Summary
The molecular mechanisms that underlie cell lineage diversification of multipotent progenitors in the pancreas are virtually unknown. Here, we show that the early fate choice of pancreatic progenitors between the endocrine and acinar cell lineage is restricted by cross-repressive interactions between the transcription factors Nkx6.1/Nkx6.2 (Nkx6) and Ptf1a. Using genetic loss- and gain-of-function approaches, we demonstrate that Nkx6 factors and Ptf1a are required and sufficient to repress the alternative lineage program and to specify progenitors toward an endocrine or acinar fate, respectively. The Nkx6/Ptf1a switch only operates during a critical competence window when progenitors are still multipotent and can be uncoupled from cell differentiation. Thus, cross-antagonism between Nkx6 and Ptf1a in multipotent progenitors governs the equilibrium between endocrine and acinar cell neogenesis required for normal pancreas development.
Keywords: Nkx6.1, Nkx6.2, Ptf1a, pancreas, development, repression, progenitor cell, exocrine, endocrine, differentiation, lineage
Introduction
At the earliest stages of pancreas development, most, if not all pancreatic epithelial cells are thought to be multipotent progenitors which are competent to develop into all pancreatic cell types, namely five different endocrine lineages as well as the exocrine acinar and ductal cells (Gu et al., 2002). During subsequent pancreas morphogenesis, cells residing in the “tips” of the branching epithelium adopt an acinar fate, while cells in the core or “trunk” become restricted to a ductal or endocrine fate (Solar et al., 2009; Zhou et al., 2007). Although significant advances have been made in defining the molecular determinants of endocrine cell subtype specification and differentiation (Oliver-Krasinski and Stoffers, 2008), little is known regarding the molecular mechanisms governing the early allocation of multipotent progenitors to either a ductal/endocrine or acinar fate.
In many tissues, the separation of lineage-restricted progenitors from a multipotent progenitor cell pool is mediated by cross-repressive interactions between two transcription factors (Briscoe et al., 2000; Laslo et al., 2006; Olguin et al., 2007). The transcriptional repressors, Nkx6.1 and Nkx6.2 (Nkx6) constitute one arm of such a cross-repressive loop during neuronal subtype specification in the spinal cord (Sander et al., 2000a; Vallstedt et al., 2001). In the pancreas, Nkx6.1 and Nkx6.2 are redundantly required for endocrine alpha- and beta-cell development (Henseleit et al., 2005), but the mechanism by which Nkx6 factors control endocrine development has remained poorly defined. The finding that co-expression of the two Nkx6 factors is only observed in multipotent progenitors, but not in endocrine lineage-committed progenitors marked by expression of the transcription factor Neurogenin 3 (Ngn3) (Henseleit et al., 2005), suggests that Nkx6 proteins play a key role in endocrine cell development prior to the initiation of Ngn3 expression. This raises the possibility that proper endocrine cell development from a multipotent progenitor cell domain first requires the specification of endocrine fate by Nkx6 factors, while Ngn3 subsequently promotes cell differentiation by promoting cell cycle exit and by inducing the expression of endocrine lineage-specific genes (Apelqvist et al., 1999; Schwitzgebel et al., 2000). The contention that Nkx6 proteins could be involved in early lineage specification is consistent with the expression domain of Nkx6.1, which becomes progressively compartmentalized to the trunk domain, as the activator of acinar-specific gene transcription, Ptf1a (Krapp et al., 1996; Rose et al., 2001), becomes exclusively restricted to cells residing in the tips (Hald et al., 2008). Interestingly, the resolution of Nkx6.1 and Ptf1a into exclusive compartments appears to coincide with the progressive restriction of trunk and tip cells to the ductal/endocrine or acinar cell lineage, respectively (Solar et al., 2009; Zhou et al., 2007).
In this study, we provide evidence that reciprocal repression between Nkx6 factors and Ptf1a enables a bistable switch in multipotent progenitors that directs progenitors to either a ductal/endocrine or acinar cell fate choice.
Results
Loss of Nkx6 activity results in an endocrine to acinar fate switch
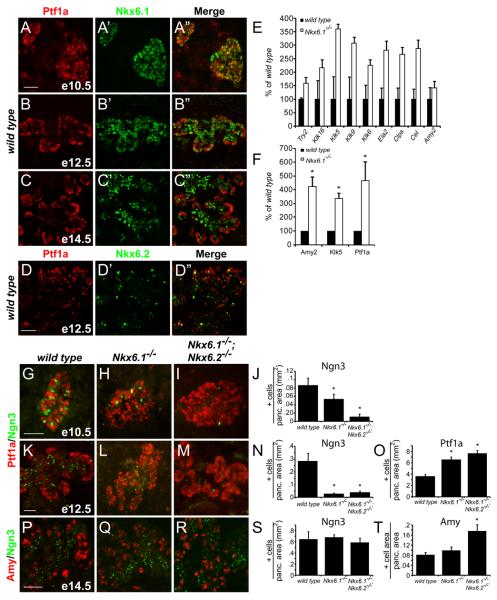
To confirm that Nkx6.1 and Ptf1a resolve into exclusive compartments (Hald et al., 2008) and to more carefully examine the extent of coexpression between both Nkx6 factors and Ptf1a, we performed co-immunofluorescence analysis for Nkx6.1 and Nkx6.2 together with Ptf1a. At embryonic day (e) 10.5, a large percentage of pancreatic epithelial cells co-expressed Nkx6.1 and Ptf1a, but cells that exclusively expressed one of the two transcription factors although scarce, were also present (Fig. 1A). At the onset of branching morphogenesis at e12.5, Nkx6.1 became progressively confined to the trunk of the developing organ, while Ptf1a marked the tips of the branches (Fig. 1B). At this stage, tip cells were additionally also largely devoid of Nkx6.2 (Fig. 1D). By e14.5, when tip cells have fully committed to an acinar fate (Zhou et al., 2007), Nkx6.1 was almost completely excluded from the tips and formed a sharp boundary with Ptf1a (Fig. 1C). As previously noted (Burlison et al., 2008), from even the earliest stages of development, Ngn3+ endocrine progenitors failed to express Ptf1a (Fig. 1G).
Figure 1. Endocrine to acinar fate conversion in Nkx6-deficient embryos.
(A-D) Immunofluorescence for Ptf1a and Nkx6.1 or Nkx6.2 in the embryonic mouse pancreas. At e12.5, Nkx6.1 and Nkx6.2 become excluded from the Ptf1a+ tip domain. (E-F) Quantification of mRNA levels for acinar markers by microarray (E) and qRT-PCR (F) in e15.5 pancreas. Nkx6.1-deficiency results in increased expression of acinar-specific genes. (G-I,K-M,P-R) Immunofluorescence staining reveals expansion of the Ptf1a+ domain into the trunk region in Nkx6.1−/− and Nkx6.1−/−;Nkx6.2−/− pancreata at e12.5. Concomitantly, Ngn3+ cells are reduced. At e14.5, Nkx6-deficient embryos show increased numbers of acinar cells. (J,N,O,S,T) Quantification of marker+ cell numbers relative to total pancreatic area. Scale bar=50 μm; Try, trypsin; Klk, kallikrein; Ela, elastase; Clps, co-lipase; Cel, carboxylester-lipase; Amy, amylase.
To assess whether loss of Nkx6 gene function affects exclusion of Ptf1a from the core of the developing pancreas, we analyzed organ patterning in Nkx6.1;Nkx6.2 (Nkx6) compound mutant mice, which exhibit normal organ size (Fig. S1M,N). In Nkx6.1- and Nkx6-deficient embryos, we detected significant ectopic expression of Ptf1a in the trunk region (Fig. 1K-N,O), which was accompanied by a 90.7% reduction in the number of Ngn3+ cells at e12.5 (Fig. 1K-N). At e10.5, when the majority of epithelial cells express both Nkx6 factors (Henseleit et al., 2005), Nkx6.1 single mutants exhibited a smaller, 38.2% reduction in Ngn3+ cells, while Nkx6 compound mutants displayed a much more extensive, 87.5% decrease compared to wild type embryos (Fig. 1G-J). This suggests that compensation by Nkx6.2 partially restores formation of Ngn3+ cells in Nkx6.1 single mutants at e10.5.
Since Ptf1a is a critical regulator of acinar cell differentiation (Esni et al., 2004), we next examined whether expansion of the Ptf1a+ domain in Nkx6 mutants manifests in increased numbers of acinar cells. Transcriptional profiling and qRTPCR analysis of pancreatic anlagen indeed confirmed an upregulation of acinar genes in Nkx6.1-deficient pancreata at e15.5 (Fig. 1E,F), which was associated with increased numbers of amylase+ acinar cells in Nkx6 mutants (Fig. 1P-R,T). However, acinar cell markers did not appear prematurely (Fig. S1A-C), indicating that the absence of Nkx6 activity does not affect the timing of acinar cell differentiation.
Significantly, in contrast to earlier developmental stages, Nkx6-deficient embryos no longer displayed a reduction in Ngn3+ progenitors at e14.5 (Fig. 1P-S). We speculate that these Ngn3+ cells arise from a residual Ptf1a−/Sox9+/Pdx1+ trunk-like progenitor cell domain that was still discernable in Nkx6 mutants (Fig. S1D-L) and might seed a partial recovery of endocrine cell differentiation during later development. The absence of the early, but presence of the later wave of Ngn3 cell genesis in Nkx6 mutants might also explain why formation of alpha- and beta-cells, but not of the later-arising delta- and pancreatic polypeptide-cells, is selectively affected in Nkx6 mutants (Henseleit et al., 2005; Johansson et al., 2007).
Nkx6 misexpression prevents acinar cell differentiation
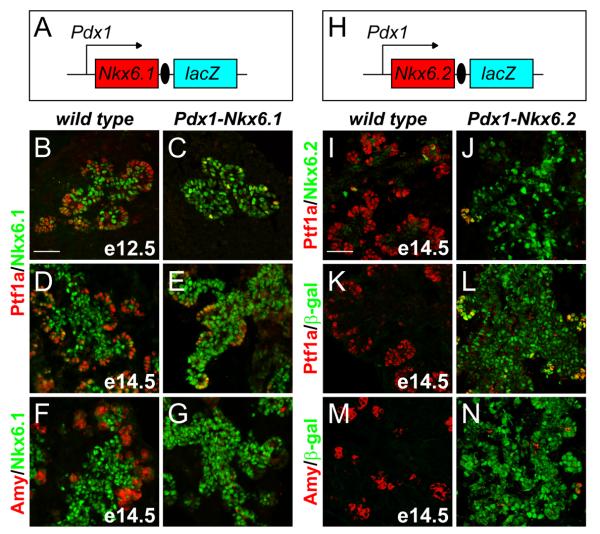
Next, we investigated whether continuous expression of Nkx6 factors throughout the entire pancreatic progenitor cell field is sufficient to repress their differentiation into acinar cells. We utilized the Pdx1 promoter to misexpress Nkx6.1 or Nkx6.2 (Fig. 2A,H). As predicted, Pdx1-Nkx6.1 or Pdx1-Nkx6.2 embryos expressed Nkx6.1 or Nkx6.2, respectively, throughout the entire pancreatic epithelium, including the presumptive tip domain (Fig. 2C,E,G,J,L,N). Organ size as well as Ngn3+ and endocrine cell numbers were unaffected in Pdx1-Nkx6.1 and Pdx1-Nkx6.2 embryos (Fig. S2, data not shown), demonstrating that Nkx6 misexpression does not induce premature endocrine cell differentiation. However, sustained expression of Nkx6.1 or Nkx6.2 significantly repressed Ptf1a expression (Fig. 2B-E,I-L) and consequently blocked acinar cell differentiation (Fig. 2F,G,M,N). Thus, the misexpression of either Nkx6.1 or Nkx6.2 in multipotent progenitors is sufficient to prevent the initiation of an acinar program.
Figure 2. Nkx6 proteins suppress acinar cell differentiation.
(A,H) Schematic of the Pdx1-Nkx6.1 and Pdx1-Nkx6.2 transgenes. Immunofluorescence staining of pancreata from Pdx1-Nkx6.1 embryos at e12.5 and e14.5 (B-G) and Pdx1-Nkx6.2 embryos at e14.5 (I-N) shows that ectopic expression of Nkx6.1 or Nkx6.2 blocks expression of Ptf1a and amylase.
Nkx6.1 specifies endocrine fate in multipotent pancreatic progenitors
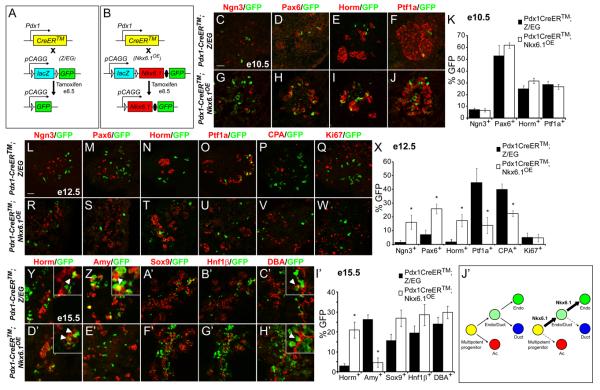
To determine whether expression of Nkx6.1 predisposes multipotent pancreatic progenitors to adopt an endocrine fate, we generated CAG-Bgeo,-Nkx6.1,-eGFP transgenic mice (hereafter abbreviated to Nkx6.1OE), in which concomitant expression of Nkx6.1 and enhanced green fluorescent protein (eGFP) can be stably and heritably induced by expression of Cre recombinase (Fig. 3B). The transgene design is analogous to that used in CAG-Bgeo,-eGFP (Z/EG) mice, in which lacZ is ubiquitously expressed in all cells unless Cre recombinase-mediated excision removes the lacZ gene and allows for expression of eGFP (Fig. 3A). Z/EG mice therefore serve as a control for the Nkx6.1OE model. To validate the Nkx6.1OE transgenic model, we intercrossed Nkx6.1OE with Pdx1-CreER™ mice and induced Cre-mediated recombination by administration of tamoxifen (TM) to pregnant dams. As expected, the majority of unrecombined cells expressed β-galactosidase (β-gal) but not GFP, while recombined cells co-expressed Nkx6.1 and GFP but were negative for β-gal (Fig. S3A). No GFP expression was observed in Nkx6.1OE mice without the Pdx1-CreER™ transgene (Fig. S3B), thus demonstrating no leakiness in our system. These results were confirmed for mouse lines established from two independent founders.
Figure 3. Nkx6.1 functions as an endocrine fate determinant.
(A,B) A tamoxifen-inducible form of Cre recombinase (CreER™) expressed under the control of the Pdx1 promoter removes a floxed Bgeo (lacZ) cassette to permanently activate expression of green fluorescent protein (GFP) from the Z/EG transgene (A) or Nkx6.1 and GFP from the Nkx6.1OE transgene (B). Immunofluorescence staining of pancreas sections from e10.5 (C-J), e12.5 (L-W) and e15.5 (Y-H′) Pdx1-CreER™;Z/EG and Pdx1-CreER™;Nkx6.1OE embryos injected with one dose of tamoxifen at e8.5. Quantification of GFP+/marker+ cells relative to the total number of GFP+ cells at e10.5 (K), e12.5 (X), or e15.5 (I′). At e10.5, similar numbers of GFP+ cells have initiated expression of the endocrine markers Ngn3, Pax6, and hormones in Pdx1-CreER™;Nkx6.1OE and Pdx1-CreER™;Z/EG control embryos (K). Similar analysis at e12.5 reveals a preference of GFP+ cells in Pdx1-CreER™;Nkx6.1OE embryos to express Ngn3, Pax6, and hormones, while showing a decreased propensity to express Ptf1a and CPA (X). Expression of the proliferation marker Ki67 does not show a difference between the two groups (X). At e15.5, GFP+ cells in Pdx1-CreER™;Z/EG embryos express endocrine hormones (Y) as well as the acinar marker amylase (Z) (arrowheads in inset, Y,Z). By contrast, in Pdx1-CreER™;Nkx6.1OE embryos, GFP+/amylase+ cells are rarely found (E′), while GFP+/hormone+ cells are abundantly detected (arrowheads in inset, D′). GFP+ cells from Pdx1-CreER™;Nkx6.1OE and Pdx1-CreER™;Z/EG embryos have a similar potential to differentiate into DBA+ ductal cells (arrowheads in inset show GFP+/DBA+ cells, C′,H′). (J′) Model of cell fate shifts induced by heritable Nkx6.1 misexpression in multipotent pancreatic progenitors. Scale bar=50 μm; triangles represent loxP sites; filled ovals, IRES; Amy, amylase; Horm, hormone (insulin, glucagon, somatostatin, pancreatic polypeptide); Endo, endocrine; Ac, acinar.
To analyze whether stable, heritable expression of Nkx6.1 biases the cell fate choice of multipotent progenitors, we injected mice with TM at e8.5, which results in nuclear translocation of CreER™ and mosaic induction of Nkx6.1 and GFP in Pdx1+ progenitors for a period of ~12-36 hours after TM administration (Zhou et al., 2007). To assess subsequent progenitor cell fate, we quantified the percentages of recombined cells that had initiated expression of specific lineage markers at different time-points following induction of Nkx6.1 expression. At e10.5, when Nkx6.1 and Ptf1a are still co-expressed in most progenitors (Fig. 1A), Nkx6.1-misexpressing cells showed the same propensity to activate the preendocrine markers Ngn3 and Pax6, hormones, or Ptf1a as normal progenitors (Fig. 3C-K). This suggests that progenitors are either not competent to repress Ptf1a in response to Nkx6.1 prior to e10.5 or that the time window between TM-mediated Nkx6.1 induction and analysis is too short to observe a response. By e12.5, however, we observed preferential activation of pre-endocrine markers in cells that heritably expressed Nkx6.1 (Fig. 3L-N,R-T,X). Crucially, we found that this increase in pre-endocrine cells was not the result of increased cell proliferation (Fig. 3Q,W,X), but was accompanied by a concomitant decrease of progenitors that had activated the pre-acinar lineage markers Ptf1a and CPA (Fig. 3O,P,U,V,X). These results provide unequivocal evidence that Nkx6.1 controls a cell lineage choice between endocrine and acinar fates in a critical time window preceding final lineage commitment of the entire tip and trunk domains.
By e15.5, most Nkx6.1-misexpressing progenitors had contributed to the endocrine cell compartment, while very few progenitors had committed to an acinar fate (Fig. 3Y,Z,D′,E′,I′; Fig. S3C-O). Nkx6.1 also increased the propensity of progenitors to contribute to the Sox9+/Hnf1β+ domain (Fig. 3A′,B′,F′G′,I′) which is bipotential and gives rise to both endocrine and ductal cells (Solar et al., 2009). Since Nkx6.1-misexpressing cells that arose from this domain preferentially activated endocrine markers and not the ductal marker DBA (Fig. 3C′,H′,I′), we conclude that expression of Nkx6.1 favors an endocrine over a ductal cell fate choice. In sum, our findings suggest a model whereby Nkx6.1 first establishes a bipotential ductal/endocrine compartment by excluding Ptf1a and subsequently promotes an endocrine fate choice in this secondary progenitor domain (Fig. 3J′). Because Nkx6.1 functions as a transcriptional repressor (Muhr et al., 2001), we tested whether Nkx6.1 is capable of directly controlling acinar gene transcription. Adenoviral misexpression of Nkx6.1 in the AR42J pancreatic acinar cell line resulted in a significant suppression of Ptf1a, amylase 2, carboxypeptidase A1, and elastase 1 mRNA and protein levels (Fig. S4A,B). Since Ptf1a is an upstream regulator of acinar gene transcription (Krapp et al., 1996; Rose et al., 2001), we next examined whether Nkx6.1 controls acinar gene expression by directly regulating Ptf1a transcription. We identified fourteen conserved Nkx6 binding motifs (Jorgensen et al., 1999; Mirmira et al., 2000) within 50 kb of the 5′-end flanking region from the Ptf1a transcriptional start site, of which eight were located in a previously-characterized 2.3 kb Ptf1a enhancer (Fig. S4C) (Masui et al., 2008). This enhancer is sufficient to initiate expression of a reporter transgene in early pancreatic progenitors and to superinduce Ptf1a expression in acinar cells. Chromatin immunoprecipitation (ChIP) analyses in βTC3 cells revealed direct and specific association of Nkx6.1 with the proximal part of the Ptf1a enhancer (E5), while no association was observed with the other Nkx6 motifs within or outside the enhancer region (Fig. S4D). Transfection of 266-6 acinar cells with an expression plasmid for Nkx6.1 and a luciferase reporter construct controlled by the Ptf1a enhancer sequence further revealed that Nkx6.1 significantly reduced reporter gene activity (Fig. S4E). This effect was blunted when the Nkx6 binding site was mutated. Likewise, Nkx6.2 also repressed the Ptf1a enhancer, albeit less potently than Nkx6.1 (Fig. S4E). These findings illustrate that Nkx6 proteins have the ability to directly repress Ptf1a gene transcription.
However, contrary to our observations in early progenitors, Nkx6.1 did not completely repress Ptf1a when misexpressed in acinar cells of embryos at e14.5 or of adult mice (data not shown; Fig. S3P,Q). This difference in responsiveness to Nkx6 repressors might be explained by the strong Ptf1a autoregulatory feedback loop that reinforces Ptf1a expression in mature acinar cells (Masui et al., 2008). We also found that induction of Nkx6.1 expression in Ptf1a+ cells at e14.5 or thereafter failed to induce acinar to endocrine fate conversion (data not shown), revealing that the ability of Nkx6 proteins to fully repress Ptf1a and to confer endocrine identity to a cell is confined to a critical early competence window, when progenitors are still multipotent.
Ptf1a represses Nkx6.1 and adoption of endocrine fate
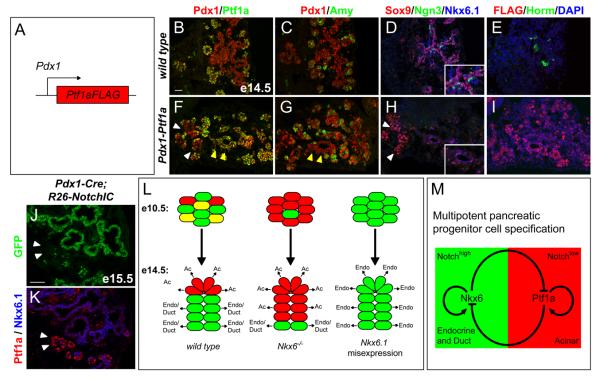
The antagonistic interplay between lineage determining transcription factors has previously been proposed as a molecular mechanism for initiating and resolving mixed lineage states in several tissues (Briscoe et al., 2000; Laslo et al., 2006; Olguin et al., 2007). To elucidate whether pancreatic endocrine versus acinar cell identity is established through a similar cross-antagonism, we tested whether uniform and sustained expression of Ptf1a in all pancreatic progenitors is also sufficient to repress Nkx6.1 and block endocrine cell differentiation. To direct Ptf1a expression ectopically to the entire pancreatic epithelium, we generated transgenic embryos that express Ptf1a under the control of Pdx1 regulatory sequences (Fig. 4A). Embryos were harvested at e14.5 and analyzed for transgene expression by immunofluorescence staining for the Ptf1a-FLAG fusion protein. Of 18 embryos exhibiting genomic integration of the transgene, four showed uniform expression of Ptf1a-FLAG throughout most of the pancreatic epithelium, including the trunk domain from which Ptf1a is normally excluded (Fig. 4B,F,E,I). Strikingly, the majority of Ptf1a+ cells in Pdx1-Ptf1a embryos were devoid of Nkx6.1 protein (Fig. 4F,H), showing that Ptf1a expression in progenitors is sufficient to suppress Nkx6.1 (4/4 embryos). Likewise, ectopic Ptf1a also repressed expression of the trunk marker Sox9 as well as Ngn3 (Fig. 4D,H) and thus completely abrogated endocrine cell differentiation (Fig. 4E,I; 4/4 embryos). The observation that not all Ptf1a+ cells in Pdx1-Ptf1a embryos activated expression of amylase (Fig. 4F,G) suggests that Ptf1a misexpression did not induce premature acinar differentiation. We conclude that the sustained expression of Ptf1a in multipotent progenitors is sufficient to block their expression of trunk markers and prevent endocrine differentiation.
Figure 4. Ptf1a is a repressor of endocrine fate.
(A) Schematic of the Pdx1-Ptf1a transgene. (B-I) Immunofluorescence staining of adjacent pancreas sections from e14.5 embryos shows ectopic expression of Ptf1a (F) and FLAG (I) in the trunk region of Pdx1-Ptf1a embryos. Ptf1a misexpression does not uniformly induce amylase expression (yellow arrow heads, F,G). Ectopic Ptf1a represses Nkx6.1, Sox9 and Ngn3 (H), and prevents endocrine cell differentiation (I). White arrowheads in (F,H) point to a region that does not misexpress Ptf1a and therefore expresses Sox9 and Nkx6.1 (H). Insets in (D,H) show higher magnifications. (J) Summary of the observed cell fate changes in Nkx6 loss- and gain-of-function models. (K,L) Immunofluorescence staining of adjacent pancreatic sections from Pdx1-Cre(mosaic);R26NotchIC embryos at e15.5 shows expression of Nkx6.1 in all cells that have recombined the R26NotchIC allele and therefore activated NotchIC/GFP expression. Ptf1a is excluded from the GFP+ domain, but is expressed in areas that have not recombined the R26NotchIC allele (arrowheads, K,L). (M) Our data support a model whereby a regulatory circuit comprised of the counter-antagonistic repressive activities of Nkx6 and Ptf1a restricts multipotent pancreatic progenitors to ductal/endocrine and acinar fates, respectively. Positive autoregulatory feedback of Nkx6 and Ptf1a creates bistability of the fate choice. High Notch activity appears to favor the Nkx6.1+ state. Note, for simplicity the redundant factors Nkx6.1 and Nkx6.2 are referred to as Nkx6. Scale bar=50 μm; Amy, amylase; Horm, hormone (insulin, glucagon, somatostatin, pancreatic polypeptide), Endo, endocrine; Ac, acinar.
A recent study has revealed that diminished Notch signaling activity results in endocrine-to-acinar cell fate conversion of Ngn3+ progenitors, suggesting that fate commitment remains plastic even in Ngn3+ cells (Cras-Meneur et al., 2009). To determine whether the Nkx6/Ptf1a switch might operate downstream of Notch, we employed a mosaic Pdx1-Cre transgene to heritably activate Notch1 in multipotent progenitors, using the R26NotchIC allele (Murtaugh et al., 2003). All cells that recombined the R26NotchIC allele and therefore expressed NotchIC and GFP were Nkx6.1+ but Ptf1a−, while cells that failed to undergo Cre-mediated recombination were Ptf1a+ but Nkx6.1− (Fig. 4K,L). These findings indicate that Notch signaling favors the expression of Nkx6.1 and the exclusion of Ptf1a in multipotent progenitors. This might be achieved either through the direct activation of Nkx6.1 by Notch or alternatively, by Notch indirectly allowing for Nkx6.1 expression by repressing Ptf1a. Taken together, our data demonstrate that a Notch-controlled cross-antagonistic switch between Nkx6 and Ptf1a restricts multipotent progenitors in the developing pancreas to either a ductal/endocrine or acinar cell fate (Fig. 4J,M).
Discussion
While reciprocal repression mechanisms are known to regulate cell lineage commitment in many tissues (Briscoe et al., 2000; Laslo et al., 2006; Olguin et al., 2007), our study demonstrates that this mechanism also operates during organogenesis of the pancreas. Paralleling their role in pancreatic endocrine cell specification described herein, the redundant activities of Nkx6.1 and Nkx6.2 also specify neuronal cell fate through the repression of alternative neuronal fate choices (Sander et al., 2000a; Vallstedt et al., 2001). An analogous role has also been demonstrated for Ptf1a in the specification of retinal progenitors (Fujitani et al., 2006). Recent analysis of zebrafish with a hypomorphic allele for Ptf1a has further shown that loss of Ptf1a activity induces an acinar-to-endocrine fate-conversion of Ptf1a+ pancreatic progenitors (Dong et al., 2008), suggesting that Ptf1a commits progenitors to an acinar cell fate. Together with the findings described in this study, these observations point to a developmentally-conserved role for Nkx6 proteins and Ptf1a as a counter-regulatory switch for specifying distinct lineages in multipotent progenitors. Notably, Nkx6.1 and Ptf1a both maintain their own expression through a direct positive feedback mechanism (Iype et al., 2004; Masui et al., 2008), which provides stability to the genetic switch and thereby reinforces a cell-specific program of gene expression.
Our findings indicate that the Nkx6 to Ptf1a arm of the repressive loop could be mediated by direct transcriptional repression of Ptf1a by Nkx6 proteins. The absence of a suitable in vitro system for primary pancreatic progenitor cells precluded experiments to test whether the direct repression of the Ptf1a enhancer by Nkx6.1 or Nkx6.2 that we observed in cell lines also occurs in embryonic progenitors. However, the observation that the Ptf1a enhancer to which Nkx6.1 binds and which it represses in acinar cell lines is active in both mature acinar cells and multipotent progenitors of the embryonic pancreas (Masui et al., 2008) favors the notion that its repression by Nkx6 factors could also be relevant during acinar cell specification in the embryo. Since Ptf1a largely functions as a transcriptional activator (Masui et al., 2008; Rose et al., 2001), it appears, however, less likely that Ptf1a represses Nkx6.1 directly.
One critical aspect of our findings is that the Nkx6/Ptf1a switch only operates during a critical competence window prior to e14, when Ptf1a+ progenitors are still multipotent. The closure of this time window coincides with the irreversible lineage commitment of tip cells to an acinar fate and trunk cells to a ductal or endocrine fate (Solar et al., 2009; Zhou et al., 2007). Our observation that Nkx6.1 induces a fate bias as early as e12.5 reveals that lineage commitment is initiated early and suggests that a substantial proportion of progenitors may have committed to an acinar or ductal/endocrine fate well before the trunk and tip domains become visually distinguishable. This early specification event mediated by cross-antagonism between Nkx6 factors and Ptf1a may therefore represent an intrinsic mechanism by which the relative numbers of newly-differentiated endocrine versus acinar cells are pre-determined during pancreas development. Neither expression of Nkx6 factors nor Ptf1a accelerated cell differentiation in our gain-of-function models. Therefore, Nkx6 proteins and Ptf1a appear to primarily function as cell fate determinants, while other factors, such as Ngn3 in the case of the endocrine lineage (Apelqvist et al., 1999; Schwitzgebel et al., 2000), are required to initiate differentiation of pre-specified progenitors into the various cell types of the mature pancreas. This notion is consistent with our observation that Nkx6 factors specify endocrine cell fate upstream of Ngn3. In case of the acinar lineage, Ptf1a itself appears to be involved in the induction of acinar cell differentiation, but its activity is refined and constrained by the availability of specific co-factors (Masui et al., 2007).
In summary, our study demonstrates the importance of a reciprocal repression mechanism in dictating cell fate choices in multipotent progenitors during pancreas development. It provides a functional framework in which to investigate the molecular mechanisms associated with the transition of progenitors from a multipotent to a lineage-committed state. Insight into the molecular underpinnings of this process may help in devising strategies to induce endocrine commitment for clinical applications in diabetes.
Experimental Procedures
Transgene construction and mice
Nkx6.1−/−, Pdx1-Nkx6.1, Pdx1-Nkx6.2, CAG-Bgeo,-eGFP, and Pdx1-CreER™ mice have been previously described (Gu et al., 2002; Nelson et al., 2007; Novak et al., 2000; Sander et al., 2000b). The generation of CAG-Bgeo,-Nkx6.1,-eGFP and Pdx1-Ptf1a mice is described in the supplemental material. All animal experiments described herein were approved by University of California, Irvine and San Diego Institutional Animal Care and Use Committees.
Tamoxifen (Sigma) was dissolved in corn oil (Sigma) at 20mg/ml and a single dose of 2mg/40g body weight administered by intraperitoneal injection at e8.5. Midday on the day of vaginal plug appearance was considered e0.5.
Immunohistochemistry, X-gal staining, morphometry, and cell quantification
Tissue preparation, immunohistochemistry, X-gal staining, TUNEL staining and morphometry were performed as previously described (Seymour et al., 2008). A detailed description of the morphometric methods is also included in the supplemental material. All values are shown as mean ± standard error of the mean (SEM); p-values were calculated using unpaired student's t-test; P<0.05 was considered significant.
mRNA quantification
Microarray analyses were performed in triplicate for each genotype. Total RNA was isolated from three Nkx6.1−/− embryos or wild type littermates at e15.5 and pooled to hybridize to an Affymetrix gene chip (MG_U74A). We used Principal Components Analysis to identify patterns of gene expression.
For qRT-PCR, RNA was isolated with the RNeasy Mini kit (Qiagen) and treated with DNAse. Synthesis of cDNA was performed with the Superscript III cDNA kit (Invitrogen). Q-PCR reactions were performed in triplicate using SYBR Green (Applied Biosystems).
Supplementary Material
Acknowledgements
We thank C. Wright, D. Melton and C. Lobe for Ptf1a-CreERT2, Pdx1-CreER™ and Z/EG mice, respectively; C. Wright for Pdx1-Ptf1aFLAG and R. MacDonald for Ptf1a-enhancer constructs; C. Newgard for AdCMV-Nkx6.1, P. Serup for anti-Nkx6.1, J. Ericson for anti-Nkx6.2, C. Kioussi for anti-GFP, B. Breant for anti-Ptf1a, and the Developmental Studies Hybridoma Bank for anti-tubulin antibodies. We thank C. Hennings for contributing experiments, L. Sussel and the Barbara Davis Center, Denver, for bioinformatics analysis, and T. Schilling as well as members of the Sander laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health Grants U19-DK072495 and RO1-DK068471 to M.S., and a CIRM postdoctoral fellowship to K.K.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A list of primer sequences for qRT-PCR analysis and of antibodies and their dilutions can be found in the supplemental material.
References
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Burlison JS, Long Q, Fujitani Y, Wright CV, Magnuson MA. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol. 2008;316:74–86. doi: 10.1016/j.ydbio.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cras-Meneur C, Li L, Kopan R, Permutt MA. Presenilins, Notch dose control the fate of pancreatic endocrine progenitors during a narrow developmental window. Genes Dev. 2009;23:2088–2101. doi: 10.1101/gad.1800209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong PD, Provost E, Leach SD, Stainier DY. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev. 2008;22:1445–1450. doi: 10.1101/gad.1663208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esni F, Ghosh B, Biankin AV, Lin JW, Albert MA, Yu X, MacDonald RJ, Civin CI, Real FX, Pack MA, et al. Notch inhibits Ptf1 function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development. 2004;131:4213–4224. doi: 10.1242/dev.01280. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Hald J, Sprinkel AE, Ray M, Serup P, Wright C, Madsen OD. Generation and characterization of Ptf1a antiserum and localization of Ptf1a in relation to Nkx6.1 and Pdx1 during the earliest stages of mouse pancreas development. J Histochem Cytochem. 2008;56:587–595. doi: 10.1369/jhc.2008.950675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henseleit KD, Nelson SB, Kuhlbrodt K, Hennings JC, Ericson J, Sander M. NKX6 transcription factor activity is required for alpha- and beta-cell development in the pancreas. Development. 2005;132:3139–3149. doi: 10.1242/dev.01875. [DOI] [PubMed] [Google Scholar]
- Iype T, Taylor DG, Ziesmann SM, Garmey JC, Watada H, Mirmira RG. The transcriptional repressor Nkx6.1 also functions as a deoxyribonucleic acid context-dependent transcriptional activator during pancreatic beta-cell differentiation: evidence for feedback activation of the nkx6.1 gene by Nkx6.1. Mol Endocrinol. 2004;18:1363–1375. doi: 10.1210/me.2004-0006. [DOI] [PubMed] [Google Scholar]
- Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, Grapin-Botton A. Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev Cell. 2007;12:457–465. doi: 10.1016/j.devcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Jorgensen MC, Vestergard Petersen H, Ericson J, Madsen OD, Serup P. Cloning and DNA-binding properties of the rat pancreatic beta-cell-specific factor Nkx6.1. FEBS Lett. 1999;461:287–294. doi: 10.1016/s0014-5793(99)01436-2. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagenbuchle O, Wellauer PK. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. Embo J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T, Swift GH, Hale MA, Meredith DM, Johnson JE, Macdonald RJ. Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol Cell Biol. 2008;28:5458–5468. doi: 10.1128/MCB.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirmira RG, Watada H, German MS. Beta-cell differentiation factor Nkx6.1 contains distinct DNA binding interference and transcriptional repression domains. J Biol Chem. 2000;275:14743–14751. doi: 10.1074/jbc.275.19.14743. [DOI] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SB, Schaffer AE, Sander M. The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting beta-cell fate specification in Pdx1+ pancreatic progenitor cells. Development. 2007;134:2491–2500. doi: 10.1242/dev.002691. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Olguin HC, Yang Z, Tapscott SJ, Olwin BB. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol. 2007;177:769–779. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose SD, Swift GH, Peyton MJ, Hammer RE, MacDonald RJ. The role of PTF1-P48 in pancreatic acinar gene expression. J Biol Chem. 2001;276:44018–44026. doi: 10.1074/jbc.M106264200. [DOI] [PubMed] [Google Scholar]
- Sander M, Paydar S, Ericson J, Briscoe J, Berber E, German M, Jessell TM, Rubenstein JL. Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 2000a;14:2134–2139. doi: 10.1101/gad.820400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000b;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel VM, Scheel DW, Conners JR, Kalamaras J, Lee JE, Anderson DJ, Sussel L, Johnson JD, German MS. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- Solar M, Cardalda C, Houbracken I, Martin M, Maestro MA, De Medts N, Xu X, Grau V, Heimberg H, Bouwens L, et al. Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev Cell. 2009;17:849–860. doi: 10.1016/j.devcel.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Vallstedt A, Muhr J, Pattyn A, Pierani A, Mendelsohn M, Sander M, Jessell TM, Ericson J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron. 2001;31:743–755. doi: 10.1016/s0896-6273(01)00412-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.