NOP58, and one molecule of fibrillarin are proposed to bind the C/D motif, while NOP56 and another copy of fibrillarin bind the C′/D′ motif. The two parts of the complex are predicted to be bridged by the interaction of the coiled-coil domains of NOP56 and NOP58 (1). It is currently unclear, however, how this model relates to the U3 snoRNP. In particular, it is not known whether the U3 snoRNP contains 2 molecules of fibrillarin and whether NOP56 contacts the analogous box B/C motif in the U3 snoRNA.
The U3 snoRNP functions as part of the small subunit (SSU) processome, a large multisubunit complex essential for the cleavages in the 5′ external transcribed spacer (5′ETS) (A′ and A0) and internal transcribed spacer (ITS1) that release the 18S rRNA from the initial precursor rRNA (pre-rRNA) transcript (15). This complex is recruited, cotranscriptionally via the tUTP proteins, to the nascent pre-rRNA transcript. It has recently been proposed that the U3 snoRNP assembles into a large, 50S complex, which is recruited into the SSU processome (41). The nucleolus contains three subcompartments, the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC) (16). Transcription occurs at the border of the FC and DFC, with the initial processing steps occurring in the DFC. The majority of snoRNAs are involved in the early processing steps and, as such, are found in the FC/DFC region of the nucleolus. The U3 snoRNP, which is involved in both early and later processing steps, is found throughout the nucleolus, and evidence suggests that this complex moves, as part of the SSU processome, from the DFC to the GC during ribosome biogenesis (14, 41).
Vertebrate U3 snoRNA expression is downregulated in response to serum starvation and during myoblast differentiation (10, 34). U3 snoRNP incorporation into the SSU processome is dependent on hU3-55K, the only U3 snoRNP-specific protein and a factor believed to be essential to the function, but not accumulation, of this complex (14, 42). Mutations in hU3-55K have been linked to breast cancers, and the expression of this protein is upregulated during tumorogenesis (23, 35, 37). Regulating U3 snoRNP levels and/or activity is likely to play an important role in controlling ribosome biogenesis. Importantly, little is known about the regulation of RNP production and activity. We therefore set out to investigate the role of hU3-55K in the structure of the U3 snoRNP and regulating the production of this complex.
MATERIALS AND METHODS
Oligonucleotides.
The following oligonucleotides (listed 5′ to 3′) were used:: 1, CACCGGATCCATGTCGGCAACAGCGGCTGCTCG; 2, GCGCGCGGCCGCTCAGGAACCAGCAGCTGGGGG; 3, CGCGGGATCCATGAAGCCAGGATTCAGTCCCG; 4, CGCGCTCGAGTCAGTTCTTCACCTTGGGGGGTGG; 5, CACCGGATCCATGGAATCCGAAATGGAAACG; 6, TCTAGATCAGAATCGATCTGCTGATCTGCTAGC; 7, CACCGGATCCATGGCGTCTCCCTCGCTGGAG; 8, TCTAGATTACTTTTTTTTCTTCTTTTTCTTTTCATCTGCC; 9, GTAGAGCACCGAAAACCACGAUGAUGAGAGGTAGCGTTTTCTCC; and 10, GGAGAAAACGCTACCTCTCATCATCGTGGTTTTCGGTGCTCTAC.
Plasmids.
The cDNAs for hU3-55K and fibrillarin were PCR amplified from pCI-neo VSVhU3-55K (13), using oligonucleotide pair 1 and 2 and pair 3 and 4, respectively, and cloned into the pcDNA5/FRT vector (Invitrogen) downstream of a FLAG tag, a PreScission (GE Healthcare) protease site, and a hexahistidine tag. For protein overexpression in Escherichia coli, it was necessary to use constructs in which the sequences encoding the RGG motif of fibrillarin, the C-terminal region of BCD1, and the KKE/KKD regions of NOP56 and NOP58 were deleted to obtain suitable amounts of soluble protein (25). The cDNAs for PNO1 (primers 5 and 6) and KRR1 (primers 7 and 8) were PCR amplified from cDNAs (Imagene) and cloned into the pET100 D/TOPO vector. The U3 snoRNA wild-type mutC constructs in the human U3 gene containing the streptomycin binding aptamer sequence (Strepto tag) were described previously (14). The C′con mutant was generated in each of these constructs by site-directed mutagenesis using oligonucleotides 9 and 10.
Cell culture, RNAi, FISH and generation of stable cell lines.
Growth of human cell lines (HeLa, HEK293, CaLu-3, and CaCo-2) involved standard cell culture techniques. For differentiation, the cells were incubated for 15 days (CaLu-3) or 21 days (CaCo-2) on polycarbonate membrane inserts. The differentiation of epithelial cells was verified by transepithelial electrical resistance (TEER) measurement (22, 39) after the cells had been incubated for 15 days (CaLu-3) or 21 days (CaCo-2) on polycarbonate membrane inserts. RNA interference (RNAi) depletions in HeLa cells were performed as previously described using small interfering RNAs (siRNAs) targeting NOP58, GL2 (a control, which targets the firefly luciferase gene), and hU3-55K (30, 44). In addition, smart pool siRNAs were used for c-Myc and Max (Thermo Scientific).
Plasmids were transfected into HEK293 Flp-In cells (Invitrogen), and stable transfectants were selected using hygromycin B. Protein expression was induced using either 1 μg/ml (for high level expression of hU3-55K and fibrillarin) or 0.5 ng/ml (1:1, tagged: endogenous hU3-55K expression) tetracycline and monitored using anti-FLAG antibodies (Sigma). Strepto-U3 snoRNA constructs were transfected into HeLa cells using Fugene 6 (Roche). To control for transfection levels, semiquantitative PCR amplification was performed using primers specific for the plasmid as described previously (26), and the product was detected by Southern blotting using a probe specific to the U3 snoRNA.
Western blotting and immunoprecipitation.
hU3-55K antipeptide antibodies were synthesized as described previously (13). NOP56, NOP58, MPP10, and UTP12 antibodies were described previously (41, 43, 45). Anti-U5-116K antibodies were kindly provided by Reinhard Luhrmann (9). Recombinant PNO1 and KRR1 were expressed in E. coli and purified using nickel affinity chromatography. The purified proteins were used to immunize rabbits to produce the anti-PNO1 and -KRR1 antibodies. Anti-FLAG (Sigma), anti-c-Myc (Santa Cruz), fibrillarin (Santa Cruz), nucleolin (Abcam), and Prp43 (Bethyl) antibodies were purchased from commercial sources. Immunoprecipitation and gradient experiments were essentially performed as previously described (14).
Extract preparation and protein-protein interaction analysis.
U3 snoRNP assembly was performed as described previously (13). The 3′ end of U3 snoRNA was PCR amplified with a T7 promoter at the 5′ end. In vitro, 32P-labeled transcripts were generated and the RNAs were incubated in mouse nuclear extract (kindly provided by Stuart Maxwell). Complexes were immunoprecipitated using wither anti-NOP58 or anti-hU3-55K antibodies, and the associated RNAs were separated by urea/PAGE and then revealed by phosphorimager analysis.
For protein-protein interaction studies, all glutathione S-transferase (GST)- and thioredoxin-tagged proteins used were prepared as described previously (13, 25). Equal amounts of GST or thioredoxin (TRX) bait proteins were immobilized on glutathione-Sepharose beads or anti-TRX antibodies coupled to protein A Sepharose, respectively, and incubated with 35S-labeled hU3-55K. The beads were washed, and the retained proteins were separated by SDS-PAGE and analyzed by autoradiography.
RESULTS
hU3-55K levels control cellular abundance of U3 snoRNA.
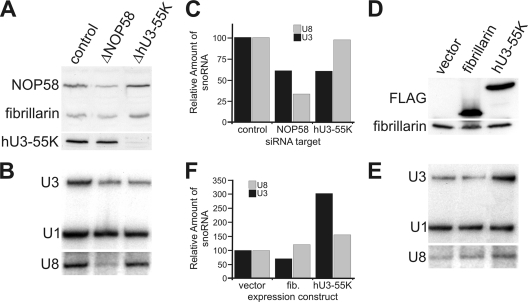
In order to investigate the role of hU3-55K in U3 snoRNP function, we used siRNAs to deplete the protein, or the general box C/D protein NOP58, from HeLa cells. The specific action of these siRNAs was monitored by Western blotting (Fig. 1A). Consistent with earlier observations (44), Northern blotting of RNA extracted from the transfected cells revealed that depletion of NOP58 specifically reduced the levels of both U3 and U8 snoRNAs relative to those seen in the control cells (Fig. 1B and C). Depletion of hU3-55K reproducibly reduced U3 snoRNA levels without affecting those of the U8 and U1 RNAs. This result was surprising, since hU3-55K was thought to be important only for U3 snoRNP function (42).
Fig. 1.
Regulation of hU3-55K controls U3 snoRNA levels in the cell. (A, B, and C) HeLa cells were transfected with either control siRNAs (targeting firefly luciferase) or siRNAs targeting NOP58 or hU3-55K. Protein levels in the transfected cells were determined by Western blot analysis (A). The protein targeted is indicated above each panel. The antibodies used are indicated on the left. Proteins derived from equal numbers of cells were loaded. RNA was extracted from an equal number of transfected cells and analyzed by Northern blotting (B). The RNA analyzed is indicated to the left of each panel. (C) Quantitation of U3 (black) and U8 (gray) snoRNA levels. The levels of the two snoRNAs were normalized to the level of the U1 snRNA. The siRNA target is indicated on the horizontal axis, and the amount of snoRNA relative to that seen with the control siRNA is indicated on the vertical axis. (D, E, and F) HEK293 cells expressing FLAG tag alone (vector), FLAG-fibrillarin, or FLAG-hU3-55K, as indicated at the top of each lane, were analyzed by Western blotting (D) or Northern blotting (E). The proteins and RNAs analyzed are indicated to the left of each panel. FLAG, anti-FLAG antibody. (F) Quantitation of U3 (black) and U8 (gray) snoRNA levels. The levels of the two snoRNAs were normalized to the level of the U1 snRNA. The expression construct used is indicated on the horizontal axis, and the amount of snoRNA relative to that seen with the vector alone is indicated on the vertical axis.
Based on this result, we next tested whether raising hU3-55K levels would increase the amount of U3 snoRNA. Stably transfected HEK293 cell lines in which FLAG-tagged hU3-55K, fibrillarin, or the FLAG tag alone were expressed from a tetracycline-regulated promoter were generated. The tagged hU3-55K and fibrillarin cell lines expressed proteins of the expected size upon the addition of tetracycline (Fig. 1D). No signal was seen for the FLAG tag alone. Immunoprecipitation analysis revealed that FLAG-fibrillarin and FLAG-hU3-55K were specifically associated with the box C/D snoRNAs and the U3 snoRNA, respectively (data not shown). Northern blot analysis of RNAs extracted from the individual cell lines revealed that expression of FLAG-hU3-55K reproducibly increased U3 snoRNA levels at least 3-fold (Fig. 1E and F) but had little or no effect on U1 snRNA levels. A slight increase in U8 snoRNA was seen when either FLAG-fibrillarin or FLAG-hU3-55K was expressed (Fig. 1E and F). Induction of the FLAG tag alone did not affect U3 snoRNA levels. A slight decrease in U3 snoRNA levels was seen when FLAG-fibrillarin was expressed, although this effect was variable.
The B/C motif is required for efficient human U3 snoRNP production.
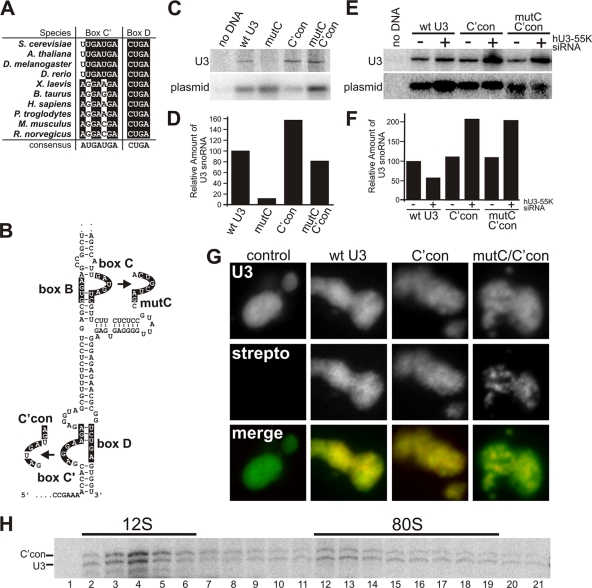
Binding of the box C/D core proteins to the C/D motif is essential for snoRNP formation and the accumulation of the snoRNA. It is therefore possible that hU3-55K controls U3 snoRNA accumulation by influencing snoRNP assembly. For this to occur, an element of the U3 snoRNA must render core box C/D snoRNP formation dependent on hU3-55K. Analysis of available U3 snoRNA sequences revealed that the C′ box (C box in other snoRNAs) in the mammalian and amphibian U3 differs significantly from the box C consensus, with the 2 Us in the C′ box being replaced by purines (Fig. 2A). Similar changes introduced into the C box of the U14 snoRNA affect box C/D snoRNP formation (43). The potentially “weaker” human C′/D motif may require hU3-55K for core snoRNP formation. We therefore tested the role of the unusual C′ box and the hU3-55K-binding site (B/C motif) in U3 snoRNP accumulation. Mutations were generated in a Strepto-tagged human U3 snoRNA construct under the control of its natural promoter (14) (Fig. 2B). The mutant constructs were expressed in HeLa cells, and the accumulation of the tagged-U3 snoRNA was determined by Northern blotting using a Strepto-tag-specific probe. Semiquantitative PCR using plasmid-specific primers was used to determine the relative transfection levels.
Fig. 2.
The box B/C motif is required for U3 snoRNP assembly and U3 snoRNA accumulation. (A) Alignment of a selection of the U3 snoRNA C′ and D box sequences from a variety of eukaryotes (S., Saccharomyces; A., Arabidopsis; D., Drosophila (melanogaster) or Danio (rerio); X., Xenopus; B., Bos; H., Homo; P., Pan; M., Mus; R., Rattus). The consensus sequence of the C/D motif is shown at the bottom. Nucleotides identical to the consensus sequence are indicated in white on a black background. Other nucleotides are shown in black on a white background. (B) Secondary structure of the 3′ end of the U3 snoRNA. The B, C, C′ and D boxes are indicated. Mutations to the C and C′ boxes are shown. The secondary structures of the C′/D and B/C motifs are drawn as described previously (13). Bars indicate base-pairing interactions. (C) HeLa cells were transiently transfected with constructs expressing wild-type or mutant U3 snoRNAs. The mutations were generated in a construct expressing a U3 snoRNA containing a Strepto tag sequence (14). RNA was extracted from the cells and analyzed by Northern blotting (upper panel). To control for transfection levels, DNA extracted from the transfected HeLa cells was analyzed by PCR using primers specific for the transfected plasmid. The product was separated by agarose gel electrophoresis and analyzed by Southern blotting using a U3-specific probe (lower panel). The U3 construct used is indicated above each lane. (D) Quantitation of tagged U3 snoRNA expression levels. The levels of tagged U3 snoRNA were normalized relative to the amount of plasmid DNA transfected into the cells. The transfected construct is indicated on the horizontal axis, and the amount of transcript relative to that of the wild type is indicated on the vertical axis. (E) HeLa cells were transfected with either control siRNAs (targeting firefly luciferase) or siRNAs targeting hU3-55K. After 48 h, cells were then transfected with constructs expressing wild-type or mutant U3 snoRNAs. RNA was extracted from the cells and analyzed by Northern blotting (upper panel). To control for transfection levels, DNA extracted from the transfected HeLa cells was analyzed by PCR, as described for panel C. The product was separated by agarose gel electrophoresis and analyzed by Southern blotting (lower panel). The U3 construct used is indicated above each lane. Use of the siRNA targeting hU3-55K (+) and the control siRNA (−) is indicated. (D) Quantitation of tagged U3 snoRNA expression levels. The levels of tagged U3 snoRNA were normalized relative to the amount of plasmid DNA transfected into the cells. The transfected construct and the presence of the control (−) and hU3-55K-targetting siRNA (+) are indicated on the horizontal axis, and the amount of transcript relative to that of the wild-type is indicated on the vertical axis. (G) HeLa cells, transiently transfected with constructs expressing wild-type or mutant Strepto-tagged U3 snoRNAs, were analyzed by fluorescence in situ hybridization (FISH) using probes specific for the endogenous U3 and the Strepto tag as indicated. In the merged images, the U3 data are shown in green and the Strepto tag in red. The transfected construct is indicated at the top of each set of images. (H) Extracts prepared from HeLa cells were transiently transfected with the construct expressing the C′con U3 snoRNA mutant and separated by glycerol gradient centrifugation. RNA was extracted from the resultant fractions and analyzed by Northern hybridization. The positions of the 12S and 80S complexes are indicated at the top. The positions of the endogenous (U3) and mutant (C′con) RNAs are indicated on the left.
Wild-type, tagged U3 snoRNA was detected in RNA derived from transfected cells, while no signal was observed in the untransfected control sample (Fig. 2C). For each construct, the Strepto-tagged U3 snoRNA signal was normalized to transfection levels (Fig. 2D). Consistent with an earlier observation (14), mutation of box C in the B/C motif (mutC) resulted in reduced U3 snoRNA expression compared to that of the wild-type RNA (Fig. 2D). Conversion of box C′ to the consensus (C′con) resulted in a slight increase in U3 snoRNA expression. Interestingly, the C′con mutation together with the mutC mutation (mutC/C′con) resulted in wild-type expression levels of U3 snoRNA. This indicates that normal U3 snoRNA accumulation is dependent on the B/C motif, and presumably hU3-55K-binding, in the presence of the weaker C′ box.
To test this hypothesis, we next investigated whether depleting hU3-55K would have an effect on the levels of the mutant U3 snoRNAs containing the C′ consensus sequence (C′con and mutC/C′con). HeLa cells were transfected with siRNAs and 48 h later were transfected with plasmids expressing the wild-type or mutant, tagged U3 snoRNAs. After a further 12 h, cells were harvested and analyzed as described above with the tagged U3 snoRNA signal (Fig. 2E) normalized to transfection levels (Fig. 2F). As expected, the wild-type U3 snoRNA levels were reduced in cells depleted of hU3-55K relative to levels in those transfected with a control siRNA. Surprisingly, the levels of mutant U3 snoRNAs containing the C′ consensus sequence (C′con and mutC/C′con) were increased about 2-fold in cells depleted of hU3-55K (Fig. 2F). This implies that, as expected, the accumulation of the C′con and mutC/C′con U3 snoRNAs is not dependent on hU3-55K but also that this protein may still, even in the case where the binding site has been mutated (mutC/C′con), influence U3 snoRNP formation (see Discussion).
It is possible that the unusual C′ box is important for U3 snoRNP function. We therefore tested whether mutating the C′ box to the consensus would affect U3 snoRNP function. In particular, we wanted to test whether the mutation would affect the ability of the U3 snoRNP to integrate into the SSU processome and localize to the GC, two key measures of U3 snoRNP function. Cells were transfected with plasmids expressing either the wild-type or mutant U3 snoRNAs and then analyzed by fluorescence in situ hybridization using probes specific for the Strepto tag sequence (inserted in the U3 snoRNA coding sequence) and the endogenous U3 snoRNA. The wild-type U3 snoRNA colocalized with the endogenous U3 snoRNA and was found throughout the nucleolus (Fig. 2G). The U3 snoRNA C′ consensus mutation (C′con) was also found throughout the nucleolus. In contrast, the U3 snoRNA containing both the C′con and box C mutations (mutC/C′con) showed a more punctuate staining pattern and was not found throughout the nucleolus, as reported previously for the mutC mutation (14). Indeed, the mutC/C′con RNA colocalized with the U8 snoRNA in the DFC/FC regions of the nucleolus (data not shown), as was reported previously for the mutC mutation alone. This therefore indicates that the unusual C′ sequence found in the human U3 snoRNA is not required for GC localization.
We next analyzed the ability of the U3 snoRNA, containing the C′con sequence, to be incorporated into the 80S SSU processome. Extracts were prepared from cells expressing the U3 snoRNA containing the C′con mutation and separated using a glycerol gradient, and the RNA content of the fractions was analyzed by Northern blotting. The endogenous U3 snoRNA was found in fractions corresponding to the free U3 snoRNP (Fig. 2H, 12S fractions 2 to 6) and the SSU processome (80S; fractions 12 to 19). The distribution of the U3 C′con RNA was almost identical to that of the endogenous snoRNA. Taken together, our data imply that the U3 C′con RNA is functional, although we cannot rule out the possibility of a subtle defect. Indeed, the key function of the unusual C′ box appears to be to provide a mechanism by which U3 snoRNA accumulation is dependent on hU3-55K binding and therefore provide a means by which to regulate U3 levels independently of all other box C/D snoRNAs.
hU3-55K facilitates U3 snoRNP assembly by directly interacting with NOP56 and NOP58.
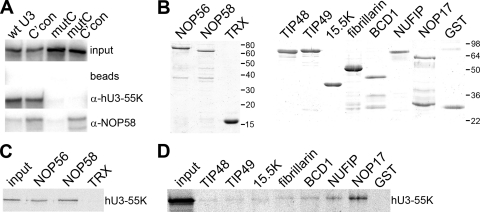
NOP56, NOP58, and fibrillarin are important in maintaining the levels of all box C/D snoRNAs, and it is believed that the recruitment of these proteins serves to stabilize the snoRNAs. hU3-55K could therefore influence U3 snoRNA accumulation by participating in the binding of the core box C/D proteins, NOP56, NOP58, and fibrillarin, to the C/D motif. To test this, wild-type and mutant 32P-labeled U3 snoRNAs (Fig. 2B) were incubated in nuclear extract, and snoRNP formation was assayed by immunoprecipitation using anti-hU3-55K and -NOP58 antibodies (13, 43). Copurifying RNAs were separated by PAGE and analyzed by autoradiography. The wild-type and C′con U3 snoRNAs but not those containing the C box mutation were efficiently coimmunoprecipitated by anti-hU3-55K antibodies after incubation in nuclear extract (Fig. 3A). Anti-NOP58 coprecipitated both the wild-type and C′con U3 snoRNA. In contrast, mutation of the B/C motif (mutC) significantly reduced copurification with anti-NOP58 antibodies. None of the RNAs were coprecipitated when no antibody was used (Fig. 3A, beads). Our data therefore indicate that the binding of the core box C/D proteins, as measured here by analyzing NOP58 recruitment, is dependent on the U3-specific box B/C motif and, presumably, hU3-55K binding. The box B/C motif, and therefore hU3-55K binding, is not required for core snoRNP formation when the C′ motif is mutated to the consensus sequence found in all other box C/D snoRNAs and plant and fungal U3 snoRNAs. Thus, the weak C′ motif in the human U3 snoRNA appears to render core box C/D snoRNP formation dependent on hU3-55K binding.
Fig. 3.
hU3-55K interacts with NOP56 and NOP58 in vitro. (A) U3 snoRNP complexes were assembled using wild-type and mutant 32P-radiolabeled RNAs in mouse nuclear extract. The mutations are indicated in Fig. 2B. RNP complexes formed during this reaction were immunoprecipitated using snoRNP-specific antibodies, and the copurifying RNAs were separated on an 8% polyacrylamide–7 M urea gel and then analyzed by autoradiography. The RNA used is indicated above each lane. Antibodies used for immunoprecipitation are indicated on the right. Input, 10% of the RNA after incubation in nuclear extract; beads, protein A Sepharose without antibodies. (B) Recombinant proteins used in protein-protein interaction studies were separated by SDS-PAGE and visualized by Coomassie staining. The protein loaded is indicated at the top of each lane. The positions of the marker proteins are indicated to the right of each panel. (C) Thioredoxin (TRX) and TRX fusion proteins of NOP56 and NOP58 were bound to anti-TRX antibody coupled to protein A Sepharose and then incubated with 35S-methionine-labeled, in vitro-translated hU3-55K. (D) Recombinant purified GST and GST-tagged proteins were bound to glutathione Sepharose and then incubated with 35S-methionine-labeled, in vitro-translated hU3-55K. Bound proteins were separated by SDS-PAGE and analyzed by autoradiography. The protein used is indicated at the top. Input: 10% of the 35S-labeled hU3-55K protein used in the interaction assay.
One mechanism by which hU3-55K could facilitate snoRNP assembly is through a direct interaction with one or more of the core box C/D proteins or assembly factors. Therefore, we tested whether hU3-55K interacted with core box C/D proteins (GST-tagged 15.5K and fibrillarin and thioredoxin-tagged NOP56 and NOP58) and/or GST-tagged snoRNP biogenesis factors (Fig. 3B). The recombinant proteins were incubated with in vitro-translated, [35S]methionine-labeled hU3-55K, and the complexes formed were purified, using either glutathione Sepharose or anti-thioredoxin antibodies bound to protein A Sepharose, as appropriate. The bound protein was then separated by SDS-PAGE and analyzed by autoradiography.
The 35S-labeled hU3-55K bound efficiently to both NOP56 and NOP58 (Fig. 3C). The labeled protein did not copurify with thioredoxin. A low level of hU3-55K also reproducibly copurified with GST-NOP17 (Fig. 3D). Very weak signals, just above the background level observed with GST alone, were seen with the other GST-tagged proteins. Our data therefore indicate that hU3-55K assists in U3 snoRNP assembly by directly binding the core box C/D proteins NOP56 and NOP58 and, to a lesser extent, the snoRNP biogenesis factor NOP17.
U3 snoRNA levels are specifically downregulated during epithelial cell differentiation.
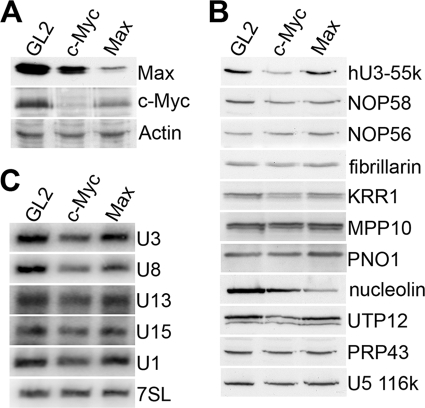
We were interested to find a situation in which hU3-55K-mediated regulation of U3 snoRNA accumulation may occur. U3 snoRNA levels are downregulated during the transition of myoblasts to myotubes (10). It is not clear, however, whether this occurs during other cellular differentiation events and whether this is specific to the U3 snoRNA. We therefore compared snoRNA and snRNA levels in undifferentiated and differentiated CaLu-3 (lung) and CaCo-2 (intestinal) epithelial cells by Northern blotting. Relative to total RNA, the levels of U1 snRNA were equal in both undifferentiated and differentiated CaCo-2 and CaLu3 cells (Fig. 4A). The amount of U15 box C/D snoRNA was also the same in both undifferentiated and differentiated cells, while there was slightly more U8 and U13 in the differentiated cells. In contrast, U3 snoRNA levels were reproducibly threefold lower in differentiated cells than in undifferentiated cells. We next compared the levels of box C/D snoRNP proteins in undifferentiated and differentiated epithelial cells. Protein loading was normalized to the spliceosomal U5-116K protein (Fig. 4B). During differentiation, hU3-55K levels were reduced and mirrored the changes in U3 snoRNA levels. In contrast, the levels of the common core box C/D proteins NOP58, NOP56, and fibrillarin were approximately the same in both differentiated and undifferentiated cells. This implies that in both CaCo-2 and CaLu-3 cells, U3 snoRNA accumulation is regulated independently of all other box C/D snoRNAs, and the similar change in U3 and hU3-55K levels is consistent with our hypothesis that controlling hU3-55K expression specifically regulates U3 snoRNA levels.
Fig. 4.
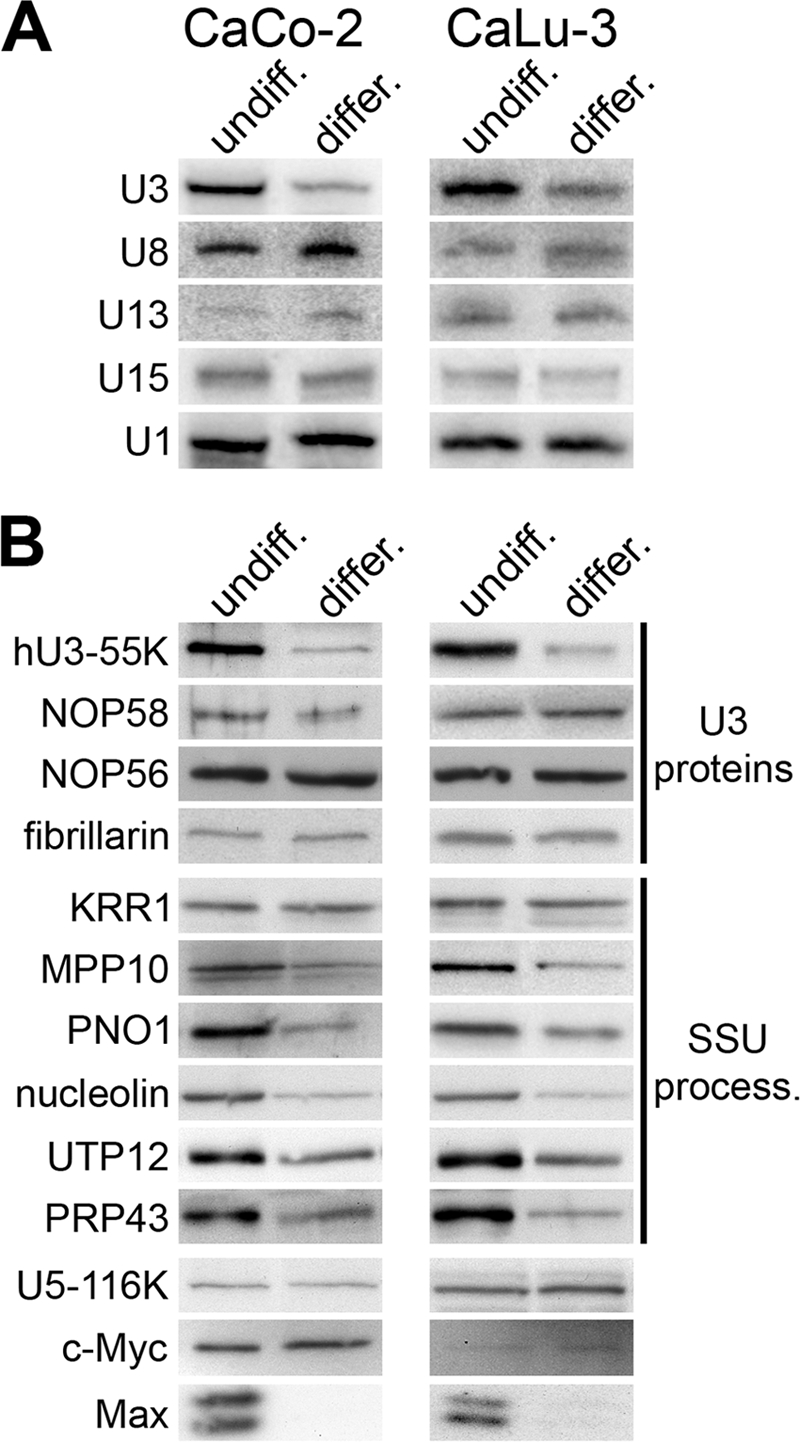
U3 snoRNA and several SSU processome components are specifically downregulated during epithelial cell differentiation. CaCo-2 and CaLu-3 epithelial cells (indicated at the top) were grown either to subconfluent levels (undiff.) or under conditions to induce differentiation (differ.). (A) RNAs were extracted from the cells, and equal amounts of RNA were separated on an 8% polyacrylamide–7 M urea gel. The RNAs were transferred to a nylon membrane and analyzed using snoRNA- and snRNA-specific probes as indicated to the left of the panels. (B) Proteins were separated using SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting using antibodies specific to U3 snoRNP or SSU processome proteins (indicated at right). The proteins detected are indicated to the left of each panel.
We next tested whether the levels of other SSU processome components were downregulated during epithelial cell differentiation. Western blot analysis revealed that the levels of MPP10, PNO1 (DIM2), nucleolin, UTP12, and PRP43 but not KRR1 were significantly lower in differentiated cells (Fig. 4B). Thus, a subset of SSU processome components is downregulated, together with the U3 snoRNP, during epithelial cell differentiation. The expression of many of these proteins is regulated by the proto-oncogene c-Myc, a protein downregulated in many differentiation events (33). Western blot analysis, however, revealed that c-Myc levels did not alter during CaCo-2 or CaLu-3 differentiation (Fig. 4B). In contrast, the levels of Max, the c-Myc cofactor, were significantly reduced in the differentiated cells relative to those seen in the undifferentiated cells. Based on these data, it is therefore not clear what role c-Myc plays in regulating U3 snoRNP and SSU processome component levels during epithelial cell differentiation.
Downregulation of c-Myc reduces hU3-55K and snoRNA levels.
The fact that the levels of ribosome biogenesis factors are downregulated during development while c-Myc levels remain unchanged raised the question of whether c-Myc is important for controlling the levels of these proteins. In particular, previous work just measured the levels of the mRNAs of ribosome biogenesis factors after c-Myc overexpression (27, 33). We therefore analyzed the levels of several ribosome biogenesis factors after RNAi-mediated depletion of c-Myc and Max. HeLa cells were transfected with siRNAs and 48 h later harvested, and protein levels were monitored by Western blotting. The c-Myc and Max siRNAs effectively depleted the levels of the target protein relative to the control siRNAs, in each case without affecting the levels of the control protein, actin (Fig. 5A). Interestingly, depletion of c-Myc resulted in a strong reduction in the levels of Max. Knockdown of Max, however, resulted in only a mild reduction in c-Myc levels.
Fig. 5.
RNAi-mediated depletion of c-Myc and Max. HeLa cells were transfected with either control siRNAs (targeting firefly luciferase) or siRNAs targeting c-Myc or Max. (A and B) Protein levels in the transfected cells were determined by Western blot analysis. The protein targeted is indicated above each panel. The antibodies used are indicated on the right. Proteins derived from equal numbers of cells were loaded. (C) RNA was extracted from an equal number of transfected cells and analyzed by Northern blotting. The RNA analyzed is indicated to the right of each panel.
Having established the knockdown of c-Myc and Max, we next analyzed the effect of depleting these two proteins on the levels of snoRNAs and ribosome biogenesis factors. Depletion of c-Myc resulted in a repeatable decrease in hU3-55K levels without a significant change in the amounts of NOP56, NOP58, and fibrillarin in the cell (Fig. 5B). Slight reductions were also seen in the levels of the SSU processome proteins UTP12, KRR1, and nucleolin, while MPP10, PNO1, PRP43, and the spliceosomal protein U5-116K remained unaffected. In contrast, depletion of Max resulted in a significant decrease in the levels of nucleolin, while all of the other proteins tested remained unaffected. Depletion of c-Myc resulted in a reduction in the levels of the U3, U8, U13, and U15 box C/D snoRNAs and the spliceosomal U1 snRNA (Fig. 5C). Interestingly, the effect of c-Myc depletion of U13 and U15 was reproducibly not as dramatic as that seen for the U3 and U8 box C/D snoRNAs. The levels of the RNA component of the signal recognition particle (SRP), 7SL, were not affected by c-Myc depletion. In contrast, depletion of Max had no effect on the levels of the RNAs tested. Therefore, c-Myc appears to affect general box C/D snoRNA levels in the cell, while Max appears not to be involved in this regulation.
DISCUSSION
We have shown that the levels of the U3 snoRNA can be regulated, independently of other box C/D snoRNAs, by controlling the level of the U3-specific protein hU3-55K. Our data imply that this protein helps recruit the core box C/D proteins to the suboptimal C′/D motif in the U3 snoRNA by directly binding to NOP56 and NOP58. This is consistent with the observation that hU3-55K is recruited early in the U3 snoRNP biogenesis pathway, along with NOP56 and NOP58 (44). The suboptimal C′/D motif appears, in the assays performed here, not to be important for U3 snoRNP function and therefore likely serves to render snoRNP formation and therefore U3 snoRNA levels dependent on hU3-55K. To our surprise, we found that depleting hU3-55K resulted in an increase in the levels of U3 snoRNAs containing the C′con mutation. We speculate that the mutant U3 snoRNAs are still being recognized as U3 snoRNAs by the snoRNP assembly machinery, even when hU3-55K binding has been abolished by mutating the C box. It is possible that other elements in the U3 snoRNA, such as elements of the U3 3′ domain, are still being recognized by the snoRNP assembly machinery, as is the case for the recruitment of the SMN complex to the spliceosomal snRNAs, where elements specific to each individual snRNA are essential (19). The U3 3′ domain has an unusual structure, with a stem structure spanning the box B/C and C′/D motifs (Fig. 6), which is essential for yeast U3 snoRNA accumulation (32). In addition, we have shown that this stem is also essential for U3 snoRNA accumulation in human cells (H. Richardson and N. J. Watkins, unpublished observation), suggesting that this double-stranded region plays an important part in U3 snoRNP formation. hU3-55K, which is limiting for U3 snoRNP formation, is likely to be part of the mechanism that recognizes the U3 snoRNA during snoRNP assembly. Depletion of hU3-55K may therefore stop the specific recognition of the U3 snoRNA, removing this level of regulation and enabling the C′con mutations to be assembled as “normal” snoRNPs.
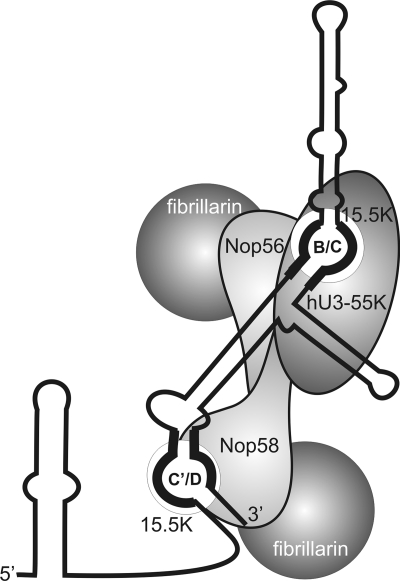
Fig. 6.
Schematic model of U3 snoRNP structure.. The secondary structure of the U3 snoRNA is shown (black line), with the predicted organization of the associated proteins indicated. The box C′/D and B/C motifs are labeled.
Our data indicate that the B/C and C′/D motifs in the U3 snoRNA function together to recruit a single protein complex. Supporting this model, the B/C motif has been previously linked to both fibrillarin binding (2) and nucleolar localization (20), functions that are normally solely attributed to the C/D motif in other snoRNAs. This raises the question of the structural organization of the core U3 snoRNP and how this relates to that of the box C/D snoRNPs that direct methylation. In the methylation guide snoRNPs, there are two copies of 15.5K and fibrillarin and one each of NOP56 and NOP58, contacting the C′/D′ and C/D motifs, respectively. Given the similar nature of the C/D and B/C motifs, it is tempting to speculate that there is a similar organization in the U3 snoRNP with 2 molecules of 15.5K and fibrillarin and one each of NOP56 and NOP58 (Fig. 6). In this case, however, the complex would be anchored to the B/C motif via the interaction between hU3-55K and NOP56/NOP58. NOP56 is predicted to make specific contacts to stem II of the C′/D′ motif. Given the different sequence of stem II in the B/C motif, it is possible that NOP56 does not directly contact the B/C motif, although evidence suggests that both NOP56 and fibrillarin contact the U3 snoRNA close to this RNA element in yeast (12).
In both epithelial cell lines analyzed, hU3-55K levels mirrored those of the U3 snoRNA, and both were reduced 3-fold during differentiation. While we cannot rule out the possibility that transcriptional regulation controls U3 snoRNA levels, it is reasonable to propose that hU3-55K regulation controls U3 snoRNA levels. Since hU3-55K is required for NOP58 association, which in turn is essential for the stability of the initial U3 snoRNA transcript (26), it is difficult to analyze transcriptional control of U3 snoRNA levels. A U3 snoRNP lacking hU3-55K is likely to be detrimental to the cell, and therefore, making U3 snoRNP accumulation dependent on hU3-55K binding avoids this problem. We found no evidence that the suboptimal C′ box in the U3 snoRNA has a functional role in ribosome biogenesis, suggesting that the main role of this unusual sequence is to enable hU3-55K-mediated regulation. The “weak” C′ box is found in mammalian and amphibian but not fish, insect, plant, or yeast U3 snoRNAs (Fig. 2A). This suggests that controlling hU3-55K expression may not regulate U3 snoRNA levels in these other eukaryotes.
Cellular differentiation results in a reduction in both cell growth and ribosome biogenesis. Upon differentiation, the adenocarcinoma cell line CaCo-2 appears to lose its tumorigenic phenotype and so represents a useful tool for examining tumor progression (38). We show that during this differentiation event, the U3 snoRNA and hU3-55K levels but not those of other box C/D snoRNAs and core snoRNP proteins are downregulated. This is consistent with hU3-55K expression controlling U3 snoRNA levels in these cells. Furthermore, several other SSU processome components are downregulated during epithelial cell differentiation. From this we conclude that many of the SSU processome components are upregulated during tumorogenesis and could be future targets for therapeutic agents. U3 is one of the few snoRNPs that is essential in both yeast and metazoans (15). This snoRNP is recruited cotranscriptionally to the pre-rRNA and could play a key role in stabilizing the nascent transcripts and therefore regulating the rate of ribosome formation. Moreover, pre-rRNA processing, rather than pre-rRNA transcription, is used to control mature ribosome levels in certain instances of tumorogenesis and cellular differentiation (5, 8, 29, 33).
The proto-oncogene c-Myc is reported to be downregulated during many differentiation events as part of the block in proliferation (21). We were therefore surprised to find that c-Myc levels were not reduced during CaCo-2 and CaLu-3 differentiation. NOP56, NOP58, fibrillarin, and hU3-55K mRNAs, and so presumably their protein products, have all been shown to be regulated by c-Myc (7, 27, 33). The fact that only hU3-55K levels are affected during differentiation is therefore consistent with c-Myc levels not changing. The levels of the c-Myc cofactor, Max, however, were significantly reduced during epithelial cell differentiation. This suggests either Max regulates a subset of ribosome biogenesis factors which are downregulated during this differentiation event or that another signaling pathway is responsible. It is important to note, however, that we cannot rule out the presence of either a posttranslationally modified or mutated c-Myc in the differentiated epithelial cells with altered activity. Depletion of c-Myc or Max only resulted in a slight reduction of a subset of the proteins, the box C/D snoRNAs and the U1 snRNA in HeLa cells. This was somewhat surprising, since many of these factors have been previously shown to be, or predicted to be, regulated by c-Myc. Based on our data, we therefore believe that it is likely that neither c-Myc nor Max plays a role in regulating the levels of ribosome biogenesis factors during epithelial cell differentiation. Other signaling pathways, such as Ras/PKA and TOR (18, 36), p53, pRB, and ARF (28), however, are key regulators in both ribosome biogenesis and cell cycle control in eukaryotes and could be responsible for the specific changes in U3 snoRNA and SSU processome protein levels we have observed during CaCo2 and CaLu3 differentiation.
ACKNOWLEDGMENTS
We thank Sabine Quitard, Brendan Kenny, James Garnett, and Mike Gray for providing CaCo-2 and CaLu-3 cells, Stuart Maxwell for the kind gift of the mouse nuclear extract, Reinhard Lührmann for providing anti-U5-116K antibodies, and Claudia Schneider, Katherine Sloan, and Jeremy Brown for critically reading the manuscript.
Footnotes
Published ahead of print on 19 April 2011.
REFERENCES
- 1. Aittaleb M., et al. 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 10: 256–263 [DOI] [PubMed] [Google Scholar]
- 2. Baserga S. J., Yang X. D., Steitz J. A. 1991. An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 10: 2645–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boulon S., et al. 2008. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 180: 579–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boulon S., et al. 2004. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell 16: 777–787 [DOI] [PubMed] [Google Scholar]
- 5. Bowman L. H., Emerson C. P., Jr 1977. Post-transcriptional regulation of ribosome accumulation during myoblast differentiation. Cell 10: 587–596 [DOI] [PubMed] [Google Scholar]
- 6. Cahill N. M., et al. 2002. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 21: 3816–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coller H. A., et al. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. U. S. A. 97: 3260–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dudov K. P., Dabeva M. D. 1983. Post-transcriptional regulation of ribosome formation in the nucleus of regenerating rat liver. Biochem. J. 210: 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fabrizio P., Laggerbauer B., Lauber J., Lane W. S., Lührmann R. 1997. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 16: 4092–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glibetic M., Larson D. E., Sienna N., Bachellerie J. P., Sells B. H. 1992. Regulation of U3 snRNA expression during myoblast differentiation. Exp. Cell Res. 202: 183–189 [DOI] [PubMed] [Google Scholar]
- 11. Granneman S., Baserga S. J. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296: 43–50 [DOI] [PubMed] [Google Scholar]
- 12. Granneman S., Kudla G., Petfalski E., Tollervey D. 2009. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U. S. A. 106: 9613–9618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Granneman S., et al. 2002. The hU3-55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in vitro. J. Biol. Chem. 277: 48490–48500 [DOI] [PubMed] [Google Scholar]
- 14. Granneman S., et al. 2004. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol. Cell. Biol. 24: 8600–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henras A. K., et al. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hernandez-Verdun D. 2006. Nucleolus: from structure to dynamics. Histochem. Cell Biol. 125: 127–137 [DOI] [PubMed] [Google Scholar]
- 17. Jastrzebski K., Hannan K. M., Tchoubrieva E. B., Hannan R. D., Pearson R. B. 2007. Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors 25: 209–226 [DOI] [PubMed] [Google Scholar]
- 18. Jorgensen P., et al. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18: 2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kolb S. J., Battle D. J., Dreyfuss G. 2007. Molecular functions of the SMN complex. J. Child Neurol. 22: 990–994 [DOI] [PubMed] [Google Scholar]
- 20. Lange T. S., et al. 1998. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol. Biol. Cell 9: 2973–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laurenti E., Wilson A., Trumpp A. 2009. Myc's other life: stem cells and beyond. Curr. Opin. Cell Biol. 21: 844–854 [DOI] [PubMed] [Google Scholar]
- 22. Lee I. J., Hom K., Bai G., Shapiro M. 2009. NMR metabolomic analysis of caco-2 cell differentiation. J. Proteome Res. 8: 4104–4108 [DOI] [PubMed] [Google Scholar]
- 23. Luo J., et al. 2001. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 61: 4683–4688 [PubMed] [Google Scholar]
- 24. Mayer C., Grummt I. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391 [DOI] [PubMed] [Google Scholar]
- 25. McKeegan K. S., Debieux C. M., Boulon S., Bertrand E., Watkins N. J. 2007. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 27: 6782–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McKeegan K. S., Debieux C. M., Watkins N. J. 2009. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol. Cell. Biol. 29: 4971–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Menssen A., Hermeking H. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. U. S. A. 99: 6274–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oskarsson T., Trumpp A. 2005. The Myc trilogy: lord of RNA polymerases. Nat. Cell Biol. 7: 215–217 [DOI] [PubMed] [Google Scholar]
- 29. Pajic A., et al. 2000. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int. J. Cancer 87: 787–793 [DOI] [PubMed] [Google Scholar]
- 30. Prieto J. L., McStay B. 2007. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 21: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ruggero D., Pandolfi P. P. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3: 179–192 [DOI] [PubMed] [Google Scholar]
- 32. Samarsky D. A., Fournier M. J. 1998. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 3431–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schlosser I., et al. 2003. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 31: 6148–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sienna N., Larson D. E., Sells B. H. 1996. Altered subcellular distribution of U3 snRNA in response to serum in mouse fibroblasts. Exp. Cell Res. 227: 98–105 [DOI] [PubMed] [Google Scholar]
- 35. Sjoblom T., et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- 36. Soulard A., Cohen A., Hall M. N. 2009. TOR signaling in invertebrates. Curr. Opin. Cell Biol. 21: 825–836 [DOI] [PubMed] [Google Scholar]
- 37. Sperger J. M., et al. 2003. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. U. S. A. 100: 13350–13355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stierum R., et al. 2003. Proteome analysis reveals novel proteins associated with proliferation and differentiation of the colorectal cancer cell line Caco-2. Biochim. Biophys. Acta 1650: 73–91 [DOI] [PubMed] [Google Scholar]
- 39. Sun H., Pang K. S. 2010. Physiological modeling to understand the impact of enzymes and transporters on drug and metabolite data and bioavailability estimates. Pharm. Res. 27: 1237–1254 [DOI] [PubMed] [Google Scholar]
- 40. Szewczak L. B., DeGregorio S. J., Strobel S. A., Steitz J. A. 2002. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9: 1095–1107 [DOI] [PubMed] [Google Scholar]
- 41. Turner A. J., Knox A. A., Prieto J. L., McStay B., Watkins N. J. 2009. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol. Cell. Biol. 29: 3007–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Venema J., Vos H. R., Faber A. W., van Venrooij W. J., Raue H. A. 2000. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA 6: 1660–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watkins N. J., Dickmanns A., Lührmann R. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22: 8342–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watkins N. J., et al. 2004. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell 16: 789–798 [DOI] [PubMed] [Google Scholar]
- 45. Watkins N. J., et al. 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466 [DOI] [PubMed] [Google Scholar]