Summary
In this issue, Katoh et al. (2011) show that the biosynthetic enzyme MATIIα binds at the HO-1 locus in mammalian cells and exhibits transcriptional corepressor activity through local production of S-adenosylmethionine, which stimulates histone methyltransferases to promote a transcriptionally repressive chromatin environment.
Regulated transcription of specific gene sets in response to cellular signals requires an extensive repertoire of highly coordinated molecular events, many of which are directed at the chromatin template. These molecular events tune the promoter chromatin environment to the required transcriptional outcome (e.g., permissive environment for enhanced transcription, or repressive environment for reduced transcription). These processes involve a wide variety of chromatin- and transcription-modulating enzymes that require small molecule cofactors or metabolites to support their catalytic activities. For example, (1) protein acetyltransferases, such as p300/CBP, PCAF/GCN5, and Tip60, require acetyl-CoA as a donor of acetyl groups in acetylation reactions; (2) poly(ADP-ribose) polymerases, such as PARP-1, require oxidized nicotinamide adenine dinucleotide (NAD+), as a donor of ADP-ribose units in poly(ADP-ribosyl) ation reactions; and (3) sirtuin family protein deacetylases, such as Sirt1, require NAD+ as a donor of ADP-ribose units to accept acetyl groups in deacetylation reactions (Table 1). The source of these metabolites in the nucleus and their use by chromatin-modifying enzymes has been receiving increased attention in the recent literature. In this issue, Katoh et al. (2011) show how the production of S-adenosylmethionine (SAM) by promoter-bound MATIIα, a methionine adenosyltransferase (MAT) isozyme, supports the activity of methyltransfersases involved in transcriptional regulation. Their results fit into an emerging paradigm of gene regulation, where chromatin-localized biosynthesis of metabolites can modulate the expression of nearby genes by providing metabolites for use by epigenetic effectors.
Table 1.
Some examples of metabolites, their synthases, and effectors involved in chromatin-dependent gene regulation.
| Metabolite | Synthase | Effector | Outcome |
|---|---|---|---|
| Acetyl-CoA | Acetyl-CoA Synthetase ATP-Citrate Lyase |
Acetyltransferases Acetyltransferases |
Histone acetylation Histone acetylation |
| ATP | ATP Synthase | Kinases Chromatin remodeling complexes |
Phosphorylation Chromatin remodeling |
| FAD+ | FMN adenylyltransferase | LSD family demethylases | Histone demethylation |
| α-Ketoglutarate | Isocitrate dehydrogenase | Jmj family demethylases | Histone demethylation |
|
NAD+ (and related cofactors) |
NMNAT-1* (and NAD+ cofactor kinases and reductases) |
Sirtuins (NAD+) PARPs (NAD+) CtBP (NAD+/NADH) Clock:BMAL1 [NAD(P)+/NAD(P)H] NPAS2:BMAL1 [NAD(P)+/NAD(P)H] |
Histone deacetylation ADP-ribosylation Transcriptional corepression Modulation of DNA binding Modulation of DNA binding |
| O-Acetyl-ADP-ribose | Sirtuins | Sir2/3/4 MacroH2A1.1 |
Regulation of enzyme activity Regulation of protein-protein interactions |
| S-adenosylmethionine | MatIIα* | Protein Methyltransferases DNA Methyltransferases |
Histone methylation DNA methylation |
Of the metabolite producers (“synthases”) listed here, only MATIIα (Katoh et al., 2011) and NMNAT-1 (Zhang et al., 2009) have been shown to be recruited to chromatin. The extent to which other metabolite-producing enzymes are recruited to target gene promoters has yet to be determined.
In their studies of gene regulation by MafK, a Maf family transcription factor, Katoh et al. (2011) used a proteomic approach to identify 289 MafK-interacting proteins. These interactors included a variety of chromatin- and transcription-related factors, such as the Maf corepressor Bach1, as well as the SAM synthetase MATIIa, which catalyzes the production of SAM by joining methionine and ATP. Interestingly, a previous genetic screen in Drosophila identified a mutation in the gene encoding a fly MAT ortholog (SamS), called Su(z)5, as a suppressor of position effect variegation, implicating SAM biosynthesis in the maintenance of heterochromatin (Larsson et al., 1996). Subsequent identification of 127 MATIIα-interacting proteins revealed 39 proteins that overlapped with the set of MafK-interacting proteins, including components of the Swi/Snf, NuRD, PARP-1, and PcG complexes, which mediate nucleosome remodeling, histone modification, and the formation of heterochromatin. Based on these results, the authors investigated a possible MafK- and MatIIα-dependent repressive chromatin network.
To understand how these various factors might function together in gene regulation, the authors used the MafK-regulated and Maf recognition element (MARE)-containing heme oxygenase-1 (HO-1) gene as a model. Using biotinylation-coupled chromatin immunoprecipitation (ChIP), the authors found that both MafK and MatIIα bound to the HO-1 locus, with MafK binding more extensively to the MARE regions and MATIIα binding more broadly, even into a neighboring gene. Upon robust stress-induced expression of the HO-1 gene in response to diethyl maleate (DEM), loss of MATIIα binding, but not MafK binding, from the HO-1 locus was observed, suggesting that MATIIα might play a critical role in repressing the expression of HO-1 in the absence of an activating signal. Using RNAi-mediated knockdown of components of the individual complexes comprising this repressive module, the authors found that Swi/Snf, NuRD, MATIIα partner MATIIβ, and PARP-1 each play a role in HO-1 gene repression. The critical observation made by the authors, however, was that introduction of a single point mutation in MATIIα, which abolishes its SAM synthetase activity, further enhanced HO-1 transcription upon DEM treatment. These results implicate MATIIα enzymatic activity - its ability to produce SAM - in MATIIα-dependent repression of the HO-1 gene.
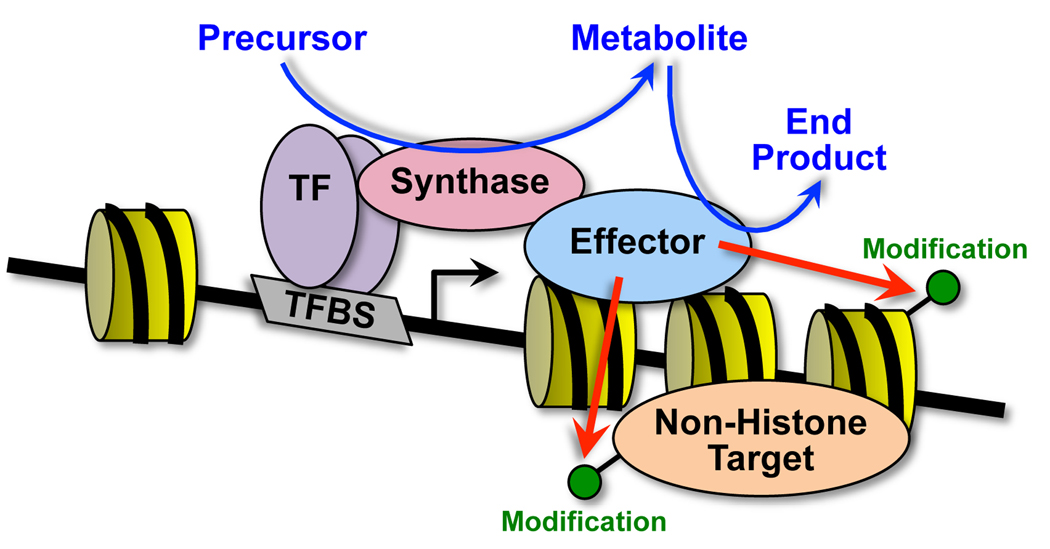
Since SAM is used by both histone and DNA methyltransferases, the authors examined which enzymes might be activated by SAM production to promote gene repression at the HO-1 locus. By using MATIIα knockdown in combination with bisulfite DNA sequencing or histone methylation ChIP, they excluded DNA methyltransferases but found evidence for histone methyltransferases as mediators of the SAM-dependent effects. In particular, H3K4me2 and H3K9me2 levels decreased upon MATIIα knockdown. The authors did not identify the specific histone methyltransferases, but they did find a number of such enzymes in their MATIIα-interacting protein set (e.g., G9a, Ehmt, and MLL1/KMT2A). Together, these results fit a model in which chromatin-localized biosynthesis of metabolites can provide chromatin-modifying enzymes with the cofactors they need to catalyze their essential chemical reactions (Figure 1).
Figure 1. Chromatin-localized biosynthesis of metabolites can provide chromatin- and transcription-modifying enzymes with the cofactors they need to catalyze their essential chemical reactions.
While bound to chromatin, metabolite producers (“synthases”; e.g., MATIIα) can synthesize small molecule cofactors (e.g., SAM) that are used by chromatin- and transcription-modifying enzymes (“effectors”; e.g., methyltransferases) to chemically modify histones and non-histone proteins to alter gene expression. The synthase may be recruited by a DNA-bound transcription factor (“TF”; e.g., MafK) or the effector. The specific examples noted here are from Katoh et al. (2011).
This model represents an emerging paradigm in gene regulation (Table 1). For example, in mammalian cells the nuclear NAD+ synthase nicotinamide mononucleotide adenylyltransferase-1 (NMNAT-1) is recruited by Sirt1 to promoters, where it produces NAD+ to support deactylation of H4K16 by Sirt1 (Zhang et al., 2009). In yeast, the Sirt1 homolog Sir2 is allosterically regulated by O-acetyl-ADP-ribose, a byproduct that it produces during deacetylation reactions (Liou et al., 2005). O-acetyl-ADP-ribose allosterically regulates the quaternary association of Sir3 and Sir2/4, which accelerates Sir2 activity on acetylhistone substrates. More broadly, ribonucleotide reductase, which synthesizes deoxyribonucleotides from ribonucleotides, is rapidly recruited by Tip60 to DNA damage foci to assist with the synthesis of new DNA, most likely due to low pools of dNTPs during quiescence (Niida et al., 2010). Beyond these specific cases of chromatin-bound biosynthetic enzymes, metabolite production in general has been shown to modulate the actions of chromatin-modifying proteins to alter gene expression. For example, histone acetyltransferases are sensitive to the availability of acetyl-CoA, produced by acetyl-CoA synthetase in yeast (Takahashi et al., 2006) and ATP-citrate lyase in mammals (Wellen et al., 2009). Moreover, the corepressor function of carboxyl-terminal binding protein (CtBP) and the DNA binding activities of the circadian transcription factors Clock and BMAL are sensitive to the redox state of NAD+ cofactors (Kumar et al., 2002; Rutter et al., 2001; Zhang et al., 2002). Whether the metabolites in these latter cases are produced locally by chromatin-bound enzymes has yet to be determined.
The results of Katoh et al. (2011) and related results in the literature raise a number of questions. First, why should metabolite production occur by a chromatin-bound enzyme as opposed to a free enzyme in the nucleoplasm? Diffusion in the nucleus is rapid and unlikely to limit the availability of metabolites for use at specific loci. Thus, stable increases in the local free pools of metabolites are unlikely to be required or even occur. Perhaps there is a more intimate and functionally important association of the metabolite producers with the metabolite consumers. Such interactions could allow for (1) substrate channeling between the producer and consumer (Srere, 1987), which would shield the metabolite from other effectors that might use it for opposing outcomes (e.g., H3K4me3 versus H3K27me3) or (2) allosteric interactions, which can stimulate the activity of either, or both, enzymes. Also, a requirement for both the producer and consumer to be present in the same place at the same time adds precision to the regulatory process. Second, how generally do metabolite producers and users associate across the genome? The answer to this question will require the application and integration of genomic, proteomic, and metabolomic approaches to the enzymes pairs described herein. Finally, what other metabolite-producing enzymes may localize to the nucleus to support the functions of transcription- and chromatin-related proteins? As more sensitive proteomic approaches yield more examples, the challenge will be to identify the functional relevance, including the specific proteins that use the metabolites, their localization across the genome, and the regulatory outcomes that they control.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Katoh Y, Ikura T, Hoshikawa Y, Tashiro S, Ohta M, Kera Y, Noda T, Igarashi K. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.02.018. [THIS ISSUE] [DOI] [PubMed] [Google Scholar]
- Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK. Mol Cell. 2002;10:857–869. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- Larsson J, Zhang J, Rasmuson-Lestander A. Genetics. 1996;143:887–896. doi: 10.1093/genetics/143.2.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Niida H, Katsuno Y, Sengoku M, Shimada M, Yukawa M, Ikura M, Ikura T, Kohno K, Shima H, Suzuki H, et al. Genes Dev. 2010;24:333–338. doi: 10.1101/gad.1863810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Srere PA. Annu Rev Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, Sauve AA, Kraus WL. J Biol Chem. 2009;284:20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]