Abstract
Histone methylations are important chromatin marks that regulate gene expression, genomic stability, DNA repair, and genomic imprinting. Histone demethylases are the most recent family of histone-modifying enzymes discovered. Here, we report the characterization of a small-molecule inhibitor of Jumonji C domain-containing histone demethylases. The inhibitor derives from a structure-based design and preferentially inhibits the sub-family of trimethyl lysine demethylases. Its methyl ester prodrug, methylstat, selectively inhibits Jumonji C domain-containing his-tone demethylases in cells and may be a useful small-molecule probe of chromatin and its role in epigenetics.
Keywords: epigenetics, histone methylation, histone demethylase, Jumonji C domain, structure-based design, small-molecule probe
INTRODUCTION
The covalent attachment of functional groups to chromatin, including DNA methylation and histone modifications, are associated with heritable changes that regulate cellular transcriptomes without altering DNA sequence. Histone methylation is one of the most important chromatin marks; these play important roles in transcriptional regulation, DNA-damage response, heterochromatin formation and maintenance, and X-chromosome inactivation.1 A recent discovery also revealed that histone methylation affects the splicing outcome of pre-mRNA by influencing the recruitment of splicing regulators.2 Histone methylation includes mono-, di-, and tri-methylation of lysines, and mono-, symmetric di-, and asymmetric di-methylation of arginines. These modifications can be activating or repressing, depending on the site and degree of methylation. Two classes of enzymes regulate the maintenance of histone methylation: histone methyltransferases (HMTs) and histone demethylases (HDMs). HDMs are the most recent family of histone-modifying enzymes discovered. Since the human HDM, LSD1, was first detected in 19983 and characterized in 2004,4 over a dozen HDMs have been discovered that modify histone H3 lysine 4 (H3K4), H3K9, H3K27, H3K36, H3R2, or H4R3 methylations.5 However, HDMs that specifically modify H3K79me3 and H4K20me3 have not yet been identified. Recent studies have shown that HDMs often display tissue-specific expression and play critical roles in gene expression, meiosis, and embryonic stem cell self-renewal.6
HDMs can be categorized into two classes based on their enzymatic mechanisms: flavin adenine dinucleotide (FAD)-dependent HDMs and Jumonji C domain-containing HDMs (JHDMs).5,7 There are two FAD-dependent HDMs, both of which are monoamine oxidases and can demethylate mono- and di-methylated H3K4 and H3K9.4,8 Compared with FAD-dependent HDMs, JHDMs appear to be more versatile in terms of their substrate specificities. These proteins are Fe2+- and α-ketoglutarate-dependent hydroxylases, and their reported substrate residues include H3K4, H3K9, H3K27, and H3K36 at all methylation states.5 As the DNA repair protein AlkB,9 JHDMs hydroxylate the C-H bond of methyl group, and the resulting hemiaminal collapses to form the demethylated product.
Small-molecule modulators of histone-modifying enzymes not only play important roles in understanding the structures and functions of these enzymes, but possibly also provide unique opportunities for treating diseases such as cancer and mental retardation.10 Small molecules specifically inhibiting FAD-dependent HDMs have been discovered recently.11 As with other Fe2+- and α-ketoglutarate-dependent hydroxylases, JHDMs are inhibited by general inhibitors such as desferoxamine (DFO, a metal-chelating agent),12 and α-ketoglutarate mimics N-oxalylglycine13 and pyridine-2,4-dicarboxylic acid.14 In addition, small-molecule inhibitors that show in vitro selectivity for JHDMs have been discovered.15 However, their cellular specificities have not been reported yet.
A number of JHDMs crystal structures have been solved, several of which are complexed with methyllysine-containing histone peptides and cofactor mimics.16 Based on these crystal structures and the enzymatic mechanism of JHDMs, we designed and synthesized potential JHDM-selective small-molecule inhibitors, each of which contains a methyllysine mimic (substrate mimic), an α-ketoglutarate mimic (cofactor mimic), and a linker combining these two (Figure 1). Herein, we characterize compound 1 (Figure 1) as a selective JHDM inhibitor in vitro, and its corresponding methyl ester prodrug 2 as a selective JHDM inhibitor in vivo.
Figure 1.
Structure-based design of a JHDM-selective inhibitor 1 and the structure of its methyl ester methylstat (2).
RESULTS AND DISCUSSION
Design and synthesis of JHDM inhibitor 1
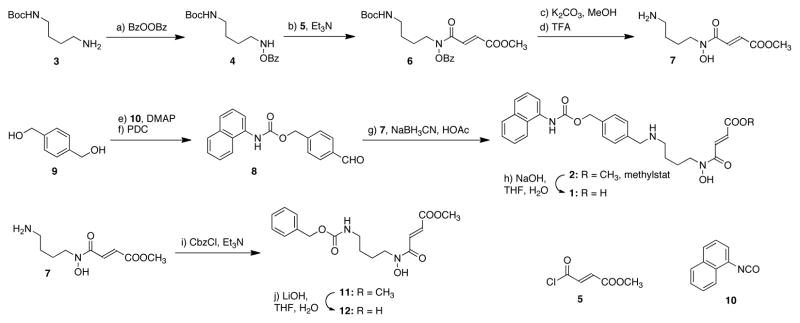
The lysine-mimicking fragment of compound 1 was derived from a well-known histone deacetylase (HDAC) inhibitor MS-275.17 The 4-carbon linker was selected based on the relative distance and geometry of α-ketoglutarate and methyl lysine in crystal structures. The synthesis of 1 began with oxidation of a commercially available amine, 3 (Scheme 1), using benzoyl peroxide to afford compound 4.18 Acylation of 4 with acyl chloride 5 gave amide 6, which was sequentially deprotected to afford amine 7 using potassium carbonate in anhydrous methanol and trifluoroacetic acid (TFA). Synthesis of the lysine-mimicking fragment 8 started with mono-carbamate formation of diol 9 with 2-naphthylene isocyanate 10. Oxidation of the remaining alcohol using pyridinium dichromate (PDC) provided an aldehyde 8, which underwent a reductive amination with amine 7 to afford methylstat (2). The corresponding acid 1 was prepared by hydrolysis of 2 using sodium hydroxide. In order to examine if under physiological conditions, the positively charged ammonium ion is an important functional group in the substrate mimicking fragment of 1, we also synthesized its analog 12 from amine 7 by a carbamate formation reaction followed by hydrolysis of the ester (Scheme 1).
Scheme 1.
Synthesis of the designed bivalent JHDM inhibitor 1 and related molecules.a
a Reagents and conditions: (a) dibenzoyl peroxide (1.2 equiv), buffer (pH = 10.5), CH2Cl2, 25 °C, 2.5 h, 75%; (b) 5 (1.3 equiv), oxalyl chloride (1.2 equiv), DMF (0.05 equiv), CH2Cl2,, 0 to 25 °C, 1 h; then Et3N (2.0 equiv), DMAP (0.01 equiv), CH2Cl2, 0 to 25 °C, 2 h, 72%; (c) K2CO3 (1.2 equiv), MeOH, 30 min; (d) TFA (20 equiv), CH2Cl2, 0 to 25 °C, 2 h; (e) 10 (0.3 equiv), DMAP (0.016 equiv), DMF, 25 °C, 12 h, 56%; (f) PDC (1.0 equiv), 4 Å molecular sieves, CH2Cl2, 25 °C, 5 h, 85%; (g) NaBH3CN (5.0 equiv), HOAc (2.0 equiv), MeOH, 25 °C, 12 h, 38% over 3 steps; (h) NaOH (2.0 equiv), THF, H2O, 25 °C, 3 h, 85%; (i) CbzCl (1.0 equiv), Et3N (2.0 equiv), CH2Cl2, 0 °C, 5 h, 82%; (j) LiOH·H2O (3.2 equiv), THF, H2O, 2 h, 87%. Abbreviations: Cbz = Carboxybenzyl, DMF = N,N-dimethylformamide, DMAP = 4-Dimethylaminopyridine, PDC = Pyridinium dichromate, TFA = trifluoroacetic acid.
In vitro profiling of compound 1
In order to evaluate the inhibitory activity of 1 in vitro, we cloned and purified a GST-fusion protein of the catalytic N-terminus of human JMJD2C(1–420).19 The JHDM, JMJD2C, predominantly uses H3K9me3 substrates, but has also been shown to demethylate H3K36me3. By using electrospray time-of-flight (ESI-TOF) mass spectrometry, we found this purified protein can remove the methyl group of a synthetic histone H3 fragment peptide substrate (H3K9me3, SI, Figure S1). Compound 1 shows a half maximal inhibitory concentration (IC50) for JMJD2C of approximately 4.3 μM determined by a dissociation-enhanced lanthanide fluorescent immunoassay (DELFIA, Figure 2a). It is noteworthy that compound 12 showed no inhibition of JMJD2C activity at up to 100 μM, the highest concentration tested. This result suggests that a positively charged substrate-mimicking fragment of our designed inhibitor 1 is important for its inhibitory activity against JHDMs.
Figure 2.

In vitro profiling of compound 1. (a) Enzyme inhibition data of compound 1 against H3K9me3 demethylases JMJD2A, JMJD2C, and JMJD2E, H3K9me2 demethylase PHF8, and H3K27me3 demethylase JMJD3. (b) Enzyme inhibition data of compound 1 against prolyl hydroxylases PHD1–3, factor inhibiting hypoxia-inducible factor FIH, an asparaginyl hydroxylase, FAD-dependent histone demethylase LSD1, and histone deacetylases HDACs.
To evaluate the enzyme selectivity of 1, we next examined its activity against several other JHDM isoforms in vitro. Compound 1 inhibits JMJD2A (H3K9me3 and H3K36me3 demethylase), JMJD2E (H3K9me3 demethylase)20 and JMJD3 (H3K27me3 demethylase)21 with IC50 values in the same order of magnitude (Figure 2a) as seen for JMJD2C; however, compound 1 shows considerably higher IC50 values against other subfamily members such as the dimethyl-specific H3K9 demethylase PHF8 (Figure 2a).22 Although comparison of IC50 values for different enzymes should be approached with caution, these data indicate that compound 1 might be in vitro a better inhibitor for the trimethyl demethylase families JMJD2 and JMJD3 compared to dimethyl demethylases such as FBXL11/PHF8 subfamily.
To further evaluate the specificity of compound 1, we tested it against a distinct Jumonji C domain-containing enzyme, the asparaginyl hydroxylase FIH,23 as well as other, even more distantly related α-ketoglutarate-dependent hydroxylases such as prolyl hydroxylases PHD1–3,24 also show higher IC50 values against compound 1 (Figure 2b). Although compound 1 contains a methyl lysine mimicking fragment, its IC50 against the FAD-dependent demethylase, LSD1, was determined as 620 μM (Figure 2b), which is over 100 folder higher than its IC50 against the JMJD2 histone demethylases. In addition, compound 1 does not inhibit class I or class II HDACs at up to 800 μM (Figure 2b), which are also transition metal-dependent histone lysine modifying enzymes.
Cellular activity and specificity of methylstat
To examine the cellular activity of the small molecules we synthesized, we chose to use JMJD2C-sensitive oesophageal carcinoma cell line KYSE150 in a growth inhibition assay.19 However, compound 1 did not show significant growth inhibition at up to 100 μM, possibly due to the poor cellular uptake arising from the high polarity of the zwitterions form in which it typically exists under physiological conditions (clogP −0.99). Therefore, we tested its methyl ester prodrug 2 (Figure 1, clogP 1.02),13,14 which we have named methylstat. Methylstat inhibited KYSE150 cell growth with a half maximal growth inhibitory concentration (GI50) at approximately 5.1 μM (Figure 3a). It should be noted that methylstat does not inhibit the enzymatic activities of JMJD2C, JMJD2A, LSD1, or HDACs at up to 50 μM, the highest concentration tested (SI, Figure S2). In order to test the stability of methylstat, it was incubated in KYSE150 cell growth media at 37 °C for 48 hours, then extracted using organic solvent (3:1 chloroform:isopropanol). We found that 88% of methylstat was recovered unchanged, the structure of which was confirmed by 1H NMR; 89% of compound 1 was recovered when this procedure was performed with 1.
Figure 3.
Methylstat as a selective JHDM inhibitor in vivo. (a) Methylstat, but not 1, inhibits KYSE150 cell growth after treatment for 48 hours. (b) Methylstat recovers H3K9me3 level in HeLa cells ectopically expressing JMJD2C(1–420)-GFP fusion protein. HeLa cells were transfected with JMJD2C(1–420)-GFP and then treated with DMSO or 10 μM methylstat. Immunostaining experiments using anti-H3K9me3 antibody (red) and DAPI nuclear stain (blue) were performed after 48 hours of incubation. Yellow arrowheads indicate transfected cells. (c) Methylstat induces hypermethylation of histone proteins in a concentration-dependent manner. KYSE150 cells were treated with DMSO, DFO at 150 μM, DMOG at 1 mM, and methylstat at 5, 10, and 15 μM for 48 hours, respectively. (d) EC50 values of methylstat for H3K4me3 and H3K9me3 in KYSE150 cells were determined by quantifying images from immunostaining assays as 10.3 μM and 8.6 μM, respectively. (e) Methylstat does not inhibit prolyl hydroxylases in cells. MCF7 cells were treated with DMSO, DFO at 150 μM, DMOG at 1 and 2 mM, methylstat at 5, 10, and 15 μM for 24 hours, respectively. Histone levels are based on coomassie stain.
To prove methylstat inhibits JMJD2C in cells, we ectopically expressed a GFP-fusion protein of the catalytic N-terminus of JMJD2C(1–420) in human epithelial carcinoma HeLa cells. Successfully transfected cells (green cells in Figure 3b) showed a significant reduction of H3K9me3 level as indicated by an immunostaining assay, and this effect was reversed by the treatment of the cells with methylstat. Next, we further evaluated the cellular effects of methylstat on a variety of histone methylation marks. DMOG and DFO were used as positive controls, since they are known cell-active inhibitors of Fe2+- and α-ketoglutarate-dependent hydroxylases. Western blotting experiments showed that methylstat induces hypermethylation of lysine residues including H3K4, H3K9, H3K27, and H3K36 at almost all methylation states investigated in a concentration-dependent manner (Figure 3c). Similar results were also observed in human breast adenocarcinoma MCF7 cells (SI, Figure S3). Of substantial interest is the observation that methylstat also induces increased cellular levels of H3K79me3 and H4K20me3. These results suggest that these histone methylation marks can also be removed by some as yet unknown Jumonji C domain-containing proteins.
To quantify the cellular activity of methylstat, we used a quantitative image-based assay. Immunostaining of KYSE150 cells treated with DMSO or methylstat showed that the cellular levels of both H3K4me3 and H3K9me3 increase in a concentration-dependent fashion (Figure 3d). Quantification of the images provided the half maximal effective concentrations (EC50) for H3K4me3 and H3K9me3 as 10.3 μM and 8.6 μM, respectively. The cellular effects of methylstat are also cell type-specific. In MCF7 cells, the EC50 values of methylstat for H3K4me3 and H3K9me3 are 6.7 μM and 6.3 μM, respectively (SI, Figure S4). It is noteworthy that methylstat induced much higher increase of the H3K9me3 mark in KYSE150 cells compared with that in MCF7 cells (10.8 fold vs. 4.5 fold), probably due to higher expression level of JMJD2C in KYSE150 cells.19
To assess the selectivity of methylstat in vivo, we examined another class of Fe2+- and α-ketoglutarate-dependent hydroxylases, prolyl hydroxylases (PHDs). PHDs have been reported to regulate normoxic inactivation of hypoxia-inducible factors (HIFs) by hydroxylating specific prolyl residues of HIFs.12,25 Inhibition of PHDs results in higher stability of HIFs and accumulation of HIFs in cells. DMOG and DFO have been used to mimic hypoxia conditions by inhibiting PHDs in vivo. MCF7 cells treated with DMOG or DFO showed significantly increased cellular levels of HIF-1α, whereas methylstat did not affect the cellular HIF-1α levels at up to 15 μM, the highest concentration tested (Figure 3e). Similar results were also observed in HeLa cells (SI, Figure S5).
Inhibition of H3K27me3 demethylase UTX-dependent myogenesis using methylstat
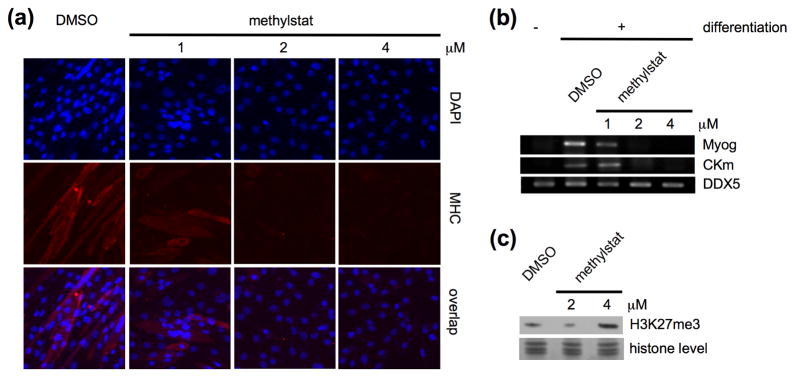
To demonstrate methylstat as a cellular probe for the studies of JHDMs further, we investigated its action during myogenesis. Recent studies have shown that UTX, an H3K27me3-specific JHDM, is recruited to the regulatory regions of muscle-specific genes Myog and CKm to remove the repressive chromatin mark H3K27me3 during myogenesis.26 Knockdown of UTX in C2C12 cells using shRNA prevents the formation of multinucleated myotubes. We found that treatment of C2C12 cells with 2 μM of methylstat is sufficient to prevent the formation myosin heavy chain (MHC)-positive myotubes (Figure 4a). Reverse transcription-polymerase chain reaction (RT-PCR) experiments also showed loss of expression of UTX-targeted genes Myog and CKm in differentiating myoblast upon treatment with methylstat (Figure 4b). It is also noteworthy that no obvious global hypermethylation of H3K27me3 was observed by Western blot in differentiating myoblast treated with 2 μM methylstat, but only at higher concentration (Figure 4c).
Figure 4.
Methylstat prevents myogenesis through inhibition of H3K27me3-demethylase UTX. (a) Methylstat prevented myotube formation in a concentration-dependent manner. C2C12 cells were treated with DMSO and methylstat at 1, 2, or 4 μM in differentiation media, respectively. Immunostaining experiments using anti-myosin heavy chain (MHC) antibody (red) and DAPI nuclear stain (blue) were performed after 6 days of incubation. (b) Methylstat inhibits the expressions of UTX-targeted genes, Myog and Ckm, during myogenesis. C2C12 cells were treated with DMSO or methylstat at 1, 2, and 4 μM in differentiation media, respectively. mRNAs were collected after 6 days of incubation and RT-PCR experiments were performed. (c) Methylstat induced hypermethylation of H3K27me3 in C2C12 cells. C2C12 cells were treated with DMSO or methylstat at 2 and 4 μM in differentiation media for 48 hours, respectively. Histone levels are based on coomassie stain.
CONCLUSIONS
We have described a cell-active selective small-molecule inhibitor of Jumonji C domain-containing histone demethylases. Compound 1 showed selective inhibitory activity against trimethyl-specific JHDMs in vitro, and its methyl ester methylstat (2) showed selective activity against a large set of JHDMs in cells. In addition, induction of hypermethylation of H3K79me3 and H4K20me3 in cells by methylstat also indicates the existence of novel JHDMs specifically targeting these important chromatin marks. This discovery provides a useful small-molecule probe that selectively targets histone demethylases. Together with other small-molecule regulators of histone-modifying enzymes,10,11,13–15,27 methylstat should find applications for studies of histone methylation dynamics in a wide range of biological processes, such as embryonic development and differentiation, germline maintenance and meiosis, and regulation of gene expression, and of disease processes, such as those leading to the development of cancer. Further studies on the structure-activity relationship and the inhibitory mechanism of methylstat are underway and will be reported in due course.
Supplementary Material
Acknowledgments
We thank S. Kato (University of Colorado) for his help with mass spectrometry and S. Fisch (Broad Institute of Harvard and MIT) and Ferran Fece de la Cruz (CeMM) for help with biochemical assays. This work was supported by the University of Colorado, Colorado Initiative of Molecular Biotechnology, the National Institute of General Medical Sciences (GM38627), European Union FP7 Marie Curie grant PIOF-GA-2008-221135 (to S.K.), and Oxford NIHR Biomedical Research Unit. The Structural Genomics Consortium is a registered charity (Number 1097737) funded by the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck and Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust.
Footnotes
Supporting Information Available. Complete ref 15b, 15c, 16b, 22a, supporting figures, materials, experimental procedures, and characterization data for all new compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Klose RJ, Zhang Y. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 2.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 5.For recent reviews on histone demethylases, see: Shi Y. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218.Loenarz C, Schofield CJ. Nat Chem Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152.Cloos PA, Christensen J, Agger K, Helin K. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908.Marmorstein R, Trievel RC. Biochim Biophys Acta. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009.
- 6.Lan F, Nottke AC, Shi Y. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Nature. 2005;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 8.(a) Wissmann M, Yin N, Müller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Günther T, Buettner R, Metzger E, Schüle R. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]; (b) Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 9.(a) Yi C, Yang CG, He C. Acc Chem Res. 2009;42:519. doi: 10.1021/ar800178j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yi C, Jia G, Hou G, Dai Q, Zhang W, Zheng G, Jian X, Yang CG, Cui Q, He C. Nature. 2010;468:330–333. doi: 10.1038/nature09497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Cole PA. Nat Chem Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M. ChemMedChem. 2009;4:1568–1582. doi: 10.1002/cmdc.200900301. [DOI] [PubMed] [Google Scholar]; (c) Natoli G, Testa G, De Santa F. Curr Opin Drug Discov Devel. 2009;12:607–615. [PubMed] [Google Scholar]
- 11.(a) Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. J Am Chem Soc. 2006;128:4536–4537. doi: 10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]; (b) Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. J Am Chem Soc. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- 12.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 13.Hamada S, Kim TD, Suzuki T, Itoh Y, Tsumoto H, Nakagawa H, Janknecht R, Miyata N. Bioorg Med Chem Lett. 2009;19:2852–2855. doi: 10.1016/j.bmcl.2009.03.098. [DOI] [PubMed] [Google Scholar]
- 14.Mackeen MM, Kramer HB, Chang KH, Coleman ML, Hopkinson RJ, Schofield CJ, Kessler BM. J Proteome Res. 2010;9:4082–4092. doi: 10.1021/pr100269b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Rose NR, Woon EC, Kingham GL, King ON, Mecinovic J, Clifton IJ, Ng SS, Talib-Hardy J, Oppermann U, McDonough MA, Schofield CJ. J Med Chem. 2010;53:1810–1818. doi: 10.1021/jm901680b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hamada S, et al. J Med Chem. 2010;53:5629–5638. doi: 10.1021/jm1003655. [DOI] [PubMed] [Google Scholar]; (c) King ON, et al. PLoS One. 2010;5:e15535. doi: 10.1371/journal.pone.0015535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, Hagman J, Hansen K, Shi Y, Zhang G. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]; (b) Ng SS, et al. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]; (c) Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Nat Struct Mol Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]; (d) Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Nat Struct Mol Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simonini MV, Camargo LM, Dong E, Maloku E, Veldic M, Costa E, Guidotti A. Proc Natl Acad Sci USA. 2006;103:1587–1592. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang QX, King J, Phanstiel IO. J Org Chem. 1997;62:8104–8108. doi: 10.1021/jo971126q. [DOI] [PubMed] [Google Scholar]
- 19.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura A, Tumber A, Rose NR, King ON, Daniel M, Oppermann U, Heightman TD, Schofield C. Anal Biochem. 2010;404:86–93. doi: 10.1016/j.ab.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]; (b) De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]; (c) Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 22.(a) Qi HH, et al. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koivunen P, Hirsilä M, Gunzler V, Kivirikko KI, Myllyharju J. J Biol Chem. 2004;279:9899–9904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- 24.(a) Hirsilä M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J. J Biol Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]; (b) Hirsilä M, Koivunen P, Xu L, Seeley T, Kivirikko KI, Myllyharju J. FASEB J. 2005;19:1308–10. doi: 10.1096/fj.04-3399fje. [DOI] [PubMed] [Google Scholar]
- 25.Asikainen TM, Schneider BK, Waleh NS, Clyman RI, Ho WB, Flippin LA, Günzler V, White CW. Proc Natl Acad Sci USA. 2005;102:10212–10217. doi: 10.1073/pnas.0504520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ. EMBO J. 2010;29:1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.