Introduction
The development of ribosomes that can read quadruplet codons could trigger a giant leap in the complexity of protein sequences. Although the practical exploration of sequence space is still limited to an infinitesimal fraction of the total volume, a full quadruplet genetic code would essentially double the information-theoretic content of proteins. Analogous studies modifying the alphabet size of ribozymes suggest that increasing the information-theoretic content of the genetic code could permit a corresponding increase in functionality. Recent work has overcome major inefficiencies in the translation of programmable quadruplet codons, paving the way for studies on fundamental questions about the origin of the genetic code and the characteristics of alternate protein “universes”.
Any protein can be thought of as occupying a specific point in sequence space, the multidimensional, discrete space in which each axis corresponds to the amino acid at a particular site in the protein [1]. The size of sequence space is limited by the number of letters in the alphabet and the length of the protein. The standard genetic code comprises 20 types of amino acid, so the number of theoretically possible proteins is 20L (where L is the number of amino acid residues in the protein sequence) although selenocysteine and pyrrolysine can increase this alphabet to 22 letters. A substantial body of work has further expanded the alphabet to include a large range of non-biological functional groups, incorporating around 70 different unnatural amino acids [2]. Like selenocysteine and pyrrolysine, artificial expansion of the genetic code usually involves translating an amber stop codon using a tRNA aminoacylated by the unnatural amino acid. However, a larger number of “blank” codons would be required to enable the incorporation of multiple different unnatural amino acids into the same protein. Therefore, quadruplet (i.e. frameshift) codons and the corresponding aminoacylated tRNA have also been explored [3–5].
In principle, a quadruplet genetic code could permit a huge increase in the volume of accessible sequence space. However, prior efforts using quadruplet or amber codons were ultimately limited by competition with native tRNAs, which are readily accepted by the native ribosome, and quadruplet codons were particularly hampered by low efficiency. Furthermore, uncontrolled incorporation of the unnatural amino acid caused widespread changes to the proteome in vivo. To circumvent these problems, Rackham and Chin designed “orthogonal” ribosomes that recognize an altered ribosome-binding site (RBS), thereby specifying that only mRNAs containing the mutant RBS would be translated by orthogonal ribosomes [6]. These ribosomes were still inefficient at incorporating unnatural amino acids, but Wang et al. [7] evolved them to reduce premature termination (ribo-X). This procedure produced orthogonal ribosomes with increased amber suppression on the desired mRNA, while native ribosomes maintained the regular level of amber suppression.
In recent work, Neumann et al. [8] evolved ribo-X even further to enhance its efficiency for translation of quadruplet codons. These ribosomes, termed ribo-Q, utilized quadruplet codons with similar efficiency and fidelity as triplets. A protein containing an azide and an alkyne was produced efficiently using a quadruplet codon and amber suppression on the orthogonal mRNA, allowing formation of an internal cross-link. In principle, ribo-Q (and perhaps its descendents) might enable even more ambitious alterations to proteins.
Information and biopolymers
How significant would a quadruplet genetic code be? One basic measure is the expansion of potential sequence space. In theory, a quadruplet code would enable an enormous increase, from 20L (or 63L, the theoretical best for a triplet code, which might be possible if the genome were heavily re-engineered) to 255L. But in practice, only a tiny fraction of this space can be explored for interesting lengths. For example, a 1 kg library of different proteins of length 100 would only constitute a fraction of 10−106 of the possible 20100 sequences, or 10−216 of the possible 255100 sequences; in both cases, sequence space is vastly larger than the number of molecules that could be explored. In other words, increasing the potential size of sequence space does not increase the library size in practice.
However, a larger alphabet would clearly include more functional groups, enabling activities that presumably could not be accessed by proteins made from the existing genetic code. This increase in functionality could be measured as simply the number of different amino acids present in a given protein (from 20 to potentially 255 in the quadruplet code), but such a measure does not capture the subtleties about the diversity of amino acids within the sequence. For example, two proteins might both contain one type of unnatural amino acid in addition to the usual 20, but if one protein contains multiple instances of the unnatural amino acid while the other contains only a single instance, the probability of novel function would presumably be higher with greater amounts of the novel amino acid. Indeed, an important technical goal of the field is the incorporation of multiple unnatural amino acids into the same protein. Orthogonal mRNA and ribosomes (like ribo-Q), which can be evolved for higher efficiencies, bring the field closer to this goal.
The Shannon information, or Shannon entropy (S), is a simple measure of the diversity of amino acids in a given protein, which takes into account the relative representation of the different amino acids. In information theory, the Shannon entropy is defined as:
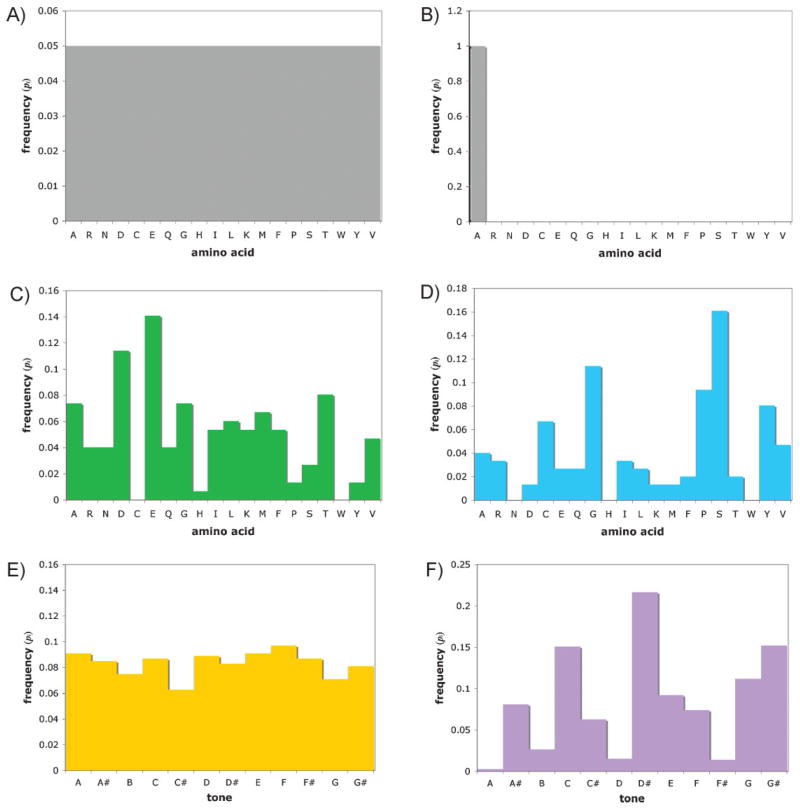
where i represents a letter of the alphabet, and pi is the frequency of i in the sequence [9]. If applied to the primary sequence of proteins, i corresponds to the 20 (or more) amino acids. The Shannon entropy would be highest for sequences containing the greatest number of different amino acids, in which the amino acids are as evenly represented as possible (Fig. 1A). The calculation of Shannon entropy does not take protein function or thermodynamic stability into account; instead, it is purely a measure of the diversity of the primary amino acid sequence. This reflects the information in the sequence in the following sense. For a very homogeneous sequence (e.g., polyalanine), knowing the identity of a particular amino acid does not contain much information, since a guess based solely on the distribution of amino acids would be correct most of the time (e.g., any given amino acid is alanine; the Shannon information is zero bits; Fig. 1B). On the other hand, in a complex protein sequence like calmodulin, knowing the identity of a particular amino acid does give quite a bit of information. In fact, we can calculate that such knowledge about a position in calmodulin would be worth about 3.9 bits of information (Fig. 1C). This is slightly less than the maximum possible value (log2 20, or ~4.3 bits), because not all amino acids are equally represented in the protein.
Figure 1.
Distribution of categorical frequencies used to calculate Shannon entropy (S). The uniform distribution A: has maximum S, while an extremely skewed sequence, like polyalanine, B: has minimum S. Among proteins, calmodulin (GenBank accession number AAD45181) has relatively high S (C; S = 3.91) while β-keratin (GenBank accession number NP_001001310 XP_424516) has lower S (D; S = 3.28). Greater information is not always “better”: Shannon information is nearly maximized in the Prelude of Schoenberg’s Suite for Piano (E; S = 3.58 ≈ log212), but this piece is relatively inaccessible compared with Beethoven’s widely beloved, yet less informative, Piano Sonata No. 23 (Appassionata) (F; S = 3.14 for the first 40 measures). Notes were counted manually from the musical score [18, 19].
How much would the Shannon entropy increase if a complete quadruplet genetic code were developed? In the best case of 255 different amino acids, knowledge of one position would be worth almost 8 bits, essentially doubling the information-theoretic content of the protein. To put this in perspective, a 4-bit leap would be similar to the difference between polyalanine and calmodulin. Using ribo-Q, Neumann et al. made a version of calmodulin containing two unnatural amino acids. We can calculate that this modification increased the Shannon entropy by 0.06 bits. This is a relatively modest gain, but the potential clearly exists for greater gains in pure information content.
Shannon information and biological function
The Shannon information of a primary sequence gives us an idea of how much information could be theoretically encoded. But how does this translate into function? Unlike functional information, which reflects the mutational robustness of a protein sequence for a specific function [10], the Shannon entropy considers a protein sequence as if it were a digital sequence in a computer, with no reference to the biological function. Thus the Shannon entropy is related to the abstract quantity of algorithmic information (the incompressibility of a sequence by a computer algorithm), which may or may not be correlated with biological activity. In human terms, increasing the Shannon entropy does increase the actual information transmitted by a code of a given length. One illustration of this general phenomenon can be found in the algorithmic complexity of songs, which was analyzed in an engaging article by computer scientist Donald Knuth [11]. A complex, internally diverse song, like Homer’s Iliad, conveys a large amount of information in a certain number of letters (high Shannon entropy). Like calmodulin, although the Iliad is very complex, its sequence actually has submaximal Shannon entropy for several reasons, including that repetitive elements served as mnemonic devices for the storyteller. When repetitive structures are taken to an extreme degree, they substantially reduce the information in the song. Knuth noted that “99 bottles of beer on the wall” (very low Shannon entropy) conveys very little information to the listener, given the same length of text.
In analogy to such everyday examples, we might intuit that Shannon information of protein sequences would also correlate with biological function, but that remains to be proven, perhaps by future comparative studies on the quadruplet versus triplet genetic code. However, the relationship between alphabet size and biological function has been studied in a different biopolymer: RNA.
More is better?
In a remarkable series of studies, Joyce and coworkers selected for RNA ligase activity among random RNA sequences in a 2-, 3-, or 4-letter alphabet, using in vitro evolution. The standard alphabet (A, C, G, U) yielded a highly active ribozyme with kcat = 20 minutes−1 [12]. One of the motivations for understanding the relationship between information and function in ribozymes is to gain insight into the RNA world, the putative early stage of life during which RNA served as both genetic repository and catalytic molecules. In reducing the alphabet, elimination of cytosine is particularly interesting for understanding the origin of life, because C is relatively unstable and degrades readily [13]. Reducing the alphabet to three letters (A, G, U) still gave a ribozyme, but with lower catalytic activity (kcat = 0.01 minute−1) [14]. Reducing the alphabet further to two letters, the smallest possible size that can convey any information at all, yielded the least active ribozyme (kcat = 0.001 minute−1, which is still 10,000 times faster than the uncatalyzed reaction) [15]. Interestingly, this ribozyme was composed of uracil and an unnatural nucleobase, diaminopurine (D). The general advantage of a D, U system over an A, U system is that the unnatural base pair contains three, rather than two, hydrogen bonds during Watson-Crick pairing, thereby stabilizing the secondary structure of the ribozyme. This example illustrates the potential benefit of large-scale substitution with unnatural subunits in biopolymers; ribo-Q may allow similar experiments in proteins. Although the discovery of a ribozyme with only two or three bases is itself quite interesting, the reduction in alphabet size did reduce activity. These studies therefore suggest that algorithmic information correlates with functional activity, confirming our intuition: perhaps more information actually is better.
However, this trend may not necessarily generalize to different functions. The optimal Shannon information may actually be less than the maximum. For example, the Shannon entropy of calmodulin is slightly lower than the maximum, and its sequence contains only 18 of the 20 canonical amino acids (cysteine and tryptophan are missing). In addition, some proteins rely heavily on repetitive elements for their function. Structural proteins, like keratin, tend to have an extremely non-uniform distribution of amino acids in their composition because of folding requirements (Fig. 1D). The arts again provide a ready analogy. In Western classical music, pitches fall into 12 categories known as tones (do, re, mi, etc.). The maximum Shannon information would be conveyed in music containing equal numbers of each tone. However, listeners generally prefer tonal music (which emphasizes some particular tones, such as the tonic and dominant notes), compared to music with very high Shannon entropies. In the early 20th century, composers such as Arnold Schoenberg did move toward very high Shannon information using the 12-tone technique and related methods, in which all notes were emphasized equally (Fig. 1E). These techniques led to some interesting innovations, but in terms of the “function” of human enjoyment, the optimum Shannon entropy appears not to be the maximum (at least for all but the most rarified circles of classical music aficionados, Fig. 1F). On the other hand, minimum Shannon entropy is also non-optimal, although there are deliberate exceptions, such as the repeating melodic note in “One Note Samba” by Antonio Carlos Jobim (the harmonic lines do change behind this melody). Altering the size and information-theoretic content of the genetic code could enable analogous studies on optimality for proteins with different functions.
Alternate universes
Quadruplet codons can be thought of in the context of expanding the existing genetic code and allowing access to additional function. But another interesting perspective is that they might enable the study of alternate protein “universes”. Orthogonal ribosomes could build proteins composed of 20 different unnatural amino acids, allowing us to examine fundamental questions about the origin of life. If early evolution had developed a different set of amino acids, how different would the resulting proteins be from existing proteins? Could they perform the same functions? Would they fold in unexpected ways? Another possibility would be to re-wire the existing genetic code, encoding the same amino acids with different codons. This could permit a direct experimental test of whether the genetic code is a “frozen accident” [16], or if it was optimized by natural selection in some way. For example, one hypothesis is that the existing code minimizes the effect of mutations by encoding similar amino acids by related codons [17]. The possibilities for studying alternate protein universes are boundless.
Conclusions
Alterations to the standard cohort of amino acids have previously been limited to one or two substitutions per protein because of problems with low efficiency and widespread proteomic disruption when incorporating unnatural amino acids. The ongoing design and evolution of orthogonal mRNA and ribosomes, so far culminating in ribo-Q, could open the floodgates to completely new protein functionalities. In principle, a quadruplet genetic code could lead to a 10-fold expansion of the amino acid alphabet, from around 20 to more than 200. Although this would not lead to the ability to explore more sequences in terms of absolute number, due to the astronomical size of sequence space, a quadruplet code could engender a large leap in the information-theoretic content of proteins. The size of this leap in information could be as large as that between a homopolymer and a typical complex protein today. In addition to the biotechnological applications, ribo-Q and related advances could eventually enable the study of deep questions about the origin of the genetic code: how optimal are the 20 amino acids? What is the relationship between abstract, algorithmic information, and biological function? Ribo-Q contains only a handful of mutations compared to the wild-type ribosome, but it could help us peer into a whole new multiverse of alternative proteomes.
Acknowledgments
We thank Julien Derr, Ramon Xulvi, and Michael Manapat for discussions about Shannon information, the Bauer Fellows program of Harvard University and NIH grant GM068763 for the National Centers of Systems Biology, and the Charles Blake Fund of the Nuttall Ornithological Club.
Abbreviations
- L
length of protein
- RBS
ribosome binding site
- ribo-X
ribosome evolved to reduce premature termination
- ribo-Q
ribosome further evolved to process quadruplet codons with high efficiency
- S
Shannon entropy
References
- 1.Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225:563–4. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 2.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010 Mar 18; doi: 10.1146/annurev.biochem.052308.105824. [Epub ahead of print] PMID: 20307192. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez EA, Lester HA, Dougherty DA. In vivo incorporation of multiple unnatural amino acids through nonsense and frame-shift suppression. Proc Natl Acad Sci USA. 2006;103:8650–5. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hohsaka T, Sisido M. Incorporation of non-natural amino acids into proteins. Curr Opin Chem Biol. 2002;6:809–15. doi: 10.1016/s1367-5931(02)00376-9. [DOI] [PubMed] [Google Scholar]
- 5.Anderson JC, Wu N, Santoro SW, et al. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci USA. 2004;101:7566–71. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat Chem Biol. 2005;1:59–66. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, Neumann H, Peak-Chew SY, et al. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007;25:770–7. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 8.Neumann H, Wang K, Davis L, et al. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–4. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 9.Shannon C. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423. [Google Scholar]
- 10.Szostak JW. Functional information: molecular messages. Nature. 2003;423:689. doi: 10.1038/423689a. [DOI] [PubMed] [Google Scholar]
- 11.Knuth D. The complexity of songs. Commun ACM. 1984;27:344–6. [Google Scholar]
- 12.Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–7. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 13.Levy M, Miller SL. The stability of the RNA bases: implications for the origin of life. Proc Natl Acad Sci USA. 1998;95:7933–8. doi: 10.1073/pnas.95.14.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers J, Joyce GF. A ribozyme that lacks cytidine. Nature. 1999;402:323–5. doi: 10.1038/46335. [DOI] [PubMed] [Google Scholar]
- 15.Reader JS, Joyce GF. A ribozyme composed of only two different nucleotides. Nature. 2002;420:841–4. doi: 10.1038/nature01185. [DOI] [PubMed] [Google Scholar]
- 16.Crick F. The origin of the genetic code. J Mol Biol. 1968;38:367–79. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 17.Freeland SJ, Hurst LD. The genetic code is one in a million. J Mol Evol. 1998;47:238–48. doi: 10.1007/pl00006381. [DOI] [PubMed] [Google Scholar]
- 18.Schoenberg A. Suite fur Klavier, op 25, plate U E 7627. Vienna: Universal Edition; 1925. [Google Scholar]
- 19.Beethoven Lv. Piano Sonata No 23, Op 57. Vienna: Bureau des Arts et d’Industrie; 1807. Plate 521. [Google Scholar]