Abstract
The overgrowth disorder Beckwith–Wiedemann syndrome (BWS) is associated with dysregulation of imprinted genes at chromosome 11p15.5. The molecular defects are heterogeneous but most of the cases are associated with defective DNA methylation at either one of two Imprinting Control Regions (IC1 and IC2) or Uniparental paternal Disomy (UPD) at 11p15.5. In rare cases, the BWS phenotype has been found associated with maternal transmission of IC1 microdeletions. We describe a family with a novel 1.8 kb deletion that is associated with hypermethylation at IC1. The mutation results from recombination between highly homologous sequences containing target sites for the zinc-finger protein CTCF (CTSs). This finding supports the hypothesis that the function of IC1 and the penetrance of the clinical phenotype depend on the spacing of the CTSs resulting from recombination in the mutant allele.
Keywords: Epigenetics, Genomic imprinting, Growth disorders
1. Introduction
The Beckwith–Wiedemann syndrome (BWS, OMIM 130650) is a congenital overgrowth disorder associated with dysregulation of imprinted genes located at chromosome 11p15.5 [1]. Children affected by BWS show with variable expressivity several characteristic features, including macroglossia, gigantism, abdominal wall defects and embryonal tumors, particularly Wilms tumor. The 11p15.5 imprinted gene cluster includes more than ten imprinted genes organized in two domains, each controlled by a separate imprinting centre region (IC1 and IC2, ref. [2]). The IGF2 and H19 genes are in the telomeric domain, and genes such as CDKN1C and KCNQ1OT1 are in the centromeric domain. Individuals with BWS have heterogeneous molecular defects affecting one or both domains of the cluster, including paternal uniparental disomy (UPD) or duplication, CDKN1C mutations and IC epimutations and microdeletions. BWS cases with alterations in the telomeric domain or UPD have a higher risk of developing Wilms tumor than cases with other molecular abnormalities.
The imprinting of the paternally expressed IGF2 and maternally expressed H19 genes is controlled by IC1, which is located upstream of the H19 transcription start site. On the maternal chromosome, IC1 binds the zinc finger protein CTCF that, by inducing higher order chromatin structures, insulates IGF2 from the effects of downstream enhancers [3]. On the paternal chromosome, the insulator function of IC1 is inactivated by DNA methylation that inhibits CTCF binding. In BWS, aberrant IGF2 activation and H19 silencing result from gain of DNA methylation at the maternal IC1. In contrast, IGF2 silencing and H19 activation are associated with loss of DNA methylation at the paternal IC1, in the growth-deficiency disorder, Silver–Russell syndrome (SRS, ref. [4]).
In the majority of BWS and SRS cases, the IC epimution is not linked with sequence changes of the region, but in a few individuals the DNA methylation defect is associated with in cis mutations [1]. In particular, microdeletions in the IC1 sequence abolishing 1–3 CTCF-target sites (CTS) have been described in BWS cases [5–7]. The clinical phenotype associated with these mutations, however, is not uniform and we have previously proposed that a critical factor may be the spacing of the remaining CTSs in the mutant allele [8].
Here we report a familial case of BWS with a novel IC1 microdeletion. The deletion is similar in size to previously reported ones, but removes different sets of CTSs. When maternally transmitted, it is associated with IC1 hypermethylation and BWS phenotype. Overall, the results are consistent with the hypothesis that the spatial organization of the CTSs is critical for the function of the imprinting regulatory element and that internal deletions resulting in long CTS clusters are generally associated with hypermethylation and fully penetrant clinical phenotype.
2. Materials and methods
2.1. Case report
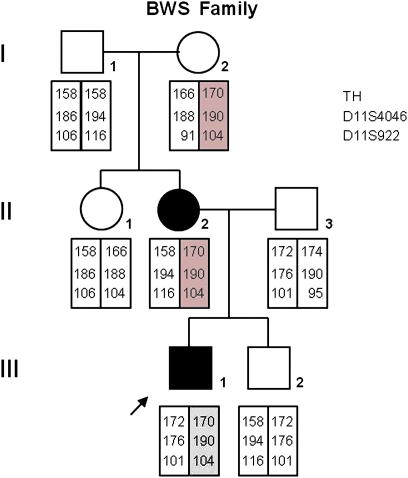
The propositus was born to unrelated parents after 37 weeks gestation and a pregnancy characterized by polyhydramnios. Birth weight was 5080 g (>95th centile), length was 59 cm (>95th centile), and OFC was 36.1 cm (75th centile). The presence of macrosomia, macroglossia, ear pits, neonatal hypoglycaemia, rectum diastasis, umbilical hernia, hepatomegaly, nephromegaly and ureteral abnormalities were consistent with the clinical diagnosis of BWS. Echocardiographic examination revealed the presence of patent foramen ovale. Feeding difficulties were observed during infancy. The propositus’ mother was a 29 year-old woman, at the time of delivery. At birth, she was macrosomic (birth weight 5400 g), displayed macroglossia and was affected by kidney disease (renal congenital renal cysts and kidney stones). No signs typical of BWS were evident in the other family members (Fig. 1).
Fig. 1.
Pedigree of the familial BWS case and segregation of chromosome 11p15.5 genotypes. The genotypes for the microsatellite markers TH, D11S922 and D11S4046 are shown in descending order; the haplotype cosegregating with the BWS phenotype is red colored.
All the genetic analyses were performed with the informed consent of the parents of the family members.
2.2. PCR and DNA sequencing
PCR amplification of the IC1 region was obtained from leukocyte DNA by using the primers: 5′-CGCTGTGGCTGATGTGTAGTAGAG-3′ and 5′-CATTTCCGTCTCCACAGCCACAAC-3′ and the Taq BIO-X-ACT Long (BIOLINE) as described [5]. The fragments generated from the allele carrying the microdeletion were gel-purified and cloned onto pCR II (Invitrogen). DNA sequencing was obtained from PRIMM (Italy).
2.3. Microsatellite analyses
11p15 microsatellite markers TH, D11S4046 and D11S922 were amplified using FAM-labeled and HEX-labeled primers and detected using an ABI 3100 capillary electrophoresis instrument. Data were analysed using GeneMapper Software. Primers and PCR conditions for the microsatellite markers were obtained from the NCBI UniSTS Database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unists).
2.4. DNA methylation analysis
The level of DNA methylation at IC1 (CTS1 and CTS6 regions) and IC2 was determined by COBRA, as previously described [7]. The allele-specific DNA methylation profile of CTS2 was determined by bisulfite genomic sequencing. DNA was treated with sodium bisulfite using EpiTect bisulfite kit (Qiagen) and PCR amplified with the following primer pairs:
In the individuals carrying the 1.8 kb B5-B2 deletion: CTS2_F 5′-AGGTGTTTTAGTTTTTTGGATGATA-3′ with CTS2∆_R 5′-CTCCAAAAATACACATATACTATACAAAAAC-3′ for the deleted allele and CTS2_F 5′-AGGTGTTTTAGTTTTTTGGATGATA-3′ with CTS2_R 5′-CAATTTCTATACCTACCAAAAACC-3′ for the wild-type allele. Annealing temperatures were 60 °C with 1.5 mM of MgCl2).
In the individual carrying the 1.4 kb B5-B3 deletion: CTS2_F 5′-AGGTGTTTTAGTTTTTTGGATGATA-3′ with CTS2∆_R2 5′-TAAATATCCTATCCCTAATAAC-3′ for the deleted allele (annealing temperature was 52 °C with 1.5 mM of MgCl2) and CTS2_F 5′-AGGTGTTTTAGTTTTTTGGATGATA-3′ with CTS2_R2 5′-CACCTAACTTAAATAACCCAAAAC-3′ for the wild-type allele (annealing temperature was 60 °C with 2.0 mM of MgCl2). The PCR products were gel-purified using QIAquick gel extraction (Qiagen), then cloned in Topo pCRII vector (Invitrogen) and individual clones sequenced by PRIMM (Italy).
3. Results
The propositus, his unaffected brother and their parents were analysed for the presence of DNA methylation defects at 11p15.5. The results obtained by COBRA demonstrated abnormal DNA methylation at IC1 and normal DNA methylation at IC2 in II2 and III1. In particular, two CTCF-target sites (CTS1 and CTS6) in the IC1 region were found hypermethylated when compared to controls (Table 1). Normal methylation was detected in the other family members.
Table 1.
Methylation-values at IC1 and IC2 in the family.
| Individual | IC1 |
IC2 | |
|---|---|---|---|
| CTS1 | CTS6 | ||
| II-2 | 7.1 | 4.3 | 1 |
| II-3 | 1 | 1 | 1 |
| III-1 | 10.2 | 3.4 | 1 |
| III-2 | 1 | 1 | 1 |
| NCa | 1 ± 0.36 | 1 ± 0.21 | 1 ± 0.19 |
Numbers represent me/unme ratios at ICs, as determined by COBRA. The ratios were normalised against the average values of 50 control samples. Numbers greater than 1 indicate gain of methylation.
NC, normal controls.
We previously demonstrated that a subset of BWS patients with IC1 hypermethylation carry microdeletions in this regulatory region [5,7]. We looked for the presence of IC1 microdeletions in this family. DNA purified from blood leukocytes of all family members was analysed by long-range PCR. A fragment carrying a 1.8 kb deletion was amplified in individuals II2 and III1, while only a PCR product of normal length was obtained from the other family members. The deletion breakpoints were identified by sequencing the PCR products and found to be located between at chr11:2021870 and chr11:2023704 (NCBI37/hg19).
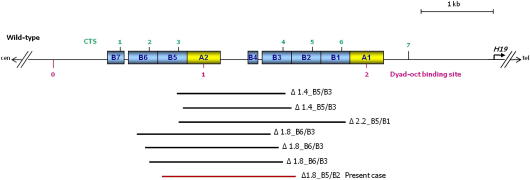
The human IC1 is organized into two large repeated units, each composed of a 450 bp A-type repeat and several highly homologous 400 bp B-type repeats containing target sites for CTCF (CTS, see Fig. 2). The novel 1.8 kb deletion eliminates most of the B5 repeat together with the entire A2, B4, B3 and a small part of the B2 repeat. Although the deletion has the same width of previously reported ones, it involves different B-repeats and CTSs. Indeed, the three previously reported 1.8 kb deletions abolish CTSs 2 and 3, while the novel deletion removes CTSs 3 and 4.
Fig. 2.
Map of the IC1 deletions. The human IC1 is organized in two large repetitive units each consisting of A- and B-type repeats. The positions of the CTCF target sites (CTSs), OCT-binding sites and H19 transcription start site are indicated. The extensions of the previously described deletions [5–7] are indicated by black bars, while the new case is in red. The OCT-binding site 1 was found mutated in BWS [11]. Note that the CTSs 2–3 are abolished in the ∆1.8 B6-B3 deletions, CTS4 in the ∆1.4 B5-B3 deletion and CTSs 3–5 in the ∆2.2 B5-B1 deletion, while CTSs 3–4 are lost in the novel ∆1.8 B5-B2.
In II2 the IC1 microdeletion originated de novo. To determine the parental origin of the chromosome carrying the deletion in II2, we analysed the segregation of the 11p15 haplotype in the family, by typing for three microsatellite markers (Fig. 1). The results showed that II2 and III1 inherited maternally the same 11p15 haplotype and that this was not shared by the unaffected II1 and III2, indicating that II2, as well as III1, carries the IC1 microdeletion on her maternal chromosome. Affected and unaffected individuals showed normal allelic balance at 11p15.5.
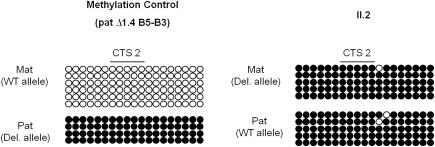
To further study the effect of the microdeletion on IC1 methylation, we determined separately the methylation status of the maternal and paternal IC1 alleles by bisulfite sequencing in II2 and III1. For this purpose, we took advantage of the deletion and used allele-specific primers to amplify selectively the maternal and paternal CTS2 sequence. As a control, we performed a similar analysis in an individual with normal phenotype previously shown to carry a 1.4 kb B5-B3 deletion on his paternal chromosome [7]. The results demonstrated that both the paternal and the maternal IC1 alleles were methylated in II2 and III1 while the paternal allele only was methylated in the control (Fig. 3 and data not shown).
Fig. 3.
DNA methylation profile of CTS2 in II2 and a control individual, as determined by bisulfite genomic sequencing. Each line corresponds to a single template DNA molecule cloned; each circle corresponds to a CpG dinucleotide. Filled circles designate methylated cytosine; open circles, unmethylated cytosines. The CpGs included in the CTS2 are indicated. The individual analysed as control carries a 1.4 kb B5-B3 deletion on its paternal chromosome. Results very similar to II2 were obtained from III1 (not shown).
4. Discussion
Familial BWS cases are rare. This study reports a novel microdeletion of the IC1 region segregating with disease in a parent of origin-dependent dominant manner. The deletion abolishes two out of the seven CTCF target sites (CTSs) and its maternal transmission is associated with hypermethylation of the residual IC1 sequence and the BWS phenotype.
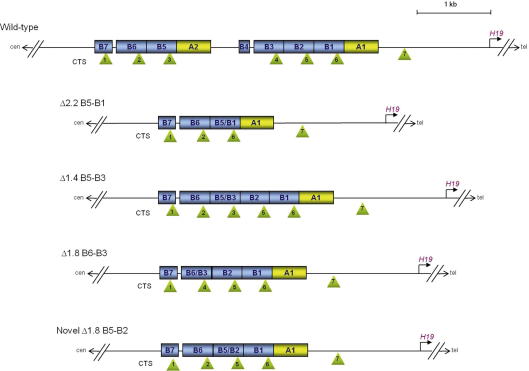
In the wildtype IC1, the CTSs are arranged in two large repetitive units of three sites each, separated by about 1000 kb. The mutation described in this study further expands the list of molecular defects derived from recombination between repeats of the IC1 region, including both microdeletions and microduplications [9,10]. Comparison with previously reported IC1 mutations supports the hypothesis that the correct spacing rather than the number or the type of CTSs is critical for IC1 function. Indeed, in the alleles carrying the microdeletions, the spacing between the CTSs changes and the two cluster of sites merge as a consequence of recombination between the repeats (Suppl. Fig. 1). We observe that the cases in which clusters of 4–5 CTSs are generated (6 mutations) are all associated with hypermethylation of the maternal IC1, while differential methylation is maintained in a case in which a longer deletion (Δ2.2 kb, ref. [6]) generates a single cluster of three sites resembling those of the two large wild-type units.
The mechanism by which some of the microdeletions result in gain of methylation of the residual IC1 sequence is unknown. Point mutations found in a couple of BWS cases implicate an OCT1/OCT4 binding site in the control of IC1 methylation [11]. However, the loss of this site does not correlate with the DNA methylation status of the deleted alleles (Fig. 2). An alternative hypothesis is that the binding of CTCF to DNA depends on the spacing of the CTSs and that this protein binds the long clusters of the Δ1.4–1.8 kb alleles less efficiently than the wild-type and the Δ2.2 kb alleles. CTS mutation results in hypermethylation of the H19 IC in the mouse, indicating that CTCF binding is required to maintain the insulator element hypomethylated and functional on the maternal chromosome [2]. The IC1 methylation status is also relevant for the clinical phenotype. Indeed, an incompletely penetrant BWS phenotype was reported for the 2.2 kb deletion that maintains the normal differential methylation of the maternal IC1 [6]. In contrast, this study demonstrated that the 1.4–1.8 kb deletions always result in IC1 hypermethylation and BWS phenotype with complete penetrance. Pre-natal genetic testing should be considered for such families, as the recurrence risk may be as high as 50% upon maternal transmission.
Acknowledgements
This work was supported by Telethon- Italia grant N. GGP07086 (AR) and Associazione Italiana Ricerca sul Cancro (AR). The authors would like to thank the patients and their family for support.
Footnotes
Supplementary data associated with this article can be found in the online version, at doi:10.1016/j.ejmg.2011.04.009.
Appendix. Supplementary data
Supplementary Fig. 1.
Comparison of the CTS spatial organization in the wild-type and mutant IC1 alleles. Note that the Δ2.2 kb deletion generates a cluster of three CTSs that resembles those of the wild-type allele, while the Δ1.4 kb and Δ1.8 kb deletions result in abnormally longer CTS clusters.
References
- 1.Weksberg R., Shuman C., Beckwith J.B. Beckwith–Wiedemann syndrome. Eur. J. Hum. Genet. 2010;18:8–14. doi: 10.1038/ejhg.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolomei M.S. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nativio R., Sparago A., Ito Y., Weksberg R., Riccio A., Murrell A. Disruption of genomic neighbourhood at the imprinted IGF2-H19 locus in Beckwith–Wiedemann syndrome and Silver-Russell syndrome. Hum. Mol. Genet. 2011;20:1363–1374. doi: 10.1093/hmg/ddr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gicquel C., Rossignol S., Cabrol S., Houang M., Steunou V., Barbu V., Danton F., Thibaud N., Le Merrer M., Burglen L., Bertrand A.M., Netchine I., Le Bouc Y. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver–Russell syndrome. Nat. Genet. 2005;37:1003–1007. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 5.Sparago A., Cerrato F., Vernucci M., Ferrero G.B., Cirillo Silengo M., Riccio A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith–Wiedemann. Nat. Genet. 2004;36:958–960. doi: 10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- 6.Prawitt D., Enklaar T., Gartner-Rupprecht B., Spangenberg C., Oswald M., Lausch E., Schmidtke P., Reutzel D., Fees S., Lucito R., Korzon M., Brozek I., Limon J., Housman D.E., Pelletier J., Zabel B. Microdeletion of target sites for insulator protein CTCF in a chromosome 11p15 imprinting center in Beckwith-Wiedemann syndrome and Wilms’ tumour. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4085–4090. doi: 10.1073/pnas.0500037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sparago A., Russo S., Cerrato F., Ferraiuolo S., Castorina P., Selicorni A., Schwienbacher C., Negrini M., Ferrero G.B., Cirillo Silengo M., Anichini C., Larizza L., Riccio A. Mechanisms causing imprinting defects in familial Beckwith–Wiedemann syndrome withWilms’ tumour. Hum. Mol. Genet. 2007;16:254–264. doi: 10.1093/hmg/ddl448. [DOI] [PubMed] [Google Scholar]
- 8.Cerrato F., Sparago A., Farina L., Ferrero G.B., Cirillo Silengo M., Riccio A. Reply to Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith–Wiedemann. Nat. Genet. 2005;37:786–787. doi: 10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- 9.Scott R.H., Douglas J., Baskcomb L., Huxter N., Barker K., Hanks S., Craft A., Gerrard M., Kohler J.A., Levitt G.A., Picton S., Pizer B., Ronghe M.D., Williams D., Cook J.A., Pujol P., Maher E.R., Birch J.M., Stiller C.A., Pritchard-Jones K., Rahman N., Factors Associated with Childhood Tumours (FACT) Collaboration Constitutional 11p15 abnormalities, including heritable imprinting center mutations, cause nonsyndromic Wilms tumor. Nat. Genet. 2008;40:1329–1334. doi: 10.1038/ng.243. [DOI] [PubMed] [Google Scholar]
- 10.Riccio A. Wilms tumor and constitutional epigenetic defects. Nat. Genet. 2008;40:1272–1273. doi: 10.1038/ng1108-1272. [DOI] [PubMed] [Google Scholar]
- 11.Demars J., Shmela M.E., Rossignol S., Okabe J., Netchine I., Azzi S., Cabrol S., Le Caignec C., David A., Le Bouc Y., El-Osta A., Gicquel C. Analysis of the IGF2/H19 imprinting control region uncovers new genetic defects, including mutations of OCT-binding sequences, in patients with 11p15 fetal growth disorders. Hum. Mol. Genet. 2009;19:803–814. doi: 10.1093/hmg/ddp549. [DOI] [PubMed] [Google Scholar]