Abstract
Objectives
This study describes parasite kinetics in the blood of visceral leishmaniasis patients treated with liposomal amphotericin B (L-AmB) or a preformed fat emulsion of amphotericin B (ApL) using real-time quantitative PCR (qPCR).
Methods
Forty-six patients were treated with a single dose (15 mg/kg of body weight) of either L-AmB (n = 13) or ApL (n = 33). qPCR was used to estimate parasite kinetics by detection of Leishmania donovani DNA using kinetoplast DNA-specific primers in peripheral blood samples using an absolute quantification method.
Results
The mean parasite load decreased from baseline (day 0) values of 894.07 and 980.48 to 71.72 and 211.52 parasite genomes/mL at day 7 in L-AmB and ApL groups, respectively, and at day 30 these further declined to 8.30 and 133.98 parasite genomes/mL, respectively. At day 30 post-treatment evaluation, the decline in parasite load was significantly greater (P = 0.024) with L-AmB compared with ApL. Four of 33 patients in the ApL group failed treatment (1 primary failure and 3 relapses) with the presence of parasites, whereas all patients in the L-AmB group were cured at 6 month follow-up.
Conclusions
qPCR can be a tool to measure parasite dynamics accurately and provide a marker to measure the efficacy of various drugs. It can be used as a test of cure, allowing us to do away with invasive and risky methods such as splenic or bone marrow aspiration.
Keywords: efficacy, fat emulsion of amphotericin B, liposomal amphotericin B, Leishmania donovani, qPCR, VL
Introduction
Visceral leishmaniasis (VL) has been treated with pentavalent antimony (Sbv) compounds in India and elsewhere for more than 70 years; however, Sbv is no longer effective in most endemic areas of India. Various alternatives, such as miltefosine, paromomycin and various lipid formulations of amphotericin, are now used in this region for the treatment of VL.1–3 Despite the initial high efficacy demonstrated by every antileishmanial drug, there is a lurking danger of the development of increasing unresponsiveness with the passage of time due to poor compliance, the inherent nature of the drugs and inefficient public health systems.4 Apart from the need to develop new therapeutic drugs that are easy to administer, economical and safe, it is important to effectively monitor the efficacy of drugs in terms of parasite clearance. Determining parasitic kinetics to assess treatment efficacy could be an important tool. Since sterile cure is never achieved in leishmaniasis after treatment,5,6 a quantitative approach [real-time quantitative PCR (qPCR)] can be employed not only for diagnostic purposes, but also for monitoring parasitaemia in patients during their treatment.7–10 In VL, the number of parasites found in blood is low compared with the spleen.11 However, in most cases it is feasible to diagnose VL by culturing either peripheral blood mononuclear cells or buffy coat/whole blood cells isolated from a small volume of blood,11 which is helpful in replacing the invasive splenic aspiration. As the Leishmania parasite is an intracellular organism, the source of the DNA detected in blood could be either live or dead parasites harboured inside phagocytic cells or DNA released from phagocytic cells.12 Kinetoplast DNA-specific primers are more suitable to detect the Leishmania parasite, as there are ∼10 000 copies in a single parasite.13–15 We used a qPCR experiment based on the SYBR Green method and targeting the minicircle kinetoplast DNA. We took a subset of patients randomized to either liposomal amphotericin B (L-AmB; AmBisome, Gilead Sciences, Foster City, CA, USA; n = 13) or a preformed fat emulsion of amphotericin B (ApL; Bharat Serum and Vaccines Limited, Mumbai, India; n = 33) to study the parasite kinetics resulting from the use of these two drugs using qPCR as a molecular tool.
Materials and methods
Study site, patients and sample collection
This study was carried out at the Infectious Diseases Research Laboratory of the Department of Medicine, Banaras Hindu University, Varanasi, India, and at its field site Kala-Azar Medical Research Centre, Muzaffurpur, Bihar, India. The experiment was approved by the Ethics Committee of the Institute of Medical Sciences, and all subjects provided written informed consent. Forty-six patients with parasitologically confirmed VL (6–55 years, males and females, and HIV negative) were enrolled in the study. Confirmation of VL was done by demonstration of amastigotes in Giemsa-stained smears of splenic aspirate. Patients were treated with L-AmB or the cheaper ApL. The drugs were administered at 15 mg/kg of body weight in a single infusion. Baseline blood samples were obtained for haematological and biochemical evaluation. For parasite quantification, 500 μL of blood was drawn from all patients on days 0, 3, 5 and 7. Patients were discharged on day 7 and post-treatment follow-up was done on day 30. Patients were followed up for a further 6 months for evaluation of definitive cure. On day 30 and at the 6 month follow-up, blood samples were drawn for parasite quantification by qPCR, as well as for haematological and biochemical evaluation. At the 6 month follow-up, clinical and laboratory evaluation was carried out in order to look for features of VL relapse. If relapse was suspected earlier, blood for parasite quantification and other haematological evaluations was drawn at that time.
DNA isolation from blood
DNA was extracted from 200 μL of blood collected in citrate tubes. Extraction was carried out using a QIAamp DNA mini kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's instructions and DNA elution was carried out in 30 μL of MiliQ water. Only DNA that had an optical density (OD) 260/280 ratio of 1.8–2.0 and an OD 260/230 ratio >1.5 by spectrophotometer measurements (ND-2000 spectrophotometer; Thermo Scientific, Waltham, MA, USA) was taken for qPCR experiments.
Standard curve
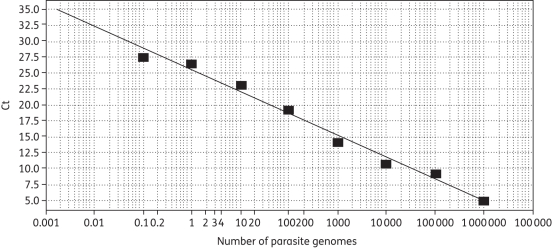
Parasite load was determined using a standard curve made with serial dilutions of Leishmania DNA. A stock solution of Leishmania donovani DNA was obtained by extraction (QIAamp DNA mini kit) from 2 × 108 promastigotes. After eluting in 200 μL of H2O, the concentration was 106 parasites/μL, assuming the extraction was nearly 100% efficient (120 Leishmania parasites have 10 pg of DNA16). Ten-fold serial dilutions of the DNA stock solution yielded eight points on a curve spanning 0.1–106 parasites (Figure 1).
Figure 1.
qPCR standard curve for quantification of Leishmania parasite. The standard curve was obtained from serial dilutions of Leishmania DNA with 0.1–106 parasites. The y-axis shows the Ct values at different parasite numbers. Efficiency = 96.775%, slope = −3.402 and r2 = 0.987.
qPCR experiment
SYBR Green-based qPCR was applied for quantification of the Leishmania parasite in patient blood. For accurate sensitivity, Leishmania-specific kinetoplast DNA was chosen as the target region. The PCR was performed in a final volume of 20 μL containing 10 μL of SYBR Green master mixture (2×) [Applied Biosystems (ABI), Carlsbad, CA, USA], 3 μL of MiliQ water and 5 μL of DNA template (20 ng/μL)16 and 1 μL (5 μM) of forward and reverse primers, 5′-CTTTTCTGGTCCTCCGGGTAGG-3′ and 5′-CCACCCGGCCCTATTTTACACCAA-3′ (Integrated DNA Technologies, Coralville, IA, USA). Amplification was conducted using a 7500 real-time PCR system (ABI). The thermal cycling conditions included an initial incubation at 50°C for 2 min, followed by a 10 min denaturation at 95°C and 40 cycles at 95°C for 15 s and 60°C for 1 min each. Each sample was tested in duplicate. Duplication was maintained both at the DNA isolation and PCR level. The generation of amplification plots, mean values and standard curves and dissociation stage analysis were carried out using ABI SDS software. The calculation of the melting temperature of each amplicon was done directly by the software provided. Appropriate negative controls (no template and healthy controls) were included in each plate to deal with contamination issues. A human tumour necrosis factor (TNF)-α primer (unpublished sequence; submitted for patent) targeting the single-copy TNF-α gene was used as a control to check for differences in human DNA amplification from samples taken at different times from the same individual. PCR conditions were the same as for the kinetoplast DNA qPCR.
Statistical analysis
Data analysis was carried out using the non-parametric Mann–Whitney test and the one-way ANOVA test using SPSS (IBM, Somers, NY, USA) and Prism (GraphPad Software, La Jolla, CA, USA) software.
Results
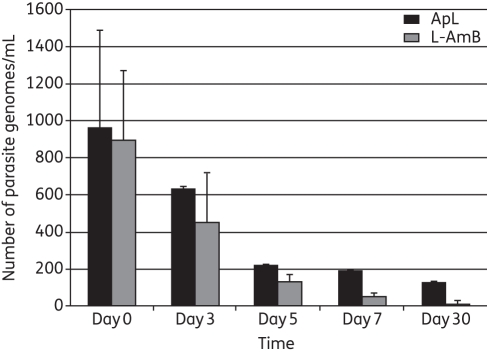
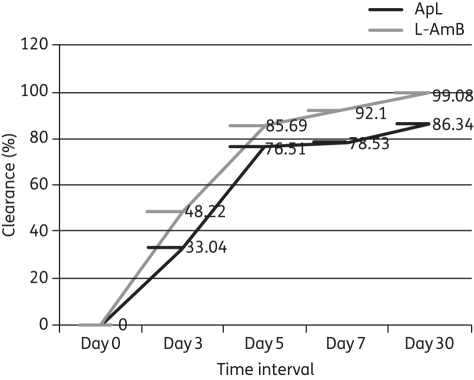
Results are expressed as the absolute number of parasite genomes/mL of blood from 200 μL of blood, and of the total DNA eluted, 100 ng was taken. Therefore, the number of Leishmania genomes found in 100 ng is further calculated in total DNA from 200 μL of blood and then per mL of blood. The difference in cycle threshold (Ct) values between two dilutions is 3.2. At the time of diagnosis, all patients were highly parasitaemic. There was good correlation between the splenic smear score and the blood parasite burden (r2 = 0.56, P ≤ 0.001). Leishmania DNA was found in all confirmed patients at day 0; the mean ± SEM parasite genomes/mL of blood in the L-AmB and ApL groups was 894.07 ± 376.99 (range = 23.8–4471.5) and 980.48 ± 528.5 (range = 20.16–6453.20), respectively, and the difference was not significant. The mean parasite load, in parasite genomes/mL, decreased to 71.72 ± 19.82 and 211.52 ± 3.47 in the L-AmB and ApL groups, respectively, at day 7. At day 30, the values were 8.30 ± 2.59 (range = 0.26–30.9) parasite genomes/mL and 133.98 ± 0.194 (range = 1.12–2681.25) parasite genomes/mL, in the L-AmB and ApL groups, respectively. At day 3, 5, 7 and 30, there was a significant decline in parasitaemia (ApL, P = 0.01; L-AmB, P = 0.045; Figure 2). However, the decline was greater with L-AmB compared with ApL at day 30 (P = 0.024). Parasite clearance in L-AmB group is significantly higher compared with ApL group (P < 0.0001) (Figure 3). At the 6 month follow-up, all the L-AmB-treated patients had a negligible parasite number (<1 parasite genomes/mL) and achieved definitive cure, whereas in the ApL group, one patient had a parasite burden of >1000 parasite genomes/mL at day 30, one patient had a parasite burden of 1266 parasite genomes/mL at 4 months and two patients had parasite burdens of 43 and 1204.13 parasite genomes/mL at the 6 month follow-up. All four of these ApL patients (after 1 month of follow-up) who failed treatment were given rescue treatment. The remaining 29 patients in the ApL group showed <2 parasite genomes/mL in their blood, and were definitively cured. Figure S1 (available as Supplementary data at JAC Online) shows the parasite load at different times in some patients in both treatment categories.
Figure 2.
Comparative graph of the Leishmania parasite load in blood at different times in VL patients of different treatment groups.
Figure 3.
Comparative parasite clearance from the blood of patients of the L-AmB and ApL groups from day 0, i.e. baseline, to day 30.
Discussion
This study demonstrates the utility of qPCR in determining an association between parasite load and clinical symptoms in patients undergoing treatment for VL. The benefits of molecular monitoring are already widely accepted in antimalarial drug trials.17,18 In leishmaniasis, however, microscopy is considered as the gold standard and splenic/bone marrow slide reports are graded as 1+ to 6+, showing different levels of parasite burden at day 0 and discharge of patients,19 but it deals only with a range of parasite loads. A great disadvantage of microscopy is the requirement for an invasive and risky splenic aspirate. PCR quantification also has the advantage of having the ability to assess the interim parasite load, and can be used as a test of cure. At diagnosis, parasite burden in peripheral blood correlated significantly with splenic score. We used the SYBR Green method for qPCR, which is less expensive than the Taqman assay. Targeting the minicircle kinetoplast DNA maximizes the sensitivity of the assay, and melting curve analysis in the SYBR Green technique confirms the specificity of the kinetoplast DNA primer (melt curve around 79°C) and also reveals the presence of primer dimers. Because of their small size, primer dimers melt at lower temperatures than the desired product. Using the melting peak of a specific product, which in our case is ∼79°C, we can be sure that the sequence of interest (amplicon) is amplified. In addition, non-specific amplification may result in PCR products that melt at temperatures above or below that of the desired product. Since readings in triplicate are highly recommended for the interpretation of results at the individual sample level, and since we estimated the parasite load in duplicate, our results should be treated with caution.
We also studied parasite kinetics in terms of burden before treatment and at different time intervals after drug administration in order to observe parasite clearance. This was primarily done to see if there was a difference in the rate of parasite clearance with the two drugs under study. We found a significant difference in the rate of parasite clearance with L-AmB and ApL in most of the patients, with significantly more rapid and greater clearance with L-AmB (Figure 3). These results correlated with clinical outcome. The cure rate with L-AmB was 100%, with every patient having <1 parasite genome/mL in peripheral blood at 6 months, which might be due to degraded residual parasite genome in the blood.20 Thus, monitoring parasite load by qPCR can indicate complete cure or early detection of treatment failure or relapse. In a few patients there was a slight increase in parasitaemia level at day 5 or 7, but it significantly decreased by day 30. The increment at this stage in some patients might be due to the release of parasites from intralesional tissues, such as the spleen, into the blood. Though this study was not designed to measure the efficacy of the two drugs, ApL performed poorly with slower and lower parasite clearance and persistence of higher levels of parasites in four patients, all of whom had to be given rescue treatment due to treatment failure. However, the parasite burden at day 30 in the 3 relapsing patients gave no indication of treatment failure, and was in a range similar to that of the 29 definitively cured patients. The resurgence of parasitaemia and, importantly, the increase in the parasite burden correlated with clinical relapses. Previous studies in human leishmaniasis in immunocompromised patients indicate that a clinical relapse is associated with a certain level of parasitaemia, and it is estimated that an increase above 10 parasites/mL of blood precedes a clinical relapse.21,22 Here, three relapsing patients in the ApL group also had >10 parasites/mL of blood. Splenic smears showed the presence of amastigotes in these patients and reconfirmed the relapse of VL. The utility of qPCR in comparing drug effectiveness is clearly demonstrated here. As qPCR helps to detect relapse cases and in India post-kala-azar dermal leishmaniasis (PKDL) appears after 3–6 years of VL, it remains to be seen whether the residual parasite burden at the end of treatment is a predictor of PKDL or not. We conclude that with the introduction of new combination therapies as well as new drugs for VL treatment, qPCR could be an important tool for monitoring their efficacy and for helping us choose the best chemotherapeutic strategies.
However, qPCR requires a high initial investment for it to be established in the laboratory and each measurement of parasite load is several times more expensive (∼$10/test) than usual parasitological examination. Highly skilled personnel are required, as well as good operating conditions, such as an uninterrupted good quality power supply, air conditioning, etc. In endemic regions of developing countries these are hard to achieve. Nevertheless, such facilities could bring about a paradigm shift in the management of VL, and reference centres equipped with qPCR facilities could guide the region about the parasite kinetics related to a particular drug. Where possible, qPCR facilities should be established, both for diagnosis and prognosis of patients.
Funding
The study was partially funded by the National Institute of Allergy and Infectious Disease (NIAID), the Division of Microbiology and Infectious Disease (DMID) funding mechanism and the Tropical Medicine Research Center (grant number P50AI074321). M. S. received financial support from the Council of Scientific and Industrial Research (CSIR), New Delhi, India.
Transparency declarations
None to declare.
Supplementary data
Figure S1 is available as Supplementary data at JAC Online (chttp://jac.oxfordjournals.org/).
Acknowledgements
We acknowledge the subjects and staff members of Kala-Azar Medical Research Centre, Muzaffarpur, for participating in the study.
References
- 1.Sundar S, Chatterjee M. Visceral leishmaniasis—current therapeutic modalities. Indian J Med Res. 2006;123:345–52. [PubMed] [Google Scholar]
- 2.Vanlerberghe V, Diap G, Guerin PJ, et al. Drug policy for visceral leishmaniasis: a cost-effectiveness analysis. Trop Med Int Health. 2007;12:274–83. doi: 10.1111/j.1365-3156.2006.01782.x. doi:10.1111/j.1365-3156.2006.01782.x. [DOI] [PubMed] [Google Scholar]
- 3.Alvar J, Croft S, Olliaro P. Chemotherapy in the treatment and control of leishmaniasis. Adv Parasitol. 2006;61:223–74. doi: 10.1016/S0065-308X(05)61006-8. doi:10.1016/S0065-308X(05)61006-8. [DOI] [PubMed] [Google Scholar]
- 4.Sundar S, Mondal D, Rijal S, et al. Implementation research to support the initiative on the elimination of kala azar from Bangladesh, India and Nepal—the challenges for diagnosis and treatment. Trop Med Int Health. 2008;13:2–5. doi: 10.1111/j.1365-3156.2007.01974.x. doi:10.1111/j.1365-3156.2007.01974.x. [DOI] [PubMed] [Google Scholar]
- 5.Francino O, Altet L, Sanchez-Robert E, et al. Advantages of real-time PCR assay for diagnosis and monitoring of canine leishmaniasis. Vet Parasitol. 2006;137:214–21. doi: 10.1016/j.vetpar.2006.01.011. doi:10.1016/j.vetpar.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Mary C, Faraut F, Drogoul MP, et al. Reference values for Leishmania infantum parasitemia in different clinical presentations: quantitative polymerase chain reaction for therapeutic monitoring and patient follow-up. Am J Trop Med Hyg. 2006;75:858–63. [PubMed] [Google Scholar]
- 7.Bell A, Ranford-Cartwright L. Real-time quantitative PCR in parasitology. Trends Parasitol. 2002;18:338. doi:10.1016/S1471-4922(02)02331-0. [PubMed] [Google Scholar]
- 8.Nicolas L, Prina E, Lang T, et al. Real-time PCR for detection and quantitation of Leishmania in mouse tissues. J Clin Microbiol. 2002;40:1666–9. doi: 10.1128/JCM.40.5.1666-1669.2002. doi:10.1128/JCM.40.5.1666-1669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy T, Bisio M, Altcheh J, et al. Accurate real-time PCR strategy for monitoring bloodstream parasitic loads in Chagas disease patients. PLoS Negl Trop Dis. 2009;3:e419. doi: 10.1371/journal.pntd.0000419. doi:10.1371/journal.pntd.0000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoun K, Chouihi E, Amri F, et al. Short report: contribution of quantitative real-time polymerase chain reaction to follow-up of visceral leishmaniasis patients treated with meglumine antimoniate. Am J Trop Med Hyg. 2009;81:1004–6. doi: 10.4269/ajtmh.2009.09-0285. doi:10.4269/ajtmh.2009.09-0285. [DOI] [PubMed] [Google Scholar]
- 11.Maurya R, Mehrotra S, Prajapati VK, et al. Evaluation of blood agar microtiter plates for culturing Leishmania parasites to titrate parasite burden in spleen and peripheral blood of patients with visceral leishmaniasis. J Clin Microbiol. 2010;48:1932–4. doi: 10.1128/JCM.01733-09. doi:10.1128/JCM.01733-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tupperwar N, Vineeth V, Rath S, et al. Development of a real-time polymerase chain reaction assay for the quantification of Leishmania species and the monitoring of systemic distribution of the pathogen. Diagn Microbiol Infect Dis. 2008;61:23–30. doi: 10.1016/j.diagmicrobio.2007.12.013. doi:10.1016/j.diagmicrobio.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Vitale F, Reale S, Vitale M, et al. TaqMan-based detection of Leishmania infantum DNA using canine samples. Ann N Y Acad Sci. 2004;1026:139–43. doi: 10.1196/annals.1307.018. doi:10.1196/annals.1307.018. [DOI] [PubMed] [Google Scholar]
- 14.Noyes HA, Reyburn H, Bailey JW, et al. A nested-PCR-based schizodeme method for identifying Leishmania kinetoplast minicircle classes directly from clinical samples and its application to the study of the epidemiology of Leishmania tropica in Pakistan. J Clin Microbiol. 1998;36:2877–81. doi: 10.1128/jcm.36.10.2877-2881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortes S, Rolao N, Ramada J, et al. PCR as a rapid and sensitive tool in the diagnosis of human and canine leishmaniasis using Leishmania donovani s.l.-specific kinetoplastid primers. Trans R Soc Trop Med Hyg. 2004;98:12–7. doi: 10.1016/s0035-9203(03)00002-6. doi:10.1016/S0035-9203(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 16.Ranasinghe S, Rogers ME, Hamilton JG, et al. A real-time PCR assay to estimate Leishmania chagasi load in its natural sand fly vector Lutzomyia longipalpis. Trans R Soc Trop Med Hyg. 2008;102:875–82. doi: 10.1016/j.trstmh.2008.04.003. doi:10.1016/j.trstmh.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felger I, Genton B, Smith T, et al. Molecular monitoring in malaria vaccine trials. Trends Parasitol. 2003;19:60–3. doi: 10.1016/s1471-4922(02)00066-1. doi:10.1016/S1471-4922(02)00066-1. [DOI] [PubMed] [Google Scholar]
- 18.Perandin F, Manca N, Calderaro A, et al. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J Clin Microbiol. 2004;42:1214–9. doi: 10.1128/JCM.42.3.1214-1219.2004. doi:10.1128/JCM.42.3.1214-1219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chulay JD, Bryceson AD. Quantitation of amastigotes of Leishmania donovani in smears of splenic aspirates from patients with visceral leishmaniasis. Am J Trop Med Hyg. 1983;32:475–9. doi: 10.4269/ajtmh.1983.32.475. [DOI] [PubMed] [Google Scholar]
- 20.Disch J, Oliveira MC, Orsini M, et al. Rapid clearance of circulating Leishmania kinetoplast DNA after treatment of visceral leishmaniasis. Acta Trop. 2004;92:279–83. doi: 10.1016/j.actatropica.2004.08.002. doi:10.1016/j.actatropica.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Bossolasco S, Gaiera G, Olchini D, et al. Real-time PCR assay for clinical management of human immunodeficiency virus-infected patients with visceral leishmaniasis. J Clin Microbiol. 2003;41:5080–4. doi: 10.1128/JCM.41.11.5080-5084.2003. doi:10.1128/JCM.41.11.5080-5084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mary C, Faraut F, Lascombe L, et al. Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J Clin Microbiol. 2004;42:5249–55. doi: 10.1128/JCM.42.11.5249-5255.2004. doi:10.1128/JCM.42.11.5249-5255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.