Abstract
The identification of genetic polymorphisms that influence the efficacy and safety of therapies for breast cancer may allow future treatments to be individualised based not only on tumour characteristics but also on host genetics. Genetic factors that affect the metabolism, efficacy and safety of tamoxifen, one of the most common drugs used for the treatment and prevention of breast cancer, have received particular attention. Cytochrome P450 2D6 (CYP2D6) is crucial in the metabolism of tamoxifen to its active metabolite endoxifen. Women with genetic variants of CYP2D6 or who take drugs that inhibit the enzyme have low endoxifen plasma concentrations and may show reduced benefits to tamoxifen treatment. CYP2D6 polymorphisms and variants in other candidate genes may also influence secondary benefits and side effects of tamoxifen. Here, we summarise data suggesting that CYP2D6 status may be an important predictor of the benefits of tamoxifen to an individual; in addition, we briefly discuss the role of variants in other candidate genes. Whether CYP2D6 status should be determined prior to initiating tamoxifen therapy is currently under debate and may be appropriate only for select women who are candidates for tamoxifen alone but for whom alternative standard options are available.
Despite the increase in breast cancer prevalence in recent years, disease-specific mortality has declined (Ref. 1). The improvement in survival is attributed both to early diagnosis and to optimal local and systemic therapies (Ref. 2). Recommendations for adjuvant systemic therapy are based on estimates of risk of subsequent recurrence for an individual as well as her risk for treatment-related toxicity. However, individual women may not encounter the same benefits from identical systemic therapies. Several tumour characteristics may predict who may benefit from specific treatments but are imperfect. For example, expression of the oestrogen (estrogen) receptor (ER) and/or progesterone receptor (PR) is essential for response to hormonal interventions but only 50% of women with ER/PR-expressing tumours will benefit from such therapies. Other specific or aggregate tumour characteristics may predict response to hormonal intervention. Host characteristics (i.e. characteristics of normal tissue of the individual rather than the tumour) also play an important role in predicting response to specific agents or treatments but have not been extensively studied (Refs 3, 4). The recent sequencing of the human genome and new high-throughput technologies have led to rapid advances in pharmacogenetics/pharmacogenomics – the study of the genetic determinants of drug responses – and to a greater appreciation of the role that host factors may play as predictors of drug response. If so, assessment of host factors prior to initiation of some medical therapies may become standard practice.
Approximately 60–70% of women with a newly diagnosed breast cancer have tumours that express ER or PR. In addition to the local treatment of their breast cancer, most of these women will be recommended endocrine manipulations. Several adjuvant endocrine treatments are currently available to postmenopausal women, including the selective ER modulator (SERM) tamoxifen, which blocks oestrogen from binding to its receptor, and the aromatase inhibitors, which block the production of oestrogen. The majority of postmenopausal women may receive an aromatase inhibitor for 5 years, or tamoxifen for 2–3 years followed by an aromatase inhibitor for a total of 5 years of hormone therapy (Ref. 5). Other postmenopausal women may receive tamoxifen for 5 years with or without 5 additional years of an aromatase inhibitor. Premenopausal women with tumours that express hormone receptors are usually recommended tamoxifen alone. Premenopausal women who cannot take tamoxifen or those enrolled in clinical trials may be offered ovarian suppression with an aromatase inhibitor (Ref. 6). Tamoxifen has been used for over three decades and its benefits and side effects have been fairly well defined.
Tamoxifen metabolism
Recently, preclinical and clinical studies have clarified the metabolic pathway of tamoxifen (Fig. 1). Tamoxifen itself has weak binding to ER and is considered a prodrug. It undergoes extensive biotransformation catalysed by both phase 1 and phase 2 liver enzymes. Tamoxifen is converted in vivo into potent anti-oestrogenic metabolites by several cytochrome P450 (CYP450) enzymes including CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4 and CYP3A5 (Ref. 7). The active metabolites of tamoxifen are metabolised further into inactive compounds by conjugation via phase 2 liver enzymes (Refs 8, 9, 10).
Figure 1. Biotransformation of tamoxifen and its metabolites.
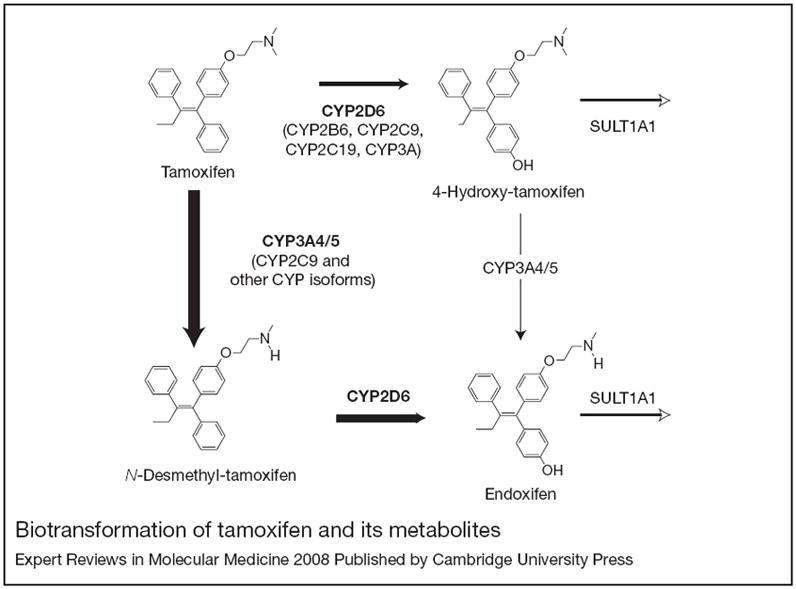
Cytochrome P450 (CYP) 3A4/5 is responsible for the formation of N-desmethyl-tamoxifen. The formation of 4-hydroxy-tamoxifen and of endoxifen are predominantly catalysed by CYP2D6. Other CYP isoforms, including CYP2C19, CYP2C9 and CYP2B6, appear to play less important roles in tamoxifen metabolism in vitro at therapeutically relevant concentrations. SULT1A1 is responsible for sulphation of 4-hydroxy-tamoxifen and may also play a role in endoxifen clearance. Reprinted, with permission from Oxford University Press, from Ref. 22 (© 2005 Oxford University Press).
Using in vitro drug metabolism models including human liver microsomes (HLMs) and expressed CYP450 isoforms, investigators have identified the primary and secondary metabolic routes of tamoxifen and the role of each enzyme in catalysing these reactions at therapeutically relevant concentrations (Ref. 7) (Fig. 1). The major primary metabolite formed is N-desmethyl-tamoxifen, a weak antioestrogen, via CYP3A4/5. The potent antioestrogen 4-hydroxy-tamoxifen is a minor primary metabolite whose formation is catalysed by multiple CYP450s, including CYP2D6. Other minor primary metabolites include α-, 3- and 4′-hydroxy-tamoxifen and an unidentified metabolite, formed by CYP3A4, CYP3A5, CYP2B6/2C19 and CYP3A4, respectively. The primary metabolites undergo further enzymatic conversion: N-desmethyl-tamoxifen is metabolised into α-hydroxy-tamoxifen, N-didesmethly-tamoxifen, and 4-hydroxy-N-desmethyl-tamoxifen (the latter also known as endoxifen); and 4-hydroxy-tamoxifen is converted to 3,4-dihydroxy-tamoxifen and also endoxifen (Refs 11, 12). The formation of every secondary metabolite is catalysed by CYP3A4/5. However, endoxifen is produced almost exclusively by CYP2D6 activity.
Although endoxifen was identified as a metabolite of tamoxifen almost two decades ago (at the time designated BX) (Ref. 13), its biological and ER-related activity was not well studied; 4-hydroxy-tamoxifen was considered the main active metabolite of tamoxifen, since it has 100-fold higher affinity for ER than tamoxifen (Ref. 14). However, evaluation of the anti-oestrogenic properties of endoxifen revealed that its affinity for the ER is equivalent to that of oestrogen and 4-hydroxy-tamoxifen, and that it inhibits in vitro oestrogen-induced breast cancer growth and induces ER-regulated gene expression changes almost identically to 4-hydroxy-tamoxifen (Refs 15, 16). Based on clinical data showing it is present at five- to sevenfold higher concentrations than that of 4-hydroxy-tamoxifen in patients with wild-type (wt) CYP2D6, endoxifen is now considered the most important metabolite of tamoxifen (Refs 12, 15).
Interindividual variation in CYPD6 activity: implications for tamoxifen metabolism
Each of the genes that encode for CYP450 enzymes implicated in tamoxifen metabolism has known genetic polymorphisms that have been shown to affect their catalytic activity; for example, single-nucleotide polymorphisms (SNPs) in CYP2C19 and CYP2D6 genes can lead to complete lack of enzymatic activity by causing the formation of truncated inactive proteins (Ref. 17). Genetic polymorphisms in genes encoding for CYP450 enzymes might explain some of the clinical variability in the plasma concentrations of tamoxifen and its metabolites. Given the importance of CYP2D6 in tamoxifen metabolism, variation in this enzyme has received particular attention.
CYP2D6, which is expressed predominately in the liver, is responsible for the metabolism of approximately 25% of drugs, including many beta-blockers, antidepressants, antipsychotics and antiarrhythmics, as well as dextromethorphan, codeine and tamoxifen (Ref. 18). The activity of CYP2D6 shows a very high degree of variability between individuals due to several genetic polymorphisms that influence protein function (Refs 18, 19, 20).
Over 80 different CYP2D6 alleles have been identified and their frequencies vary among ethnic groups (Ref. 20). The most common variant allele in Caucasians, the null allele CYP2D6*4, has an allelic frequency of ~25%, whereas CYP2D6*10 is most common in Asian populations (Ref. 18). Different designations can be ascribed to individual patients based on their CYP2D6 genotype and thus genetic testing can be used to categorise patients into one of four phenotypic groups (Table 1): poor metaboliser (PM), intermediate metaboliser (IM), extensive metaboliser (EM) and ultrarapid metaboliser (UM). In Caucasian and Western European populations, ~1–2% are UMs (carrying more than two copies of CYP2D6 in their genome), ~70% are EMs (carrying two wt alleles), ~20% are IMs (heterozygous for null or partial function alleles) and ~8% are PMs (homozygous for variant alleles).
Table 1.
Classification of CYP2D6 phenotype
| Metaboliser status | Activity | Genotype and CYP2D6 inhibitor statusa |
|---|---|---|
| Poor metabolisers | No activity | Two null alleles (CYP2D6*3,*4,*5,*6,*11), or one null allele and a strong inhibitor |
| Intermediate metabolisers | Reduced activity | One null allele (CYP2D6*3,*4,*5,*6,*11) or one or two variants *9,*10,*17,*41, and/or a moderate inhibitor |
| Extensive metabolisers | Normal activity | Two ‘wild-type’ or normal alleles (CYP2D6*1,*2,*35) and no inhibitor |
| Ultrarapid metabolisers | Excess activity | *1xN, *2xN, *35xN, *41xN and no inhibitor |
Inhibitor staus refers to the coadministration of drugs that may affect CYP2D6 activity (see Table 2).
Abbreviation: N, number of gene duplication alleles.
In addition to genetic factors that influence CYP2D6 activity, enzymatic activity can be influenced by several common drugs. Paroxetine, fluoxetine and bupropion are strong inhibitors; duloxetine, thioridazine, amiodarone, diphenhydramine and cimetidine are moderate inhibitors (Ref. 21) (Table 2).
Table 2.
Partial list of drugs that are commonly administered with tamoxifen, and CYP2D6 inhibition
| Inhibitor Stronga | Ref. |
|---|---|
| Paroxetine (Paxil) | 22 |
| Fluoxetine (Prozac) | 22 |
| Bupropion (Wellbutrin) | 45 |
| Quindine (Cardioquin) | 46 |
| Moderate | |
| Duloxetine (Cymbalta) | 47 |
| Thioridazine (Mellaril) | 48 |
| Amiodarone (Cordarone) | 49 |
| Diphenhydramine (Benadryl) | 50 |
| Cimetidine (Tagamet) | 51 |
| Weak or Noninhibitorb | |
| Sertraline (Zoloft) | 22 |
| Venlafaxine (Effexor) | 22 |
| Citalopram (Celexa) | 52 |
| Escitalopram (Lexapro) | 53 |
Should be avoided in women taking tamoxifen.
Probably safe in women taking tamoxifen.
Initial prospective studies demonstrated that tamoxifen metabolism, and perhaps efficacy, is greatly affected by lack of CYP2D6 as a result of polymorphisms or interactions with drugs that inhibit the enzyme (Refs 12, 22, 23). However, these studies were not designed to answer questions regarding long-term efficacy of tamoxifen use based on CYP2D6 status. Several recent investigations have attempted to correlate CYP2D6 genotype with long-term outcomes. Here we review studies reported to date suggesting that CYP2D6 may be an important predictor of tamoxifen’s benefits. In addition, we briefly discuss the role of variants in other candidate genes that may affect tamoxifen’s efficacy.
Pharmacogenetic studies of tamoxifen metabolism
In the mid 1990s, emerging data suggested that selective serotonin-reuptake inhibitors (SSRIs) or serotonin–noradrenergic-reuptake inhibitors (SNRIs) may alleviate hot flushes (flashes) (Ref. 24). Because many breast cancer survivors taking tamoxifen suffer hot flushes, nonhormonal remedies such as the SSRI paroxetine and the SNRI venlafaxine have been commonly prescribed. However, many SSRIs are potent CYP2D6 inhibitors at therapeutic concentrations (Ref. 15). A pilot study designed to evaluate the effects of paroxetine on tamoxifen metabolite plasma concentrations showed that women on chronic tamoxifen who had genetic variants in CYP2D6 or who were coprescribed paroxetine had very low plasma concentrations of the active tamoxifen metabolite endoxifen (Ref. 12).
The Consortium On Breast Cancer Pharmacogenomics (COBRA) investigators initiated a larger prospective clinical trial to confirm early results and to investigate other pharmacogenetic effects of tamoxifen. This prospective trial included 300 women with a primary breast cancer who started tamoxifen in the adjuvant setting or for prevention of a new primary tumour in one of three academic centres. Baseline samples were collected to evaluate candidate gene variants for their association with tamoxifen-related side effects and secondary benefits. Additional samples were collected following 1, 4, 8 and 12 months of treatment to determine tamoxifen and metabolite concentrations. The women completed hot-flush diaries and quality of life questionnaires, and provided a complete medication list at baseline and at several time points during the first year of tamoxifen therapy. An analysis of the first 80 patients confirmed the pilot study results that women with one or two null CYP2D6 alleles (IMs or PMs, respectively) had significantly lower endoxifen plasma concentrations compared with women with normal CYP2D6 (EMs). In addition, women with normal CYP2D6 who were taking concomitant medications that are known strong inhibitors of CYP2D6 (e.g. paroxetine) had marked reductions in their endoxifen plasma concentrations, similar to that seen in women with two null alleles (PMs) (Ref. 22). Women prescribed moderate inhibitors of CYP2D6, such as duloxetine, had moderate reductions in endoxifen plasma concentrations, while women who were administered a weak inhibitor, such as venlafaxine or citalopram, had very little if any change in endoxifen concentrations. These results have been further evaluated with a larger sample size of 130 women genotyped for 27 genetic variants in CYP2D6 using a chip-based approach (Ref. 23). Together, all available data show that the CYP2D6 activity of a patient is the major determinant of plasma concentration of endoxifen.
Pharmacogenetic studies of tamoxifen efficacy
CYP2D6
The studies demonstrating the crucial role of CYP2D6 in converting tamoxifen to its active metabolite endoxifen generated much interest and concern among clinical breast oncologists, and attempts have been made to validate CYP2D6 genotype as a biomarker for predicting tamoxifen response. Although most prospective trials of single-agent tamoxifen did not include a collection of blood for DNA analysis, paraffin tumour blocks are often available on a majority of participants. Indeed, DNA extraction from formalin-fixed, paraffin-embedded archival tumour specimens, and analysis of genetic variant candidate genes of interest is feasible (Ref. 25). Several investigators have evaluated genetic variants in CYP2D6 and other genes important in tamoxifen metabolism and long-term outcomes of tamoxifen-treated women. To date, there have been nine reports that included CYP2D6 variants among the genes tested for an association between genotype and benefit from tamoxifen therapy (Table 3).
Table 3.
Summary of main studies that evaluated genetic variants and tamoxifen-associated long-term outcomes
| Study | Study overview | Main results |
|---|---|---|
| Goetz et al., 2005 (Refs 27, 28) | 256 postmenopausal women with ER+/PR+ tumours; tamoxifen (20 mg/day) for 5 yearsa | CYP2D6*4/*4 vs other groups |
| Higher risk of recurrence: | ||
| RFS HR = 1.85, P = 0.176 | ||
| DFS HR = 1.86, P = 0.089 | ||
| Schroth et al., 2007 (Ref. 32) | 486 pre- and postmenopausal women; 206 adjuvant tamoxifen (100% ER+), 280 chemotherapy or nothing (54% ER+) | Tamoxifen-treated PMs vs other groups |
| Higher risk of recurrence: | ||
| RFS HR = 2.24, P = 0.02 | ||
| EFSR HR = 1.89, P = 0.02 | ||
| Wegman et al., 2005 (Ref. 29) | 226 postmenopausal women; 112 (70% ER+) tamoxifen (40 mg/day) for 2 years | CYP2D6*4 carrier |
| Lower risk of recurrence: RR = 0.28, P = 0.0089 | ||
| SULT1A1*1/*1 carrier | ||
| Lower risk of recurrence: RR = 0.48, P = 0.0074 | ||
| CYP2D6*4 and /or SULT1A1*1/*1 carrier | ||
| Lower risk of distant recurrence: RR = 0.38, P = 0.0041 | ||
| Wegman et al., 2007 (Ref. 30) | 677 peri-and postmenopausal ER+ women; 238 randomised to tamoxifen (40 mg/day or 20 mg/day) for 2 or 5 years, 439 tamoxifen (40 mg/day) for 2 or 5 years (before 1994) or 20 mg/day for 5 years (after 1994) | CYP2D6*4/*4 vs CYP2D6*1/*1 or CYP2D6*1: |
| Improved DFS: P = 0.05, and 0.04, respectively | ||
| CYP3A5*3/*3 | ||
| Improved RFS: HR = 0.20, P = 0.002 | ||
| Lim et al., 2007 (Ref. 33) | 211 pre- and postmenopausal women (12 with metastatic breast cancer; 190 adjuvant); tamoxifen (20 mg/day) | CYP2D6*10/*10 carriers vs other groups |
| Lower concentration of tamoxifen metabolites: P < 0.0001 | ||
| CYP2D6*10 | ||
| More often found among nonresponders: | ||
| 100% vs 50%; P = 0.0186; Median TTP = 5 vs 21.8 months; P = 0.0032 | ||
| Nowell et al., 2005 (Ref. 31) | 337 pre- and postmenopausal women (retrospective);160 tamoxifen, 177 no tamoxifen | CYP2D6*4 vs other |
| Lower risk of recurrence: | ||
| DFS HR = 0.67, P = NS | ||
| OS HR = 0.79, P = NS | ||
| Kiyotani et al., 2008 (Ref. 34) | 67 pre- and postmenopausal women (retrospective); tamoxifen (20 mg/day) for 5 years | CYP2D6*10/*10 vs *1v*1 |
| Higher risk of recurrence: | ||
| RFS HR = 10.94, P = 0.036 | ||
| Bonanni et al., 2006 (Ref. 35) | 46 women from a prospective chemoprevention study; tamoxifen (20 mg/day) for 5 years vs placebo for 5 years | Higher frequency of breast cancer in CYP2D6*4/*4 carrier: |
| 8.7% vs 0.7% in other |
Women who received 5 years of tamoxifen and 1 year of fluoxymesterone were not included in the pharmacogenetics study.
Abbreviations: DFS, disease-free survival; EFSR, event-free survival rate; EM, extensive metaboliser; ER, estrogen (estrogen) receptor; HR, hazard ratio, IM, intermediate metaboliser; NS, statistically not significant, OS, overall survival; PM, poor metaboliser; PR, progesterone receptor; RFS, relapse-free survival; RR, relative risk; TTP, time to progression; UM, ultrarapid metaboliser; vs, versus.
In the first report, samples were collected from Caucasian postmenopausal women with resected ER-positive breast cancer who were previously enrolled in the North Central Cancer Treatment Group (NCCTG) randomised Phase III clinical trial designed to test the value of adding 1 year of fluoxymesterone to 5 years of tamoxifen adjuvant therapy (NCCTG 89-30-52). Difference in the long-term outcomes were not detected between the two treatment arms (Ref. 26). Tumour blocks from initial breast surgery were analysed for CYP2D6 genotype, and patient information regarding concomitant medication use was extracted from medical records. Of the 256 women enrolled to the tamoxifen-only arm, genotyping was successful from samples from 190 patients for CYP2D6*4, the most common CYP2D6-null variant in Caucasians. The primary objectives of the study were to determine the relationship between genotype and relapse-free time, disease-free survival and overall survival. Women who were PMs with the CYP2D6*4/*4 genotype (n = 13) had significantly worse recurrence-free time and disease-free survival but not overall survival compared with IMs (*4/wt; n = 40) or EMs (wt/wt; n = 137) (log rank P = 0.030, P = 0.020 and P = 0.360, respectively) (Ref. 27) (Table 3). Cox proportional hazard modelling demonstrated that nodal status and tumour size were significantly associated with recurrence-free time, disease-free survival, and overall survival. Once nodal status and tumour size were accounted for, PMs still had worse recurrence-free time and disease-free survival compared with IMs and EMs.
A follow-up study by the same investigators considered coprescription of CYP2D6 inhibitors with CYP2D6 genotype to determine CYP2D6 activity. Indeed, CYP2D6 activity was an independent predictor of breast cancer outcome in postmenopausal women receiving tamoxifen (Ref. 28). The results of these studies were consistent with the hypothesis that patients with CYP2D6-null variants will exhibit lower response to tamoxifen. However, the study was limited by a small sample size, a homogeneous patient population, and determination of a limited number of CYP2D6 alleles.
Swedish investigators reported a retrospective genetic association study in 112 women who were prescribed adjuvant tamoxifen (Ref. 29). The authors compared the outcomes of patients with at least one CYP2D6*4 allele who received tamoxifen versus no tamoxifen. Contrary to the NCCTG study results, they found that patients with the CYP2D6*4 genotype did significantly better than patients not treated with tamoxifen. However, of the 112 women receiving tamoxifen, only 70% were ER-positive. The same group has recently published a second retrospective study in a different and larger cohort of 677 tamoxifen-treated, ER-positive, postmenopausal women. Of those, 238 women were included in one study in which they were randomised to tamoxifen for 2 or 5 years (Ref. 30). The other 439 women received either tamoxifen at 40 mg/day for 2 or 5 years (before 1994) or tamoxifen at 20 mg/day (after 1994). The authors reported that patients homozygous for CYP2D6*4 had significantly improved disease-free survival compared with patients with wild-type CYP2D6. These results are consistent with a study conducted by Nowell et al. (Ref. 31) using formalin-fixed, paraffin-embedded samples collected from a hospital tumour registry. After adjusting for age, stage of disease, ethnicity and hormone receptor status, Cox proportional hazard modelling suggested a trend towards better overall survival in patients with CYP2D6*4 (Ref. 31). These results are contradictory to the NCCTG data and may be explained in part by the nature of these latter studies. Most samples were collected retrospectively from an existing database, the duration of treatment and dose of tamoxifen were variable, women may have received chemotherapy, and hormone receptor status was not tested centrally. While the authors did not collect information regarding CYP2D6 inhibitors, it is likely that at the time of collection only a few women were taking such agents.
A nonrandomised cohort study where women received either tamoxifen or no treatment in a German cohort evaluated genes encoding for five different cytochrome enzymes, with a total of sixteen different variants (Ref. 32). A total of 486 women were included: 206 were ER-positive and received tamoxifen, and 280, who were either ER-positive or -negative, received chemotherapy or no systemic therapy. Tamoxifen-treated women who were carriers for a variant allele CYP2D6*4 were more likely to suffer recurrences and have shorter relapse-free survival compared with women with functional alleles. Women with one or two reduced-activity enzymes who were treated with tamoxifen had a worse breast cancer recurrence-free survival [hazard ratio (HR) = 2.24; P = 0.02] and event-free survival (HR = 1.89; P = 0.02) compared with those who did not receive tamoxifen.
More recently, two Asian studies evaluated the role of CYP2D6*10 and tamoxifen-related outcomes. Asian populations are less likely to carry CYP2D6*4 alleles but have a greater likelihood of carrying the CYP2D6*10 alleles, which are associated with reduced enzyme activity. The status of CYP2D6 and outcome was investigated in 202 patients who received tamoxifen in a study from Korea. Of those, most women received the treatment in an adjuvant setting but 12 women received the treatment for metastatic disease. In the metastatic patients, 57% carried the *10 allele compared with 24% of women in the adjuvant setting. However, it is not clear whether patients in the metastatic setting were previously treated with tamoxifen. Patients who had two *10 alleles had a statistically significantly lower concentration of the tamoxifen metabolites 4-hydroxy-tamoxifen and endoxifen compared with patients who had one or two normal alleles (Ref. 33). In the second part of the study the investigators included the 12 patients with metastatic breast cancer and nine additional patients with metastatic breast cancer and reported that patients with two *10 alleles had a shorter time to progression compared with those with one allele. Finally, a study of 67 women who received tamoxifen monotherapy in Japan revealed that those homozygous for the CYP2D6*10 alleles had a higher incidence of recurrence (odds ratio = 16.63; P = 0.0057), compared with those with normal alleles (Ref. 34).
The influence of CYP2D6 variants on tamoxifen effects have also been examined in the prevention setting. In a ‘letter to the editor’, Bonanni et al. reported a correlation of CYP2D6*4 genotype in a nested case–control study derived from the Italian Chemoprevention trial. The authors demonstrated that the frequency of the CYP2D6*4/*4 genotype was higher in 46 tamoxifen-treated women who developed breast cancer (cases) than in 136 tamoxifen-treated women who did not develop breast cancer (controls) (Ref. 35). Although a relatively small study, these data provide a strong incentive to examine the effect in larger datasets to establish more robustly the relationship between CYP2D6 genotype and outcomes in women treated with tamoxifen in the prevention setting.
Current status of CYP2D6 genotype and tamoxifen response
Taken together, several retrospective analyses suggest that tamoxifen-treated women with CYP2D6-null alleles have worse prognosis compared with those with one or two normal alleles, but data are not consistent among trials. These preliminary studies suggest that CYP2D6 activity may be an important factor determining long-term outcomes of women taking tamoxifen. Information regarding the metabolism of tamoxifen was not available in the studies that reported outcome data. All studies to date on the association between CYP2D6 genotype and tamoxifen response have been limited by the small numbers of patients and small number of CYP2D6 alleles examined. Indeed, none of the studies shown in Table 3 has conducted a comprehensive CYP2D6 genotyping approach and therefore all may have been confounded by misclassification of the true CYP2D6 phenotype of patients.
Recently, Blake et al. (Ref. 36) developed a method that takes into account all genetic polymorphisms in CYP2D6 with significant frequency and uses this information to assign a CYP2D6 activity ‘score’ that is directly related to the ability of a patient to metabolise CYP2D6-dependent drugs. The CYP2D6 activity score method has been validated as a genotypic measure of phenotype (Ref. 37). Ideally, future prospective studies should include a comprehensive analysis of CYP2D6 variants and other candidate genes and will utilise novel scoring methods.
Additional genetic variants affecting tamoxifen metabolism
PMs have very low endoxifen plasma concentrations, while EMs have on average five- to sevenfold higher endoxifen concentrations (Ref. 22). However, a great deal of variability in endoxifen remains within the CYP2D6 EM group, suggesting that factors other than CYP2D6 influence endoxifen plasma concentrations. Indeed, while production of endoxifen is almost entirely dependent on CYP2D6, elimination of the metabolite from the plasma requires other enzymes, including the phase 2 drug-metabolising enzymes sulphotransferase isoform 1A1 (SULT1A1) and UDP-glucuronosyltransferase isoform 2B15. Therefore, genetic variants in genes other than CYP2D6 could be responsible for variability in endoxifen concentrations and affect benefit from tamoxifen therapy. Indeed, six of the nine studies shown in Table 3 examined multiple gene variants, some of which correlated with benefit from tamoxifen.
For example, Swedish investigators analysed the role of other gene variants and reported that there was no prognostic significance for CYP3A5, SULT1A1 and UGT2B15 (Ref. 30). However, for women who were prescribed 2 years of tamoxifen, those homozygous for CYP3A5*3 were at increased risk of recurrence (HR = 2.84, 0.68–11.99; P = 0.15). By contrast, women who received 5 years of tamoxifen and were homozygous for CYP3A5*3 had improved recurrence-free survival (HR = 0.20, 0.07–0.55, P = 0.002) (Ref. 30). In the German cohort, patients with a CYP2C19*17 allele had a more favourable clinical outcome than carriers of *1, *2 and *3 alleles (HR = 0.45; P = 0.03) (Ref. 32).
Genetic variation affecting tamoxifen secondary benefits and side effects
In postmenopausal women, secondary benefits of tamoxifen include increase in bone mineral density, and reduction in plasma concentrations of total cholesterol and low-density lipoprotein (and possibly reduction of cardiovascular events) (Ref. 38). However, tamoxifen is also associated with frequent and bothersome menopausal symptoms such as hot flushes and atrophic vaginitis. Rare effects include thromboembolic events, thrombocytopaenia or leukopaenia, endometrial hyperplasia, polyps or cancer (Ref. 38). Tamoxifen-associated side effects and secondary benefits are likely mediated through the effect of tamoxifen on its targets – ERα and ERβ – in non-breast-cancer tissues; thus it is reasonable to assume that genetic variants in ERα and ERβ may be associated with tamoxifen clinical outcomes.
Although well-studied genetic variants in the ERα gene (ESR1 polymorphisms PυuII and XbaI) do not cause amino acid changes, they have been associated with several clinical phenotypes, presumably by inducing changes in protein levels. In the observational study conducted by COBRA investigators, tamoxifen was associated with a statistically significant reduction in total cholesterol in postmenopausal women with the ERα ESR1 XbaI GG genotype and in triglycerides in women with the ERβ ESR2-02 GG genotypes (Ref. 39). In premenopausal women, the ESR1 XbaI genotypes were associated with tamoxifen-induced changes in triglycerides and high-density lipoprotein. Preliminary data have also suggested that bone mineral density may be influenced by ER genotypes (Ref. 40).
In addition, preliminary data suggest that tamoxifen-induced hot flushes are influenced by ER genotypes. Postmenopausal women with ESR1 PυuII CC and ESR2-02 GG genotypes reported the greatest increase in hot-flush scores (P = 0.0007) compared with women with other alleles. By contrast, women with ESR2-02 AA genotype were less likely to experience tamoxifen-induced hot flushes than women who carried at least one ESR-02 G allele (HR = 0.26, 95% confidence interval = 0.10–0.63; P = 0.001) (Ref. 41).
One report suggested that women with normal CYP2D6 genotype were more likely to report hot flushes compared with women who were IMs or PMs (Ref. 27). Among the 223 patients included in the NCCTG study, a total of 61% (n = 136) reported having hot flushes, with 40% (n = 90) reporting mild (grade 1), 15% (n = 34) reporting moderate (grade 2), and 5% (n = 12) reporting severe (grade 3) hot flushes. By contrast, none (0/13) of the PMs had moderate or severe hot flushes compared with 20% (36/177) for IMs and EMs (one-sided P = 0.06). At the same time, others did not observe a correlation between CYP2D6 and hot flushes (Ref. 35). It is possible that EMs are more likely to suffer the anti-oestrogenic effects due to an increase in endoxifen concentrations compared with other groups. Indeed, a recent report suggested that EMs are more likely to discontinue tamoxifen during the first year of treatment compared with women who are IMs or PMs (Ref. 42).
Conclusions and future directions
Several studies have provided an initial correlation between variants in CYP2D6 and tamoxifen metabolism, efficacy and safety. More-preliminary studies suggest a role for genetic variants in other genes in tamoxifen efficacy. Overall, the data reported to date suggest that polymorphisms in ER genotypes and CYP2D6 may be useful in selecting women who would gain the greatest benefit from tamoxifen and those who may be more susceptible to adverse effects. However, prospective studies are needed to validate preliminary findings and to further delineate the magnitude of effect. Such investigations will help provide definitive answers to the question of CYP2D6 activity and tamoxifen response. In addition, the role of other genes that influence tamoxifen metabolism and elimination and its target should be further investigated; indeed, it is possible that a multigene assay will be required.
Because many SSRI/SNRI agents are available, it is recommended that women receiving tamoxifen should be prescribed a weak CYP2D6 inhibitor such as venlafaxine or citalopram. Women who have benefited from long-term use of drugs that are strong inhibitors of CYP2D6 such as paroxetine or fluoxetine may need to either continue such use if it has been the best treatment for their underlying disease or consider transitioning into another agent under the direction of a primary care physician or a psychiatrist.
Aromatase inhibitors are also commonly used in the treatment of postmenopausal breast cancer, but pharmacogenetic studies on these are few. The role of genetic polymorphism in the aromatase gene (CYP19) has been extensively investigated as a determinant of breast cancer risk. In one study, time to progression was significantly improved in patients with one CYP19 variant (rs4646), compared with those with a wild-type gene (17.2 versus 6.4 months; P = 0.02) (Ref. 43). Ongoing studies are evaluating the effects of pharmacogenetics of aromatase and drug-metabolising enzymes on several aromatase inhibitors such as anastrozole, letrozole and exementane. Such studies will also consider variation in aromatase inhibitor activity on oestrogen concentrations, breast density, bone density, lipids and other factors.
It is hoped that the results from the tamoxifen and aromatase inhibitor studies may be useful in the future to predict response to individual agents and may help individualise treatments based on host genetic factors. In addition to hormone therapies, many women with breast cancer will be prescribed chemotherapy. Pharmacogenetics may be important in determining response or resistance to different agents. However, little work has been done to evaluate such effects. Agents that are commonly used in the treatment of breast cancer include the anthracyclines doxorubicin and epirubicin and the taxanes paclitaxel and docetaxel; other agents include cyclophosphamide, vinorelbine, gemcitabine and 5-fluorouracil. Potential pharmacogenetics of response and side effects to such agents have been reviewed elsewhere (Ref. 44).
In summary, data available to date suggest that reduced CYP2D6 activity leads to low concentration of an active metabolite and possibly to poor response to tamoxifen treatment. While women receiving tamoxifen treatment who have CYP2D6 polymorphisms leading to reduced activity or who take drugs that inhibit CYP2D6 may have worse long-term outcome, a direct correlation between change in the metabolic profile of tamoxifen and long-term outcome has not been evaluated. More data are needed to determine pharmacogenetic effects on aromatase inhibitor action or on specific chemotherapy agents. Finally, additional information is required to assess pharmacogenetic effects on treatment-related toxicity. Until prospective data are available we do not recommend routine CYP2D6 testing for women prescribed tamoxifen. However, it is reasonable to consider testing in select women for whom tamoxifen is considered but who may have alternative treatment options; for example, a postmenopausal woman with osteoporosis may be treated with an aromatase inhibitor and a bisphosphonate if she is a poor metaboliser, but may be better served with tamoxifen if she is an extensive metaboliser. Indeed, the promise of pharmacogenetic testing is that the most effective yet least toxic agent will be prescribed to an individual.
Acknowledgments
Acknowledgements and funding The authors are supported in part by Pharmacogenetics Research Network grants U-01 GM61373. We thank Drs Desta, Flockhart and Hayes for helpful discussions, and the anonymous reviewers for constructive suggestions.
Footnotes
Citation details for this article Vered Stearns and James M. Rae (2008) Pharmacogenetics and breast cancer endocrine therapy: CYP2D6 as a predictive factor for tamoxifen metabolism and drug response?. Expert Rev. Mol. Med. Vol. 10, e34, November 2008, doi:10.1017/S1462399408000896
References
- 1.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Berry DA, et al. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 3.Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348:529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 4.Evans WE, McLeod HL. Pharmacogenomics–drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 5.Winer EP, et al. American Society of Clinical Oncology technology assessment on the use of aromatase inhibitors as adjuvant therapy for postmenopausal women with hormone receptor-positive breast cancer: status report 2004. J Clin Oncol. 2005;23:619–629. doi: 10.1200/JCO.2005.09.121. [DOI] [PubMed] [Google Scholar]
- 6.Brown RJ, Davidson NE. Adjuvant hormonal therapy for premenopausal women with breast cancer. Semin Oncol. 2006;33:657–663. doi: 10.1053/j.seminoncol.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Desta Z, et al. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 8.Sun D, et al. Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab Dispos. 2007;35:2006–2014. doi: 10.1124/dmd.107.017145. [DOI] [PubMed] [Google Scholar]
- 9.Nishiyama T, et al. Reverse geometrical selectivity in glucuronidation and sulfation of cis- and trans-4-hydroxytamoxifens by human liver UDP-glucuronosyltransferases and sulfotransferases. Biochem Pharmacol. 2002;63:1817–1830. doi: 10.1016/s0006-2952(02)00994-2. [DOI] [PubMed] [Google Scholar]
- 10.Nowell S, Falany CN. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene. 2006;25:1673–1678. doi: 10.1038/sj.onc.1209376. [DOI] [PubMed] [Google Scholar]
- 11.Dehal SS, Kupfer D. Cytochrome P-450 3A and 2D6 catalyze ortho hydroxylation of 4-hydroxytamoxifen and 3-hydroxytamoxifen (droloxifene) yielding tamoxifen catechol: involvement of catechols in covalent binding to hepatic proteins. Drug Metab Dispos. 1999;27:681–688. [PubMed] [Google Scholar]
- 12.Stearns V, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 13.Lien EA, et al. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989;49:2175–2183. [PubMed] [Google Scholar]
- 14.Borgna JL, Rochefort H. Hydroxylated metabolites of tamoxifen are formed in vivo and bound to estrogen receptor in target tissues. J Biol Chem. 1981;256:859–868. [PubMed] [Google Scholar]
- 15.Johnson MD, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 16.Lim YC, et al. Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J Pharmacol Exp Ther. 2006;318:503–512. doi: 10.1124/jpet.105.100511. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006;25:1679–1691. doi: 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- 18.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 19.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]
- 20.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 21.Alfaro CL, et al. CYP2D6 inhibition by fluoxetine, paroxetine, sertraline, and venlafaxine in a crossover study: intraindividual variability and plasma concentration correlations. J Clin Pharmacol. 2000;40:58–66. doi: 10.1177/00912700022008702. [DOI] [PubMed] [Google Scholar]
- 22.Jin Y, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 23.Borges S, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Stearns V. Clinical update: new treatments for hot flushes. Lancet. 2007;369:2062–2064. doi: 10.1016/S0140-6736(07)60959-3. [DOI] [PubMed] [Google Scholar]
- 25.Rae JM, et al. Genotyping for polymorphic drug metabolizing enzymes from paraffin-embedded and immunohistochemically stained tumor samples. Pharmacogenetics. 2003;13:501–507. doi: 10.1097/00008571-200308000-00008. [DOI] [PubMed] [Google Scholar]
- 26.Ingle JN, et al. Randomized trial of tamoxifen alone or combined with fluoxymesterone as adjuvant therapy in postmenopausal women with resected estrogen receptor positive breast cancer. North Central Cancer Treatment Group Trial 89-30-52. Breast Cancer Res Treat. 2006;98:217–222. doi: 10.1007/s10549-005-9152-1. [DOI] [PubMed] [Google Scholar]
- 27.Goetz MP, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 28.Goetz MP, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 29.Wegman P, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wegman P, et al. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowell SA, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 32.Schroth W, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 33.Lim HS, et al. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol. 2007;25:3837–3845. doi: 10.1200/JCO.2007.11.4850. [DOI] [PubMed] [Google Scholar]
- 34.Kiyotani K, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008;99:995–999. doi: 10.1111/j.1349-7006.2008.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonanni B, et al. Polymorphism in the CYP2D6 tamoxifen-metabolizing gene influences clinical effect but not hot flashes: data from the Italian Tamoxifen Trial. J Clin Oncol. 2006;24:3708–3709. doi: 10.1200/JCO.2006.06.8072. author reply 3709. [DOI] [PubMed] [Google Scholar]
- 36.Blake MJ, et al. Ontogeny of dextromethorphan O- and N-demethylation in the first year of life. Clin Pharmacol Ther. 2007;81:510–516. doi: 10.1038/sj.clpt.6100101. [DOI] [PubMed] [Google Scholar]
- 37.Gaedigk A, et al. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83:234–242. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- 38.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 39.Ntukidem NI, et al. Estrogen receptor genotypes, menopausal status, and the lipid effects of tamoxifen. Clin Pharmacol Ther. 2008;83:702–710. doi: 10.1038/sj.clpt.6100343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henry NL, et al. Association of chemotherapy and estrogen receptor genotype with change in bone mineral density after one year of tamoxifen therapy. Breast Cancer Res Treat. 2007;106:S118. abstract. [Google Scholar]
- 41.Stearns V, et al. Tamoxifen-induced hot flashes are associated with estrogen receptor polymorphisms. J Clin Oncol. 2007;25:18S. abstract. [Google Scholar]
- 42.Rae JM, et al. Cytochrome P450 2D6 activity predicts adherence to tamoxifen therapy. Breast Cancer Res Treat. 2007;106:S21. abstract. [Google Scholar]
- 43.Colomer R, et al. A single-nucleotide polymorphism in the aromatase gene is associated with the efficacy of the aromatase inhibitor letrozole in advanced breast carcinoma. Clin Cancer Res. 2008;14:811–816. doi: 10.1158/1078-0432.CCR-07-1923. [DOI] [PubMed] [Google Scholar]
- 44.Stearns V, Davidson NE, Flockhart DA. Pharmacogenetics in the treatment of breast cancer. Pharmacogenomics J. 2004;4:143–153. doi: 10.1038/sj.tpj.6500242. This review details common therapies for breast cancer and potential genetic variants that may influence efficacy or safety. An excellent link to detailed clinical information on drug interactions, including references, is provided via the website of the Indiana University School of Medicine, Department of Medicine, Division of Clinical Pharmacology: http://medicine.iupui.edu/flockhart/ [DOI] [PubMed]
- 45.Kotlyar M, et al. Inhibition of CYP2D6 activity by bupropion. J Clin Psychopharmacol. 2005;25:226–229. doi: 10.1097/01.jcp.0000162805.46453.e3. [DOI] [PubMed] [Google Scholar]
- 46.Branch RA, et al. In vivo modulation of CYP enzymes by quinidine and rifampin. Clin Pharmacol Ther. 2000;68:401–411. doi: 10.1067/mcp.2000.110561. [DOI] [PubMed] [Google Scholar]
- 47.Skinner MH, et al. Duloxetine is both an inhibitor and a substrate of cytochrome P4502D6 in healthy volunteers. Clin Pharmacol Ther. 2003;73:170–177. doi: 10.1067/mcp.2003.28. [DOI] [PubMed] [Google Scholar]
- 48.Baumann P, et al. Dextromethorphan and mephenytoin phenotyping of patients treated with thioridazine or amitriptyline. Ther Drug Monit. 1992;14:1–8. doi: 10.1097/00007691-199202000-00001. [DOI] [PubMed] [Google Scholar]
- 49.Fukumoto K, et al. Effect of amiodarone on the serum concentration/dose ratio of metoprolol in patients with cardiac arrhythmia. Drug Metab Pharmacokinet. 2006;21:501–505. doi: 10.2133/dmpk.21.501. [DOI] [PubMed] [Google Scholar]
- 50.Hamelin BA, et al. Significant interaction between the nonprescription antihistamine diphenhydramine and the CYP2D6 substrate metoprolol in healthy men with high or low CYP2D6 activity. Clin Pharmacol Ther. 2000;67:466–477. doi: 10.1067/mcp.2000.106464. [DOI] [PubMed] [Google Scholar]
- 51.Ruderman J, et al. Control of the cell cycle in early embryos. Cold Spring Harb Symp Quant Biol. 1991;56:495–502. doi: 10.1101/sqb.1991.056.01.056. [DOI] [PubMed] [Google Scholar]
- 52.Preskorn SH. Clinically relevant pharmacology of selective serotonin reuptake inhibitors. An overview with emphasis on pharmacokinetics and effects on oxidative drug metabolism. Clin Pharmacokinet. 1997;32(Suppl 1):1–21. doi: 10.2165/00003088-199700321-00003. [DOI] [PubMed] [Google Scholar]
- 53.Preskorn SH, et al. Comparison of duloxetine, escitalopram, and sertraline effects on cytochrome P450 2D6 function in healthy volunteers. J Clin Psychopharmacol. 2007;27:28–34. doi: 10.1097/00004714-200702000-00005. [DOI] [PubMed] [Google Scholar]


