Abstract
Background
CYP2C9 and VKORC1 are two major genetic factors associated with inter-individual variability in warfarin dose. Additionally, genes in the warfarin metabolism pathway have also been associated with dose variance. We analyzed Single Nucleotide Polymorphisms (SNPs) in these genes to identify genetic factors that might confer warfarin sensitivity in Indonesian patients.
Methods
Direct sequencing method was used to identify SNPs in CYP2C9, VKORC1, CYP4F2, EPHX1, PROC and GGCX genes in warfarin-treated patients. Multiple linear regressions were performed to model the relationship warfarin daily dose requirement with genetic and non-genetic variables measured and used to develop a novel algorithm for warfarin dosing.
Results
From the 40 SNPs analyzed, CYP2C9 rs17847036 and VKORC1 rs9923231 showed significant association with warfarin sensitivity. In our study population, no significant correlation could be detected between CYP2C9*3, CYP2C9C-65 (rs9332127), CYP4F2 rs2108622, GGCX rs12714145, EPHX1 rs4653436 and PROC rs1799809 with warfarin sensitivity.
Conclusions
VKORC1 rs9923231 AA and CYP2C9 rs17847036 GG genotypes were associated with low dosage requirements of most patients (2.05 ± 0.77 mg/day and 2.09 ± 0.70 mg/day, respectively). CYP2C9 and VKORC1 genetic variants as well as non-genetic factors such as age, body weight and body height account for 15.4% of variance in warfarin dose among our study population. Additional analysis of this combination could allow for personalized warfarin treatment in ethnic Indonesians.
Keywords: Warfarin, SNP, CYP2C9, VKORC1, Indonesia
Background
Warfarin is the most widely used oral anticoagulant in the world. It is usually prescribed for treatment of atrial fibrillation, heart valve prosthesis, recurrent stroke, deep vein thrombosis and pulmonary embolism [1]. Although warfarin is indispensable for treatment of thromboembolism and for prophylaxis of stroke, due to the large inter-individual variation in the requirement for this drug the appropriate dose to each patient is not easily adjustable. An insufficient dose will result in failure to prevent thrombosis, while overdose increases the risk of unexpected bleeding. Pharmacogenetic differences are believed to cause the variation in individual response to warfarin [1,2].
Warfarin is primarily metabolized to the 7-hydroxylated form in humans, principally by cytochrome P450 2C9. So far, more than 30 variant alleles in the CYP2C9 gene have been described (http://www.cypalleles.ki.se/cyp2c9.htm). Two common allelic variants CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910) are among the most well characterized of the CYP2C9 alleles; both alleles have been associated with reduced enzymatic activity, and thus with reduced warfarin metabolism. Individuals bearing variant alleles CYP2C9*2 and *3 are reported to require a lower maintenance dose of warfarin and a longer time to achieve stable dosing. These individuals are also reported to have a higher proportion of prothrombin-time measurements above therapeutic range, and to experience more frequent bleeding events as compared to individuals with the CYP2C9*1 wild-type allele [3].
Variability in warfarin response is also attributable to variants in the vitamin K-related gene. Warfarin makes use of its anticoagulant effects by interfering with regeneration of vitamin K by reduction of its 2,3-epoxide in the vitamin K cycle, leading to inhibition of gamma-carboxylation of vitamin K-dependent clotting factor II (prothrombin), VII, IX and X. Several reports have demonstrated that non-coding SNPs in the vitamin K related gene influenced warfarin sensitivity. These data strongly suggest that differences in genetic variations in both CYP and vitamin K-related genes could explain the diversity in warfarin sensitivity and dose requirement [2-4].
Many reports indicated an additive effect on warfarin dose requirement in combinations of variant forms of VKORC1 and CYP2C9 genes in Caucasian patients. Furthermore, it is well known that there are significant differences in warfarin dose requirement in different ethnic groups; for example, Chinese patients were reported to require a warfarin dose nearly 40% lower than that required by Caucasian patients. The therapeutic maintenance dose of warfarin for Japanese was also 31% lower than that for Caucasians [5].
Recent genome wide association studies have confirmed known polymorphisms in CYP2C9 and VKORC1 as the primary genetic determinants of stabilized warfarin dose. Variants in both genes have been reported to cause ~40% of the variability in warfarin dose. In addition SNP rs2108622 of CYP4F2 was reported to contribute to 1%-2% of the variability [6,7].
In this study we investigate genetic variants previously identified to associate with warfarin metabolism in other populations, in order to identify genetic variations that might confer sensitivity to warfarin in Indonesian patients. Based on the gene variants, which are significantly associated with warfarin sensitivity, we aim to develop a prediction system that will be able to determine the dose appropriate to each patient in Indonesia.
Methods
Study population
All patients were unrelated ethnic Indonesians treated at the National Cardiovascular Centre, Harapan Kita Hospital, Jakarta, Indonesia for thromboembolic diseases, such as atrial fibrillation, valve replacement, atherothrombotic diseases or congestive heart failure from October 2007 to November 2008. All patients had a stable dosage of warfarin for minimum 3 months and an International Normalized Ratio (INR) of the Thrombo Test within the range of 1.5-3.0. Patients with incomplete history of warfarin medication were excluded in this study. Information about gender, age (missing in two patients), bodyweight, body height (missing in two patients), treatment indication and other diseases was taken from the patients' medical records. The ethical committee of Harapan Kita Hospital, Jakarta, approved the study. Prior to their participation in this study all patients have given their written informed consents. For genotyping purposes 122 subjects were included in this study; of which 85 samples with complete medical information were used in the statistical analysis for association with warfarin dose requirement.
DNA isolation and genotyping
Genomic DNA was extracted from 200 μL whole blood using Illustra Blood GenomicPrep Mini Spin Kit (GE Healthcare, Buckinghamshire, UK) and QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's standard protocol. The genomic DNA concentration was measured using NanoDrop™ Spectrophotometer (Thermo Fisher Scientific, USA) and adjusted to 10 ng/μl.
The initial screening of SNPs was performed using 122 samples for partial CYP2C9 locus, which included Promoter regions, Exon 1, Exon 2, Exon 3, Exon 4, Exon 5, Exon 6, Exon 7, Exon 8 and Exon 9. Genotyping was also performed for VKORC1 (rs9923231); EPHX1 (rs4653436); GGCX (rs12714145); PROC (rs2069920; rs1799808; rs1799809); and CYP4F2 (rs2108622). Primers used for CYP2C9 and VKORC1 genotyping were designed according to Ref. [7,8]. The nucleotide position for the primers were designed according to corresponding published sequence in the NCBI database using Primer3 software (GenBank accession number NG_008385 for CYP2C9, NG_011564 for VKORC1, NG_011811.1 for GGCX, NG_009776 for EPHX1, M11228 for PROC and NG_007971 for CYP4F2; Primer3 is available at http://frodo.wi.mit.edu/primer3/).
The primer sequences were listed in Additional file 1: Table S1. Approximately 30 ng of total DNA were subjected to Polymerase Chain Reaction (PCR) amplification in a final volume of 25 μL. Each PCR reaction contained 1× PCR buffer (AmpliTaq® Gold, Applied Biosystems, Foster City, CA), 1.5 mM MgCl2, 200 μM dNTPs, 0.4 μM forward and reverse primers, and 0.625 U HotStart Taq Polymerase (Applied Biosystems, Foster City, CA). The PCR were carried out in a Bio-Rad PTC-200 DNA Engine Cycler with an initial denaturation step of 10 min at 95°C, followed by 45 cycles of denaturation at 94°C for 30 s, annealing for 30 s (annealing temperature of each primer were listed in Additional file 1: Table S1), and extension at 72°C for 1 min, with a final extension for 10 min at 72°C. The purity and mobility of each PCR product were confirmed in 2% agarose gel electrophoresis; each PCR product was either purified using Multi Screen PCR96 Filter Plate (Millipore, Billerica, MA) or subjected to incubation with 1 U of Shrimp Alkaline Phosphatase (New England Biolabs, Ipswich, MA) and 1 U of Exonuclease I (New England Biolabs, Ipswich, MA). Subsequently each PCR product was sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit and ABI Prism® 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA) according to manufacturer's standard protocol. Each sample was sequenced for both strands. All sequencing reactions were performed at least twice with DNA amplified from at least two independent PCR. Sequencing results were aligned and analyzed for SNPs using BioEdit 7.0.0 program (Applied Biosystems, USA). All SNPs detected in this study were located within the high quality region of the chromatogram.
Statistical Analysis
Each polymorphism was tested for Hardy Weinberg equilibrium in the study population. An adjustment for multiple tests by Bonferroni correction was not applied in our analysis. The Bonferroni correction is commonly used by assuming that all tests are independent of each other. However in practical applications, such as our study; that is often not the case. The Bonferroni correction can be extremely conservative and leads to a high rate of false negatives, which may also contribute to publication bias [9]. Therefore Bonferroni correction in which the P-values are multiplied by the number of comparisons was not used in our analysis. Correlations between warfarin dosage and clinical and genetic factors were performed using the categoric Spearman-rank correlation with 5% two-tailed significance level. Warfarin dosage was log-transformed to normalize their distribution. Differences in the daily maintenance dose of warfarin in the different genotype groups were calculated using ANOVA with post hoc comparison using least significant difference (LSD) analysis. Multiple linear regressions were performed to model the relationship warfarin daily dose requirement with other variables measured and used to develop a novel algorithm for warfarin dosing. P-value <0.05 was considered statistically significant. All statistical analyses were carried out using SPSS 15 (SPSS Inc., Chicago, IL).
Results
This is a one-centre study on inter-individual variability of warfarin dose requirements. Anticoagulant response is measured by INR. Inclusion criteria were INR readings between 1.5 and 3.0 during a minimum series of three consecutive INR measurements for a period of 3 months. The collected clinical data included gender, age, bodyweight, height and indication for anticoagulation. Clinical characteristics of the patients were listed in Table 1.
Table 1.
Clinical characteristics of subjects
| Variable | Subjects (n = 85) |
|---|---|
| Gender | |
| Male | 47 (55.3%) |
| Female | 38 (44.7%) |
| Age (yrs) | |
| Male | 58 ± 12 (26-78) |
| Female | 56 ± 10 (32-71) |
| Bodyweight (kg) | |
| Male | 65.25 ± 10.41 (41-83) |
| Female | 57.64 ± 14.27 (35-88) |
| Body height (cm) | |
| Male | 165.74 ± 6.74 (151-180) |
| Female | 156.10 ± 5.06 (144-167) |
| Body Mass Index (kg/m2) | |
| Male | 23.93 ± 3.57 (15.72-36.52) |
| Female | 23.46 ± 5.07 (15.55-32.55) |
| Indication for anticoagulation | |
| Atrial fibrillation | 24 (28.91%) |
| Heart valve replacement | 13 (15.66%) |
| Atrial fibrillation and heart valve replacement | 6 (7.23%) |
| Atherothrombotic diseases | 8 (9.64%) |
| Other | 32 (38.55%) |
| INR | 2.11 ± 0.39 (1.48-3.00) |
We screened for polymorphisms in partial CYP2C9 locus including promoter region and regions surrounding exons 1 to 9, in addition to genetic variants in VKORC1 (rs9393231) and CYP4F2 (rs2108622). Since some reports have showed that genes other than CYP2C9, VKORC1 and CYP4F2 might also contribute to warfarin sensitivity, screening was also extended for PROC promoter region and intron 3, EPHX1 (rs4653436), GGCX (rs1271415) [3,8,10]. A total of 40 SNPs were evaluated in our study; 22 SNPs with Minor Allele Frequency (MAF) greater than 1% and their genotype distributions were listed in Table 2. The allele frequencies were in the Hardy Weinberg Equilibrium (HWE), except the polymorphism in GGCX rs12714145, which was deviated from the HWE (P ≤ 0.05).
Table 2.
Allele frequencies and genotype distribution of SNPs evaluated in this study
| Gene | SNP | Position relative to transcription start site | Allele | n | % | MAF | Genotype | n | % | P-Value HWE |
|---|---|---|---|---|---|---|---|---|---|---|
| CYP2C9** | rs17847036 | 3259 | G | 265 | 98.15 | 0.9815 | GG | 130 | 96.30 | 0.8265 |
| A | 5 | 1.85 | 0.0185 | GA | 5 | 3.70 | ||||
| AA | 0 | 0.00 | ||||||||
| Novel SNP | 3164 | G | 224 | 91.80 | 0.9180 | GG | 102 | 83.61 | 0.3240 | |
| Intron 2 | A | 20 | 8.20 | 0.0820 | GA | 20 | 16.39 | |||
| AA | 0 | 0.00 | ||||||||
| rs9332120 | 3435 | T | 264 | 97.78 | 0.9778 | TT | 129 | 95.56 | 0.7917 | |
| C | 6 | 2.22 | 0.0222 | TC | 6 | 4.44 | ||||
| CC | 0 | 0.00 | ||||||||
| rs2860905 | 3880 | G | 224 | 91.80 | 0.9180 | GG | 104 | 85.25 | 0.1556 | |
| A | 20 | 8.20 | 0.0820 | GA | 16 | 13.11 | ||||
| AA | 2 | 1.64 | ||||||||
| rs28371675 | 3922 | C | 198 | 81.15 | 0.8115 | CC | 79 | 64.75 | 0.4291 | |
| T | 46 | 18.85 | 0.1885 | CT | 40 | 32.79 | ||||
| TT | 3 | 2.46 | ||||||||
| rs28371676 | 3948 | T | 234 | 95.90 | 0.9590 | TT | 112 | 91.80 | 0.6369 | |
| C | 10 | 4.10 | 0.0410 | TC | 10 | 8.20 | ||||
| CC | 0 | 0.00 | ||||||||
| rs28371677 | 4057 | A | 224 | 91.80 | 0.9180 | AA | 104 | 85.25 | 0.1556 | |
| G | 20 | 8.20 | 0.0820 | AG | 16 | 13.11 | ||||
| GG | 2 | 1.64 | ||||||||
| rs9332127 | 9056 | G | 234 | 95.90 | 0.9590 | GG | 112 | 91.80 | 0.6369 | |
| (CYP2C9C-65) | C | 10 | 4.10 | 0.0410 | GC | 10 | 8.20 | |||
| CC | 0 | 0.00 | ||||||||
| rs9332129 | 10322 | A | 234 | 96.69 | 0.9630 | AA | 112 | 92.56 | 0.6696 | |
| G | 9 | 3.72 | 0.0370 | AG | 9 | 7.44 | ||||
| GG | 0 | 0.00 | ||||||||
| rs1057910 | 42625 | A | 235 | 96.31 | 0.9631 | AA | 113 | 92.62 | 0.6723 | |
| (CYP2C9*3) | C | 9 | 3.69 | 0.0369 | AC | 9 | 7.38 | |||
| CC | 0 | 0.00 | ||||||||
| Novel SNP | 47635 | C | 240 | 98.36 | 0.9836 | CC | 118 | 96.72 | 0.8539 | |
| Intron 8 | T | 4 | 1.64 | 0.0164 | CT | 4 | 3.28 | |||
| TT | 0 | 0.00 | ||||||||
| rs2298037 | 47650 | C | 199 | 81.56 | 0.8156 | CC | 79 | 64.75 | 0.1957 | |
| T | 45 | 18.44 | 0.1844 | CT | 41 | 33.61 | ||||
| TT | 2 | 1.64 | ||||||||
| rs9332238 | 50064 | G | 233 | 95.49 | 0.9549 | GG | 111 | 90.98 | 0.6020 | |
| A | 11 | 4.51 | 0.0451 | GA | 11 | 9.02 | ||||
| AA | 0 | 0.00 | ||||||||
| rs1934969 | 50067 | A | 173 | 70.90 | 0.7090 | AA | 60 | 49.18 | 0.5595 | |
| T | 71 | 29.10 | 0.2910 | AT | 53 | 43.44 | ||||
| TT | 9 | 7.38 | ||||||||
| rs1057911 | 50309 | A | 234 | 95.90 | 0.9590 | AA | 112 | 91.80 | 0.6369 | |
| T | 10 | 4.10 | 0.0410 | AT | 10 | 8.20 | ||||
| TT | 0 | 0.00 | ||||||||
| VKORC1 | rs9923231 | -1639 | A | 208 | 77.04 | 0.7704 | AA | 78 | 57.78 | 0.3027 |
| G | 62 | 22.96 | 0.2296 | AG | 52 | 38.52 | ||||
| GG | 5 | 3.70 | ||||||||
| EPHX1 | rs4653436 | -2586 | G | 203 | 87.50 | 0.8750 | GG | 91 | 78.45 | 0.0633 |
| A | 29 | 12.50 | 0.1250 | GA | 21 | 18.10 | ||||
| AA | 4 | 3.45 | ||||||||
| GGCX | rs12714145 | 1291 | C | 145 | 59.43 | 0.5943 | CC | 34 | 27.87 | 0.0006 |
| T | 99 | 40.57 | 0.4057 | CT | 77 | 63.11 | ||||
| TT | 11 | 9.02 | ||||||||
| PROC | rs2069920 | 3643 | C | 140 | 57.38 | 0.5738 | CC | 36 | 29.51 | 0.1232 |
| T | 104 | 42.62 | 0.4262 | CT | 68 | 55.74 | ||||
| TT | 18 | 14.75 | ||||||||
| PROC | rs1799808 | -141 | C | 99 | 40.57 | 0.4057 | CC | 15 | 12.30 | 0.0563 |
| T | 145 | 59.43 | 0.5943 | CT | 69 | 56.56 | ||||
| TT | 38 | 31.15 | ||||||||
| PROC | rs1799809 | -128 | A | 141 | 57.79 | 0.7747 | AA | 72 | 59.02 | 0.3009 |
| G | 41 | 16.80 | 0.2253 | AG | 46 | 37.70 | ||||
| GG | 4 | 3.28 | ||||||||
| CYP4F2 | rs2108622 | 18454 | C | 198 | 81.15 | 0.8115 | CC | 82 | 67.21 | 0.3248 |
| T | 46 | 18.85 | 0.1885 | CT | 34 | 27.87 | ||||
| TT | 6 | 4.92 |
** SNPs in CYP2C9 with MAF ≤ 0.01: CYP2C9*2 (rs1799853); CYP2C9*4 (rs56165452); CYP2C9*5 (rs28371686); rs17847032; rs2017319; CYP2C9*6 (rs9332131); rs9332173; CYP2C9*7; CYP2C9*8 (rs7900194); rs5031019; CYP2C9*9 (rs2256871); CYP2C9*10 (rs9332130); CYP2C9*11 (rs28371685); rs1057909; CYP2C9*12 (rs9332239); rs9332100; CYP2C9*13; rs12414460
Two reported alleles for CYP2C9, CYP2C9*2 and *3 have been identified with decreased hydroxylation activity of the enzyme, which in turn can effect the efficacy of anticoagulation [1,3]. Similar to earlier reports on other Asian populations [7,8,10,11], CYP2C9*2 carriers were not detected in our population (Table 2). We searched for additional SNPs in CYP2C9 locus in the promoter region of the gene, up to -700 bp upstream of the transcriptional start site, and regions surrounding exons 1 to 9. A total of 33 SNPs in CYP2C9 were genotyped in our study. Twelve known CYP2C9 allelic variants were identified in our screen (CYP2C9*2 to CYP2C9*13). CYP2C9*3 was detected with MAF greater than 1%, whereas CYP2C9*2 as well as CYP2C9*4 to CYP2C9*13 were found to be homozygous in our study population. The percentage frequencies for genotype CYP2C9*1/*1 and CYP2C9*1/*3 were 92.62% and 7.38% respectively (Table 2).
Additionally seven SNPs in CYP2C9, rs9332100 in promoter region, rs12414460 and rs5031019 in exon 3, rs9332173 in exon 6, rs1057909 in exon 7, rs17847032 in exon 8 and rs2017319 in exon 9, were identified as homozygous SNPs in our population (data not shown). Two novel CYP2C9 variants were detected in introns 1 and 8 with MAF of 8.2% and 1.64% respectively (Table 2). The two novel SNPs were detected in the position 3164 and 47635 relative to transcription start site.
Two SNPs in VKORC1, rs9934438 and rs9923231, have been reported to correlate with pair-wise Linkage Disequilibrium (LD) r2 values of 1.0 [7], therefore only one SNP, rs9923231, was selected as candidate SNP in our study. For rs9923231 or VKORC1 (-1639G > A) allelic variation, 57.78% of our subjects were homozygous for the wild-type A allele, 38.52% were heterozygous and 3.7% were homozygous for the variant G allele.
Genetic variants in EPHX1 (rs4653436), GGCX (rs12714145), PROC (rs2069920, rs1799808, rs1799809) and CYP4F2 (rs2108622) were detected in our screen with MAF of 12.50%, 40.57%, 42.62%, 40.57%, 22.53% and 18.85% respectively (Table 2).
Together with non-genetic indicators such as gender, age, body weight and height, all 22 identified SNPs with MAF > 1% (Table 2) were evaluated as indicators for warfarin dose association. The non-parametric Spearman-rank correlation of warfarin dose based on the non-genetic and genetic factors is shown in Table 3. Our results indicated that among the non-genetic indicators included in the analysis, age and height were significantly correlated with warfarin dose with a P-value of 0.0232 and 0.0183 respectively. Bodyweight exhibited a less significant association with daily warfarin dose (P-value 0.0521). Among the 22 genetic indicators analyzed, only rs17847036 in CYP2C9 and rs9923231 in VKORC1 showed significant correlation with warfarin dose with a P-value of 0.0058 and 0.0248 respectively (Table 3).
Table 3.
Spearman correlation of warfarin dose based on different indicators
| Indicators | Spearman Correlation | P-value (Sig 2. tailed) | Total Samples |
|---|---|---|---|
| Gender | 0.1435 | 0.1900 | 85 |
| Age | -0.2490 | 0.0232 | 83 |
| Bodyweight | -0.2140 | 0.0521 | 83 |
| Body height | -0.2584 | 0.0183 | 83 |
| CYP2C9 rs17847036 | -0.2971 | 0.0058 | 85 |
| VKORC1 rs9923231 | 0.2433 | 0.0248 | 85 |
Comparisons of age, weight, height, maintenance INR and warfarin dose were performed across the different genotypes of CYP2C9 and VKORC1 using t-test. Patients with GA genotype for CYP2C9 rs17847036 received significantly higher doses of warfarin (3.67 ± 0.88 mg/day; P-value 0.005) than patients with GG genotype (2.09 ± 0.70 mg/day). For VKORC1, patients carrying AA genotype for rs9923231 received lower dose albeit insignificantly (2.05 ± 0.77 mg/day; P-value 0.080) than patients with AG or GG genotype (2.32 ± 0.73 mg/day).
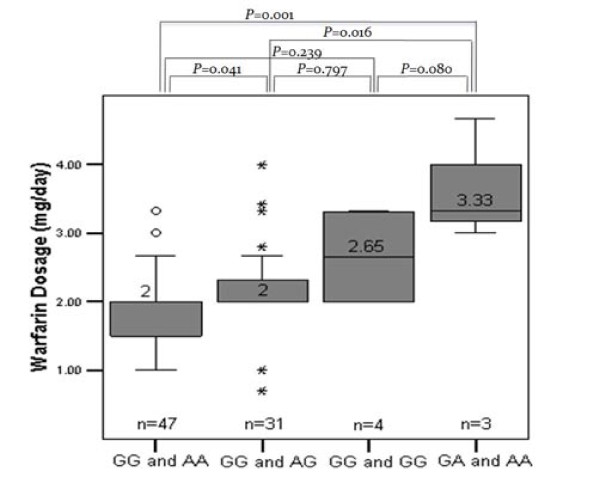
The variability in warfarin stable dose for patients grouped by the VKORC1 variants (AA, AG or GG for rs9923231) and CYP2C9 (AG or GG for rs17847036) is shown in Figure 1. All patients were separated into four categories according to the genotypes identified in VKORC1 and CYP2C9. The median warfarin dose for patients carrying heterozygous GA genotype for CYP2C9 and AA genotype for VKORC1 was 3.33 mg/day; higher than for patient group carrying homozygous GG genotype for CYP2C9 and AA genotype for VKORC1 with 2 mg/day (P-value 0.001).
Figure 1.
Box plot of the distribution for warfarin dose by CYP2C9 rs17847036 (GG and GA) of CYP2C9 and VKORC1 rs9923231 (GG, AG and AA) genotypes. Number indicates the median warfarin dose. P-values for the distributions in each genotype are indicated.
Using the non-genetic and genetic indicators, we analyzed the effect of these indicators by multiple regression model analysis as shown in Table 4. The R2 value varied depending on the indicators included in the five different models used in our analysis. The model 5 showed the highest adjusted R2 value of 0.154; thus, according to our analysis this model might be used to explain 15.4% of the variance in the warfarin dose. Figure 2 showed the correlation between warfarin dose and the proposed model 5. Out of 85 patients included in our analysis, our model could be used to predict the dosage of 83 patients.
Table 4.
Multiple linear regression for estimation of warfarin daily dose requirements based on age, bodyweight, body height, CYP2C9 rs17847036 and VKORC1 rs9923231
| Model | Predictors (include constant) | R2 | Adjusted R2 | P-value |
|---|---|---|---|---|
| 1 | Age, Bodyweight and body height | 0.094 | 0.059 | 0.053 |
| 2 | CYP2C9 rs17847036 and VKORC1 rs9923231 | 0.106 | 0.084 | 0.010 |
| 3 | Age, Bodyweight, body height and CYP2C9 rs17847036 | 0.187 | 0.145 | 0.003 |
| 4 | Age, Bodyweight, body height and VKORC1 rs9923231 | 0.107 | 0.060 | 0.069 |
| 5 | Age, Bodyweight, body height, CYP2C9 rs17847036 and VKORC1 rs9923231 | 0.207 | 0.154 | 0.003 |
Figure 2.
Correlation between warfarin dose and model 4 predicted value.
Discussion
Therapy with warfarin requires frequent monitoring due to its narrow therapeutic index and inter-individual variability in the dose necessary to reach a therapeutic INR [1,12]. In this study we assessed the contribution of previously reported genetic variants to inter-individual variability in ethnic Indonesians [3,5-7,12,13]. This is the first report on the effect of genetic polymorphisms on warfarin dose requirement for this population. Based on the analysis we conclude that age, body weight, body height, and genetic variants of CYP2C9 and VKORC1 could explain for approximately 15.4% of the variability in warfarin daily dose requirement of our study population.
CYP2C9 metabolizes S-warfarin, the more active enantiomer of warfarin, in the liver [14,15]. In our screen for candidate SNPs in CYP2C9, we found that among the known CYP2C9 variants identified, only CYP2C9*3 was present in warfarin treated Indonesian patients. To some extent this is in agreement with published reports from other Asian populations including Chinese, Japanese, Malaysian and Korean patients [10,16,17]. Nevertheless other CYP2C9 genetic variants (e.g. CYP2C9*8, *11 and *13) have been reported with minor allele frequencies (MAF) as low as 1.02% in certain population [18-21]. Our analysis revealed that despite reports from other populations [3,13,22-24], CYP2C9*3 was not significantly associated with warfarin dose requirement in our study (P-value 0.9308, data not shown). The probable reason for this might be the low number of subjects carrying a single CYP2C9*3 allele and the absence of homozygous CYP2C9*3 individuals in our study; further analysis with a greater sample size will be required to confirm these findings.
Our study suggested a genetic variant in the exon 2 of CYP2C9, rs17847036, as a genetic factor that contribute to warfarin sensitivity with a P-value of 0.005. Together with non-genetic indicators such as patient age, body weight and body height, this genetic variant could explain about 14.5% of the inter-individual variability. To the best of our knowledge this is the first time that this polymorphism is linked with warfarin sensitivity. G/A polymorphism in rs17847036 results in a synonymous amino acid change in V76. Synonymous SNPs are not expected to alter the function of the protein; however the functional significance of these SNPs cannot be overlooked. It has been reported that synonymous mutations in coding regions may act alone or in combination with other mutations in the same transcript to influence mRNA stability and translation, thereby causing functional effects [25]. Indeed, a "silent" polymorphism in the Multi Drug Resistance (MDR1) gene has been attributed to alteration in the structure of substrate and inhibitor interaction sites and thus to a change in substrate specificity [26].
Recent genome-wide association studies for genetic determinants of warfarin dose have verified the involvement of CYP2C9*2 and *3 in warfarin sensitivity, and additional genetic variants of CYP2C9 were not significantly correlated with warfarin dose requirement [6,7]. Smaller studies however indicated that both genetic variants might not be the major predictors for warfarin dosing in African American population [27,28]. Recently, Scott et al. identified CYP2C9*8 genetic variant as the most frequent variant in African Americans, and suggested the incorporation of CYP2C9*8 into genotyping panels to improve dose prediction of CYP2C9-metabolized drugs, including warfarin in this particular ethnics [29]. It is therefore tempting to suggest, that CYP2C9 variants other than CYP2C9*2 and *3 might be associated with CYP2C9 enzymatic activity, and subsequently with warfarin dosing in different populations. Nevertheless additional studies in larger populations are required to confirm these findings.
Various publications have demonstrated the significant role of VKORC1 -1639G/A polymorphism (rs9923231) in warfarin requirements. Recent studies on VKORC1 suggested that SNPs in VKORC1 might contribute more to dose variance than SNPs in CYP2C9 [3,11,15,23,29]. VKORC1 variants have been suggested to play a role on transcriptional regulation and warfarin dose determination as haplotypes [30,31]. Indeed, Lee et al. reported that individuals carrying VKORC1 H1 haplotype require lower warfarin maintenance dose than individuals with VKORC1 H7, H8 or H9 haplotypes [32]. Moreover, these VKORC1 haplotypes were identified with variable frequency in Chinese, Malayan and Indian populations. Our result suggested that rs9923231 in VKORC1 contributed less significantly than CYP2C9 genetic variant rs17847036 (Table 4). From common VKORC1 variants only rs9923231 was analyzed in this study, this SNP was in LD with rs9934438 [13]. It is therefore conceivable that other VKORC1 polymorphisms might additionally contribute to dose variance in ethnic Indonesians; and this warrants further investigation. Similar to other studies, our study also suggested that patients with AA genotype in VKORC1 rs9923231 require lower doses of warfarin than those with AG or GG genotype [11,33].
CYP4F2 genetic variant rs2108622 was associated with a clinically relevant effect on warfarin requirement in Caucasian populations [34,35]. This result was confirmed by a genome wide association study, which reported that this CYP4F2 genetic variant might contribute to 1%-2% of the variability in warfarin dose in Caucasian populations [7]. This genetic variant was detected in our study population with a MAF of 18.85%; however this polymorphism was not significantly correlated with warfarin dose as indicated by a P-value of 0.9394 (data not shown). In a UK study, CYP4F2 rs2108622 was not associated with stable warfarin dose, but the authors reported an association between CYP4F2 rs2189784 and time-to-therapeutic INR, which confirmed the role of CYP4F2 in vitamin K metabolism [36]. A functional regulatory CYP4F2 haplotype has also been associated with increased susceptibility to hypertension and myocardial infarction in Chinese and Japanese population, respectively [37,38]. It is therefore tempting to suggest that additional CYP4F2 variants or haplotypes might affect warfarin dose requirement; and this needs further clarifications.
Similarly, unique SNPs, such as rs9332127 (CYP2C9C-65) and rs4653436 in EPHX1, were not strongly correlated with warfarin dose. These SNPs were reported to correlate with warfarin sensitivity in Chinese, Taiwanese and Caucasian populations [3,13,24]. The observed P-values for these genetic variants were 0.7304 and 0.6960 respectively (data not shown). Similar to Wang et al. [13], we were not able to detect any correlation between genetic variants in GGCX and PROC with warfarin dose requirement, despite report on other populations [3,24]. This further confirms that polymorphisms in major warfarin metabolizing genes might contribute not only to inter-individual difference within a particular population but also to inter-population variability among different populations [24].
Taken together, the present study indicated that SNPs in two major warfarin-metabolizing genes, CYP2C9 and VKORC1, contributed to the variability in warfarin dose of ethnic Indonesians. This study demonstrated the inter-individual variability in warfarin maintenance dose was due to VKORC1 -1639G/A polymorphism (rs9923231), CYP2C9 rs17847036, age, bodyweight and height in Indonesian patients. Clinical factors such as age, bodyweight and body height contributed to 5.9% of warfarin reactivity. Interestingly, the two genetic factors, VKORC1 -1639G/A polymorphism (rs9923231), CYP2C9 rs17847036, were associated with a greater contribution to warfarin sensitivity (8.4%). This clearly indicated that genetic factors play an important role in warfarin dosing in ethnic Indonesians similar to other population. Analysis of the warfarin dosing algorithm by multiple regression analysis showed that together with the non-genetic factors, the genetic predictors accounted for 15.4% of warfarin reactivity; thus this suggests that additional genetic determinants might possibly contribute to warfarin sensitivity in our study population. Genetic variants in Apolipoprotein E (APOE), for example, have been associated with warfarin metabolism. Polymorphisms in APOE gene were found to influence warfarin dose requirement differently in Caucasians, African Americans and Asians [39,40]. Recent study also identified a polymorphism in calumenin (CALU) (vitamin K reductase regulator) as genetic factor for warfarin requirement in African Americans [41]. Identification of additional genetic variants and further evaluation of common genetic predictors associated with warfarin sensitivity might substantially improve warfarin dose prediction for ethnic Indonesians.
Conclusions
The study confirmed that the impacts of genetic variants on warfarin dosage requirement might vary in different population. Together with non-genetic predictors the genetic variants might be used to improve warfarin dose prediction for ethnic Indonesians.
Abbreviations
CYP2C9: cytochrome P450, family 2, subfamily C, polypeptide 9; VKORC1: vitamin K epoxide reductase complex, subunit 1; PROC: Protein C; EPHX1: Epoxy hydrolase 1; GGCX: γ-glutamyl carboxylase; ORM: Orcosumucoid; INR: international normalized ratio; CYP4F2: cytochrome P450, family 4, subfamily F, polypeptide 2; APOE: Apolipoprotein E; SNP: Single Nucleotide Polymorphism; MAF: Minor allele frequency; LD: Linkage Disequilibrium.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
The study was conceived and designed by TW and IS. IS drafted the manuscript. WT carried out the molecular genetic studies and assisted in interpreting the results. AU, SR, YY and ST participated in its design and coordination. All authors have read and have approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table S1 - List of primers used in this study.
Contributor Information
Ivet M Suriapranata, Email: isuriapranata@mrinstitute.org.
Wen Ye Tjong, Email: wenye@mrinstitute.org.
Tingliang Wang, Email: wangtingliang@hotmail.com.
Andi Utama, Email: autama@mrinstitute.org.
Sunu B Raharjo, Email: sunu.b.raharjo@gmail.com.
Yoga Yuniadi, Email: yogayun@yahoo.com.
Susan SW Tai, Email: stai@mrinstitute.org.
Acknowledgements
The authors would like to thank all patients who participated in this study. We acknowledge Elizabeth Sarah Salmon, Elok Ekawati, Lasmaria Sitorus, Ivan Chandra, Rama Dhenni, and Widyartini Made Sudania for their assistance in subjects' enrollment and data management. The authors would like to thank Dr. rer. nat. Elke Schaeffeler and Michael Flynn, EdM. for critical reading of this manuscript and Etih Sudarnika for her help in statistical analysis. This work was supported by Mochtar Riady Institute Foundation Grant (Budget no. cc301/2008).
References
- Daly AK, King BP. Pharmacogenetics of oral anticoagulants. Pharmacogenetics. 2003;13:247–252. doi: 10.1097/00008571-200305000-00002. [DOI] [PubMed] [Google Scholar]
- Wadelius M. Use of pharmacogenetics in guiding treatment with warfarin. Clin Chem. 2009;55:709–711. doi: 10.1373/clinchem.2008.115964. [DOI] [PubMed] [Google Scholar]
- Wadelius M, Chen LY, Eriksson N, Bumpstead S, Ghori J, Wadelius C, Bentley D, McGinnis R, Deloukas P. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121:23–34. doi: 10.1007/s00439-006-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadelius M, Chen LY, Downes K, Ghori J, Hunt S, Eriksson N, Wallerman O, Melhus H, Wadelius C, Bentley D. et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5(4):262–720. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- Xie HG, Kim RB, Wood AJJ, Stein CM. Molecular Basis of Ethnic Differences in Drug Disposition and Response. Annual Review of Pharmacology and Toxicology. 2001;41:815–850. doi: 10.1146/annurev.pharmtox.41.1.815. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD. et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112(4):1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W. et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5(3):e100043. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao LH, Zhang H, Lai MP, Lau KW, Lai AK, Zhang JH, Wang Q, Wei W, Chai JH, Lung ML. et al. The Association of CYP2C9 gene polymorphisms with colorectal carcinoma in Han Chinese. Clin Chim Acta. 2007;380:191–196. doi: 10.1016/j.cca.2007.02.033. [DOI] [PubMed] [Google Scholar]
- Nakagawa S. A farewell to Bonferroni: the problems of low statistical power and publication bias. Behavioral Ecology. 2005;15(6):1044–1045. [Google Scholar]
- Yuan HY, Chen JJ, Lee MT, Wung JC, Chen YF, Charng MJ, Lu MJ, Hung CR, Wei CY, Chen CH. et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Gen. 2005;14(13):1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- Schelleman H, Chen Z, Kealey C, Whitehead AS, Christie J, Price M, Brensinger CM, Newcomb CW, Thorn CF, Samaha FF. et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007;81:742–747. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- Moyer TP, O'Kane DJ, Baudhuin LM, Wiley CL, Fortini A, Fisher PK, Dupras DM, Chaudhry R, Thapa P, Zinsmeister AR. et al. Warfarin Sensitivity Genotyping: A Review of the Literature and Summary of Patient Experience. Mayo Clin Proc. 2009;84(12):1079–1094. doi: 10.4065/mcp.2009.0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TL, Li HL, Tjong WY, Chen QS, Wu GS, Zhu HT, Hou ZS, Xu S, Ma SJ, Wu M. et al. Genetic factors contribute to patient-specific warfarin dose for Han Chinese. Clin Chim Acta. 2008;396:76–79. doi: 10.1016/j.cca.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Rettie AE, Korzekwa KR, Kunze KL, Lawrence RF, Eddy AC, Aoyama T, Gelboin HV, Gonzalez FJ, Trager WF. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol. 1992;5(1):54–59. doi: 10.1021/tx00025a009. [DOI] [PubMed] [Google Scholar]
- Kaminsky LS, Zhang ZY. Human P450 metabolism of warfarin. Pharmacol Ther. 1997;73(1):67–74. doi: 10.1016/S0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- Veenstra DL, You JH, Rieder MJ, Farin FM, Wilkerson HW, Blough DK, Cheng G, Rettie AE. Association of Vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics. 2005;15:687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- Ngow HA, Wan Khairina WM, Teh LK, Lee WL, Harun R, Ismail R, Salleh MZ. CYP2C9 polymorphism: prevalence in healthy and warfarin-treated Malay and Chinese in Malaysia. Singapore Med J. 2009;50(5):490–493. [PubMed] [Google Scholar]
- Bae JW, Kim HK, Kim JH, Yang SI, Kim MJ, Jang CG, Park YS, Lee SY. Allele and genotype frequencies of CYP2C9 in a Korean population. Br J Clin Pharmacol. 2005;60:418–422. doi: 10.1111/j.1365-2125.2005.02448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell J, Jorge-Nebert LF, Coulter S, Ferguson SS, Lee SJ, Chanas B, Xi T, Mohrenweiser H, Ghanayem B, Goldstein JA. Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics. 2004;14:527–537. doi: 10.1097/01.fpc.0000114759.08559.51. [DOI] [PubMed] [Google Scholar]
- King BP, Khan TI, Aithal GP, Kamali F, Daly AK. Upstream and coding region CYP2C9 polymorphisms: correlation with warfarin dose and metabolism. Pharmacogenetics. 2004;14:813–822. doi: 10.1097/00008571-200412000-00004. [DOI] [PubMed] [Google Scholar]
- Si D, Guo Y, Zhang Y, Yang L, Zhou H, Zhong D. Identification of a novel variant CYP2C9 allele in Chinese. Pharmacogenetics. 2004;14:465–469. doi: 10.1097/01.fpc.0000114749.08559.e4. [DOI] [PubMed] [Google Scholar]
- Kimura R, Miyashita K, Kokubo Y, Akaiwa Y, Otsubo R, Nagatsuka K, Otsuki T, Okayama A, Minematsu K, Naritomi H. et al. Genotypes of vitamin K epoxide reductase, gamma-glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese patients. Thromb Res. 2007;120(2):181–186. doi: 10.1016/j.thromres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Shennan M, Reynolds KK, Johnson NA, Herrnberger MR, Valdes R Jr, Linder MW. Estimation of warfarin maintenance dose based on VKORC1 (-1639 G > A) and CYP2C9 genotypes. Clin Chem. 2007;53(7):1199–1205. doi: 10.1373/clinchem.2006.078139. [DOI] [PubMed] [Google Scholar]
- Lee MT, Chen CH, Chou CH, Lu LS, Chuang HP, Chen YT, Saleem AN, Wen MS, Chen JJ, Wu JY. et al. Genetic determinants of warfarin dosing in the Han-Chinese population. Pharmacogenomics. 2009;10(12):1905–1913. doi: 10.2217/pgs.09.106. [DOI] [PubMed] [Google Scholar]
- Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12(3):205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Limdi N, Goldstein J, Blaisdell J, Beasley T, Rivers C, Acton R. Influence of CYP2C9 Genotype on warfarin dose among African American and European Americans. Per Med. 2007;4(2):157–169. doi: 10.2217/17410541.4.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kealey C, Chen Z, Christie J, Thorn CF, Whitehead AS, Price M, Samaha FF, Kimmel SE. Warfarin and cytochrome P450 2C9 genotype: possible ethnic variation in warfarin sensitivity. Pharmacogenomics. 2007;8(3):217–25. doi: 10.2217/14622416.8.3.217. [DOI] [PubMed] [Google Scholar]
- Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10(8):1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352(22):2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- Limdi NA, Beasley TM, Crowley MR, Goldstein JA, Rieder MJ, Flockhart DA, Arnett DK, Acton RT, Liu N. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-Americans. Pharmacogenomics. 2008;9(10):1445–1458. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Ng SS, Oldenburg J, Chong PY, Rost S, Guo JY, Yap HL, Rankin SC, Khor HB, Yeo TC. et al. Interethnic variability of warfarin maintenance requirement is explained by VKORC1 genotype in an Asian population. Clin Pharmacol Ther. 2006;79(3):197–205. doi: 10.1016/j.clpt.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Miao L, Yang J, Huang C, Shen Z. Contribution of age, body weight, and CYP2C9 and VKORC1 genotype to the anticoagulant response to warfarin: proposal for a new dosing regimen in Chinese patients. Eur J Clin Pharmacol. 2007;63(12):1135–1141. doi: 10.1007/s00228-007-0381-6. [DOI] [PubMed] [Google Scholar]
- Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C. et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgiani P, Ciccacci C, Forte V, Sirianni E, Novelli L, Bramanti P, Novelli G. CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics. 2009;10(2):261–266. doi: 10.2217/14622416.10.2.261. [DOI] [PubMed] [Google Scholar]
- Zhang JE, Jorgensen AL, Alfirevic A, Williamson PR, Toh CH, Park BK, Pirmohamed M. Effects of CYP4F2 genetic polymorphisms and haplotypes on clinical outcomes in patients initiated on warfarin therapy. Pharmacogenet Genomics. 2009;19(10):781–789. doi: 10.1097/FPC.0b013e3283311347. [DOI] [PubMed] [Google Scholar]
- Fu Z, Nakayama T, Sato N, Izumi Y, Kasamaki Y, Shindo A, Ohta M, Soma M, Aoi N, Sato M. et al. A haplotype of the CYP4F2 gene associated with myocardial infarction in Japanese men. Mol Genet Metab. 2009;96(3):145–147. doi: 10.1016/j.ymgme.2008.11.161. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhao Y, Nie D, Shi J, Fu L, Li Y, Yu D, Lu J. Association of a functional cytochrome P450 4F2 haplotype with urinary 20-HETE and hypertension. J Am Soc Nephrol. 2008;19(4):714–721. doi: 10.1681/ASN.2007060713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel SE, Christie J, Kealey C, Chen Z, Price M, Thorn CF, Brensinger CM, Newcomb CW, Whitehead AS. Apolipoprotein E genotype and warfarin dosing among Caucasians and African Americans. Pharmacogenomics J. 2008;8(1):53–60. doi: 10.1038/sj.tpj.6500445. [DOI] [PubMed] [Google Scholar]
- Lal S, Sandanaraj E, Jada SR, Kong MC, Lee LH, Goh BC, Lee SC, Chowbay B. Influence of APOE genotypes and VKORC1 haplotypes on warfarin dose requirements in Asian patients. Br J Clin Pharmacol. 2008;65(2):260–264. doi: 10.1111/j.1365-2125.2007.03053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voora D, Koboldt DC, King CR, Lenzini PA, Eby CS, Porche-Sorbet R, Deych E, Crankshaw M, Milligan PE, McLeod HL. et al. A Polymorphism in the VKORC1 Regulator Calumenin Predicts Higher Warfarin Dose Requirements in African Americans. Clin Pharmacol Ther. 2010;87(4):445–51. doi: 10.1038/clpt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 - List of primers used in this study.