Abstract
Cardiac infections caused by the foodborne bacterium Listeria monocytogenes represent a significant but poorly studied facet of disease. It is not known whether L. monocytogenes cardiac infections stem solely from host susceptibility, or whether bacterial isolates exist that exhibit a tropism for cardiac tissue. Here we examine the cardio-invasive capacity of a recent L. monocytogenes cardiac case strain (07PF0776) as well as nine additional outbreak and clinical isolates. Mice infected with the cardiac isolate 07PF0776 had 10-fold more bacteria recovered from heart tissue than those infected with L. monocytogenes strain 10403S, a well-characterized clinical isolate originally obtained from a human skin lesion. Additional L. monocytogenes isolates exhibited varied capacities to colonize the hearts of mice; however, those with the highest efficiency of mouse cardiac invasion also demonstrated the highest levels of bacterial invasion in cultured myoblast cells. Our findings strongly suggest that subpopulations of L. monocytogenes strains have acquired an enhanced ability to target and invade the myocardium.
INTRODUCTION
Listeria monocytogenes is a foodborne, facultative, intracellular bacterial pathogen that infects humans and causes disease following translocation across the gut epithelial barrier (Swaminathan & Gerner-Smidt, 2007). While healthy individuals usually exhibit mild forms of disease such as gastroenteritis, immunocompromised individuals and elderly patients often suffer more severe forms of illness, which include meningoencephalitis and septicaemia, with 20–30 % of these patients succumbing to infection despite antibiotic treatment (Mead et al., 1999; Vázquez-Boland et al., 2001). A significant but much less documented sequela of L. monocytogenes infections involves the heart. This type of infection is estimated to occur in at least 7–10 % of those with systemic disease; however, it is exceedingly rare in HIV-AIDS (acquired immunodeficiency syndrome) patients (0.5 %) (Berenguer et al., 1991; Brouqui & Raoult, 2001; Brusch, 2001; Kales & Holzman, 1990). Cardiac infection with L. monocytogenes most often manifests as endocarditis or invasive myocarditis (Antolín et al., 2008; Haddad et al., 2007; Hill et al., 2006; Lindholm, 2008; Llanwarne et al., 2007; Pocar et al., 2009; Rua Galisteo et al., 2007). Approximately 60 % of endocarditis cases are associated with individuals who have prior valvular abnormalities (Antolín et al., 2008), and 50 % of those infected require valve replacement due to severe damage and failure (Brouqui & Raoult, 2001).
The death rate from cardiac illness is estimated to be up to 35 % despite treatment, and yet very little is known regarding L. monocytogenes colonization of cardiac tissue and its resultant pathologies (Antolín et al., 2008). Although less common in comparison to Staphylococcus aureus or viridans streptococci-associated heart infections, bacterial endocarditis due to L. monocytogenes infection still requires careful consideration for diagnosis and treatment in susceptible individuals due to its high mortality (Brouqui & Raoult, 2001; Hill et al., 2006). Additionally, cases of bacterial myocarditis and abscess formation due to L. monocytogenes infection have been described (Haddad et al., 2007; Hood & Baxter, 1999; Makaryus et al., 2004; McCue & Moore, 1979). Although myocarditis and abscess formation represent a smaller proportion of L. monocytogenes cardiac cases, these cases are noteworthy in that the organism colonizes what had previously been assumed to be a non-permissive site for bacterial replication (Luo et al., 2003). While the majority of patients with endocarditis or invasive myocarditis due to L. monocytogenes can be considered immunocompromised (Brusch, 2001), at least two documented cases of Listeria-associated cardiac illness have occurred in patients who were apparently completely healthy prior to infection (Adler et al., 2009; Haddad et al., 2007).
It is not known whether L. monocytogenes cardiac manifestations occur as a result of patient predisposition or whether they result from strains that possess enhanced colonization capacities. A study by Luo et al. (2003) suggested that the heart is not normally a tissue that is permissive for L. monocytogenes colonization. Here we describe the examination of a series of clinical and food outbreak L. monocytogenes isolates for cardiac infection, including a recent isolate (07PF0776) cultured from an HIV-infected patient who had a non-resuscitatable asystolic arrest due to invasive L. monocytogenes infection of the heart. Our studies indicate that a subpopulation of L. monocytogenes strains (that includes 07PF0776) exhibit an enhanced capacity to colonize cardiac tissue.
METHODS
Bacterial strains and culture conditions.
All bacterial strains used are described in Table 1 along with serotype and source. J4403, J5043 and J4901 were obtained courtesy of Dr Lewis Graves at the CDC. Clinical isolate 07PF0776 was obtained from both the Massachusetts Public Health Laboratory and from the CDC (where it is classified as J4533). Isolate NWI4b was obtained courtesy of the Children's Memorial Hospital and Northwestern Memorial Hospital Chicago, IL, USA, from Dr Stanley Schulman (Adler et al., 2009). All other strains were derived from laboratory stocks, and the original sources from which they were isolated are indicated. Prior to experiments, all strains were grown statically overnight from a single colony in 2 ml Brain Heart Infusion (BHI) broth (Difco Laboratories) at 37 °C.
Table 1.
Clinical and outbreak isolates used in this study
| Strain no. | Designation | Isolate type | Serotype |
|---|---|---|---|
| NF-L100 | 10403S | Virulent laboratory strain (skin lesion) | 1/2a |
| NF-L101 | EGDe | Virulent laboratory strain (guinea pig) | 1/2a |
| NF-L725 | ScottA | Virulent laboratory strain (foodborne) | 4b |
| NF-L828 | F2379 | Foodborne – Los Angeles (Jalisco cheese) | 4b |
| NF-L831 | OIS4b | Foodborne – Sweden (Brie cheese) | 4b |
| NF-L832 | OIJ4b | Outbreak – Japan | 4b |
| NF-L1403 | 07PF0776 | Clinical isolate – cardiac interventricular septal abscess | 4b |
| NF-L1590 | J4403 | Clinical isolate – pericardial fluid | 1/2b |
| NF-L1591 | J4901 | Clinical isolate – heart tissue | nt |
| NF-L1592 | J5043 | Clinical isolate – patient blood | 4b |
| NF-L1776 | NWI4b | Clinical isolate – cardiac pseudotumour | nd |
nt, Not typable; nd, not determined.
Case report and isolation of L. monocytogenes 07PF0776.
A 51-year-old HIV-infected man with dyslipidaemia and hypertension presented with chest pain, dyspnoea, transient fevers and chills for 3 weeks, and evidence of inferior ischaemia by electrocardiogram on admission. Cardiac markers indicated that his myoglobin B fraction was elevated. The patient's CD4+ T-cell count was 207 cells mm−3 with an HIV RNA level of 310 765 copies ml−1, and he had not been on any antiretroviral or preventative treatment for opportunistic infection for over 1 year. Intermittent chest pain continued, and on the second hospital day his temperature rose to 39.2 °C transiently, but he was not started on antibiotics. On day three, his vital signs were normal that morning, but 1 h later he became asystolic and could not be resuscitated. Five of six blood cultures later grew L. monocytogenes.
The organism (from here on referred to as 07PF0776) was isolated from blood cultures from the patient by the Baystate Health Microbiology Laboratory. The isolate was biochemically characterized as L. monocytogenes using standard procedures by the Massachusetts Public Health Department and the CDC Foodborne and Diarrheal Diseases Branch, Division of Bacterial and Mycotic Diseases, National Center for Infectious Diseases, and confirmed by AccuProbe (GenProbe) (Weaver, 1989). Serological typing was by the method of Seeliger & Höhne (1979). PFGE was performed using AscI and ApaI restriction endonuclease digests of bacterial DNA in accordance with the CDC PulseNet standardized protocol (Graves & Swaminathan, 2001). Computer-assisted analysis of PFGE patterns and comparison with patterns in the PulseNet national database was performed using Molecular Analyst Fingerprinting Plus with Data Sharing Tools Version 1.6 (Bio-Rad Laboratories) (Graves & Swaminathan, 2001; Graves et al., 2005). The CDC pattern identifiers for the cardiac isolate are GX6A.1019 for the AscI pattern and GX6A.12.0088 for the ApaI pattern.
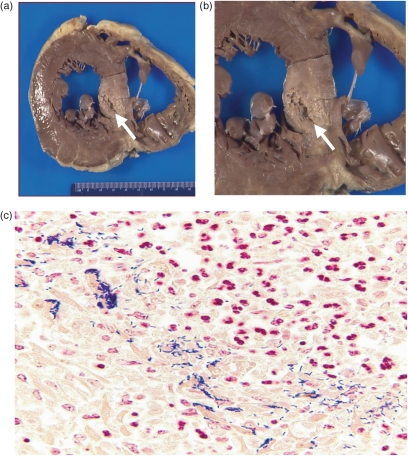
Gross and microscopic autopsy analysis was performed by the Baystate Health Center Department of Pathology (Springfield, MA, USA) on the cardiovascular, respiratory, gastrointestinal, hepatobiliary systems and pancreas, urogenital, endocrine, haematopoietic, musculoskeletal and central nervous systems of the deceased. Haematoxylin, Brown and Brenn, acid-fast bacilli and Gomori methenamine–silver stains were used for analysis. Gross examination revealed an enlarged heart with left ventricular hypertrophy, and an interventricular septal abscess of 3.5×3.5×3.0 cm (Fig. 1a, b). There were no valvular or other lesions in the heart or great vessels, and the coronary arteries were free of obstruction. Clusters of L. monocytogenes were seen by Brown and Brenn stain along with a neutrophilic infiltrate (Fig. 1c). It was difficult to discern from the specimen whether the bacteria were intracellular, extracellular, or both. There were also clusters of L. monocytogenes within the hepatic sinusoids (data not shown). There was no sign of infection in the other organs. The cause of death was likely to be a fatal arrhythmia due to death of myocardial cells in the interventricular septum and disruption of the main impulse conduction pathway of the heart from the invasive infection.
Fig. 1.
Gross and microscopic examination reveals the presence of an abscess within the interventricular septum (IVS) of the heart as well as bacillary micro-organisms in and around cardiac tissue. (a) Cross-section of patient's heart. The white arrow denotes the location of an abscess within the IVS. (b) Enlarged view of the abscess shown in (a). (c) Brown and Brenn stain of the cardiac septal abscess showing both the presence of many bacillary organisms and a pronounced neutrophilic infiltrate.
Mouse intravenous infections.
All animal procedures were IACUC approved and performed in the Biological Resources Laboratory at the University of Illinois at Chicago. Intravenous inoculation of mice via tail vein was performed as previously described (Alonzo et al., 2009) with some modifications. Six to eight week old female ND4 Swiss Webster mice (Harlan) were injected with 0.2 ml PBS [0.144 g KH2PO4 ml−1, 9.00 g NaCl ml−1, 0.795 g Na2HPO4 (anhydrous) ml−1] containing 2×104 c.f.u. of each strain of L. monocytogenes. For time-course experiments, 15 mice were inoculated and at 24, 48 and 72 h post-inoculation, five mice were sacrificed and livers, spleens and hearts were dissected. Organs were homogenized and dilutions of the tissue homogenate were spread onto BHI agar plates to enumerate bacterial burdens to each organ. For the multistrain comparisons of bacterial burden to the heart, mice were inoculated as described, but were sacrificed at only the 24 h time point. All animal experiments were repeated a minimum of three times. Statistics were calculated using a one-way ANOVA with Tukey's multiple comparison test, and P<0.05 was considered significant.
Intracellular growth in tissue culture cells.
The H9C2 cell line (ATCC CRL-1446) was kindly provided by Dr David Engman (Northwestern University, Chicago, IL, USA) and serves as a rat myoblast cell line routinely used for cardiac-related pathogenesis studies (Hyland et al., 2008; Lee et al., 2006; Tyler et al., 2005). H9C2 and J774 mouse macrophage-like cells were maintained in DMEM supplemented with penicillin (100 μg ml−1), streptomycin (100 μg ml−1), l-glutamine (292 μg ml−1) and 10 % fetal bovine serum (Hyclone) and grown at 37 °C with 5 % CO2. Monolayers of cells were grown on acid-washed coverslips in DMEM without antibiotics. Cells were infected with L. monocytogenes at an m.o.i. of 100 bacteria to 1 myoblast (H9C2) or 0.1 bacteria to 1 macrophage (J774). After 1 h, cells were washed three times with 37 °C PBS followed by the addition of 37 °C DMEM containing gentamicin (15 μg ml−1). At the time points indicated, coverslips were removed and lysed in 5 ml H2O with vigorous vortexing, or were processed for fluorescence microscopy. Dilutions of lysates were spread onto LB agar for enumeration of bacterial c.f.u. Fluorescence staining was performed as previously described (Mueller & Freitag, 2005) and images were obtained on a DeltaVision microscope (Applied Precision). Images are representative of at least ten independently viewed fields and were captured using SoftWorx image software (Applied Precision).
Invasion assays.
H9C2 cells were infected at an m.o.i. of 100 : 1. After 1 h, cells were washed three times with 37 °C PBS followed by removal of coverslips for enumeration of total bacterial c.f.u. 37 °C DMEM containing gentamicin (15 μg ml−1) was then added to the remaining cells and allowed to incubate for an hour at 37 °C. Additional coverslips were then removed and intracellular bacterial c.f.u. were enumerated. The ratio of intracellular bacteria to that of the total inoculum was calculated and indicated as per cent invasion. Assays were conducted three times in triplicate.
L2 plaque assays.
Plaque assays were conducted as previously described (Sun et al., 1990). A monolayer of L2 fibroblast cells was infected with L. monocytogenes at an m.o.i. of 30 : 1 for 1 h followed by the addition of gentamicin (10 μg ml−1) in a DMEM/0.7 % agarose overlay. After 72 h, plaques were visualized by staining with Neutral Red and measured with a micrometer. A minimum of three plaque assays were performed with approximately 25 plaques measured per experiment.
Haemolytic activity.
Haemolytic activity and phospholipase activity were measured as described by Alonzo et al. (2009). Briefly, overnight cultures of L. monocytogenes in LB were diluted 1 : 10 into 10 ml fresh LB and grown for 5 h at 37 °C with shaking. After 5 h, OD600 readings were measured and ∼1.2 ml bacterial culture was centrifuged at maximum speed in a microcentrifuge. After centrifugation, supernatants were removed and bacterial pellets were discarded. The supernatants from cultures with higher optical densities after 5 h of growth were normalized by diluting into fresh LB, thus accounting for differences in bacterial abundance between strains. Serial dilutions of culture supernatants were made into PBS+1 mM DTT (pH 5.0) followed by the addition of sheep red blood cells (RBCs). After incubation at 37 °C for 30 min, supernatant/RBC mixtures were centrifuged at maximum speed for 1 min and the dilution resulting in 50 % lysis of RBCs based on visual examination of RBC pellets was recorded. Haemolytic units are defined as the reciprocal of the culture supernatant dilution required for 50 % RBC lysis. Experiments were conducted a minimum of three times in triplicate.
Phospholipase activity.
Phospholipase activity was assessed by streaking single colonies of each L. monocytogenes strain onto egg yolk agar plates containing 25 mM glucose 6-phosphate, 2.5 % egg yolk and 0.2 % activated charcoal (to enhance PrfA-dependent plcB expression) (Alonzo et al., 2009). After 72 h, the white precipitates surrounding the bacterial streaks were examined. Strain 10403S was considered the standard to which all other strains were compared. Thus, a strain with increased phospholipase activity had a larger zone of opacity than 10403S, while a strain with decreased phospholipase activity exhibited a smaller zone.
Bacterial swimming motility.
Motility of isolates was measured in a soft agar assay as described by Shetron-Rama et al. (2003). One microlitre of an overnight culture of each isolate was injected into a BHI soft agar plate (0.3 % agar) and incubated at 37 °C for 48 h. Motile distance was determined by measuring the diameter of the zones of bacterial growth for each strain tested. Measurements are from a minimum of three independent experiments.
DNA sequencing and construction of cladogram relationship trees.
Bacterial genomic DNA was isolated using a DNeasy Blood/Tissue kit (Qiagen). Oligonucleotides used for sequencing hly were as follows: 1SEQllo, GAGTAATAAAACTAATGTGCG; 2SEQllo, AACAATTTCGTTACCTTCAGG; 3SEQllo, GAGAAAAAGAAGAAATCCATC; 4SEQllo, AAAAGCTATTACCATGATGAT; 5SEQllo, AATTCAATTTCATCCATGGCA; 6SEQllo, ATTTCTATTTTTCACAAGTGG. Oligonucleotides for sequencing inlA and inlB were designed as previously described (Van Stelten et al., 2010). Sequencing was carried out by the DNA Services Facility at the University of Illinois at Chicago and analysed using the ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html). DNA sequence data for previously unpublished hly, inlA and inlB genes have been submitted to GenBank and assigned accession numbers HQ703556–HQ703578.
RESULTS
L. monocytogenes human cardiac isolate 07PF0776 exhibits an enhanced capacity for colonization of heart tissue in mice
Although host physiology and immunity are critical components of infectious disease progression, pathogens have also been observed to acquire traits that increase their pathogenic capacity. Given that L. monocytogenes cardiac infections are rare in HIV-AIDS individuals (Berenguer et al., 1991; Brouqui & Raoult, 2001; Brusch, 2001; Kales & Holzman, 1990), we investigated whether a recent HIV-AIDS patient cardiac infection isolate, 07PF0776, exhibited an enhanced ability to target cardiac tissue. Mice were intravenously infected with 07PF0776 and at various time points post-infection the bacterial burdens in hearts, livers and spleens were compared to those of animals infected with an independent and well-characterized L. monocytogenes human skin lesion isolate, strain 10403S (Bishop & Hinrichs, 1987; Edman et al., 1968).
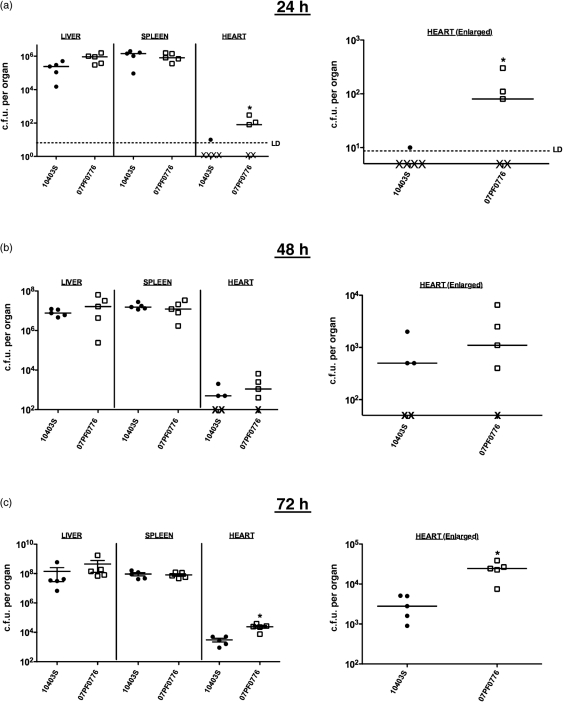
At 24 h post-infection, strains 10403S and 07PF0776 produced strikingly similar bacterial burdens in the livers and spleens of infected mice, and both strains established infection in these tissues with similar efficiencies (Fig. 2). In contrast to the recovery of bacteria from liver and spleen, mice infected with 07PF0776 had increased bacterial burdens in the heart as well as an increased efficiency of heart colonization when compared to animals infected with 10403S (Fig. 2a). Whereas only one of five mice infected with 10403S contained detectable numbers of bacteria within the heart, three of five mice infected with 07PF0776 had bacteria recovered from cardiac tissue. Animals infected with 07PF0776 were observed to have 10–15-fold more bacteria in the heart than the animals for which 10403S could be recovered. At 48 h post-infection, the bacterial burdens in the livers and spleens of animals infected with either 07PF0776 or 10403S were nearly identical, and differences in bacterial burdens in the hearts were less pronounced and not of statistical significance (Fig. 2b). The 07PF0776 case isolate still exhibited a greater efficiency of heart infection than 10403S with four out of five 07PF0776-infected mice having detectable bacterial c.f.u. recovered from the hearts in comparison with three of five mice infected with 10403S (Fig. 2b). At 72 h post-infection, however, the difference in bacterial burdens in the hearts of animals infected with 07PF0776 was 10-fold greater than observed for those infected with 10403S, while the numbers of bacteria recovered from the livers and spleens remained nearly identical (Fig. 2c). These data indicate an enhanced proclivity of strain 07PF0776 for cardiac infection in comparison to 10403S.
Fig. 2.
Strain 07PF0776 exhibits an enhanced ability to colonize cardiac tissue of infected mice. Bacterial burdens present within livers, spleens and hearts at 24 h (a), 48 h (b) and 72 h (c) following intravenous inoculation of 2×104 c.f.u. of L. monocytogenes. X below the dashed limit of detection (LD) line represents mice that either contained no bacteria within the heart or whose bacterial burdens were below the limit of detection. Statistics were calculated using a one-way ANOVA with Dunnett's post-test. *, P<0.05. •, 10403S; □, 07PF0776.
Additional L. monocytogenes isolates can be grouped based on their capacity for cardiac colonization
Based on the enhanced ability of 07PF0776 to colonize the hearts of infected mice, it seemed of interest to determine whether cardiac infection by L. monocytogenes represents a trait shared by many strains or alternatively one that is limited to a few strains or even unique to 07PF0776. Nine additional clinical and outbreak strains of L. monocytogenes were thus compared for their capacity to establish cardiac infections (Table 1). Included in this comparison were strains isolated from patients with cardiac-associated Listeria infections (J4403, J4901 and NWI4b) and a blood isolate (J5043) as well as the first genome sequenced strain (EDGe) and a variety of strains isolated from independent foodborne outbreaks (ScottA, OIS4b and OIJ4b). The comparison included strains of serotypes 1/2a, 1/2b and 4b, and one strain designated non-typable (Table 1). This sampling of isolates thus represented the two serotypes most commonly associated with systemic disease (1/2a and 4b), as well as isolates directly associated with cardiac disease and isolates obtained from food as well as blood samples. Each isolate was examined for its ability to colonize the hearts of intravenously infected mice. Consistent with the results obtained for strains 07PF0776 and 10403S, similar numbers of bacteria were recovered from the livers and spleens of infected animals for each isolate at 24 h post-infection (Fig. 3a, b). No statistically significant differences in bacterial c.f.u. were detected for isolates recovered from either organ.
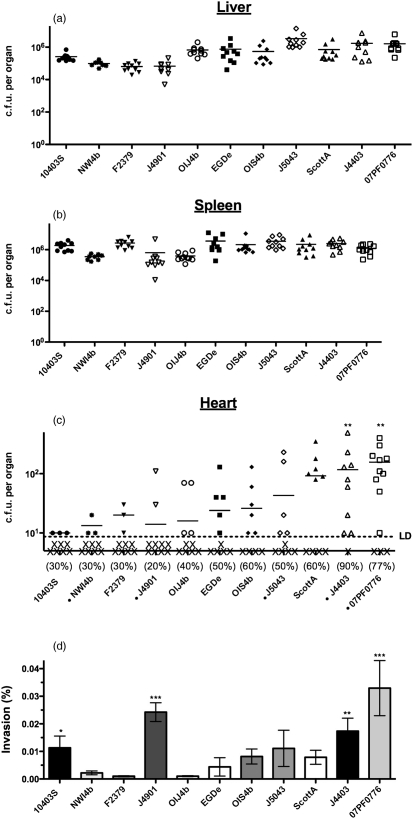
Fig. 3.
Multistrain comparisons reveal a subset of strains with enhanced invasion characteristics. Mice were intravenously inoculated with 2×104 c.f.u. of 1 of 11 isolates of L. monocytogenes. Bacterial burdens to the liver (a), spleen (b) and heart (c) were quantified 24 h post-inoculation. The percentage of mice containing quantifiable bacteria within the heart is indicated beneath the scatter plot for each individual strain. X below the dashed limit of detection (LD) line represents mice that either contained no bacteria within the heart or whose bacterial burdens were below the limit of detection. (d) Measurement of bacterial invasion efficiency by gentamicin protection of H9C2 cardiomyoblasts. Per cent invasion is the ratio of total intracellular bacteria to the bacterial inoculum. Statistics were calculated using a one-way ANOVA with Tukey's multiple comparison test. Statistically significant increases in burden compared to the five least invasive strains are indicated: *, P<0.01; **, P<0.001; ***, P<0.0001. •, 10403s; ✶, NW4b; ▾, F2379; ▿, J4901; ○, OIJ4b; ▪, EGDe; ⧫, OIS4b; ◊, J5043; ▴, ScottA; ▵, J4403; □, 07PF0776. Strains marked with a small black circle are of cardiac or patient blood origin.
Interestingly, striking differences were observed when strains were examined for colonization of mouse hearts (Fig. 3c). Isolate 07PF0776 had a high efficiency of heart infection (11 out of 14 mice with detectable c.f.u. within the heart) as well as the largest numbers of bacterial c.f.u. recovered. An additional cardiac isolate, J4403, was also recovered with high efficiency from the hearts of infected mice (nine out of ten animals infected) and in numbers that resembled those of 07PF0776 (Fig. 3c). The remaining strains were found to vary in their ability to colonize heart tissue. Indeed, there were no detectable bacteria in the hearts of the majority of infected animals for several strains, including the cardiac-associated patient isolates NWI4b and J4901 (Fig. 3c). It is of interest to note that isolates recovered most abundantly from the hearts of infected animals were not recovered from the livers or the spleens in increased numbers in comparison to other strains. In contrast, the isolates recovered in the largest numbers from the livers and/or spleens of mice had the lowest numbers of bacteria recovered from the heart (F2379, EGDe, J5043 and 10403S) (Fig. 3a–c). Taken together, these data indicate that cardiac isolates J4403 and 07PF0776 appeared better able to target, infect and/or replicate within cardiac tissue than the other examined isolates, including isolates associated with other patient cardiac infections.
A majority of L. monocytogenes isolates replicate within the cytosol of infected cardiac tissue culture cells
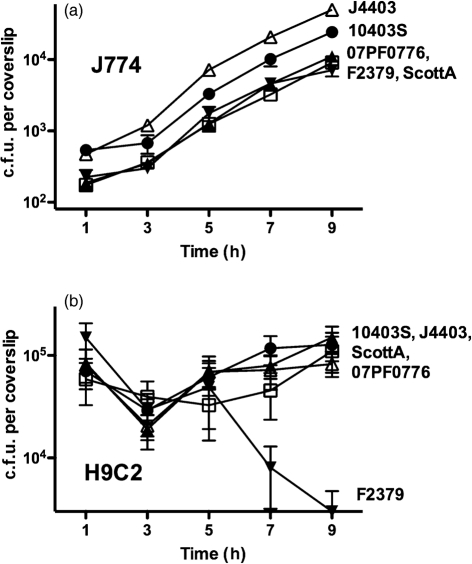
To compare the ability of selected L. monocytogenes isolates to invade host cells, escape the phagosome and replicate within the cytosol, bacterial replication was examined in murine J774 macrophage-like tissue culture cells as well as in H9C2 rat myoblast tissue culture cells. Five strains were selected as representative of bacteria with differing efficiencies of heart colonization (Fig. 3). Strains 10403S and F2379 were selected as strains displaying low-level infection of cardiac tissue in vivo, and J4403, ScottA and 07PF0776 were selected as three strains exhibiting a higher efficiency of cardiac infection. All five isolates had similar rates of replication in J774 cells although modest differences in the efficiency of bacterial uptake into macrophages were observed (Fig. 4a).
Fig. 4.
L. monocytogenes strains invade and replicate within cardiomyoblasts. (a) Intracellular growth in J774 macrophage-like cells (m.o.i. 0.1 : 1) of representative cardio-invasive/non-invasive strains of varied origin and serotype. (b) Intracellular growth in H9C2 rat myoblasts infected at an m.o.i. of 100 : 1. •, 10403S; ▵, J4403; ▴, ScottA; □, 07PF0776; ▾, F2379.
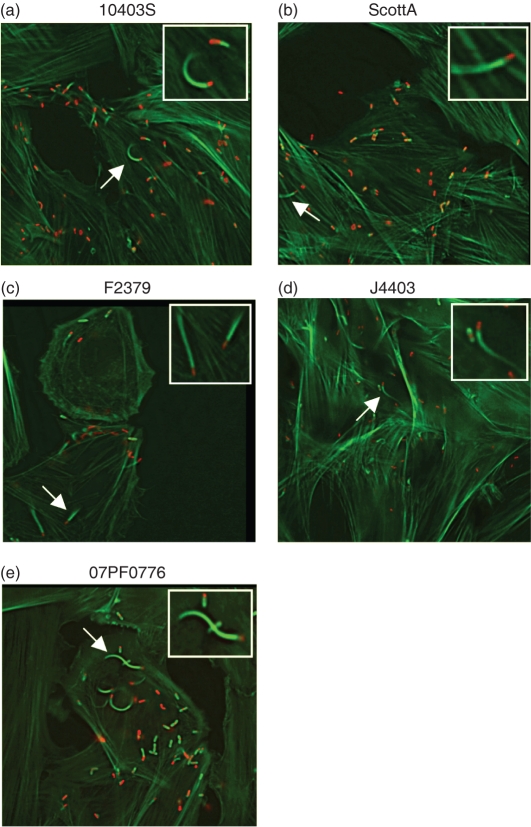
In contrast to the results observed for the infection of macrophage-like cells, differences in bacterial intracellular replication were observed for the selected isolates in rat myoblast H9C2 cells (Fig. 4b). All strains were capable of cell entry (albeit with varied efficiencies) with reduced rates of intracellular bacterial replication in comparison with the rates of growth observed in J774 cells (Fig. 4b). In J774 macrophages, the doubling times for all strains were between approximately 60 and 75 min. In the rat myoblast cells, the doubling times were increased approximately two- to threefold with ScottA having the shortest doubling time (120 min) and 07PF0776 the longest (195 min). One strain, F2379, exhibited a dramatic decrease in bacterial numbers beginning at 5 h post-infection, potentially reflective of bacterial cytotoxicity (Fig. 4b). All strains of L. monocytogenes were capable of phagosome lysis and entry into the myoblast cytosol based on bacterial association with host cell actin (Fig. 5a–e).
Fig. 5.
L. monocytogenes mediates phagosome escape and entry into the cytosol of infected cardiomyoblasts. (a–e) L. monocytogenes 10403S, J4403, ScottA, 07PF0776 and F2379 were used to infect H9C2 cells at an m.o.i. of 100 : 1 for 6.5 h. Cells were then dually stained with 7-nitrobenz-2-oxa-1,3-diazole–phallacidin (actin – green) and a secondary antibody conjugated to rhodamine (for all Listeria strains – red). Bacterial association with host cell actin is indicative of cytosolic bacteria.
L. monocytogenes strains that target the heart in vivo exhibit enhanced invasion of cardiac cells in vitro
The L. monocytogenes isolates were examined for several activities associated with virulence, including secreted haemolytic and phospholipase activities, efficiency of cell-to-cell spread and the presence of listeriolysin S (LLS) as well as flagella-mediated swimming motility (an activity that has been associated with enhanced colonization of the intestinal epithelium) (O'Neil & Marquis, 2006) (Table 2). Strains were additionally examined for their ability to invade a H9C2 rat myoblast cell line in tissue culture (Table 2 and Fig. 3d). While variability in plaque size, haemolytic activity, phospholipase activity, swimming motility and the presence/absence of LLS was observed for all strains tested, no phenotype appeared to directly correlate with the ability of a strain to establish cardiac colonization in animals (Table 2). In contrast, the efficiency of bacterial invasion of H9C2 cells was found to be significantly increased for both cardiac colonizers 07PF0776 and J4403 (0.033 % and 0.017 %, respectively) compared to that of the other isolates (ranging from 0.001 % to 0.008 %, with the exception of J4901, and to a lesser degree, 10403S) (Fig. 3d). In fact, a striking correlation appeared to exist between the efficiency of H9C2 invasion for nearly all strains tested and the overall bacterial burdens in the hearts of infected mice. These findings suggest that the L. monocytogenes isolates that efficiently target the heart for replication may do so as a result of an increased capacity for cardiac cell invasion.
Table 2.
Virulence characteristics of the selected clinical and outbreak isolates used in this study
| Strain | Haemolytic activity (HU) | Phospholipase activity | Motility (cm) | Plaque diameter (%) | LLS | Invasivity (H9C2) | Heart colonization |
|---|---|---|---|---|---|---|---|
| 10403S | + (84.4) | + | + (16.2) | + (100) | − | + | − |
| EGDe | + (100.0) | + | +/− (9.75) | + (89 %) | − | + | +/− |
| ScottA | ++ (120.0) | − | + (17.5) | + (102 %) | − | ++ | ++ |
| F2379 | + (70.0) | + | + (13.0) | + (90 %) | + | + | − |
| OIS4b | ++ (120.0) | + | +/− (10.75) | + (94 %) | + | ++ | +/− |
| OIJ4b | +++ (320.0) | +++ | + (15.75) | ++ (120 %) | + | + | +/− |
| 07PF0776 | ++ (160.0) | + | + (12.2) | ++ (117 %) | + | +++ | +++ |
| J4403 | ++ (120.0) | + | + (14.75) | ++ (121 %) | − | +++ | +++ |
| J4901 | ++ (140.0) | + | + (16.25) | − (65 %) | + | +++ | − |
| J5043 | − (37.5) | + | + (14.75) | + (102 %) | + | ++ | +/− |
| NWI4b | + (70.0) | − | + (13.25) | − (70 %) | + | + | − |
Amino acid sequence alignments of key invasion proteins reveal extensive conservation for cardio-invasive and non-invasive strains, and suggest potential polymorphisms that might contribute to altered tissue tropism
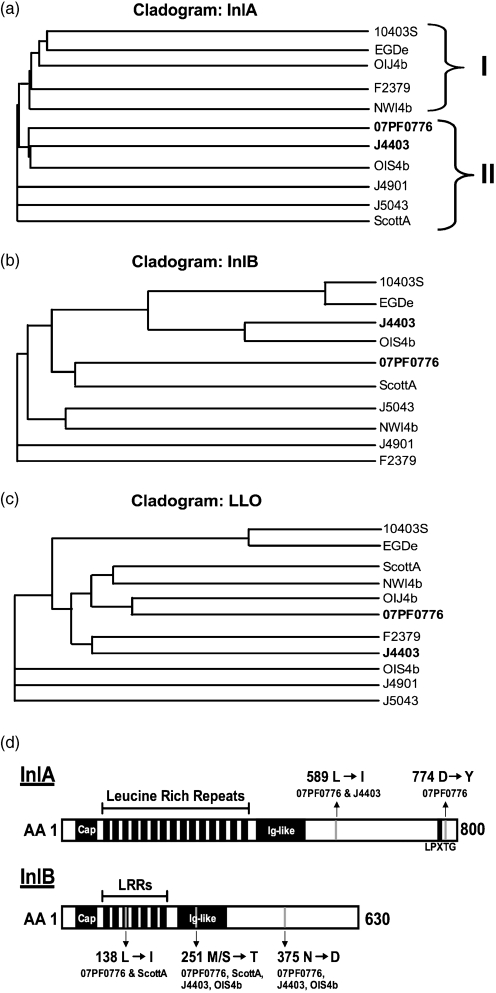
InlA and InlB are the two most well-characterized bacterial invasion proteins associated with L. monocytogenes and they directly facilitate bacterial uptake into specific host cell types (Bierne et al., 2007). As few as two amino acid substitutions within InlA have been shown to dramatically alter species-specific invasive capacity by enhancing bacterial binding to host cell receptors (Wollert et al., 2007). To determine whether there were detectable amino acid polymorphisms in either InlA or InlB that could potentially correlate with cardiac cell invasion, each gene was amplified from the L. monocytogenes isolates and amino acid sequences were compared along with those of the conserved virulence protein listeriolysin O. The construction of cladogram relationship trees suggested a potential phylogenetic link between strains with increased cardio-invasive capacity, particularly for InlA, where J4403 and 07PF0776 were most closely linked (Fig. 6a). Additionally, phylogenetic linkage based on InlA sequence suggested relationships between strains with the highest and lowest levels of cardiac invasion (compare Fig. 6a to Fig. 3c and d). It should be noted that isolate OIJ4b contained both a stop codon in inlA and significant rearrangements of inlB sequence that complicated alignment and comparison (Fig. 6b). Amino acid sequence comparisons based on cladogram relationships revealed five unique amino acid polymorphisms associated with cardio-invasive isolates for either InlA (L589I and D744Y) or InlB (L138I, M/S251T and N375D) (Fig. 6d). Overall, the isolates of Listeria that efficiently target cardiac tissue in vivo appear more closely related than those that do not, and these isolates share amino acid polymorphisms that may indicate a link of InlA and/or InlB to enhanced cardiac cell invasion.
Fig. 6.
Amino acid sequence alignment and cladogram grouping suggest that amino acid polymorphisms within InlA and InlB may contribute to the cardio-invasive phenotypes of highly invasive strains. Cladograms of the amino acid sequence alignments from (a) InlA, (b) InlB and (c) listeriolysin O (LLO). Alignments and cladograms were constructed using the ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html). For InlA comparisons, brackets indicate strains with low-level in vivo cardiac colonization (I) or mid- to high-level colonization (II). For InlB, strain OIJ4b was omitted due to substantial rearrangement of the internalin sequence resulting in insufficient sequence coverage and misalignment. (d) Schematic illustrating the unique amino acid polymorphisms present in both InlA and InlB that may influence cardiac cell invasive capacity.
DISCUSSION
Although systemic L. monocytogenes infections in humans have primarily been associated with the central nervous system, the placenta and the developing fetus, a significant percentage of invasive disease cases involve cardiac tissue (Brouqui & Raoult, 2001; Brusch, 2001). Despite this association, cardiac disease resulting from L. monocytogenes infections remains poorly defined. While host physiology and immune status influence the course and extent of L. monocytogenes infection, it is possible that some bacterial isolates possess an enhanced ability to colonize alternative tissue sites. Here we have presented evidence to suggest that subpopulations of L. monocytogenes exhibit an enhanced capacity for establishing cardiac disease in vivo resulting from increased bacterial invasion of cardiac cells, a trait potentially linked to amino acid polymorphisms in InlA and/or InlB.
While isolates 07PF0776 and J4403 appeared to efficiently target the hearts of infected mice, it remains possible that these isolates are capable of invading other host sites in addition to the heart, liver and spleen. Work by Hardy et al. (2004, 2006) has shown that L. monocytogenes 10403S colonizes the mouse gall bladder, a location with the potential to serve as a reservoir for reactivation of illness. Listeria has also been reported in the bone marrow of infected mice and humans (de Bruijn et al., 1998; Hardy et al., 2009; Khan et al., 2001). It would thus appear that the number of body sites that have the potential to serve as foci for Listeria replication may be more diverse than originally appreciated. Efforts to determine the progression and distribution of 07PF0776 and J4403 in animal infection models are currently under way.
The apparent correlation between the ability of L. monocytogenes to invade cardiac cells in tissue culture and its capacity to colonize the hearts in infected mice suggests that L. monocytogenes might establish infections at novel body sites by acquiring cell type-specific invasive capacities. As L. monocytogenes has the capacity to spread intracellularly from one cell type to another, the colonization of a tissue would not be expected to be limited by whether a strain is capable of direct bacterial invasion at that location. We speculate that some organs such as the heart, which moves blood rapidly through its chambers, might have limited contact with L. monocytogenes-infected cells present within the circulation. Bacteria free in the blood, however, may more efficiently target the heart if they are capable of direct cardiac cell attachment and invasion. One notable exception to the apparent link between myoblast invasive capacity and cardiac infection was isolate J4901, which exhibited high efficiency invasion of H9C2 cells but was unable to establish cardiac infections in mice. This strain was, however, isolated from a human cardiac infection, indicating that it is capable of targeting cardiac tissue in other circumstances.
Amino acid comparisons of the bacterial invasion proteins InlA and InlB suggest that the cardio-invasive strains form phylogenetically similar subsets, particularly when grouped based upon InlA sequence. A closer comparison of amino acid polymorphisms between strains revealed differences that may individually or in combination influence cardiac cell invasion. As an example, the InlA L589I polymorphism was present in both 07PF0776 and J4403; while this represents a conservative amino acid substitution, it is interesting that this InlA polymorphism has not been previously identified as a natural variant. Additionally, the InlB polymorphisms could potentially influence invasion independently or in concert with one another and/or with InlA to increase cardiac cell invasive capacity. It is worth noting that the most cardio-invasive isolate (07PF0776) contains all five amino acid polymorphisms, while less invasive isolates contain a subset. We are currently investigating whether these amino acid polymorphisms serve to alter the invasive capacity of L. monocytogenes; however, it remains possible that other yet to be defined bacterial factors contribute to cardiac disease. An enhanced understanding of bacterial factors that contribute to tissue tropism could reduce cardiac morbidity and mortality through rapid diagnostic and therapeutic intervention.
Acknowledgments
We thank Dr Lewis Graves for L. monocytogenes clinical isolates 07PF0776 (also known as J4533), J4403, J5043 and J4901; Dr Stanley Schulman for the L. monocytogenes clinical isolate NWI4b; Dr David Engman for providing the H9C2 rat myoblast cell line (ATCC CRL-1446); and members of the Freitag lab for helpful discussions. This work was supported by NIAID with Public Health Service grant AI41816 (N. E. F.), an American Heart Association Predoctoral Fellowship, 0910080G (F. A.), and by Institutional Training Grant T32-AI007172 and post-doctoral training grant UL1 RR024992 (L. D. B.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the funding sources.
Abbreviations
LLS, listeriolysin S
RBC, red blood cell
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the DNA sequence data for the previously unpublished hly, inlA and inlB genes are HQ703556–HQ703578.
References
- Adler, A., Fimbres, A., Marcinak, J., Johnson, A., Zheng, X., Hasegawa, S. & Shulman, S. T. (2009). Inflammatory pseudotumor of the heart caused by Listeria monocytogenes infection. J Infect 58, 161–163. [DOI] [PubMed] [Google Scholar]
- Alonzo, F., III, Port, G. C., Cao, M. & Freitag, N. E. (2009). The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect Immun 77, 2612–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolín, J., Gutierrez, A., Segoviano, R., López, R. & Ciguenza, R. (2008). Endocarditis due to Listeria: description of two cases and review of the literature. Eur J Intern Med 19, 295–296. [DOI] [PubMed] [Google Scholar]
- Berenguer, J., Solera, J., Diaz, M. D., Moreno, S., López-Herce, J. A. & Bouza, E. (1991). Listeriosis in patients infected with human immunodeficiency virus. Rev Infect Dis 13, 115–119. [DOI] [PubMed] [Google Scholar]
- Bierne, H., Sabet, C., Personnic, N. & Cossart, P. (2007). Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect 9, 1156–1166. [DOI] [PubMed] [Google Scholar]
- Bishop, D. K. & Hinrichs, D. J. (1987). Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J Immunol 139, 2005–2009. [PubMed] [Google Scholar]
- Brouqui, P. & Raoult, D. (2001). Endocarditis due to rare and fastidious bacteria. Clin Microbiol Rev 14, 177–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusch, J. L. (2001). Cardiac infections in the immunosuppressed patient. Infect Dis Clin North Am 15, 613–638, xi. [DOI] [PubMed] [Google Scholar]
- de Bruijn, M. F., van Vianen, W., Ploemacher, R. E., Bakker-Woudenberg, I. A., Campbell, P. A., van Ewijk, W. & Leenen, P. J. (1998). Bone marrow cellular composition in Listeria monocytogenes infected mice detected using ER-MP12 and ER-MP20 antibodies: a flow cytometric alternative to differential counting. J Immunol Methods 217, 27–39. [DOI] [PubMed] [Google Scholar]
- Edman, D. C., Pollock, M. B. & Hall, E. R. (1968). Listeria monocytogenes L forms. I. Induction maintenance, and biological characteristics. J Bacteriol 96, 352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves, L. M. & Swaminathan, B. (2001). PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int J Food Microbiol 65, 55–62. [DOI] [PubMed] [Google Scholar]
- Graves, L. M., Hunter, S. B., Ong, A. R., Schoonmaker-Bopp, D., Hise, K., Kornstein, L., DeWitt, W. E., Hayes, P. S., Dunne, E. & other authors (2005). Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J Clin Microbiol 43, 2350–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad, F., Berry, G., Doyle, R. L., Martineau, P., Leung, T. K. & Racine, N. (2007). Active bacterial myocarditis: a case report and review of the literature. J Heart Lung Transplant 26, 745–749. [DOI] [PubMed] [Google Scholar]
- Hardy, J., Francis, K. P., DeBoer, M., Chu, P., Gibbs, K. & Contag, C. H. (2004). Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303, 851–853. [DOI] [PubMed] [Google Scholar]
- Hardy, J., Margolis, J. J. & Contag, C. H. (2006). Induced biliary excretion of Listeria monocytogenes. Infect Immun 74, 1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, J., Chu, P. & Contag, C. H. (2009). Foci of Listeria monocytogenes persist in the bone marrow. Dis Model Mech 2, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, E. E., Herijgers, P., Herregods, M. C. & Peetermans, W. E. (2006). Evolving trends in infective endocarditis. Clin Microbiol Infect 12, 5–12. [DOI] [PubMed] [Google Scholar]
- Hood, S. & Baxter, R. H. (1999). Listeria endocarditis causing aortic root abscess and a fistula to the left atrium. Scott Med J 44, 117–118. [DOI] [PubMed] [Google Scholar]
- Hyland, K. V., Asfaw, S. H., Olson, C. L., Daniels, M. D. & Engman, D. M. (2008). Bioluminescent imaging of Trypanosoma cruzi infection. Int J Parasitol 38, 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kales, C. P. & Holzman, R. S. (1990). Listeriosis in patients with HIV infection: clinical manifestations and response to therapy. J Acquir Immune Defic Syndr 3, 139–143. [PubMed] [Google Scholar]
- Khan, K. M., Pao, W. & Kendler, J. (2001). Epidural abscess and vertebral osteomyelitis caused by Listeria monocytogenes: case report and literature review. Scand J Infect Dis 33, 714–716. [DOI] [PubMed] [Google Scholar]
- Lee, S. D., Wu, C. C., Kuo, W. W., Lin, J. A., Hwang, J. M., Lu, M. C., Chen, L. M., Hsu, H. H., Wang, C. K. & other authors (2006). Porphyromonas gingivalis-related cardiac cell apoptosis was majorly co-activated by p38 and extracellular signal-regulated kinase pathways. J Periodontal Res 41, 39–46. [DOI] [PubMed] [Google Scholar]
- Lindholm, A. C. (2008). Prosthetic valve Listeria endocarditis caused septic cerebral embolism. Lakartidningen 105, 2670–2671. [PubMed] [Google Scholar]
- Llanwarne, N., Badic, B., Delugeau, V. & Landen, S. (2007). Spontaneous splenic rupture associated with Listeria endocarditis. Am J Emerg Med 25, 1086.e3–1086.e5. [DOI] [PubMed] [Google Scholar]
- Luo, Y., Lee, A., Shen, H. & Radice, G. L. (2003). Altering tissue tropism of Listeria monocytogenes by ectopically expressing human E-cadherin in transgenic mice. Microb Pathog 35, 57–62. [DOI] [PubMed] [Google Scholar]
- Makaryus, A. N., Yang, R., Cohen, R., Rosman, D., Mangion, J. & Kort, S. (2004). A rare case of Listeria monocytogenes presenting as prosthetic valve bacterial endocarditis and aortic root abscess. Echocardiography 21, 423–427. [DOI] [PubMed] [Google Scholar]
- McCue, M. J. & Moore, E. E. (1979). Myocarditis with microabscess formation caused by Listeria monocytogenes associated with myocardial infarct. Hum Pathol 10, 469–472. [DOI] [PubMed] [Google Scholar]
- Mead, P. S., Slutsker, L., Dietz, V., McCaig, L. F., Bresee, J. S., Shapiro, C., Griffin, P. M. & Tauxe, R. V. (1999). Food-related illness and death in the United States. Emerg Infect Dis 5, 607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, K. J. & Freitag, N. E. (2005). Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect Immun 73, 1917–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil, H. S. & Marquis, H. (2006). Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect Immun 74, 6675–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocar, M., Passolunghi, D., Moneta, A. & Donatelli, F. (2009). Fulminant prosthetic valve endocarditis caused by Listeria monocytogenes. Eur J Cardiothorac Surg 36, 1077. [DOI] [PubMed] [Google Scholar]
- Rua Galisteo, O., Keituqwa Yáñez, I. & López Sánchez, L. (2007). Listeria induced ventriculitis. Med Intensiva 31, 50–51. [DOI] [PubMed] [Google Scholar]
- Seeliger, H. P. R. & Höhne, K. (1979). Serotyping of Listeria monocytogenes and related species. Methods Microbiol 13, 31–49. [Google Scholar]
- Shetron-Rama, L. M., Mueller, K., Bravo, J. M., Bouwer, H. G., Way, S. S. & Freitag, N. E. (2003). Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol Microbiol 48, 1537–1551. [DOI] [PubMed] [Google Scholar]
- Sun, A. N., Camilli, A. & Portnoy, D. A. (1990). Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect Immun 58, 3770–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan, B. & Gerner-Smidt, P. (2007). The epidemiology of human listeriosis. Microbes Infect 9, 1236–1243. [DOI] [PubMed] [Google Scholar]
- Tyler, K. M., Luxton, G. W., Applewhite, D. A., Murphy, S. C. & Engman, D. M. (2005). Responsive microtubule dynamics promote cell invasion by Trypanosoma cruzi. Cell Microbiol 7, 1579–1591. [DOI] [PubMed] [Google Scholar]
- Van Stelten, A., Simpson, J. M., Ward, T. J. & Nightingale, K. K. (2010). Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl Environ Microbiol 76, 2783–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domínguez-Bernal, G., Goebel, W., González-Zorn, B., Wehland, J. & Kreft, J. (2001). Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14, 584–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, R. (1989). Morphological, physiological and biochemical characterization. In Isolation of and Identification of Listeria monocytogenes, 1st edn, pp. 39–43. Edited by G. L. Jones. CDC Laboratory Manual.
- Wollert, T., Pasche, B., Rochon, M., Deppenmeier, S., van den Heuvel, J., Gruber, A. D., Heinz, D. W., Lengeling, A. & Schubert, W. D. (2007). Extending the host range of Listeria monocytogenes by rational protein design. Cell 129, 891–902. [DOI] [PubMed] [Google Scholar]