Summary
All living organisms communicate with the external environment for their survival and existence. In prokaryotes, communication is achieved by two-component systems (TCS) comprising histidine kinases and response regulators. In eukaryotes, signalling is accomplished by serine/threonine and tyrosine kinases. Although TCS and serine/threonine kinases coexist in prokaryotes, direct cross-talk between these families was first described in Group B Streptococcus (GBS).Aserine/threonine kinase (Stk1) and a TCS (CovR/CovS) co-regulate toxin expression in GBS. Typically, promoter binding of regulators like CovR is controlled by phosphorylation of the conserved active site aspartate (D53). In this study, we show that Stk1 phosphorylates CovR at threonine 65. The functional consequence of threonine phosphorylation of CovR in GBS was evaluated using phosphomimetic and silencing substitutions. GBS encoding the phosphomimetic T65E allele are deficient for CovR regulation unlike strains encoding the non-phosphorylated T65A allele. Further, compared with wild-type or T65A CovR, the T65E CovR is unable to bind promoter DNA and is decreased for phosphorylation at D53, similar to Stk1-phosphorylated CovR. Collectively, we provide evidence for a novel mechanism of response regulator control that enables GBS (and possibly other prokaryotes) to fine-tune gene expression for environmental adaptation.
Introduction
Post-translational modification of proteins is crucial for regulation of biological processes in prokaryotes and eukaryotes. Although 200 different types of post-translational modifications are known, reversible modifications are most important for control of cellular functions (Krishna and Wold, 1998; Raggiaschi et al., 2005). Of these, reversible protein phosphorylation is fundamental to signal transduction and enables organisms to respond to changes in the environment. Phosphorylation can occur at multiple sites and affect protein function. O-phosphorylation is common in eukaryotes and occurs on serine, threonine and tyrosine amino acids. These phosphorylation events require the function of serine/threonine and tyrosine kinases and their cognate phosphatases of which as many as 500 are encoded in the human genome (Manning et al., 2002). Deviation from normal phosphorylation events mediated by these signalling cascades can lead to changes in cellular function and disease (Cohen, 2002).
In contrast to the serine/threonine and tyrosine signalling cascades in eukaryotes, signal transduction in prokaryotes is primarily accomplished by two-component systems (TCS). TCS regulate gene expression in response to external signals and play a critical role in bacterial disease pathogenesis (Hoch and Silhavy, 1995; Beier and Gross, 2006). A typical TCS comprises a membrane-associated sensor histidine kinase and its cognate response regulator (often a DNA binding protein). Environmental signals are detected by the histidine kinase, which then phosphorylates its cognate response regulator at a conserved aspartate residue. Aspartate phosphorylation usually alters the effector activity which is most often DNA binding affinity and control of gene expression (Hoch and Silhavy, 1995; Stock et al., 2000). As a consequence of altering gene expression, the organism can respond to the initiating external signal, and the altered environment.
Although previously thought to be mutually exclusive, recent advances in genome analyses have indicated the presence of serine/threonine kinases (STK) and their cognate phosphatases in prokaryotes (Kennelly, 2002; Deutscher and Saier, 2005). Similarly, the existence of TCS has been described in eukaryotes such as Saccharomyces cerevisiae, Oryza sativa and Arabidopsis thaliana (Chang et al., 1993; Chang and Meyerowitz, 1995; Zhang et al., 2004; Stepanova and Alonso, 2005; Pareek et al., 2006). Interestingly, environmental adaptation in S. cerevisiae and A. thaliana is accomplished in part, through heterologous TCS and STK signalling systems. Responses to ethylene in A. thaliana and osmotic stress in S. cerevisiae involve TCS that sense external signals and regulate downstream mitogen-activated protein kinase (STK) signalling pathways (Chang et al., 1993; Chang and Meyerowitz, 1995; Zhang et al., 2004; Stepanova and Alonso, 2005). Despite the importance of STKs in virulence of bacterial pathogens such as Streptococci, Enterococci, Yersinia, Pseudomonas, and Mycobacterial sp., their role in prokaryotic signalling is not completely understood (Hakansson et al., 1996; Barz et al., 2000; Juris et al., 2000; Koul et al., 2001; Chaba et al., 2002; Madec et al., 2002; Rajagopal et al., 2003; Cowley et al., 2004; Jin and Pancholi, 2006; Kristich et al., 2007; Mougous et al., 2007). Examples of cross-talk between STK and bacterial signalling molecules in regulation of gene expression for environmental adaptation are minimal, suggesting that these signalling pathways are discrete. Although STKs in Myxo-coccus (Pkn14) and Mycobacteria (PknH) were described to phosphorylate transcriptional regulators such as MrpC of the cAMP receptor protein family and EmbR of the SARP (Streptomyces antibiotic regulatory protein) family (Molle et al., 2003; Nariya and Inouye, 2005; 2006; Sharma et al., 2006), the role of phosphorylation on DNA binding is not completely understood. Our previous studies established that an STK (Stk1) and a classical TCS (CovR/CovS) co-regulate toxin expression in the human pathogen Group B Streptococci (GBS; Rajagopal et al., 2006).
Group B Streptococci or Streptococcus agalactiae are Gram positive bacteria that can cause invasive infections in human newborns and in diabetic, immunocompromised and elderly adults. However, GBS commonly reside as commensal organisms in the genital and gastrointestinal tracts of healthy adult women (Baker and Edwards, 1995, Doran and Nizet, 2004). Because of its ability to transition from a commensal organism to an invasive pathogen in the human host, regulation of gene expression in response to the external environment is essential for GBS disease pathogenesis. Signalling systems play a crucial role in adaptive responses of GBS; in their absence, inappropriate expression of virulence factors can severely compromise its existence as a commensal or as a pathogen. Signal transduction systems in GBS comprise approximately 20 TCS, a single STK Stk1 and its cognate phosphatase Stp1, and a tyrosine kinase CpsD and its cognate phosphatase CpsB (for genome sequences, see Glaser et al., 2002; Tettelin et al., 2002; 2005). Although 17 TCS are conserved among GBS serotypes (Glaser et al., 2002; Tettelin et al., 2002; Jiang et al., 2005), the role of only four systems in regulation of gene expression has been described (DltR/DltS, CovR/CovS, RfgA/RgfC and CiaR/CiaH; Poyart et al., 2001; 2003; Spellerberg et al., 2002; Lamy et al., 2004; Jiang et al., 2005; Quach et al., 2008). Relevant to this study, the TCS known as CovR/CovS (also called CsrR/CsrS) and a STK known as Stk1 regulate GBS virulence and cytotoxin expression (Rajagopal et al., 2003; 2006; Lamy et al., 2004; Jiang et al., 2005).
Expression of the GBS cytotoxins known as β-haemolysin/cytolysin (β-H/C) and CAMP factor is regulated by CovR/CovS and Stk1. The response regulator CovR represses the expression of β-H/C and activates the expression of CAMP factor in GBS (Lamy et al., 2004; Jiang et al., 2005). Studies have shown that CovS, the sensor histidine kinase of CovR, enhances CovR-mediated repression of β-H/C and activation of CAMP factor through phosphorylation of the conserved aspartate residue at position 53 (D53) (Jiang et al., 2005; 2008). Our previous work indicates that Stk1 positively regulates expression of β-H/C and negatively regulates expression of CAMP factor in a CovR-dependent manner (Rajagopal et al., 2006). Although these studies suggested that Stk1 can phosphorylate CovR in vitro, the in vivo relevance and functional consequence of the post-translational modification was not established. The interactions between different signalling systems triggered our interest in understanding the functional consequence of serine/threonine phosphorylation of the response regulator CovR on GBS gene expression. In this report, we demonstrate that a threonine residue at position 65 (T65) is required for Stk1 phosphorylation. GBS encoding the phosphomimetic T65E CovR allele is deficient for CovR function, whereas the strain encoding the non-phosphorylated T65A CovR allele is not. Furthermore, we show that the T65E CovR protein is unable to bind promoter DNA due to decreased phosphorylation at D53, unlike either wild-type (WT) or T65A CovR. Taken together, our results indicate that threonine phosphorylation of CovR reduces its activation at the conserved aspartate and consequently diminishes its ability to bind DNA and control GBS gene expression. The existence of two diverse phosphorylation events that are linked to control the function of a response regulator is likely to be beneficial for environmental adaptation and GBS disease pathogenesis.
Results
Stk1 phosphorylates the N-terminus of CovR
The response regulator CovR was shown to repress expression of β-H/C and activate expression of CAMP factor (Lamy et al., 2004). Our previous studies showed that Stk1 alleviates CovR repression of β-H/C and activation of CAMP factor and can also phosphorylate CovR in vitro (Rajagopal et al., 2006). Thus, we hypothesized that phosphorylation at a serine or threonine (S/T) may decrease CovR activity in GBS. To test this hypothesis, we first sought to identify the serine and/or threonine (S/T) residues that are phosphorylated in CovR. We subsequently compared cytotoxin expression and CovR regulation in GBS that express WT or a substitution which mimics the constitutively phosphorylated state of CovR (also known as phosphomimetic and are represented by S/T to glutamate or aspartate substitutions S/T: E/D). Because phosphorylation of polar, uncharged amino acids such as S/T and tyrosine (Y) confers a negative charge to these residues, their substitution with negatively charged amino acids such as E or D are considered to function as phosphomimetic substitutions. In contrast, substitution of phosphorylated S/T and Y residues with neutral or basic amino acids are considered to function as silencing substitutions (Wang and Klemke, 2008). Consequently, these two classes of mutations are often used to evaluate the role of S/T and Y protein phosphorylation in eukaryotes (for examples, see Fillebeen et al., 2003; Wagner et al., 2004; Lauberth et al., 2007; Onischenko et al., 2007).
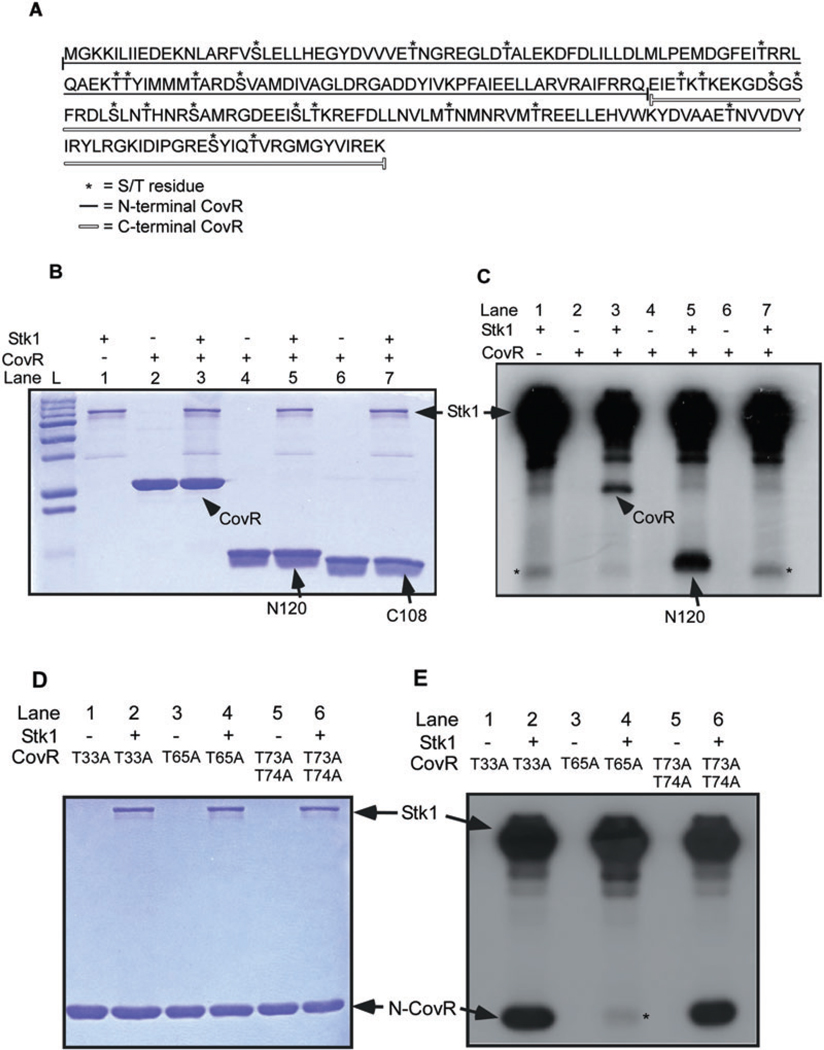
In order to identify the S/T amino acids that are phosphorylated in CovR, we initially examined whether Stk1 preferentially phosphorylated CovR at the N- or C-terminus. Truncated CovR proteins encoding either the first 120 amino acids (M1-Q120, hereafter referred to as N120) or the C-terminal 120 amino acids (E121-K229, hereafter referred to as C108) were constructed and purified as described in Experimental procedures. The amino acid sequence of the entire CovR protein and amino acids encoded in the N- and C-terminal truncations are shown in Fig. 1A. In vitro phosphorylation reactions were performed with full-length, N120 and C108 CovR in the presence or absence of Stk1 as described in Experimental procedures. The reaction products were analysed on 15% SDS-PAGE followed by autoradiography. Figure 1B shows the Coomassie-stained gel and Fig. 1C depicts the autoradiograph of the same gel. As observed previously (Rajagopal et al., 2006), we confirmed that Stk1 auto-phosphorylates in vitro (Fig. 1C, lanes 1, 3, 5 and 7), and phosphorylates full-length CovR (Fig. 1C, lane 3). Auto-phosphorylation was not observed with the full-length or truncated forms of CovR in the presence of [γ-32P]-ATP (Fig. 1C, lanes 2, 4, and 6). The addition of Stk1 to N120 CovR produced a robust phosphorylated band, whereas Stk1 did not substantially phosphorylate C108 (Fig. 1C, compare lanes 5 and 7). The faint band denoted by a asterisk (*) in C108 (Fig. 1C, lane 7) is also observed in the lane with Stk1 alone (Fig. 1C, lane 1). Taken together, these data indicate that S/T phosphorylation occurs within the first 120 amino acids of CovR.
Fig. 1.
Stk1 phosphorylates CovR at a threonine residue in position 65.
A. The entire CovR coding sequence comprising 229 amino acids is shown. The amino acids encoded in the N- (N120) and C-terminal (C108) CovR are indicated by the filled and unfilled line respectively. The N120 CovR contains two serine and six threonine residues, while the C108 CovR (which was constructed to have a methionine start codon) has six serine and eight threonine residues.
B and C. In vitro phosphorylation reactions were performed with approximately 0.5–1 µg of GST-Stk1 protein and WT, N120 or C108 CovR as described in the Experimental procedures. All samples were analysed on 15% SDS-PAGE, stained with Coomassie and exposed to autoradiography. Note that Stk1 phosphorylates full-length CovR-His6 and N-terminal CovR-His6 (see lanes 3 and 5 in ‘C’). The band denoted by asterisk (*) observed with C108 (see lane 7 in ‘C’) is also seen with Stk1 alone (see lane 1 in ‘C’). Lane ‘L’ is the pre-stained Protein Standards (Bio-Rad).
D and E. In vitro phosphorylation reactions were performed with approximately 0.5–1 µg of GST-Stk1 protein and CovR substitution mutants as described in the Experimental procedures. All samples were analysed on 15% SDS-PAGE, stained with Coomassie and exposed to autoradiography. Autoradiograph shows that Stk1 phosphorylation is drastically reduced or abolished in T65A N-terminal CovR-His6 (Fig. 1E, lane 4) in contrast to T33A (see lane 2 in ‘E’) or T73/74A CovR (see lane 6 in ‘E’). Autophosphorylation of T65A, T33A and T73/74A was not observed (see lanes 1, 3 and 5 in ‘E’).
Stk1 phosphorylates CovR at threonine 65
We next constructed site-directed mutants in the threonine and/or serine residues located within N120. CovR mutants were constructed to replace threonine residues at position 33, 41, 65, 73, 74, 80 and serine residues at position 19 and 84 with alanine respectively. These mutants were derived using both N120 and full-length CovR plasmids as templates (see Experimental procedures). Subsequently, mutant CovR proteins were purified and in vitro phosphorylation reactions were performed in the presence and absence of Stk1 as described in Experimental procedures. The reaction products were analysed on 15% SDS-PAGE followed by autoradiography. Among all CovR mutants tested, we observed decreased Stk1 phosphorylation only with the CovR T65A substitution (shown in Fig. 1E). Figure 1D is the Coomassie-stained gel and Fig. 1E is the autoradiograph of the same gel. Note that Stk1-dependent phosphorylation was not affected in the T33A or the double T73A/T74A CovR (Fig. 1E, lanes 2 and 6). In contrast, Stk1-mediated phosphorylation was severely reduced in T65A CovR (Fig. 1E, lane 4), indicating that T65 is the predominant amino acid necessary for Stk1 phosphorylation. The faint band denoted by a asterisk (*) in lane 4 was similar to that observed previously with Stk1 alone as shown in Fig. 1C. Similar results were observed with full-length T65A CovR (data not shown). Further, decreased Stk1-mediated phosphorylation was not observed with other site-directed CovR mutants T41A, S19A, S84A, T124A, T126A, T157A, S132A, S134A and S155A (data not shown). Thus, T65 is critical for Stk1 phosphorylation of CovR.
T65E CovR does not complement ΔcovR for cytotoxin expression
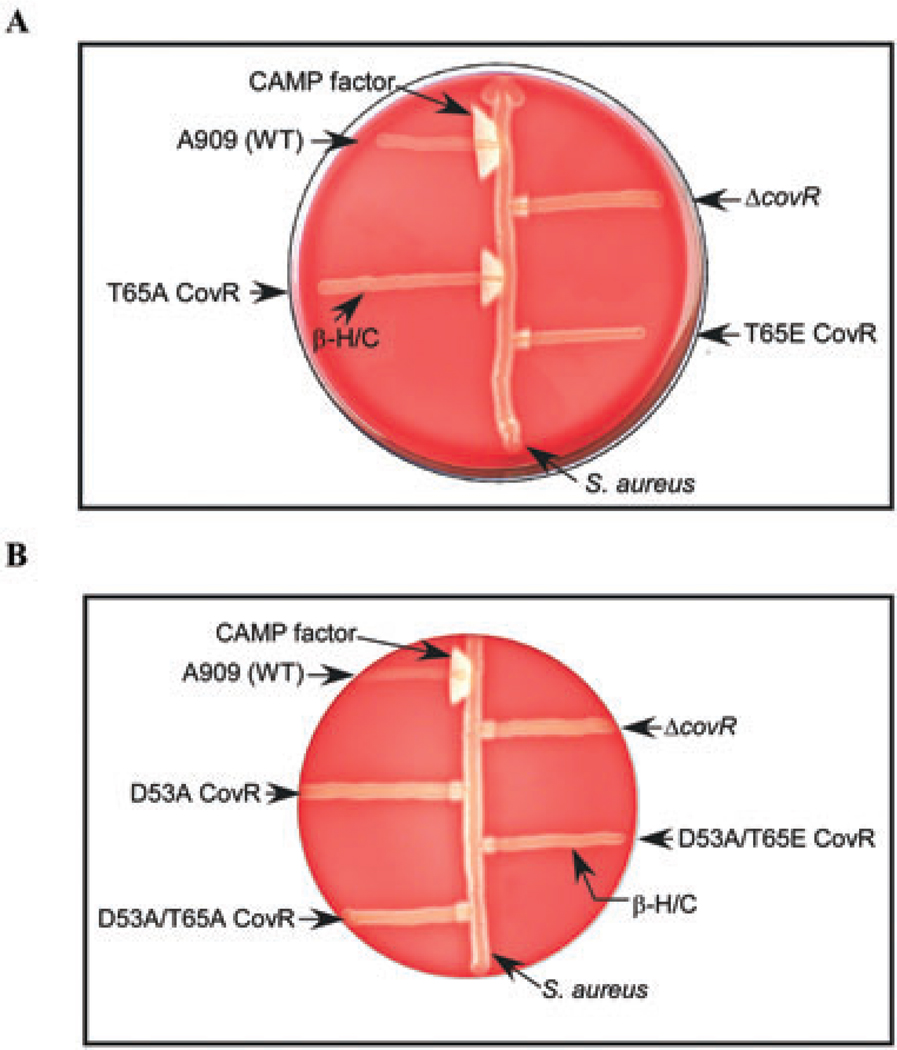
To test our hypothesis that phosphorylation at T65 decreases CovR activity in vivo, we compared cytotoxin expression in GBS strains that encoded either the T65A or T65E CovR allele. Our previous studies have shown that CovR-mediated repression of β-H/C and activation of CAMP factor was increased in the Δstk1 strains (Rajagopal et al., 2006). Therefore, we predicted that the negatively charged T65E CovR would not complement the ΔcovR strain for cytotoxin expression and CovR function. Conversely, substitution of T65 with a neutral amino acid such as alanine (T65A) should restore cytotoxin expression and CovR function to the ΔcovR strain. Using ΔcovR as the parent strain, we derived ΔGBS strains that carried either the T65E or the T65A CovR alleles on the chromosome as described in Experimental procedures. Because the mutants are not plasmid integrants and are exact allelic replacements of covR in GBS, these alleles are stable and provide us with the advantage of analysing gene expression without plasmid-associated problems such as (i) copy number effects; (ii) addition of antibiotics for retention of the plasmid integrant strains; and (iii) the presence of non-endogenous promoter elements. We initially analysed expression of CAMP factor and β-H/C on blood agar plates and subsequently performed quantitative reverse transcription polymerase chain reaction (qRT-PCR) to examine transcription of these cytotoxins. Consistent with our hypothesis, we observed increased expression of CAMP factor in the presence of T65ACovR, but not in the presence of T65E CovR (compare the triangular zone of lysis at the junction between GBS and Staphylococcus aureus in Fig. 2A). Likewise, repression of β-H/C was restored in the presence of T65A CovR and was similar to the Δstk1 strain. In contrast, β-H/C expression in the presence of T65E CovR was similar to the ΔcovRstrain (Fig. 2A). qRT-PCR confirmed that transcription of cylE (β-H/C) was 10–12-fold higher compared with WT in both ΔcovR and T65E CovR (Table 1). In contrast, cylE transcription was lower in the presence of T65A CovR compared with ΔcovR and T65E CovR and was similar to the Δstk1 strain (see Table 1). Transcription of cfb (CAMP factor) was also restored in the ΔcovR strain by T65ACovR (see Table 1). In contrast, cfb transcription was ≤ 10-fold compared with WT in the presence of T65E CovR (Table 1). These results confirm that the T65A CovR substitution is functional in GBS unlike the T65E CovR substitution (see Table 1).
Fig. 2.
Cytotoxin expression in GBS strains encoding T65A and T65E CovR.
A. T65A CovR complements the ΔcovR strain for repression of β-H/C and activation of CAMP factor expression unlike the T65E CovR. CAMP factor activity is represented by the triangular zone of lysis at the junction between GBS and β-lysin producing Staphylococcus aureus on blood-agar plates. β-H/C activity is represented by the zone of clearing around GBS. A909 represents wild-type GBS. The strains encoding the T65A and T65E CovR alleles on the GBS chromosome are isogenic to A909. The ΔcovR is a covR deletion in A909. Note that CAMP factor activity is greater in T65A when compared with T65E or ΔcovR.
B. Complementation of the ΔcovR strain by T65A CovR requires D53. GBS strains encoding D53A, D53A/T65A and D53A/T65E CovR are compared with WT and the isogenic ΔcovR strain. Note that the decrease in β-H/C and increase in CAMP factor expression seen in T65A (shown in A) is abolished in D53A/T65A CovR.
Table 1.
Expression of CovR-regulated genes in GBS T65A, T65E, D53A and Δstk1 strains.
| Gene expression | |||||
|---|---|---|---|---|---|
| qRT-PCRa |
|||||
| Locus | ΔcovRb | CovRD53Ab | CovRT65Eb | CovRT65Ac | Δstk1c |
| CovR-repressed genes | |||||
| cylE (β-H/C) | 12.5 ± 2 | 11 ± 4 | 10 ± 0.5 | 0.35 ± 0.15 | 0.19 ± 0.15 |
| SAK_0838 | 35 ± 10 | 44 ± 10 | 44 ± 10 | 0.4 ± 0.2 | 0.3 ± 0.2 |
| SAK_0899 | 3.8 ± 0.9 | 3.7 ± < 0.01 | 3.5 ± 1 | 0.19 ± 0.1 | 0.3 ± < 0.01 |
| SAK_1097 | 21 ± 14 | 20 ± 10 | 18.6 ± 10 | 0.3 ± 0.2 | 0.3 ± 0.1 |
| SAK_1142 | 30 ± 4 | 22 ± 4 | 22 ± 2 | 0.4 ± 0.1 | 0.4 ± < 0.01 |
| SAK_1574 | 180 ± 100 | 134 ± 95 | 142 ± 100 | 0.4 ± < 0.01 | 0.4 ± < 0.01 |
| CovR-activated genes | |||||
| cfb (CAMP factor) | 0.14 ± <0.01 | 0.02 ± <0.01 | 0.1 ± <0.01 | 2.0 ± 0.2 | 1.9 ± 0.2 |
| SAK_1777 | 0.3 ± 0.1 | 0.3 ± <0.01 | 0.4 ± 0.2 | 4 ± 0.4 | 4.5 ± 0.6 |
| SAK_1777 | 0.3 ± < 0.01 | 0.3 ± < 0.01 | 0.3 ± < 0.01 | 3.3 ± 1 | 2.1 ± 0.5 |
| SAK_0255 | 0.5 ± < 0.01 | 0.5 ± < 0.01 | 0.4 ± < 0.01 | 1.9 ± 0.4 | 2.3 ± < 0.01 |
qRT-PCR was performed as described in Experimental procedures. Gene expression is denoted as fold difference relative to the WT GBS strain A909.
No significant difference in gene expression was observed between these sets of strains. Standard error is indicated. Choice of CovR-regulated genes other than cylE and cfb were based on the CovR core regulon described in Jiang et al. (2008).
Aspartate at position 53 is required for CovR regulation of GBS cytotoxins
Previous studies have indicated that the cognate sensor kinase CovS enhances CovR activity through phosphorylation at the conserved aspartate (D53) similar to CovR/S in Group A Streptococcus (GAS) (Lamy etal., 2004; Jiang et al., 2005; 2008; Gusa et al., 2006). To examine whether the presence of D53 (or its phosphorylation state) altered the reactivity of T65, we analysed cytotoxin expression in GBS strains that had an alanine substitution of aspartate 53 (D53A) in the presence and absence of T65E and T65A CovR respectively. The CovR allele containing an alanine substitution of the predicted active site D53 failed to complement the ΔcovR strain (Fig. 2B). Transcription of β-H/C and CAMP factor in the GBS strain encoding D53A CovR was similar to the ΔcovR strain (Table 1). Interestingly, cytotoxin expression in GBS strains containing the double substitutions (i.e. D53A/T65A or D53A/T65E CovR) was also similar to the ΔcovR strain (Fig. 2B). Transcription of cylE and cfb in these strains was also similar to the ΔcovR strain (data not shown). Taken together, these data indicate that D53 is essential for CovR function, including its regulation at T65 in GBS.
Other CovR-regulated genes are affected by the T65E CovR substitution
We examined whether the T65E and T65A substitutions differentially affect the expression of other CovR-regulated GBS genes. A recent study showed that although CovR/S regulates 150 genes in GBS serotypes Ia, III and V, 39 genes comprise the core regulon (Jiang et al., 2008). A subset of genes that represent both CovR-repressed and -activated genes from this core regulon were chosen for verification by qRT-PCR. The results shown in Table 1 indicate that genes that are negatively regulated by CovR are repressed in the GBS strain encoding T65A CovR, in contrast to the strains encoding either T65E or D53A CovR. Likewise, increased transcription of CovR activated genes was observed in the strain encoding T65A CovR, but not strains encoding T65E or D53A CovR (Table 1). Despite the observation that CovR family members differentially regulate expression of their promoters (Lamy et al., 2004; Jiang et al., 2005; 2008) [as does CovR in GAS as well as the archetype member OmpR (Gao et al., 2005; Gusa et al., 2006; Rhee et al., 2008)], our results confirm that T65A and T65E substitutions have similar effects on CovR function. Thus, T65A but not T65E CovR is functional in GBS and requires D53. We also predict that in the absence of Stk1, CovR activity might be further increased in the presence of (as yet unidentified) environmental cues that enhance CovS activation of CovR.
T65E CovR does not bind promoter DNA
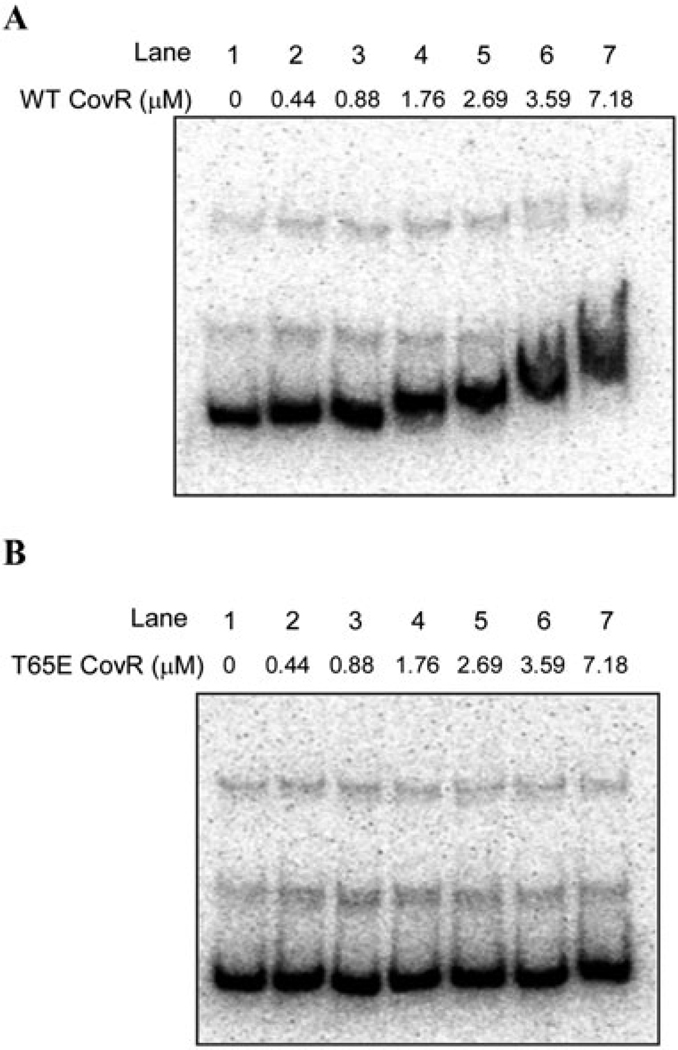
Previous studies have shown that CovR binds to the promoter of β-H/C and represses transcription (Lamy et al., 2004; Jiang et al., 2008). Aspartate phosphorylation (CovR~P) increases CovR affinity and enhances repression at this locus. We hypothesized that phosphorylation of CovR at T65 modulates CovR activity by decreasing the DNA binding affinity to target promoters. In particular, as T65E CovR did not restore CovR function (Fig. 2), we predicted that the T65E CovR protein would demonstrate decreased promoter binding. To test this possibility, we compared the ability of WT and T65E CovR to bind to the β-H/C promoter, PcylX. WT and CovR mutant proteins were pretreated with the acetyl phosphate (denoted as CovR~P) prior to binding as described in Experimental procedures. The results shown in Fig. 3A indicate that CovR~P binds to the PcylX promoter as shown previously (Lamy et al., 2004). The concentration of CovR~P required and the pattern of retardation of probe PcylX in the electrophoretic mobility shift assay (EMSA) was similar to that previously observed (Lamy et al., 2004; Jiang et al., 2008). CovR-mediated retardation of DNA was competitively inhibited by excess unlabelled PcylX DNA (data not shown). Most notably, the T65E mutant was unable to bind PcylX at any of the concentrations employed (compare lanes 4, 5, 6 and 7 between Fig. 3A and B). The lowest [CovR~P] where a mobility shift was observed was 1.76 µM, yet at fourfold higher concentrations of T65E CovR, no significant shift was apparent, indicating the lower affinity of T65E CovR at the β-H/C promoter.
Fig. 3.
T65E CovR does not bind promoter DNA. Electrophoretic mobility shift assay was performed using the PcylX promoter DNA as probe. The 176 bp PcylX promoter was amplified using the oligos PcylxL and PcylxR (see Table S1) and incubated with equimolar concentrations of acetyl phosphate treated WT CovR (CovR~P) (A) and T65E CovR~P (B). In both cases, lane 1 represents the probe-only control and lanes 2–7 represent increasing amounts of CovR from 0. 44 to 7.18 µ. Note that retardation in mobility of the probe DNA was observed with increasing concentrations of WT CovR~P and not with the T65E CovR~P.
T65E CovR does not protect PcylX promoter from DNase I cleavage
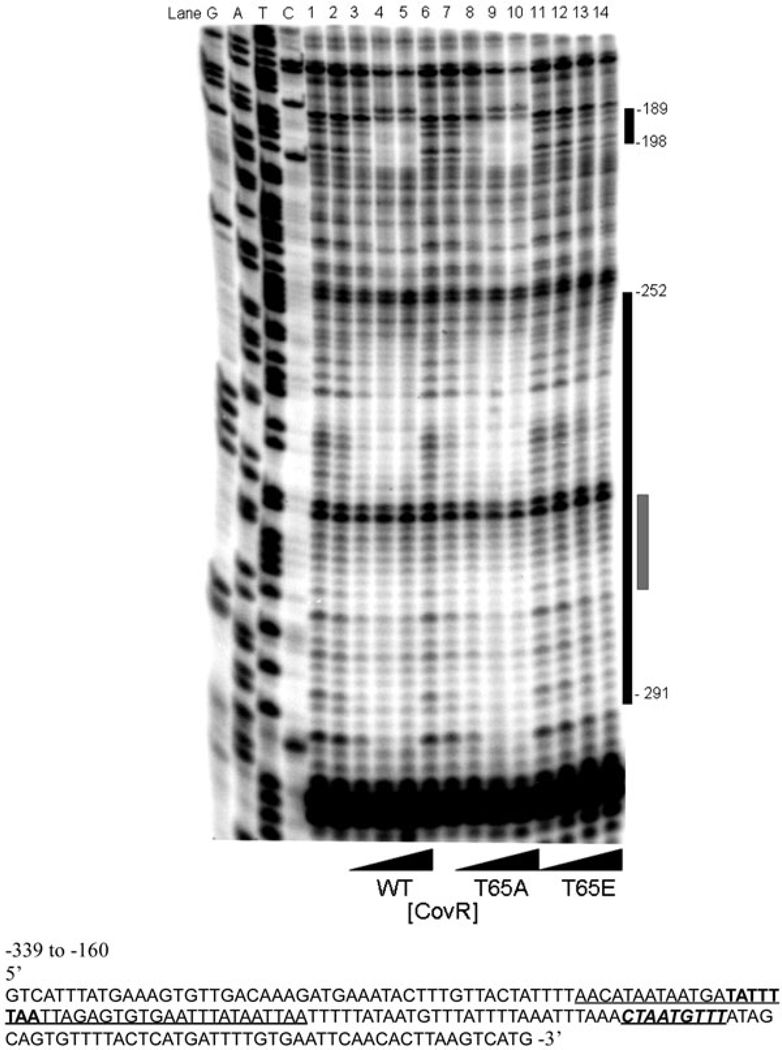
Due to the relatively modest decrease in DNA mobility observed with CovR~P in the EMSA (see Fig. 3A and Lamy et al., 2004; Jiang et al., 2005; 2008), we performed DNase I footprinting of the PcylX promoter region in the presence of WT, T65E and T65A CovR~P respectively (Fig. 4). In the presence of WT (lanes 2–5) and T65A CovR~P (lanes 7–10), two regions of protection are evident. One covers approximately 40 bp of PcylX DNA from −291 to −252 relative to the translational start site of cylX, as reported previously (Lamy et al., 2004). An additional downstream promoter region (from −198 to −189) is also protected by WT and T65A CovR~P. CovR~P appears to have lower affinity for this downstream site, as it takes slightly higher concentrations (about twofold) for protection to be evident. In contrast, in the presence of T65E CovR~P, no significant protection of promoter DNA was observed (lanes 11–14). The concentrations of CovR~P employed in the footprinting assay were similar to those in the EMSA (Fig. 3). Of note, incubation of CovR with the phosphodonor was required to observe protection from DNase I (data not shown). Taken together, these results suggest that modification of T65 to confer a negative charge (e.g. substitution with glutamate or phosphorylation of the threonine) substantially reduces the ability of CovR~P to bind to target promoters and likely accounts for the increased CovR function observed previously in the Δstk1 strains (Rajagopal et al., 2006).
Fig. 4.
T65E CovR does not protect promoter PcylX from DNase I cleavage. Equimolar concentrations of phosphoramidate-treated WT, T65A and T65E CovR~P were used in DNase I protection assays of the PcylX promoter. The lanes G, A, T and C represent the DNA sequencing ladder. Lanes 1 and 6 represent DNase I-only controls. Lanes 2–5, 7–10 and 11–14 represent WT, T65A and T65E CovR~P at 1, 2, 4 and 8 µM respectively. The co-ordinates of CovR protection from −291 to −252 of the predicted cylX translational start site was described previously (Lamy et al., 2004). These footprinting analyses also indicate CovR protection of PcylX from −198 to −189 which was not previously observed. Importantly, in all cases, the T65A CovR~P demonstrated DNA protection similar to WT CovR, whereas no significant DNA protection is observed with T65E CovR. The black bar to the right of the panel delineates the extent of CovR-mediated protection from DNase I cleavage. The grey bar depicts the predicted CovR DNA sequence motif (Lamy et al., 2004).
T65E CovR decreases phosphorylation of D53 by acetyl phosphate
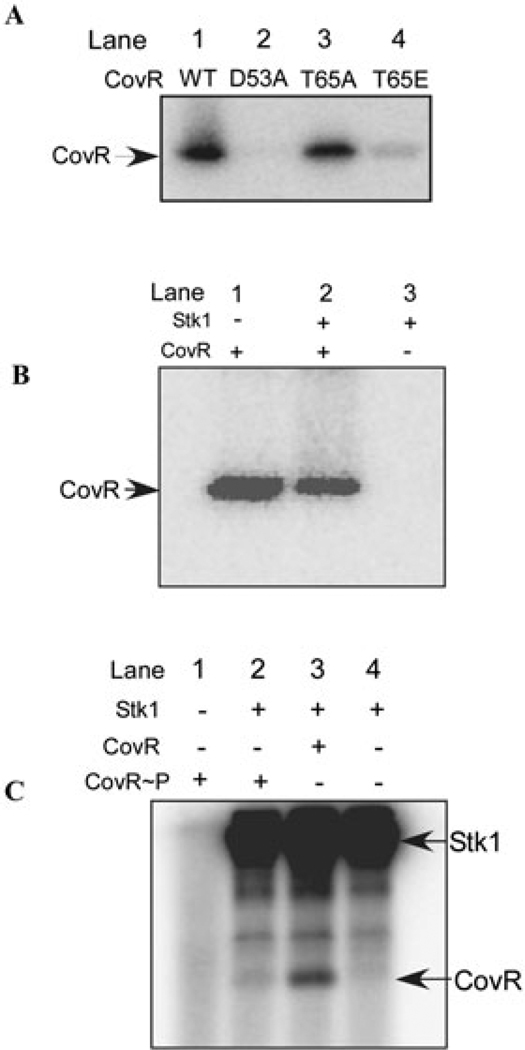
One explanation for the diminished ability of T65E CovR to bind promoter DNA and its lack of function in GBS (Figs 3 and 4; Table 1) is that the presence of this substitution decreases or prevents phosphorylation at the conserved aspartate residue in the active site (D53). To test this hypothesis, we compared phosphorylation of WT, T65E, D53A and T65A CovR using [32P]-acetyl phosphate as the phosphodonor. Low-molecular-weight compounds such as acetyl phosphate have been described to serve as phosphodonors for aspartate phosphorylation of response regulators both in vitro and in vivo (Lukat et al., 1992; Kenney et al., 1995; Klein et al., 2007). Synthesis of [32P]-acetyl phosphate and phosphorylation of CovR were performed as described in Experimental procedures and the results are shown in Fig. 5A. Our results indicate that both WT and T65A CovR were phosphorylated in the presence of acetyl phosphate at similar efficiencies (see lanes 1 and 3). As expected, alanine substitution of D53 abolished acetyl phosphate phosphorylation (lane 2). Importantly, T65E CovR exhibited a dramatic decrease (≥ 3-fold) in phosphorylation by acetyl phosphate (lane 4) suggesting that substitution of T65 with a negative charge reduces phosphorylation at the conserved aspartate residue at position 53. This result suggests that T65E CovR is deficient for CovR function because it reduces or prevents phosphorylation at D53, thus preventing DNA binding (Figs 3 and 4).
Fig. 5.
A. T65E CovR decreases phosphorylation of D53 by [32P]-acetyl phosphate. Equal concentrations of WT, T65A, T65E and D53A CovR were incubated with 32 mM [32P]-acetyl phosphate (specific activity 8.5 mCi mmol−1) at 37°C for 90 min. The samples were subsequently analysed on a 12% SDS-PAGE followed by autoradiography. Note that [32P]-acetyl phosphate-mediated phosphorylation of T65E CovR is significantly reduced (≥ 3-fold) compared with T65A and WT CovR. The experiment was repeated at least three times.
B. Stk1 phosphorylation of CovR decreases D53 phosphorylation by [32P]-acetyl phosphate. In vitro phosphorylation of CovR was performed with non-radioactive ATP in the presence and absence of Stk1 prior to incubation with 32 mM [32P]-acetyl phosphate (specific activity 8.5 mCi mmol−1) at 37°C for 90 min. Controls included a reaction containing only Stk1 (lane 3) and no phosphorylation by acetyl phosphate was observed. The samples were subsequently analysed on a 12% SDS-PAGE followed by autoradiography. The figure shown represents one of three independent experiments. Note that [32P]-acetyl phosphate-mediated phosphorylation of CovR is significantly reduced (≥ 2-fold) when phosphorylated by Stk1 (compare lane 2 with lane 1).
C. CovR phosphorylation at D53 reduces phosphorylation by Stk1. Equal concentrations of CovR were pre-incubated in the presence and absence of 32 mM acetyl phosphate prior to the addition of Stk1 that was phosphorylated with radioactive ATP (see Experimental procedures). All samples were analysed on 12% SDS-PAGE and exposed to autoradiography. The figure shown represents one of three independent experiments. Lane 4 contains only Stk1 and lane 1 contains only CovR~P. CovR phosphorylation by Stk1 was reduced ≥ 4-fold when pretreated with acetyl phosphate (lane 2) compared with the absence of acetyl phosphate (lane 3).
Phosphorylation by Stk1 decreases CovR phosphorylation at D53
As the phosphomimetic T65E CovR exhibited decreased phosphorylation at D53 (Fig. 5A), we next examined whether Stk1 phosphorylation of CovR also reduced phosphorylation at D53. CovR was first phosphorylated by Stk1 in the presence of non-radioactive ATP and subsequently [32P]-acetyl phosphate was added to the reaction. The results shown in Fig. 5B indicate that CovR phosphorylation by acetyl phosphate decreased ≥ 2-fold when it was first phosphorylated by Stk1 (compare lane 2 with lane 1). As expected, we did not observe auto-phosphorylation of Stk1 in the presence of [32P]-acetyl phosphate (lane 3). These data confirm that Stk1 phosphorylation of CovR at T65 decreases the reactivity of D53 and reduces its phosphorylation. Given that the presence of the phosphodonor was essential for protection against DNase I, decreasing CovR~P at D53 would lead to a reduction in DNA binding and a relief of repression of PcylX.
CovR phosphorylation at D53 decreases Stk1 phosphorylation at T65
As phosphorylation by Stk1 at T65 decreased CovR phosphorylation at D53, we predicted that CovR phosphorylation at D53 would exclude Stk1 phosphorylation at T65. To test this hypothesis, we incubated CovR in the presence and absence of non-radioactive acetyl phosphate prior to addition of 32P-phosphorylated Stk1. The results shown in Fig. 5C indicate that Stk1 phosphorylation of CovR was reduced ≥4-fold when CovR was phosphorylated at D53 by acetyl phosphate (compare lane 2 with lane 3). Significant Stk1 phosphorylation of CovR~P was not observed even on prolonged exposure (data not shown). Autophos-phorylation of CovR~P is not observed in the presence of [γ-32P]-ATP (lane 1). These data confirm that phosphorylation at D53 decreases CovR phosphorylation at T65 and vice versa. This result suggests that once CovR is phosphorylated at D53, T65 is inaccessible for phosphorylation by Stk1.
Phosphorylation by Stk1 decreases CovR promoter binding
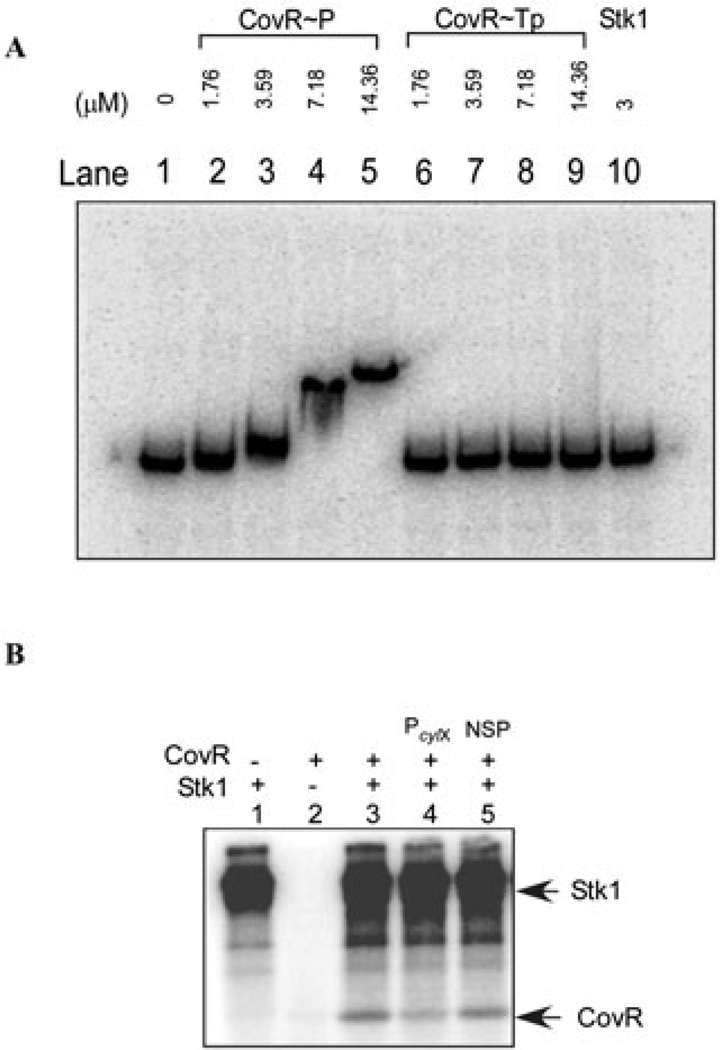
We next examined the effect of Stk1 (threonine) phosphorylation on promoter binding of CovR. To this end, CovR was first phosphorylated by Stk1 that was retained on glutathione sepharose and subsequently the threonine-phosphorylated CovR was treated with acetyl phosphate as described in the Experimental procedures. Equimolar concentrations of threonine phosphorylated CovR (CovR~Tp) and aspartate-phosphorylated CovR (CovR~P) were used in promoter binding assays that employed the PcylX probe. The results shown in Fig. 6A indicate that promoter binding of CovR~P to PcylX (lanes 2–5) was similar to that observed previously (Fig. 3A). In contrast, promoter binding of CovR was eliminated when CovR was first phosphorylated by Stk1 (CovR~Tp, lanes 6–9). Taken together, these data demonstrate that Stk1 phosphorylation of CovR prevents promoter DNA binding. As expected, incubation of PcylX probe with Stk1 alone indicated that Stk1 does not bind DNA because it does not contain canonical DNA binding domain/s.
Fig. 6.
A. Stk1 phosphorylation prevents promoter binding of CovR. In vitro phosphorylation of CovR was performed using Stk1 that was bound to GST-sepharose. Electrophoretic mobility shift assay was then performed using PcylX DNA as probe. The 176 bp PcylX promoter was amplified using the oligos PcylxL and PcylxR (see Table S1) and incubated with equimolar concentrations of threonine-phosphorylated CovR (CovR~Tp) and non-threonine-phosphorylated CovR (see Experimental procedures). Lane 1 represents the probe-only control and lanes 2–5 represent increasing amounts of CovR~P from 1.76 to 14.36 µM and lanes 6–9 represent increasing amounts of CovR~Tp from 1.76 to 14.36 µM. Note that retardation in mobility of the probe DNA was observed with increasing concentrations of CovR~P and not with the CovR~Tp. As an additional control, lane 10 contained 3 µM of Stk1 alone (i.e. no CovR) and demonstrates that Stk1 itself does not bind to PcylX DNA.
B. CovR phosphorylation by Stk1 is reduced in the presence of CovR-specific promoter DNA. Equal concentrations of unphosphorylated CovR were pre-incubated with either a CovR-specific promoter, i.e. PcylX (lane 4) or non-specific DNA (lane 5) prior to in vitro phosphorylation reaction with Stk1 (see Experimental procedures). All samples were analysed on 12% SDS-PAGE and exposed to autoradiography. The figure shown is one of three independent experiments. In lane 3, a control reaction that did not contain DNA is shown. CovR phosphorylation was reduced to 60% in the presence of PcylX DNA (lane 4) compared with the absence of DNA (lane 3).
DNA binding decreases CovR phosphorylation by Stk1
In the OmpR subfamily of response regulators, previous studies have shown that DNA binding stimulates aspartate phosphorylation and vice versa (Ames et al., 1999; Tran et al., 2000; Mattison et al., 2002; Gao et al., 2005; Gusa et al., 2006). It was therefore of interest to determine whether CovR-specific promoter DNA can affect the reactivity of T65 to the Stk1 kinase. To this end, we pre-incubated unphosphorylated CovR with either the CovR-specific PcylX promoter or non-specific DNA prior to in vitro phosphorylation reactions in the presence of Stk1 (see Experimental procedures for details). As observed previously, Stk1 phosphorylates CovR (lane 3, Fig. 6B). However, in the presence of PcylX, a 40% decrease in phosphorylation is observed (lane 4). As this decrease was not observed when CovR was pre-incubated with non-specific DNA (lane 5), these data together with the results in Fig. 5C suggest that Stk1 preferentially phosphorylates free and unphosphorylated CovR rather than DNA-bound CovR or free CovR~P.
Discussion
Signalling systems are essential for organisms to respond to their dynamic external environment. The importance of TCS in bacterial signalling and their role in expression of virulence factors has been described for many pathogenic organisms (for recent reviews, see Beier and Gross, 2006; Laub and Goulian, 2007; Bourret, 2008). Control of the function of two-component response regulators by cognate histidine kinases through phosphorylation at the conserved aspartate residue has been extensively studied in both Gram negative and Gram positive bacteria. Our studies on the link between a TCS (CovR/CovS) and a S/T kinase (Stk1) in the human pathogen GBS provides evidence for a novel method of control of a response regulator that is important for toxin expression.
We previously showed that Stk1 regulates toxin expression in a manner that is opposite to but dependent on the response regulator CovR and that it can phosphorylate CovR in vitro (Rajagopal et al., 2006). To elucidate the role of CovR phosphorylation by Stk1, we identified the amino acid residue that is modified and subsequently compared the function of a negatively charged or phosphomimetic allele with a corresponding non-phosphorylated allele. Because reversible phosphorylation is a substoichiomet-ric reaction, only a small fraction of a protein is in the phosphorylated state at any given time (Raggiaschi et al., 2005; Thomas et al., 2008). This complexity is further accentuated for CovR because in addition to threonine phosphorylation by Stk1, it is also subject to aspartate phosphorylation by CovS and/or low-molecular-weight phosphodonors (Lamy et al., 2004; Jiang et al., 2008). Therefore, the phosphomimetic allele enables us to selectively define whether the negative charge conferred by the phosphate on the threonine residue due to Stk1 had a role in CovR function.
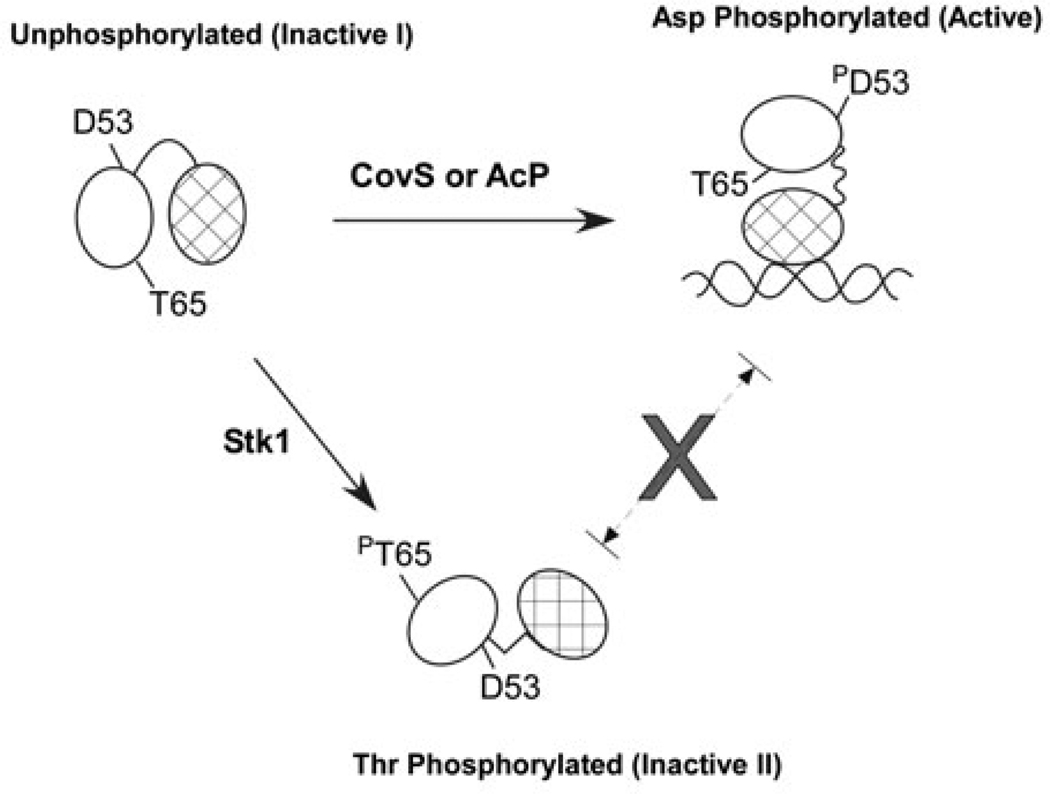
In this study, we have extended our previous observations and identified that Stk1 phosphorylates CovR at threonine 65. Consistent with the phenotypes of the Δstk1 strains (Rajagopal et al., 2006) and our hypothesis that Stk1 phosphorylation decreases CovR activity, GBS encoding the T65E CovR are deficient for CovR function in contrast to T65A CovR. Interestingly, the T65E CovR and Stk1-phosphorylated CovR exhibit dramatically reduced phosphorylation of D53. As the substitution of T65 with an alanine did not negatively affect D53 phos-phorylation, DNA binding or CovR function in vivo, this suggests that the negative charge from the phosphate group is responsible for allosteric hindrance at position 65. Based on these data, we predict that Stk1-mediated phosphorylation of CovR at T65 induces a conformational change that decreases phosphorylation at D53 and reduces promoter DNA binding and function (see Fig. 7). Conversely, as Stk1 phosphorylation is diminished in the presence of CovR~P and CovR-specific promoter DNA, this suggests that when phosphorylated at D53 and bound to DNA, accessibility of T65 is decreased for Stk1 phosphorylation and CovR inactivation (see Fig. 7). Detailed mechanistic insights into how phosphorylation at T65 affects CovR phosphorylation at D53 and the subsequent effect on promoter binding and interaction with the transcriptional machinery are not completely understood and require further study.
Fig. 7.
Proposed model of Stk1 and CovS regulation of CovR. CovR requires D53 in order to bind DNA and repress or activate transcription of β-H/C and CAMP factor respectively. Phosphorylation by small molecule phosphodonors like acetyl phosphate (AcP) or its cognate kinase CovS converts CovR to the active form. Stk1 phosphorylates the inactive form of CovR, as phosphorylation at D53 or DNA binding decrease the reactivity of T65 to Stk1. As a consequence of T65 phosphorylation, CovR is prevented from binding to DNA and D53 exhibits reduced reactivity.
Homology modelling of CovR with the related response regulator RegX3 of Mycobacterium tuberculosis indicates that T65 is located in the a helix 3 in the N-terminal receiver domain of CovR (see Fig. S1). Previous studies have described that residues in α helix 3 of response regulators can function in allosteric interactions. Fluorescent labelling of OmpR at its lone cysteine at position 67 in α helix 3 was reported to inhibit phosphorylation of the conserved aspartate at position 55 and vice versa (Ames et al., 1999). Our studies provide in vivo relevance that phosphorylation of a threonine residue in α helix 3 by a STK can regulate aspartate phosphorylation and CovR function.
As T65 is conserved in CovR among GBS serotypes (Tettelin et al., 2001; 2002; 2005; Glaser et al., 2002; Lamy et al., 2004; Jiang et al., 2008) and is important for its regulation by Stk1, we also examined whether T65 was conserved in response regulators that are similar to CovR and are present in bacteria that harbour a STK. Of note, the role of CovR/S in GBS was elucidated based on its homology to the extensively characterized CovR/S system in GAS (Levin and Wessels, 1998; Federle et al., 1999; Heath et al., 1999; Federle and Scott, 2002; Graham et al., 2002; Kreikemeyer et al., 2003; Dalton and Scott, 2004; Lamy et al., 2004; Jiang et al., 2005). Although the GAS CovR has a threonine residue at position 65, the GBS Stk1 enzyme did not phosphorylate GAS CovR (W.-J. Lin et al., unpublished data). Because the amino acids immediately flanking the target S or T residue play a critical role in recognition by the STK (Yang and Huang, 1994; Aitken, 2003; Huang et al., 2003; Schwartz and Gygi, 2005; Remenyi et al., 2006), we speculate that the valine residue at position 64 in GAS CovR (in contrast to the isoleucine in GBS, Fig. S2) contributed to the absence of phosphorylation. Whether GAS CovR is regulated by its cognate STK (SP-STK) (Jin and Pancholi, 2006) remains to be investigated.
CovR homologues from other Streptococcal sp. that encode STK such as S. pneumoniae and S. mutans contain either an alanine or an isoleucine residue at position 65 (see Fig. S2). Thus, these regulators are not subject to STK phosphorylation at this residue. While T65 is present in only three of the 21 other response regulators in the GBS serotype Ia, the amino acids that flank T65 in these regulators are not similar to CovR (data not shown, see Tettelin et al., 2005 for genome sequence). These observations suggest that CovR is the predominant regulator controlled by Stk1 phosphorylation in GBS. Recently, the S/T kinase StkP of S. pneumoniae was described to phosphorylate the response regulator RitR (Ulijasz et al., 2008). These studies showed that StkP phosphorylated RitR at the C-terminal DNA binding domain. However, the amino acid residue/s in RitR that is phosphorylated was not identified and the functional consequence of StkP phosphorylation of RitR is not understood. As an asparagine residue replaces the conserved aspartate at the active site of RitR (see Fig. S2) and StkP phosphorylates the C-terminal domain of RitR (Ulijasz et al., 2008), these observations suggest that S/T phosphorylation does not affect RitR similar to the GBS CovR. Mechanistic insight on how StkP phosphorylation affects RitR function remains to be elucidated and will provide information on how this interaction affects virulence factor expression and pneumococcal pathogenesis.
It is well known that bacteria modulate expression of their virulence factors because constitutive expression can be as detrimental as the absence of virulence gene expression. This phenomenon is well established in Salmonella wherein constitutive expression of virulence factors by TCS causes attenuation in disease similar to the lack of virulence factors (Miller and Mekalanos, 1990; Mouslim et al., 2004). Thus, it is advantageous for pathogens to regulate gene expression in response to the host environment. As CovR is a global regulator that affects the expression of at least 150 genes in the GBS serotypes 1a, III and V (Lamy et al., 2004; Jiang et al., 2005; 2008), modulation of its activity is critical for disease pathogenesis. Cognate sensor kinases play a crucial role in control of response regulator function. Although the sensor kinases Stk1 and CovS only moderately regulate CovR during GBS growth in laboratory media (Lamy et al., 2004; Rajagopal et al., 2006; Jiang et al., 2008), we predict that GBS may preferentially use either Stk1 or CovS to regulate CovR during disease pathogenesis. This additional level of regulation can be particularly advantageous for control of toxins such as β-H/C and CAMP factor.
The pluripotent toxin β-H/C is critical for various facets of GBS disease including entry and invasion of host cells and also in evasion of host defences (Nizet et al., 1996; 1997; Gibson et al., 1999; Spellerberg et al., 2000; Puliti et al., 2000; Ring et al., 2000; Pritzlaff et al., 2001; Doran et al., 2002; 2003; Nizet, 2002; Spellerberg, 2003). Interestingly, the normal human pulmonary surfactant Dipalmitoylphosphatidylcholine (DPPC) has antagonistic properties to this toxin (Liu et al., 2004; Hensler et al., 2008a), indicating that expression of β-H/C in a normal human lung can be detrimental to GBS. Furthermore, a recent study showed that although previously presumed important for many facets of GBS disease (Skalka and Smola, 1981; Jurgens et al., 1985; 1987; Tapsall and Phillips, 1987; Podbielski et al., 1994; Takaisi-Kikuni et al., 1997; Lang and Palmer, 2003), CAMP factor is dispensable for systemic virulence (Hensler et al., 2008b). It is therefore conceivable that opposing regulation of the two cytotoxins achieved through CovR/S and Stk1 is important for GBS pathogenesis. We predict that in certain host niches (e.g. during invasion of host tissues), expression of CovR-repressed genes like β-H/C and repression of CovR-activated genes like CAMP factor are important for GBS. Under these conditions, Stk1 can sense environmental cues to phosphorylate CovR at T65; phosphorylation at T65 will diminish CovR phosphorylation and activation at D53 by CovS or low-molecular-weight phosphodonors. Thus, Stk1 alleviates CovR-mediated repression and activation of GBS genes respectively (see Fig. S3). Conversely, in other host niches (e.g. as a colonizer on epithelial or mucosal surfaces and in DPPC environments), enhanced CovR-mediated repression and activation might be advantageous for survival of GBS. Consequently, GBS can use CovS to sense these environments to enhance CovR activity (see Fig. S3). Such co-ordinated regulation of gene/virulence factor expression will be beneficial to the pathogen during its disease cycle. Whether or not Stk1 preferentially affects CovS phosphorylation of CovR or CovR phosphorylation by low-molecular-weight phosphodonors in vivo is under investigation and will enhance our understanding on the link between STK and TCS signalling.
It is important to note that despite the plethora of information available on genes regulated by TCS, little is known about the extracellular signals that trigger the sensor kinases of TCS. Extracellular signals that have been identified include chemical gradients, pH and quorum sensing or antimicrobial peptides (for reviews, see Lyon and Novick, 2004; Mascher et al., 2006; Gunn and Richards, 2007). Although the GAS sensor kinase CovS responds to the extracellular Mg2+ concentration (Gryllos et al., 2003; 2007), the GBS CovS does not (Jiang et al., 2005). External signals that trigger STK in prokaryotes are not known. Identification of the signals that specifically trigger CovS or Stk1 to regulate CovR function will provide important clues on intertwined signalling networks and their role in the GBS virulence programme.
In summary, the results presented in this study indicate that phosphorylation at a threonine residue can decrease aspartate phosphorylation and DNA binding of the GBS response regulator CovR. Conversely, aspartate phosphorylation and DNA binding reduce threonine phosphorylation of CovR. These results suggest that two different phosphorylation events can affect promoter DNA binding of a response regulator with opposite outcomes and is likely to be beneficial for environmental adaptation of GBS and possibly other organisms that encode both STK and response regulators.
Experimental procedures
Bacterial strains and growth conditions
All strains, plasmids and primers used in this study are listed in Table S1. GBS strains were cultured in Tryptic Soy Broth (Difco Laboratories, Detroit, MI) at 37°C in the presence of 5% CO2 unless indicated otherwise. The WT GBS strain used in this study is A909 (Madoff et al., 1991). A909 is a clinical isolate of GBS that belongs to the capsular polysaccharide serotype Ia (Madoff et al., 1991). LR128 is isogenic to A909 where an allelic replacement of covR with sp. abolished CovR expression (Rajagopal et al., 2006). Routine cultures of Escherichia coli were performed in Luria–Bertani broth (Difco Laboratories, Detroit, MI) at 37°C. Cell growth was monitored by reading optical density at 600 nm. Antibiotics were added at the following concentrations when necessary: for E. coli, ampicillin 100 µg ml−1, kanamycin 50 µg ml−1, erythromycin 300 µg m−1 and spectinomycin 50 µg ml−1; for GBS, kanamycin 1000 µg ml−1, erythromycin 1 µg ml−1 and spectinomycin 300 µg ml−1. All chemicals were purchased from Sigma-Aldrich (St Louis, MO) and molecular biology reagents were purchased from New England Biolabs or Promega Corporation, USA.
Construction of truncated and site-directed CovR mutants
The plasmid pLR136 encoding the full-length CovR with a C-terminal His-tag (Rajagopal et al., 2006) was used as the template. Inverse PCR was performed with primers CovR NF and CovR NR to generate a plasmid pSL1 (see Table S1) that encodes a N-terminal truncated CovR protein encoding amino acid residues from M1-Q120 (also called N120). Similarly, inverse PCR was performed with primers CovR CF and CovR CR to generate the plasmid pSL2 that encodes a C-terminal truncated CovR encoding amino acids from E121-K229 (also called C108). The plasmid pSL2 was designed to contain a methionine start codon directly upstream of E121. All primers were designed to generate an in-frame HindIII restriction site at the junction that did not alter the coding sequence of CovR in either N120 or C108. Inverse PCR was performed using the following conditions: 98°C 2 min; 30 cycles each at 98°C 10 s, 55°C 15 s, 72°C 5 min 36 s, one cycle at 72°C for 7 min. Site-directed mutants were derived using either pSL1 or pLR136 (Rajagopal et al., 2006) as the template. The QuickChange II Site-Directed Mutagenesis Kit from Stratagene was used to generate CovR mutants following the manufacturer’s instructions. Plasmids pSL3 to pSL8 that contain CovR amino acid substitutions are listed in Table S1. All plasmids were sequenced to confirm the substitution mutation, and the absence of other mutations in the constructs.
Protein purification and in vitro phosphorylation assays
Purification of recombinant glutathione S transferase (GST) Stk1 fusion protein and WT and mutant CovR proteins were performed as described previously (Rajagopal et al., 2006). In vitro phosphorylation reactions were also carried out as described (Rajagopal et al., 2006). Briefly, approximately 0.5–1 µg of purified Stk1 was incubated with equal amounts of CovR in kinase buffer (25 mM Tris-HCl pH 8.0, 10 mM MgCl2, 3 µM ATP) containing 10 µCi [γ-32P]-ATP (PerkinElmer, Waltham, MA) for 15 min. Controls included reactions not containing Stk1 and those containing Stk1 alone. Subsequently, all reactions were heated to 100°C for 5 min as phosphoserine and phosphothreonine are heat stable unlike phosphohistidine and phosphoaspartate (Rosenberg, 1996; Ritte et al., 2002). The samples were then analysed either on 12% or 15% SDS-PAGE, stained with Coomassie and exposed to autoradiography. All experiments were repeated three times for reproducibility. For in vitro phosphorylation in the presence of DNA, CovR was incubated with either CovR specific or non-specific DNA as described for EMSA below. In vitro phosphorylation was then performed as described above.
Construction of GBS chromosomal substitution mutations
Primers comprising either the T65E or T65A alleles were used to amplify a segment of covR and 1Kb of flanking DNA from the 5′ and 3′ end respectively. A909 genomic DNA was used as the template and primer pairs for the T65A construct were VectorF and T65AR and T65AF and VectorR respectively. Likewise, the primers pairs for the T65E construct were VectorF and T65ER and T65EF and VectorR respectively. The resulting PCR fragments that overlapped at position 65 of CovR were ligated using SOEing PCR (Horton, 1995) to generate one large DNA fragment that comprised (i) the entire covR gene; (ii) the desired mutation in covR; and (iii) 1 kb of flanking DNA on both ends of covR. The PCR products had unique restriction sites engineered at the 5′ and 3′ ends of the fragment (NotI and XhoI) and were digested and cloned into NotI and XhoI-linearized pBluescript II KS+ to generate pSL10 encoding T65A CovR and pSL11 encoding T65E CovR respectively. These plasmids were sequenced to confirm the desired covR mutation and the insert DNA from pSL10 and pSL11 were subsequently cloned into NcoI and XhoI-linearized pJRS233 to generate pSL12 and pSL13 respectively. The plasmid pJRS233 is temperature sensitive and is routinely used for construction of mutants in Streptococci (Perez-Casal et al., 1993; Jiang et al., 2005). The plasmids pSL12 and pSL13 were also sequenced to confirm the presence of T65A and T65E substitutions in CovR and were then transformed into the GBS strain LR128; LR128 has an allelic replacement of covR with a gene that confers resistance to spectinomycin (Rajagopal et al., 2006). Selection for single cross-overs and screening for the double cross-over mutants were performed as described (Jiang et al., 2005) with a few modifications. Briefly, at least four independent transformants of each covR allele (T65E or T65A) in LR128 were grown in TSB containing erythromycin (encoded on the plasmid) at 30°C. The cultures were diluted 1:20 in liquid broth in the presence of erythromycin and grown overnight at 37°C. Overnight cultures were serially diluted and plated for single colonies on solid media at 37°C, i.e. TSA containing erythromycin. The single cross-overs or plasmid integrants were re-streaked on TSA containing erythromycin at 37°C. Five serial passages were performed for each integrant at 30°C in the absence of antibiotics as described previously (Jiang et al., 2005). The cultures were then diluted 1:20 in TSB and grown at 37°C till the OD600 reached ~0.6; erythro-mycin was included to the medium and the cultures were incubated for a period of 1 h to promote the growth of any remaining plasmid integrants. Subsequently, D-cycloserine was added to the cultures at a concentration of 100 µg ml−1 and incubated overnight at 37°C. The bacteria were serially diluted and plated on TSA without antibiotics. Colonies that were sensitive to both erythromycin and spectinomycin were selected as potential T65E (LR161) and T65A (LR160) strains. PCR was performed to confirm the presence of the covR gene and DNA sequencing was used to confirm the presence of the desired mutation/s. Subsequently, qRT-PCR was also performed to confirm covR and covS expression in these strains.
The D53A mutant LR159 was also derived as described above. Primer pairs used to construct the D53A mutation were VectorF and D53AR and D53AF and VectorR. The PCR fragments were ligated into pBluescript to generate pSL14. Plasmid pSL14 was used as a template to generate the double D53A\T65A and D53A\T65E substitutions by site-directed mutagenesis as described previously. Selection for single integrants and screening for the double cross-over events in GBS were performed as described above. The strains LR162 and LR163 encode D53A/T65E and D53A/T65A CovR respectively. PCR and DNA sequencing were used to confirm the presence of the desired mutation in CovR and qRT-PCR was used to confirm covR/S transcription.
Isolation and purification of total RNA
Total RNA was isolated using the RNeasy Mini kit (QIAGEN, Valencia, CA). In brief, GBS strains were grown to an OD600 of 0.6, centrifuged and washed in 1:1 mixed RNA Protect and TE buffer (QIAGEN, Valencia, CA). The cells were then resuspended in RLT buffer (QIAGEN, Valencia, CA) and lysed using a FastPrep FP101 bead beater (Bio 101) with 3 × 30 s bursts at a power setting of 6, followed by centrifugation. RNA was isolated from the supernatants using the RNeasy Mini kit as described by the manufacturer (QIAGEN, Valencia, CA). RNA concentration and integrity was determined using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA) and a NanoDrop 1000 (NanoDrop, Wilmington, DE) for subsequent use in qRT-PCR.
qRT-PCR
qRT-PCR was performed using a one-step QuantiTect SYBR Green RT-PCR kit (QIAGEN, Valencia, CA) following the manufacturer’s instructions for a total reaction volume of 20 µl (iCycler thermocycler; Bio-Rad, Hercules, CA). Amplification was performed using the following cycling protocol: denature at 95°C for 15 min, hold at 50°C for 20 min, followed by 40 cycles of a three-step profile consisting of denaturing at 94°C for 15 s, annealing at 52°C for 25 s, and extension at 72°C for 20 s.All runs were immediately followed by a melting curve analysis to evaluate PCR specificity, and showed single primer-specific melting temperatures. All assays were performed in triplicate with three independent biological replicates and three technical replicates for each biological replicate. All primers were calculated using Primer3 services (http://frodo.wi.mit.edu/), purchased from Integrated DNA Technologies (Coralville, IA) and are listed in Table S1. Relative expression was calculated according to the Bio-Rad Real-Time PCR Applications Guide section on the ΔCT method using a reference gene (Bio-Rad, 2006). The reference gene used for all runs was the housekeeping ribosomal protein S12 gene rpsL (Jiang et al., 2005) and statistical significance was determined as described previously (Rajagopal et al., 2006).
Electrophoretic mobility shift assay
Primers to amplify the CovR-regulated β-H/C promoter PcylX for EMSA and DNase I footprinting are listed in Table S1. The promoter region was PCR amplified using A909 genomic DNA as the template and Pfu DNA Polymerase (Stratagene, La Jolla, CA). The PCR fragment was purified using the QIAquick PCR Purification Kit (QIAGEN, Valencia, CA). End-labelling of the PCR fragment was performed using T4 Polynucleotide Kinase (New England Biolabs, Ipswich, MA) and 125 µCi [γ-32P]-ATP (PerkinElmer, Waltham, MA) as described (Ausubel et al., 1997). Unincorporated nucleotides were removed using a ProbeQuant G-50 Micro Column (Amersham Bioscience, Piscataway, NJ) and specific activity was determined (SA, unit: c.p.m. µl−1) using a Beckman-Coulter LS6500 scintillation counter. Binding reactions were carried out in a final volume of 10 ml in binding buffer that contained 2.5 mM sodium phosphate buffer, 50 mM NaCl, 2 mM MgCl2, 0.05 µg ml−1 poly DI-DC, 0.3 µg µl−1 BSA, 10% glycerol as described (Lamy et al., 2004). Binding reaction mixtures were added to a constant amount of probe DNA (20 000 c.p.m. µl−1) and incubated at 37°C for 15 min. Subsequently, the samples were resolved on 5% PAGE with TBE for 1.4 h at 4°C and 120 V. All TBE-5% PAGE gels were pre-run for 30 min at 4°C immediately prior to use. Dried gels were exposed to the phosphorimager Typhoon (Amersham Bioscience, Piscataway, NJ). To increase specificity of the EMSA, all binding reactions also contained 0.5 pmole of unlabelled, non-specific (non-CovR) binding DNA amplified from GBS using the primers NSPF and NSPR (see Table S1). For acetyl phosphate treatment, equimolar concentrations of WT, T65E and T65A CovR were incubated with 32 mM acetyl phosphate for 90 min at 37°C as described (Jiang et al., 2008) prior to the binding reactions. For threonine phospho-rylation of CovR in the presence of Stk1, Stk1 was bound to the GST sepharose column as described previously (Rajagopal et al., 2006). Subsequently 10 µg of CovR protein was phosphorylated by Stk1 that remained bound to the GST column in the presence of 10 mM ATP in kinase buffer (see above). The sample was centrifuged and the supernatant that is enriched for threonine phoshorylated CovR was then treated with acetyl phosphate and subsequently used in the EMSA as described previously.
DNase I footprinting
Templates for DNase I footprinting and DNA sequencing ladders were prepared as described (Walthers et al., 2007). The primers used for amplification of the PcylX promoter are listed in Table S1 and GBS genomic DNA was used as the template. Purified CovR was incubated in 50 mM Tris-HCl pH 7.5, 50 mM KCl, 20 mM MgCl2 (Gusa and Scott, 2005) and 25 mM phosphoramidate for 1 h at room temperature. After incubation in the presence of phosphodonor, CovR was added to binding reactions (final volume 20 µl) containing 50 000 c.p.m. of labelled DNA, 1 µg poly[d(I-C)], 10% glycerol, 10 mM MgCl2, 25 mM Tris-HCl pH 8.0 and 0.3 µg µl−1 of BSA. The reactions were incubated for 20 min at 37°C. Subsequently, 0.025 U DNase I (Roche) in a volume of 5 µl of 1× reaction buffer was added and incubated for another 30 s at room temperature prior to the addition of 5 ml of stop buffer [1.95 M NaAc pH 5.7, 125 mM EDTA and 2.5 µg of glycogen (Roche)]. The samples were ethanol precipitated and resuspended in loading dye containing 95% formamide, 25 mM EDTA and 0.05% each of xylene cyanol and bromophenol blue. Reactions were resolved on an 8% polyacrylamide gel containing 1× TBE and 7.5 M urea and analysed using a phosphorimager.
Phosphorylation of CovR with [32P]-acetyl phosphate
Synthesis of [32P]-acetyl phosphate was accomplished using carrier-free [32P]-orthophosphate (10 mCi ml−1, Perkin Elmer) as described (McCleary and Stock, 1994) with the minor modification that the entire protocol was reduced by half. The precipitated [32P]-acetyl phosphate was dissolved in a final volume of 100 µl of 100 mM Tris-HCl, pH 7.0 and the concentration was estimated using a modification of the colorimetric assay described (Lipmann and Tuttle, 1945). Briefly, neutralized hydroxylamine solution was prepared fresh by combining an equal volume of 28% hydrolyamine hydrochloride and 14% NaOH. Lithium potassium acetyl phosphate (Sigma) was used as the standard for the assay. Various concentrations of acetyl phosphate (Sigma) or synthesized [32P]-acetyl phosphate in a final volume of 50 µl were combined with an equal volume of neutralized hydroxylamine and the reaction was incubated at room temperature for 10 min. Subsequently, the samples were acidified by the addition of 50 µl of 1:3 solution of concentrated HCl : dH20 followed by the addition of 50 µl of 5% FeCl3.6H2O in 0.1 N HCl. The absorbance of the purple iron complex was read at 540 nm. The specific activity of the synthesized [32P]-acetyl phosphate was determined (SA, c.p.m. µl−1) using a Beckman-Coulter LS6500 scintillation counter. For CovR phosphorylation, 1 µg of either WT or CovR substitution proteins was incubated for 90 min at 37°C with 32 mM acetyl phosphate (specific activity 8.5 mCi mmol−1) in a final volume of 20 µl. Subsequently, SDS-PAGE sample buffer was added to a final concentration of 1× and the samples were analysed on a 12% SDS-PAGE and exposed to the phosphorimager as described for other response regulators (Lukat et al., 1992).
Supplementary Material
Acknowledgements
This work was supported by funding from the National Institutes of Health, Grant # RO1 AI070749 to L.R. and MCB-0613014 from the NSF and NIH GM058746 to L.J.K. We thank Dr Lakshminarayan M. Iyer (NCBI, NLM, NIH), Donald Chaffin (Seattle Children’s Hospital Research Institute (SCHRI) and Joyce Karlinsey (University of Washington) for their advice and support. We are grateful to Kristy Seidel for statistical assistance (SCHRI) and Thao Tran for technical support. We thank Drs June Scott and Gordon Churchward (Emory University) for helpful discussions on CovR binding and the kind gift of GAS CovR.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aitken A. Protein consensus sequence motifs. Methods Mol Biol. 2003;211:465–485. doi: 10.1385/1-59259-342-9:465. [DOI] [PubMed] [Google Scholar]
- Ames SK, Frankema N, Kenney LJ. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc Natl Acad Sci USA. 1999;96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. [Google Scholar]
- Baker CJ, Edwards MW. Group B streptococcal infections. In: Remington JS, Klein JO, editors. Infectious Diseases of the Fetus and Newborn Infant. Philadelphia, PA: W.B. Saunders; 1995. pp. 980–1054. [Google Scholar]
- Barz C, Abahji TN, Trulzsch K, Heesemann J. The Yersinia Ser/Thr protein kinase YpkA/YopO directly interacts with the small GTPases RhoA and Rac-1. FEBS Lett. 2000;482:139–143. doi: 10.1016/s0014-5793(00)02045-7. [DOI] [PubMed] [Google Scholar]
- Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol. 2006;9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Bio-Rad. Real-Time PCR Applications Guide. Hercules, CA: Bio-Rad Laboratories Inc. Life Science Group; 2006. [Google Scholar]
- Bourret RB. Signal transduction meets systems biology: deciphering specificity determinants for protein-protein interactions. Mol Microbiol. 2008;69:1336–1340. doi: 10.1111/j.1365-2958.2008.06379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaba R, Raje M, Chakraborti PK. Evidence that a eukaryotic-type serine/threonine protein kinase from Mycobacterium tuberculosis regulates morphological changes associated with cell division. Eur J Biochem. 2002;269:1078–1085. doi: 10.1046/j.1432-1033.2002.02778.x. [DOI] [PubMed] [Google Scholar]
- Chang C, Meyerowitz EM. The ethylene hormone response in Arabidopsis: a eukaryotic two-component signaling system. Proc Natl Acad Sci USA. 1995;92:4129–4133. doi: 10.1073/pnas.92.10.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Cohen P. Protein kinases – the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- Cowley S, Ko M, Pick N, Chow R, Downing KJ, Gordhan BG, et al. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol. 2004;52:1691–1702. doi: 10.1111/j.1365-2958.2004.04085.x. [DOI] [PubMed] [Google Scholar]
- Dalton TL, Scott JR. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J Bacteriol. 2004;186:3928–3937. doi: 10.1128/JB.186.12.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Saier MH., Jr Ser/Thr/Tyr protein phosphorylation in bacteria – for long time neglected, now well established. J Mol Microbiol Biotechnol. 2005;9:125–131. doi: 10.1159/000089641. [DOI] [PubMed] [Google Scholar]
- Doran KS, Nizet V. Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol Microbiol. 2004;54:23–31. doi: 10.1111/j.1365-2958.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- Doran KS, Chang JC, Benoit VM, Eckmann L, Nizet V. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J Infect Dis. 2002;185:196–203. doi: 10.1086/338475. [DOI] [PubMed] [Google Scholar]
- Doran KS, Liu GY, Nizet V. Group B streptococcal beta-hemolysin/cytolysin activates neutrophil signaling pathways in brain endothelium and contributes to development of meningitis. J Clin Invest. 2003;112:736–744. doi: 10.1172/JCI17335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ, Scott JR. Identification of binding sites for the group A streptococcal global regulator CovR. Mol Microbiol. 2002;43:1161–1172. doi: 10.1046/j.1365-2958.2002.02810.x. [DOI] [PubMed] [Google Scholar]
- Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillebeen C, Chahine D, Caltagirone A, Segal P, Pantopoulos K. A phosphomimetic mutation at Ser-138 renders iron regulatory protein 1 sensitive to iron-dependent degradation. Mol Cell Biol. 2003;23:6973–6981. doi: 10.1128/MCB.23.19.6973-6981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Gusa AA, Scott JR, Churchward G. Binding of the global response regulator protein CovR to the sag promoter of Streptococcus pyogenes reveals a new mode of CovR-DNA interaction. J Biol Chem. 2005;280:38948–38956. doi: 10.1074/jbc.M506121200. [DOI] [PubMed] [Google Scholar]
- Gibson RL, Nizet V, Rubens CE. Group B streptococcal beta-hemolysin promotes injury of lung microvascular endothelial cells. Pediatr Res. 1999;45:626–634. doi: 10.1203/00006450-199905010-00003. [DOI] [PubMed] [Google Scholar]
- Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, Msadek T, et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol. 2002;45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, et al. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryllos I, Levin JC, Wessels MR. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+ Proc Natl Acad Sci USA. 2003;100:4227–4232. doi: 10.1073/pnas.0636231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryllos I, Grifantini R, Colaprico A, Jiang S, Deforce E, Hakansson A, et al. Mg(2+) signalling defines the group A streptococcal CsrRS (CovRS) regulon. Mol Microbiol. 2007;65:671–683. doi: 10.1111/j.1365-2958.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- Gunn JS, Richards SM. Recognition and integration of multiple environmental signals by the bacterial sensor kinase PhoQ. Cell Host Microbe. 2007;1:163–165. doi: 10.1016/j.chom.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Gusa AA, Scott JR. The CovR response regulator of group A streptococcus (GAS) acts directly to repress its own promoter. Mol Microbiol. 2005;56:1195–1207. doi: 10.1111/j.1365-2958.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- Gusa AA, Gao J, Stringer V, Churchward G, Scott JR. Phosphorylation of the group A Streptococcal CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. J Bacteriol. 2006;188:4620–4626. doi: 10.1128/JB.00198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson S, Galyov EE, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- Heath A, DiRita VJ, Barg NL, Engleberg NC. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect Immun. 1999;67:5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensler ME, Miyamoto S, Nizet V. Group B streptococcal beta-hemolysin/cytolysin directly impairs cardiomyocyte viability and function. PLoS ONE. 2008a;3:e2446. doi: 10.1371/journal.pone.0002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensler ME, Quach D, Hsieh CJ, Doran KS, Nizet V. CAMP factor is not essential for systemic virulence of Group B Streptococcus. Microb Pathog. 2008b;44:84–88. doi: 10.1016/j.micpath.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J, Silhavy T. Two-Component Signal Transduction. Washington, DC: American Society for Microbiology; 1995. [Google Scholar]
- Horton RM. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- Huang B, Zeng G, Ng AY, Cai M. Identification of novel recognition motifs and regulatory targets for the yeast actin-regulating kinase Prk1p. Mol Biol Cell. 2003;14:4871–4884. doi: 10.1091/mbc.E03-06-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SM, Cieslewicz MJ, Kasper DL, Wessels MR. Regulation of virulence by a two-component system in group B streptococcus. J Bacteriol. 2005;187:1105–1113. doi: 10.1128/JB.187.3.1105-1113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SM, Ishmael N, Hotopp JD, Puliti M, Tissi L, Kumar N, et al. Variation in the group B Streptococcus CsrRS regulon and effects on pathogenicity. J Bacteriol. 2008;190:1956–1965. doi: 10.1128/JB.01677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Pancholi V. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J Mol Biol. 2006;357:1351–1372. doi: 10.1016/j.jmb.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Jurgens D, Shalaby FY, Fehrenbach FJ. Purification and characterization of cAMP-factor from Streptococcus agalactiae by hydrophobic interaction chromatography and chromatofocusing. J Chromatogr. 1985;348:363–370. doi: 10.1016/s0021-9673(01)92474-4. [DOI] [PubMed] [Google Scholar]
- Jurgens D, Sterzik B, Fehrenbach FJ. Unspecific binding of group B streptococcal cocytolysin (CAMP factor) to immunoglobulins and its possible role in pathogenicity. J Exp Med. 1987;165:720–732. doi: 10.1084/jem.165.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juris SJ, Rudolph AE, Huddler D, Orth K, Dixon JE. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc Natl Acad Sci USA. 2000;97:9431–9436. doi: 10.1073/pnas.170281997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ. Protein kinases and protein phos-phatases in prokaryotes: a genomic perspective. FEMS Microbiol Lett. 2002;206:1–8. doi: 10.1111/j.1574-6968.2002.tb10978.x. [DOI] [PubMed] [Google Scholar]
- Kenney LJ, Bauer MD, Silhavy TJ. Phosphorylation-dependent conformational changes in OmpR, an osmoregulatory DNA-binding protein of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:8866–8870. doi: 10.1073/pnas.92.19.8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol. 2007;189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A, Choidas A, Tyagi AK, Drlica K, Singh Y, Ullrich A. Serine/threonine protein kinases PknF and PknG of Mycobacterium tuberculosis: characterization and localization. Microbiology. 2001;147:2307–2314. doi: 10.1099/00221287-147-8-2307. [DOI] [PubMed] [Google Scholar]
- Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- Krishna RG, Wold . Posttranslational modifications. In: Angeletti RH, editor. Protein Analysis and Design. San Diego, CA: Academic Press; 1998. pp. 121–206. [Google Scholar]
- Kristich CJ, Wells CL, Dunny GM. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci USA. 2007;104:3508–3513. doi: 10.1073/pnas.0608742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamy MC, Zouine M, Fert J, Vergassola M, Couve E, Pellegrini E, et al. CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence. Mol Microbiol. 2004;54:1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x. [DOI] [PubMed] [Google Scholar]
- Lang S, Palmer M. Characterization of Streptococcus agalactiae CAMP factor as a pore-forming toxin. J Biol Chem. 2003;278:38167–38173. doi: 10.1074/jbc.M303544200. [DOI] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Lauberth SM, Bilyeu AC, Firulli BA, Kroll KL, Rauchman M. A phosphomimetic mutation in the Sall1 repression motif disrupts recruitment of the nucleosome remodeling and deacetylase complex and repression of Gbx2. J Biol Chem. 2007;282:34858–34868. doi: 10.1074/jbc.M703702200. [DOI] [PubMed] [Google Scholar]
- Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- Lipmann F, Tuttle CF. A specific micromethod for the determination of acyl phosphates. J Biol Chem. 1945;159:21–28. [Google Scholar]
- Liu GY, Doran KS, Lawrence T, Turkson N, Puliti M, Tissi L, Nizet V. Sword and shield: linked group B streptococcal beta-hemolysin/cytolysin and carotenoid pigment function to subvert host phagocyte defense. Proc Natl Acad Sci USA. 2004;101:14491–14496. doi: 10.1073/pnas.0406143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon GJ, Novick RP. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides. 2004;25:1389–1403. doi: 10.1016/j.peptides.2003.11.026. [DOI] [PubMed] [Google Scholar]
- McCleary WR, Stock JB. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- Madec E, Laszkiewicz A, Iwanicki A, Obuchowski M, Seror S. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol Microbiol. 2002;46:571–586. doi: 10.1046/j.1365-2958.2002.03178.x. [DOI] [PubMed] [Google Scholar]
- Madoff LC, Michel JL, Kasper DL. A monoclonal antibody identifies a protective C-protein alpha-antigen epitope in group B streptococci. Infect Immun. 1991;59:204–210. doi: 10.1128/iai.59.1.204-210.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K, Oropeza R, Byers N, Kenney LJ. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J Mol Biol. 2002;315:497–511. doi: 10.1006/jmbi.2001.5222. [DOI] [PubMed] [Google Scholar]
- Miller SI, Mekalanos JJ. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Kremer L, Girard-Blanc C, Besra GS, Cozzone AJ, Prost JF. An FHA phosphop-rotein recognition domain mediates protein EmbR phosphorylation by PknH, a Ser/Thr protein kinase from Mycobacterium tuberculosis. Biochemistry (Mosc) 2003;42:15300–15309. doi: 10.1021/bi035150b. [DOI] [PubMed] [Google Scholar]
- Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- Mouslim C, Delgado M, Groisman EA. Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence. Mol Microbiol. 2004;54:386–395. doi: 10.1111/j.1365-2958.2004.04293.x. [DOI] [PubMed] [Google Scholar]
- Nariya H, Inouye S. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol Microbiol. 2005;58:367–379. doi: 10.1111/j.1365-2958.2005.04826.x. [DOI] [PubMed] [Google Scholar]
- Nariya H, Inouye S. A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol Microbiol. 2006;60:1205–1217. doi: 10.1111/j.1365-2958.2006.05178.x. [DOI] [PubMed] [Google Scholar]
- Nizet V. Streptococcal beta-hemolysins: genetics and role in disease pathogenesis. Trends Microbiol. 2002;10:575–580. doi: 10.1016/s0966-842x(02)02473-3. [DOI] [PubMed] [Google Scholar]
- Nizet V, Gibson RL, Rubens CE. The role of group B streptococci beta-hemolysin expression in newborn lung injury. Adv Exp Med Biol. 1997;418:627–630. doi: 10.1007/978-1-4899-1825-3_146. [DOI] [PubMed] [Google Scholar]
- Nizet V, Gibson RL, Chi EY, Framson PE, Hulse M, Rubens CE. Group B streptococcal beta-hemolysin expression is associated with injury of lung epithelial cells. Infect Immun. 1996;64:3818–3826. doi: 10.1128/iai.64.9.3818-3826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko EA, Crafoord E, Hallberg E. Phosphomimetic mutation of the mitotically phosphorylated serine 1880 compromises the interaction of the transmem-brane nucleoporin gp210 with the nuclear pore complex. Exp Cell Res. 2007;313:2744–2751. doi: 10.1016/j.yexcr.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Pareek A, Singh A, Kumar M, Kushwaha HR, Lynn AM, Singla-Pareek SL. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 2006;142:380–397. doi: 10.1104/pp.106.086371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Casal J, Price JA, Maguin E, Scott JR. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- Podbielski A, Blankenstein O, Lutticken R. Molecular characterization of the cfb gene encoding group B streptococcal CAMP-factor. Med Microbiol Immunol (Berl) 1994;183:239–256. doi: 10.1007/BF00198458. [DOI] [PubMed] [Google Scholar]
- Poyart C, Lamy MC, Boumaila C, Fiedler F, Trieu-Cuot P. Regulation of D-alanyl-lipotechoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J Bacteriol. 2001;183:6324–6334. doi: 10.1128/JB.183.21.6324-6334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart C, Pellegrini E, Marceau M, Baptista M, Jaubert F, Lamy MC, Trieu-Cuot P. Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol Microbiol. 2003;49:1615–1625. doi: 10.1046/j.1365-2958.2003.03655.x. [DOI] [PubMed] [Google Scholar]
- Pritzlaff CA, Chang JC, Kuo SP, Tamura GS, Rubens CE, Nizet V. Genetic basis for the beta-haemolytic/cytolytic activity of group B Streptococcus. Mol Microbiol. 2001;39:236–247. doi: 10.1046/j.1365-2958.2001.02211.x. [DOI] [PubMed] [Google Scholar]
- Puliti M, Nizet V, von Hunolstein C, Bistoni F, Mosci P, Orefici G, Tissi L. Severity of group B streptococcal arthritis is correlated with beta-hemolysin expression. J Infect Dis. 2000;182:824–832. doi: 10.1086/315773. [DOI] [PubMed] [Google Scholar]
- Quach D, van Sorge NM, Kristian SA, Bryan JD, Shelver DW, Doran KS. The CiaR response regulator in Group B Streptococcus promotes intracellular survival and resistance to innate immune defenses. J Bac-teriol. 2008 doi: 10.1128/JB.01216-08. [Epub ahead of print] PMID: 19114476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggiaschi R, Gotta S, Terstappen GC. Phosphoproteome analysis. Biosci Report. 2005;25:33–44. doi: 10.1007/s10540-005-2846-0. [DOI] [PubMed] [Google Scholar]
- Rajagopal L, Clancy A, Rubens CE. Aeukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J Biol Chem. 2003;278:14429–14441. doi: 10.1074/jbc.M212747200. [DOI] [PubMed] [Google Scholar]
- Rajagopal L, Vo A, Silvestroni A, Rubens CE. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol Microbiol. 2006;62:941–957. doi: 10.1111/j.1365-2958.2006.05431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr Opin Struct Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Rhee JE, Sheng W, Morgan LK, Nolet R, Liao X, Kenney LJ. Amino acids important for DNA recognition by the response regulator OmpR. J Biol Chem. 2008;283:8664–8677. doi: 10.1074/jbc.M705550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring A, Braun JS, Nizet V, Stremmel W, Shenep JL. Group B streptococcal beta-hemolysin induces nitric oxide production in murine macrophages. J Infect Dis. 2000;182:150–157. doi: 10.1086/315681. [DOI] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. The starch-related R1 protein is an alpha -glucan, water dikinase. Proc Natl Acad Sci USA. 2002;99:7166–7171. doi: 10.1073/pnas.062053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg IM. Protein Analysis and Purification. Boston, MA: Birkhäuser; 1996. [Google Scholar]
- Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- Sharma K, Gupta M, Pathak M, Gupta N, Koul A, Sarangi S, et al. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J Bacteriol. 2006;188:2936–2944. doi: 10.1128/JB.188.8.2936-2944.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka B, Smola J. Lethal effect of CAMP-factor and UBERIS-factor – a new finding about diffusible exo-substances of Streptococcus agalactiae and Streptococcus uberis. Zentralbl Bakteriol A. 1981;249:190–194. [PubMed] [Google Scholar]
- Spellerberg B. CylI and group B streptococcal hemolysis. Trends Microbiol. 2003;11:497–498. doi: 10.1016/j.tim.2003.09.006. author reply 498. [DOI] [PubMed] [Google Scholar]
- Spellerberg B, Martin S, Brandt C, Lutticken R. The cyl genes of Streptococcus agalactiae are involved in the production of pigment. FEMS Microbiol Lett. 2000;188:125–128. doi: 10.1111/j.1574-6968.2000.tb09182.x. [DOI] [PubMed] [Google Scholar]
- Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Lutticken R. rgf encodes a novel two-component signal transduction system of Streptococcus agalactiae. Infect Immun. 2002;70:2434–2440. doi: 10.1128/IAI.70.5.2434-2440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Alonso JM. Arabidopsis ethylene signaling pathway. Sci STKE. 2005:cm4. doi: 10.1126/stke.2762005cm4. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Takaisi-Kikuni NB, Jurgens D, Wecke J, Fehren-bach FJ. Immunochemical localization of CAMP factor (protein B) in Streptococcus agalactiae. Microbios. 1997;89:171–185. [PubMed] [Google Scholar]
- Tapsall JW, Phillips EA. Presumptive identification of group B streptococci by rapid detection of CAMP factor and pigment production. Diagn Microbiol Infect Dis. 1987;7:225–228. doi: 10.1016/0732-8893(87)90010-1. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, Wessels MR, et al. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc Natl Acad Sci USA. 2002;99:12391–12396. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SA, Brewster JA, Bourret RB. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol. 2008;69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran VK, Oropeza R, Kenney LJ. A single amino acid substitution in the C terminus of OmpR alters DNA recognition and phosphorylation. J Mol Biol. 2000;299:1257–1270. doi: 10.1006/jmbi.2000.3809. [DOI] [PubMed] [Google Scholar]
- Ulijasz AT, Falk SP, Weisblum B. Phosphorylation of the RitR DNA-binding domain by a Ser-Thr Phosphokinase: Implications for global gene regulation in the streptococci. Mol Microbiol. 2008;71:382–390. doi: 10.1111/j.1365-2958.2008.06532.x. [DOI] [PubMed] [Google Scholar]
- Wagner LE, Li WH, Joseph SK, Yule DI. Functional consequences of phosphomimetic mutations at key cAMP-dependent protein kinase phosphorylation sites in the type 1 inosital 1,4,5-Trisphosphate receptor. J Biol Chem. 2004;279:46242–46252. doi: 10.1074/jbc.M405849200. [DOI] [PubMed] [Google Scholar]
- Walthers D, Carroll R, Navarre WW, Libby S, Fang FC, Kenney LJ. The resonse regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol. 2007;65:477–493. doi: 10.1111/j.1365-2958.2007.05800.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klemke RL. PhosphoBlast, a computational tool for comparing phosphoprotein signatures among large datasets. Mol Cell Proteomics. 2008;7:145–162. doi: 10.1074/mcp.M700207-MCP200. [DOI] [PubMed] [Google Scholar]
- Yang SD, Huang TJ. Identification of -R-X-(X)-S/T-X3-S/T- as consensus sequence motif for autophosphorylation-dependent protein kinase. J Biol Chem. 1994;269:29855–29859. [PubMed] [Google Scholar]
- Zhang ZG, Zhou HL, Chen T, Gong Y, Cao WH, Wang YJ, et al. Evidence for serine/threonine and histidine kinase activity in the tobacco ethylene receptor protein NTHK2. Plant Physiol. 2004;136:2971–2981. doi: 10.1104/pp.103.034686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.