Abstract
Rab31, a protein that we cloned from an oligodendrocyte cDNA library, is required for transport of mannose 6-phosphate receptors (MPRs) from the TGN to endosomes and for the Golgi/TGN organization. Here we extend the knowledge of the mechanism of action of Rab31 by demonstrating its interaction with OCRL-1, a phosphatidylinositol 4, 5 diphosphate 5-phosphatase (PI(4,5)P2 5-phosphatase) that regulates the levels of PI(4,5)P2 and PI(4)P, molecules involved in transport and Golgi/TGN organization. We show that Rab31 interacts with OCRL-1 by yeast two hybrid system, GST-Rab31 pull-down experiments and coimmunoprecipitation of OCRL-1 using oligodendrocyte culture lysates. Rab31 and OCRL-1 colocalize in the TGN, post TGN carriers and endosomes. Cation-dependent-MPR (CD-MPR) is sorted to OCRL-1-containing carriers, but CD63 and vesicular stomatitis virus G (VSVG) are not. siRNA-mediated depletion of endogenous Rab31 causes collapse of the TGN apparatus and markedly decreases the levels of OCRL-1 in the TGN and endosomes. Our observations indicate that the role of Rab31 in the Golgi/TGN structure and transport of MPRs depends on its capability to recruit OCRL-1 to domains of the TGN where the formation of carriers occurs. The importance of our observations is highlighted by the fact that mutation of OCRL-1 causes demyelination in humans.
Keywords: Rab proteins, vesicle transport, oligodendrocytes, signaling, myelin, endosomes, trans Golgi network and phosphatidylinositol 4, 5 diphosphate 5-phosphatase
INTRODUCTION
Myelin, the highly specialized membrane that surrounds axons, is generated in the CNS by oligodendrocytes (Morell, 1994). Multiple processes from these cells wrap around the axon in a spiral manner, generating the complex multilamellar structure of the myelin membrane (Raine C.S, 1984). The myelin sheath is not only essential for normal nerve conduction, but also plays a crucial role in maintaining axonal integrity. In multiple sclerosis and other demyelinating conditions, disruption of the myelin membranes has irreparable consequences.
Myelin biogenesis is a highly regulated process that requires coordination of several oligodendrocytic events including lipid and protein synthesis, cytoskeletal reorganization and intracellular trafficking of membranes (Morell, 1994;Larocca and Rodriguez-Gabin, 2002). Membrane trafficking is essential to maintain both the structural and functional organization of the oligodendrocytes and myelin (Larocca and Rodriguez-Gabin, 2002;Huber et al., 1994). However, the oligodendrocyte membrane transport pathways and the signaling mechanisms involved in their regulation remain largely unknown. Studies from others and our laboratories showed the expression of several members of the Rab family of proteins in oligodendrocytes (Bouverat et al., 2000;Madison et al., 1996;Burcelin et al., 1997;Rodriguez-Gabin et al., 2001). Rab proteins are essential components of the mechanisms that regulate intracellular protein transport (Armstrong, 2000). Each member of this family of proteins is specifically localized to a particular membrane transport pathway (Pfeffer, 2005). We showed that Rab31, a Rab protein cloned from an oligodendrocyte cDNA library and originally identified as rRab22b, is located at the Golgi/TGN and endosomes in oligodendrocyte processes(Rodriguez-Gabin et al., 2001). Recently, we showed that Rab31 is involved in both the Golgi/TGN organization and the transport of mannose phosphate receptors (MPRs) from TGN to endosomes(Rodriguez-Gabin et al., 2009). This transport connects biosynthetic pathways to endocytic pathways, and plays an essential role in the biology of the cells, since it is involved in the biogenesis of endosomes, lysosomes and plasma membrane (Mullins and Bonifacino, 2001;Rouille et al., 2000). The transport of MPRs is also implicated in the trafficking of myelin proteins (Rodriguez-Gabin et al., 2001;Larocca and Rodriguez-Gabin, 2002).
The molecular mechanisms underlying the involvement of Rab31 in both maintenance of the Golgi/TGN structure and in the transport of MPRs to endosomes remain largely unknown. However, our results showed that Rab31 regulates the formation of MPRs containing carriers at the TGN(Rodriguez-Gabin et al., 2009). This observation is in agreement with reports that Rab proteins participate in each of the four principal events in membrane traffic: carrier budding, carrier delivery, carrier tethering, and fusion of the carrier with the target compartment (Bonifacino and Glick, 2004;Zerial and McBride, 2001). The ability of Rab proteins to regulate several cellular events depends on their capacity to switch between two conformational states and to interact with a number of proteins located in the membrane donor compartment, in the traffic vesicle and in the membrane acceptor compartment (Zerial and Stenmark, 1993). Indeed, the interaction of Rab proteins with effector proteins leads to specificity in membrane traffic by defining specific membrane domains. Thus, localized clustering of Rab proteins ensures specific recruitment and regulation of proteins required for membrane traffic (Zerial and McBride, 2001;Allan et al., 2000). Therefore, to define the role of Rab31 in both the regulation of MPRs transport from TGN to endosomes and in the Golgi/TGN organization it is necessary to identify the proteins that interact with Rab31, and to determine their localization in cellular compartments.
In this study, we set to identify proteins that interact with Rab31 by molecular cloning using the yeast two hybrid system. A mouse brain cDNA library was screened using the Rab31 full sequence as bait. Our results showed that Rab31 interacts with OCRL-1, a phosphatidyl inositol 4,5 diphosphate 5 phosphatase (PI(4,5)P2 5-phosphatase) (Dressman et al., 2000) that plays a key role in the regulation of the levels of PI(4)P and PI(4,5)P2 (Zhang et al., 1995), two signaling molecules involved in membrane trafficking and Golgi/TGN organization (De Matteis et al., 2002). The formation of Rab31-OCRL-1 complex was confirmed by both GST-Rab31 pull-down experiments and coimmunoprecipitation of oligodendrocyte lysates. Analysis by double fluorescence microscopy of HeLa cells co-expressing Rab31-EYFP and OCRL-1-ECFP showed that Rab31 and OCRL-1 colocalizes in both the TGN and endosomes. Time lapse double fluorescent microscopy showed that Rab31 and OCRL-1 are present in carriers budding from the TGN. Our results also showed that CD-MPR is specifically sorted to carriers containing OCRL-1. In contrast, newly synthesized CD63-EYFP and VSVG-EYFP are not sorted to OCRL-1-containing carriers.
MATERIALS AND METHODS
Recombinant DNA Procedures
The cloning of Rab31 and OCRL-1 tagged with EYFP or ECFP was carried out as described previously(Rodriguez-Gabin et al., 2001). Rab31(S19N) and Rab31(Q64L)mutants were obtained by site–directed mutagenesis according to the method of Ho et al.(Ho et al., 1989). The CD63-ECFP, CD63-EYFP and CD-MPR-ECFP expression plasmids were provided by Dr. Juan Bonifacino (Cell Biology and Metabolism Branch, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA). The VSVG-Venus expression plasmid was provided by Dr. Erik Snapp (Anatomy and Structural Biology Department, Albert Einstein College of Medicine, Yeshiva University).
Oligodendrocyte cultures
Primary cultures of oligodendrocytes were generated from newborn rat cerebral hemispheres by the method of McCarthy and de Vellis (McCarthy and de Vellis, 1980), as described in our previous reports (Larocca and Almazan, 1997;Burcelin et al., 1997). The lineage composition of the cellular preparations was determined by immunocytochemical analysis as described previously (Rodriguez-Gabin et al., 2001). Oligodendrocyte cultures were more than 90% GalC- and MBP-positive cells, whereas 5% were A2B5-positive and the rest were complement receptor C3b (OX-42) or GFAP-positive cells.
Coimmunoprecipication
The formation of the Rab31-OCRL-1 complex was assessed by the capacity of antibodies against Rab31 to coimmunoprecipitate OCRL-1 from cell lysates. These experiments were carried out as previously described (Larocca and Almazan, 1997). Briefly, the cells were lysed in ice-cold immunoprecipitation buffer which contained 20 mM Tris-HCl (pH 7.4), 1 mM EDTA, 137 NaCl, 50 mM NaF, 1 mM Na3VO4, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 1 μg/ml aprotinin, 100 μM PMSF, 10 μg/ml leupeptin. The lysates were incubated with rabbit polyclonal antibodies against Rab31 or rabbit non specific polyclonal antibodies, followed by protein A/G-agarose. The immune complexes were washed twice with lysis buffer and once with Tris-buffered saline containing 1 mM Na3VO4, 100 μM PMSF, 10 μg/ml leupeptin and 1 μg/ml aprotinin. They were resuspended in 30 μl of Laemmli’s sample buffer and heated at 100°C for 3 min. The mixture was subjected to SDS-PAGE on 10% polyacrylamide gels. Proteins were transferred to nitrocellulose membranes by electroblotting. Nitrocellulose membranes were overlaid with media containing monoclonal against OCRL-1 or polyclonal antibodies against Rab31. Bound antibodies were detected by enhanced chemiluminescence using: goat anti rabbit antibodies or rabbit anti mouse antibodies, conjugate with horseradish peroxidase.
Reverse transcriptase and DNA polymerase chain reactions
Complementary DNA (cDNA) from oligodendrocyte total RNA was generated using the Moloney Murine Leukemia Virus reverse transcriptase (MMLV-RT), as described in Burcelin et al. 1997. Oligo dT was used to prime the reaction. PCR was then carried out using a 5′ primer containing the first 20 nucleotides of the OCRL-1 coding region, and a 3′ primer containing nucleotides complementary to the last 20 nucleotides of the OCRL-1 coding sequence, including the stop codon. PCR was carried out according to Burcelin et al. (Burcelin et al., 1997). PCR products were purified by agarose gel electrophoresis and subcloned into pGEMT vector. The inserts were sequenced from the plasmid in the DNA Sequencing Facility at Albert Einstein College of Medicine using the ABI Prism Dye Terminator Kit with AmpliTaq DNA polymerase. Similar studies were performed using total RNA isolated from HOG cells. The isolation of total RNA was performed as described in Burcelin et al. (Burcelin et al., 1997)
Cell line cultures
HeLa cells (Epitheloid carcinoma, from human cervix, obtained from American Type Culture Collection) were maintained in MEM supplemented with 10% FCS, 1% essential amino acids, 2 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin at 37° C in a 5% CO2 incubator.
For transfection experiments, HeLa cells were plated on 35-mm glass-bottom cultures dishes (MatTek Corporation, Ashland, MA) at a density of 0.4-1.6 × 105 cells per dish for 24 hours before transfection.
Transfection
HeLa cells were transfected using Lipofectamine Plus (GIBCO BRL, Gaithersburg, MD) according to the instruction of the manufacturer. Briefly, cultures were treated with 1.2 ml of growth medium containing 3 μg DNA, 12.5 μg of Lipofectamine and 10 μl of Plus reagent at 37° C for 3-6 hr. To select HeLa cells permanently transfected with pGolgi-ECFP, CD63-ECFP, MPR-ECFP and Rab31-ECFP, G418 (2 mg/ml) was added to the culture media, 48 hours after transfection.
Intranuclear injection of DNA
HeLa cells were pressure micro-injected intranuclearly with cDNAs prepared in KCl micro-injection buffer (10 mM Hepes, 140 mM potassium chloride at pH 7.4); 20 μg ml-1 for CD63-ECFP and 20 μg ml-1 for CD63-EYFP, using back-loaded glass capillaries and FenmtoJet micro pump and InjectMan micromanipulator (Eppendorf, Hamburg, Germany). After injection, cells were maintained at 37 °C in a humidified CO2 environment for 60 min to allow for expression of injected cDNAs. Newly synthesized protein was accumulated in the TGN by incubating cells at 20 °C in bicarbonate-free MEM supplemented with 10% FCS, 1% essential amino acids, 2 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin, 20 mM Hepes and 100 μg ml-1 cycloheximide (Sigma, St Louis, MO). Cells were transferred to recording medium (bicarbonate and phenol-red -free MEM supplemented with 1% FBS, 20 mM Hepes, 1% essential amino acids, 2 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin and 100 μg ml-1 cycloheximide) and the transport and fusion of post-Golgi carriers was monitored by time-lapse fluorescence microscopy after shifting to the permissive temperature for transport out of the Golgi (32° C).
Fluorescence microscopic analysis of living cells
Cells were imaged in recording medium (MEM without bicarbonate and phenol-red, and supplemented with 1% FBS, 20 mM Hepes, 1% essential amino acids, 2 mM glutamine, and 100 U/ml penicillin and 100 μg/ml streptomycin) at the temperatures indicated in the figure legends using an Olympus IX70 inverted microscope with a 60X Olympus Planapo oil immersion objective NA 1.4 and equipped with a cooled CCD camera (Photometrics coolSNAP HQ, Roper Scientific) with a 2 × 2 binning. All imaging hardware was controlled by a workstation running I.P. Lab Spectrum imaging software (Scanalytics, Inc. Fairfax, VA). Cells expressing ECFP chimeras were excited using a 436 ± 20 nm filter (Chroma Technology) and imaged with a filter 480 ± 40 nm (Chroma Technology). The exciter filter 500 ± 20 nm, and the emitter filter 535 ± 30 nm (Chroma Technology) were used to visualize EYFP chimeras. Cells expressing both ECFP and EYFP chimeras were excited using a double band filter 436 ± 10 nm and 500 ± 10 nm (Chroma Technology), the emitted fluorescence was splinted using a bean splitter (Dual-View, Roper Scientific) and ECFP fluorescence imaged with a filter 465 ± 30 nm and EYFP fluorescence imaged with a filter 535 ± 30 nm. Out of focus haze was removed from the images by deconvolution (Scanalytics’ IPLab image processing and analysis software; Scanalytics, Inc. Fairfax, VA.
Time lapse studies were carried out to analyze the dynamics of organelles; images were collected at the intervals indicated in the figure legend. At least 40 cells were analyzed per each experimental condition. Kymograph analysis was carried out using the ImageJ 1.36 software (Rasband, 1997-2007). Kymograph analysis of carrier formation and carrier movement was carried out by first defining the area of the cell that included the budding site in the TGN and the carrier trajectory. Second the fluorescence detected in the defined area was graphed as a function of the time and space. To evaluate quantitatively the level of colocalization, the Pearson’s correlation coefficient (r) (Manders et al., 1992) was calculated, with one indicating complete positive correlation and zero indicating no correlation. Values were calculated using the image analysis software ImageJ 1.36b.
RNA-mediated Interference (RNAi)
RNAi was performed using siRNAs (Ambion, Austin, Texas) that target three different regions of the human Rab31 sequence: 1) 5’-GGAUCACUUUGACCACCACAAC-3’, 2) 5’-GGAUGCUAAGGAAUACGCU-3’, and 3) 5’-GCAGGAUUCAUUUUAUACC-3’). Control experiments were performed with non targeting siRNA (Negative Control #1, Ambion). HeLa cells were transfected using siPORT™ NeoFX™ reagent (Ambion) according to the instructions of the supplier, using a reverse transfection protocol. Briefly, cultures were trypsinaized and the cells collected by centrifugation. The pellet was resuspended in medium without FCS at a density of 2 × 105 cells/ml. Aliquots of 1.5 ml of cell suspension were added to a 35 mm Petri dish containing siRNA/siPORT NeoFX complexes (30 nM siRNA, final concentration). Cells were analyzed 48 hours after transfection. The expression of Rab31 was determined by immunoblot analysis using mouse monoclonal antibodies against Rab31 (ABCAM, Cambridge, MA, USA). Expression of GAPDH was determined using mouse monoclonal antibody (Ambion). Immunoblot analysis was carried out according to Larocca and Almazan (Larocca and Almazan, 1997) using the SuperSignal West Dura reagent (Pierce, Rockford, IL, USA). The signals were quantified by densitometry.
Yeast two hybrid system
The MATCHMAKER two-hybrid system (Clontech) was used to screen a mouse brain cDNA library. All the experimental procedures were carried out following the instructions for the Matchmaker two-hybrid system (Clontech). Saccharomyces cerevisiae AH109 host strain were transfected with pGBKT7 vector encoding BD/Rab31. The transfected AH109 host strain were mated with the Y187 host strain transfected with pGADT7 vector encoding AD/library. The plating and selection of the diploid containing proteins that interact with Rab31 were carried out as described in the RESULTS section. Positive experimental controls were carried out by mating AH109 host strain transfected with pGBKT7-53 and Y187 transfected with pGADT7-T. pGBKT7-53 encode fusions between the GAL4 DNA-BD and murine p53, while pGBKT7-T -encode fusions between AD and SV40 large T-antigen; p53 and large T-antigen interact in a yeast two-hybrid assay. Negative experimental controls were carried out by mating AH109 host strain transfected with pGBKT7-Lam with Y187 transfected with pGADT7-T. pGBKT7-Lam encode a fusion of the DNA-BD with human lamin C; large T-antigen does not interact with lamin C in a yeast two-hybrid assay.
GST-Rab31 pull down experiments
The formation of Rab31-OCRL-1 complex was assessed by GST-Rab31 pull-down experiments. A GST-Rab31 fusion protein was used to pull down in vitro translated OCRL-1. Glutathione S-transferase (GST)-Rab31 was produced in Escherichia coli (E. coli) using the GST Gene Fusion System (Amersham Biosciences, Piscataway, NJ). The Rab31 cDNA was cloned in the pGEX plasmid vector following the method described by Rodriguez-Gabin et al. (Rodriguez-Gabin et al., 2004). GST-Rab31 was expressed in E. coli, extracted from the bacterial lysate, and purified by affinity chromatography on a glutathione Sepharose 4B column. OCRL-1 was in vitro translated using Promega’s TnT T7 coupled Reticulocyte Lysate System. Linearize AD/OCRL-1 pGADT7 (0.5 μg) vector was used as template, and the reaction mixture contained [35S]Methionine (20 μCi). [35S] -labeled OCRL-1 protein was incubated with GST-Rab31 in a media containing 100 μM GTP or with GST for 1 h at room temperature. Agarose-glutathione beads were added to the reaction mixtures and the suspension was incubated for 1 h at room temperature. The beads were centrifuged and washed three times with PBS. Then they were resuspended in Laemmli’s buffer and heated at 100° C for 3 min. The proteins were subjected to SDS-PAGE and the band containing the [35S] -labeled OCRL-1 fusion protein was revealed using a fluographic reagent and exposing the gel overnight to X-ray film.
RESULTS
We searched for proteins that interact with Rab31 using the yeast two hybrid system. Rab31 was expressed as a fusion to the Gal4 DNA-binding domain (Rab31-BD), while a mouse brain cDNA library was expressed as fusions to the Gal4 activation domain (brain protein-AD).
Interaction of Rab31-BD with brain protein-AD was detected by mating Saccharomyces cerevisiae AH109 host strain expressing BD/Rab31 with Saccharomyces cerevisiae host Y187 strain expressing AD/library. After mating, the diploid containing the proteins that interact with Rab31 were selected by first plating the mating mixtures on agar containing low stringency media (to select colonies expressing the HIS3 reporter gene), and then a replica plate onto agar containing high stringency media was made (to select colonies expressing both the ADE2 and the HIS3 reporter genes). Additionally, the expression of the lacZ gene was assessed. We detected 400 colonies that grew in high stringency conditions and expressed the lacZ gene.
Three consecutive sets of studies were carried out to rule out false positives and to confirm the two hybrid interaction. First, the AD/library plasmids were isolated from the positive clones, and they were transformed again into AH109 yeast strain. The transformed AH109 were mated with Y187 strain transformed with: BD/Rab31 plasmid, BD/empty vector plasmid, BD/unrelated protein plasmid. Additionally, Y187 strain transformed with BD/Rab31 plasmid were mated with the AH109 strain transformed with: AD/empty plasmid and AD/unrelated protein plasmid. Only hundred fifty AD/library fusion proteins of the original 400 hundred were found to interact with BD/Rab31 fusion protein by this test (only diploids containing AD/plasmid library and BD/Rab31 plasmid should grow in high stringency conditions).
Second, the nucleotide sequence of the AD/library plasmids was determined. A large number of cDNA insert were not in frame with the GAL4 AD sequence while other encoded proteins of unknown function. However, at least 35 plasmids contained cDNA inserts corresponding to mRNA encoding the a isoform of OCRL-1 fused in frame with the GAL4 AD sequence (Accession Number, NP_796189) (Blewitt et al., 2008). OCRL-1 is a member of the group II inositol polyphosphate 5-phosphatases. There are two alternatively-spliced forms of OCRL-1, a and b (Nussbaum et al., 1997;Johnson et al., 2003); the a isoform contains an additional exon lacking in isoform b that encodes a segment of 8 amino acids present in the C terminal region of OCRL-1.
Third, the OCRL-1 cDNA was cloned by PCR and subcloned into BD/plasmid, and the BD/OCRL-1 plasmid was transformed into Y187 yeast strain. The cDNA encoding Rab31 was cloned into AD/plasmid, and the AD/Rab31 plasmid was transformed into AH109 strain. Subsequently, the AH109 transformed with BD/OCRL-1 plasmid was mated with the Y187 strain transformed with the following plasmid: a) AD/Rab31, b) AD/unrelated protein, c) AD/empty. Additionally, Y187 strain transformed with AD/Rab31 was mated with the AH109 strain transformed with following plasmid: d) BD/unrelated protein and e) BD/empty plasmid. As shown in Figure 1 A only the diploid containing the AD/Rab31 and BD/OCRL-1 plasmids grew in high stringency conditions and expressed the lacZ gene, confirming that Rab31 and OCRL-1 form a complex.
Figure 1.
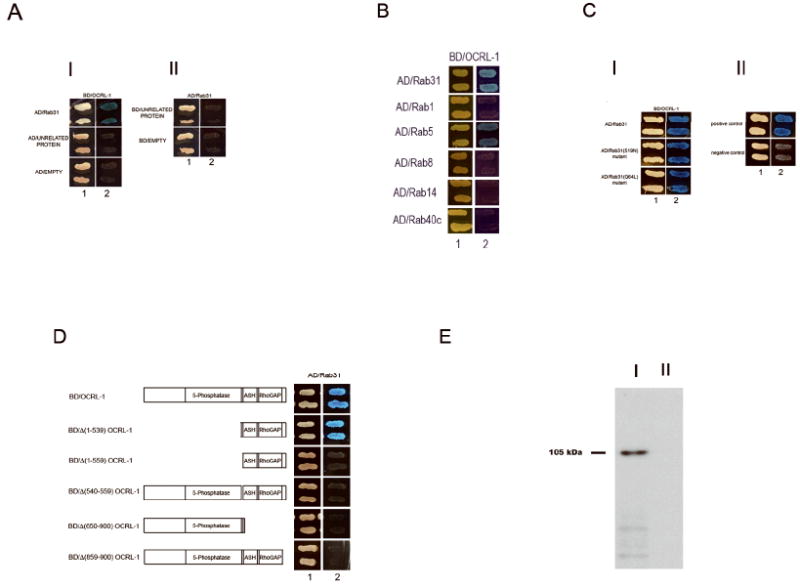
A) Rab31 interacts with OCRL-1. (I) Y187 yeast strain transformed with the BD/OCRL-1 plasmid was mated with the AH109 yeast strain transformed with the following plasmid: a) AD/Rab31, b) AD/unrelated protein, c) AD/empty. (II) Y187 strain transformed with AD/Rab31 was mated with the AH109 strain transformed with following plasmid: d) BD/unrelated protein and e) BD/empty plasmid. The mating mixtures were first plated on agar containing double drop out (DO) media (1), to assess for mating efficiency. Then, a replica plate onto agar containing quadrate drop out media (QO, high stringency media) was made (2), to identify the diploids expressing fusion proteins that interact. Additionally, the expression of the lacZ gene was assessed; X-α-gal was used as α-galactosidase substrate (interaction results in growth of blue colonies). B) Interaction of OCRL-1 with Rab proteins. The capacity of OCRL-1 to interact with other Rab proteins was assessed using the yeast two hybrid system. The AH109 yeast strain transformed with BD/OCRL-1 plasmid was mated with the Y187 yeast strain transformed with the following plasmid: a) AD/rRab22b, b) AD/ Rab1, c) AD/Rab5, d)AD/Rab8, e) AD/Rab14 and f) AD/Rab40c g) positive control and h) negative control. C) Formation of OCRL-1-Rab31 complex does not depend on the conformational state of Rab31. (I) The AH109 yeast strain transformed with BD/OCRL-1 plasmid was mated with the Y187 yeast strain transformed with the following plasmid: a) AD/Rab31, b) AD/ Rab31(S19N), c) AD/Rab31(Q64L). (II) Positive control and negative control. Positive experimental controls were carried out by mating AH109 host strain transfected with pGBKT7-53 and Y187 transfected with pGADT7-T. pGBKT7-53 encode fusions between the GAL4 DNA-BD and murine p53, while pGBKT7-T -encode fusions between AD and SV40 large T-antigen. Negative experimental controls were carried out by mating AH109 host strain transfected with pGBKT7-Lam with Y187 transfected with pGADT7-T. pGBKT7-Lam encode a fusion of the DNA-BD with human lamin C. D) Mapping of the Rab31 binding site in OCRL-1. Full-length and truncated OCRL-1 constructs were tested for interaction with Rab31 using the yeast two-hybrid system. The Y187 yeast strain transformed with AD/Rab31 plasmid was mated with the AH109 yeast strain transformed with the following plasmids: a) BD/OCRL-1, b) BD/Δ (1-539) OCRL-1, c) BD/Δ(1-559) OCRL-1, d) BD/Δ(540-559) OCRL-1, e) BD/Δ (650-900) OCRL-1 and f) BD/Δ (858-900) OCRL-1. E) GST-Rab31 pull-down experiments. [35S]-labeled OCRL-1 protein was incubated in a media containing 100 μM GTP with either GST-Rab31 (I) or GST (II). The GST-Rab31-OCRL-1 complex was pulled down with agarose-glutathione beads. The proteins were separated by SDS-PAGE. The band containing the [35S]-labeled protein was revealed using a fluographic reagent and exposing the gel overnight to X-ray film. Number to the left indicates the molecular weight of [35S]-labeled OCRL-1 protein.
These yeast two hybrid system studies showed that Rab31 interacts with the a isoform of OCRL-1.
Analysis of OCRL-1 interaction with Rab proteins
The capacity of OCRL-1 to interact with other Rab proteins was assessed using the yeast two hybrid system. The AH109 strain transformed with BD/OCRL-1 plasmid was mated with the Y187 strain transformed with the following plasmid: a) AD/Rab31, b) AD/ Rab1, c) AD/Rab5, d)AD/Rab8, e) AD/Rab14 and f) AD/Rab40c. As shown in Figure 1 B, only the diploid containing BD/OCRL-1 and AD/Rab31, or BD/OCRL-1 and AD/Rab5 grew in high stringency conditions and expressed the lacZ gene (Figure 1 B). These results showed that OCRL-1 interact with Rab31 and Rab5, but not with Rab1, Rab8, Rab14 and Rab40c, suggesting that the Rab31 and Rab5 interactions with OCRL-1 are specific.
GST-Rab31 pull-down studies
The formation of Rab31-OCRL-1 complex was also assessed by GST-Rab31 pull-down experiments. A GST-Rab31 fusion protein was used to pull down in vitro translated [35S]-labeled OCRL-1. in 100 uM GTP containing medium. Unfuse GST was used as control. As is shown in Figure 1 E, [35S] -labeled OCRL-1 protein is present in the pull-down experiments carried out with GST-Rab31, but not when the pull down experiment is carried out with GST. These results confirmed the interaction between of OCRL-1 and Rab31.
Formation of the OCRL-1-Rab31 complex does not depend on the conformational state of Rab31
We determined whether interaction of OCRL-1 with Rab31 depends on the conformational state (GTP-bound or GDP-bound), of Rab31. To this end we assessed the interaction of OCRL-1 with two Rab31 mutants, Rab31(S19N) that preferentially binds GDP, and Rab31(Q64L) that is locked in the GTP-bound state because it lacks GTPase activity. The formation of the OCRL-1-Rab31 mutant complexes was assessed by yeast two hybrid system. As shown in Figure 1 C, the diploid containing the AD/Rab31, AD/ Rab31(S19N) or AD/Rab31(Q64L) and BD/OCRL-1 plasmids grow in high stringency conditions and express the lacZ gene, indicating that the formation of the OCRL-1-Rab31 complex does not depend on the Rab31 conformational state. Similarly, previous reports showed that the interaction of Rac with OCRL-1 does not dependent on the conformational state of Rac (Faucherre et al., 2003).
Mapping of the Rab31 binding site in OCRL-1
The OCRL-1 structure encompasses several domains including a central 5-phosphatase domain, followed by a sequence identified as an ASH domain and a C terminal catalytically inactive RhoGAP domain (Figure 1 D). Thus, we carried out studies to assess the involvement of the different regions of OCRL-1 in the binding to Rab31. Several truncated mutants of OCRL-1 were tested for binding to Rab31 using the yeast two hybrid system (Figure 1 D). The N terminal region containing the phosphatase domain is not involved in the formation of the Rab31-OCRL-1 complex, as demonstrated by the fact that an OCRL-1 mutant lacking this region interacted with Rab31. Further analysis of the importance of the C terminus indicated that at least two separate sequences are essential for the formation of the OCRL-1-Rab31 complex. The first includes residues 540 to 559, while the second encompasses residues 858 to 900. Deletion of either the first segment (residues 540-559) (Δ(540-559)OCRL-1) or the second (residues 858 to 900) (Δ(858-900)OCRL-1) resulted in deletion OCRL-1 mutants that did not bind Rab31 (Figure 1 D).
In summary, this analysis showed that both the N-terminus and 5-phosphatase domain of OCRL-1 (residues 1-539) are not involved in the formation of the Rab31-OCRL-1 complex. Instead the Rab31 binding domain located at the C-terminus of OCRL-1 and it is formed by at least two separate regions including 540 to 559, and residues 858 to 900.
OCRL-1 is expressed in oligodendrocytes
Since we had previously cloned Rab31 from oligodendrocyte cDNA library, we analyzed whether OCRL-1 is expressed in these cells. To this end we used primary cultures of oligodendrocytes and the human oligodendroglioma cell line HOG. Differentiated oligodendrocytes were generated from newborn rat cerebral hemispheres by the method of McCarthy and de Vellis. Immunocytochemistry analysis showed that between 85-90% of the cells present had the phenotype of mature oligodendrocytes (galacto cerebrosides positive and MBP positive cells (results not shown). The expression of OCRL-1 was assessed by two independent methods: reverse transcriptase polymerase chain reaction (RT-PCR) and immunoblot analysis. RT-PCR was performed using total RNA isolated from rat oligodendrocytes and HOG cells as template and primers directed against the C terminus and N terminus of OCRL-1. The products of the reaction were cloned in pGEMT vector and sequenced. The results showed a band of approximately 2,700 bp (Figure 2 A, lanes 1 and 2), and its sequence corresponded to the a isoform of OCRL-1 (results not shown). The possibility that the positive RT-PCR is caused by the presence of DNA in the RNA preparations was discarded because no reaction products were obtained when the PCR was performed using total RNA as template (Figure 2 A, lanes 3 and 4).
Figure 2. OCRL-1 is expressed in oligodendrocytes and HOG cells.

A) RT-PCR analysis of the expression of OCRL-1 in rat oligodendrocytes and HOG cells. RT reaction was performed using 10 μg of total RNA and primers targeting the C terminus of OCRL-1 (rat and human). PCR reactions were carried out using the cDNA obtained in the RT reaction and primers targeting the C- and the N- termini (lanes 1 and 2). To rule out DNA contamination, PCR was carried out using as templates total RNA (lane 3 and 4). PCR products were separated by electrophoresis on 1 % agarose gels. Oligodendrocytes, lanes 1 and 3; HOG cells, lanes 2 and 4. B) Western blot analysis of OCRL-1 expression in differentiated oligodendrocyte cultures and HOG cells with monoclonal antibody against OCRL-1. Lysate samples containing 20 μg of protein (Oligodendrocytes, lanes 1 and 3; HOG cells, lanes 2 and 4) were resolved by SDS-PAGE on 6 % gels. Proteins were transferred to nitrocellulose membranes by electroblotting. Nitrocellulose membranes were overlaid with media containing monoclonal antibody against OCRL-1 (lanes 1 and 2). Negative control consisted in membranes incubated in the absence of antibody against OCRL-1 (lanes 3 and 4). Bound antibodies were detected by enhanced chemiluminescence.
The presence of OCRL-1 in oligodendrocytes and HOG cells was confirmed by immunoblot analysis using a mouse monoclonal antibody against OCRL-1 (provided by Dr. Robert Nussbaum, NIH). A polypeptide of 105 kDa reacted with specific antibody against OCRL-1 (Figure 2 B, lane 1). When the antibody against OCRL-1 was excluded from the reaction media the 105 kDa was not observed (Figure 2 B, lane 3). Similar results were obtained by immunoblot analysis of protein from HOG cells proteins (Figure 2 A, lane 2 and 4).
Coimmunoprecipitation of Rab31 and OCRL-1
The formation of the Rab31-OCRL-1 complex was assessed by coprecipitation of OCRL-1 by antibodies against rat Rab31 using oligodendrocyte culture lysates. In controls, to detect the formation of nonspecific complexes, non specific rabbit IgG was used. OCRL-1 was present in the immune complexes obtained by adding antibodies against Rab31 (Figure 3 A). In contrast, the immune complexes formed in the control experiments did not contain OCRL-1 (Figure 3 B). These observations confirm the interaction of Rab31 with OCRL-1 in living cells, in particular demonstrate the formation of complexes containing Rab31 and OCRL-1 in oligodendrocytes.
Figure 3. Coimmunoprecipitation of Rab31 and OCRL-1.
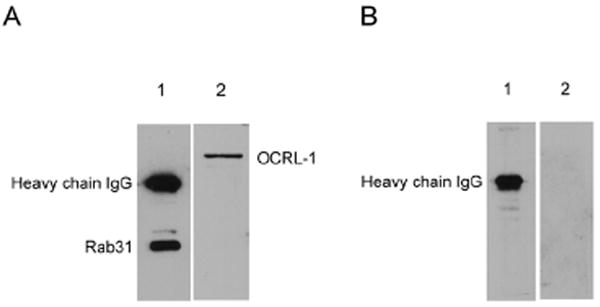
A) Immune complexes formed by treatment of oligodendrocyte lysates with rabbit IgG against Rab31. B) Immune complexes formed by treatment of the lysates with non specific rabbit IgG. Samples were resolved by SDS-PAGE on 10 % gels. Proteins were transferred from gel to nitrocellulose membranes by electroblotting. Nitrocellulose membranes were overlaid with media: containing rabbit IgG against Rab31 (lane 1) or mouse monoclonal anti- OCRL-1 (lane 2). Bound antibodies were detected by enhanced chemiluminescence using goat anti-rabbit antibodies (lane 1) or rabbit antimouse antibodies (lane 2) conjugated to horseradish peroxidase.
OCRL-1 and Rab31 co-localized at the TGN and in post-TGN carriers
Our previous studies showed that Rab31 localized along the pathway that transports MPRs from TGN to endosomes. Certainly, our studies showed the presence of Rab31 in the TGN and carriers that bud from TGN and fuse with endosomes, demonstrating that the transport of MPRs from TGN to endosomes is carried out by carriers containing Rab31. These observations together with the data presented here demonstrating the interaction of Rab31 with OCRL-1, led us to hypothesize that both proteins co-localize along the pathway that transports MPRs from the TGN to endosomes.
First, to demonstrate the presence of OCRL-1 in Golgi/TGN and post-TGN carriers we analyzed by time-lapse fluorescence microscopy HeLa cells co-expressing OCRL-1 and pGolgi-ECFP (a chimera constructed with ECFP and an 81-amino acid fragment of the N terminal region of human 1,4 beta galactosyl transferase, a protein that localizes at the Golgi/TGN). The time-lapse experiments were carried out at 32° C, a temperature widely used in the study of post-Golgi transport, which facilitated the visualization of the carriers budding from the TGN. OCRL-1-EYFP exhibited a perinuclear localization in structures of the Golgi/TGN, as shown by its co-localization with pGolgi (r=0.850 ± 0.057) (Figure 4). OCRL-1-EYFP was also present in small vesiculo-tubular compartments that are dispersed all through the cell cytoplasm including the cell periphery. Kymographic analysis of data obtained in the time-lapse experiments showed that some of these structures break up from the TGN and move toward the cell periphery along linear or curvilinear trajectories, at speeds ranging from 0.40 to1.20 μm/sec. As shown in Figure 4 B and C movements were intermittent, with frequent changes in direction (see also Video Figure 4). As they move toward the cell periphery, the majority drop out of the focal plane. However, some seem to interact and fuse with small vesicles, which are probably part of the endosomal system (Video Figure 4).
Figure 4. OCRL-1 localizes at the Golgi/TGN and in carriers budding from the TGN.
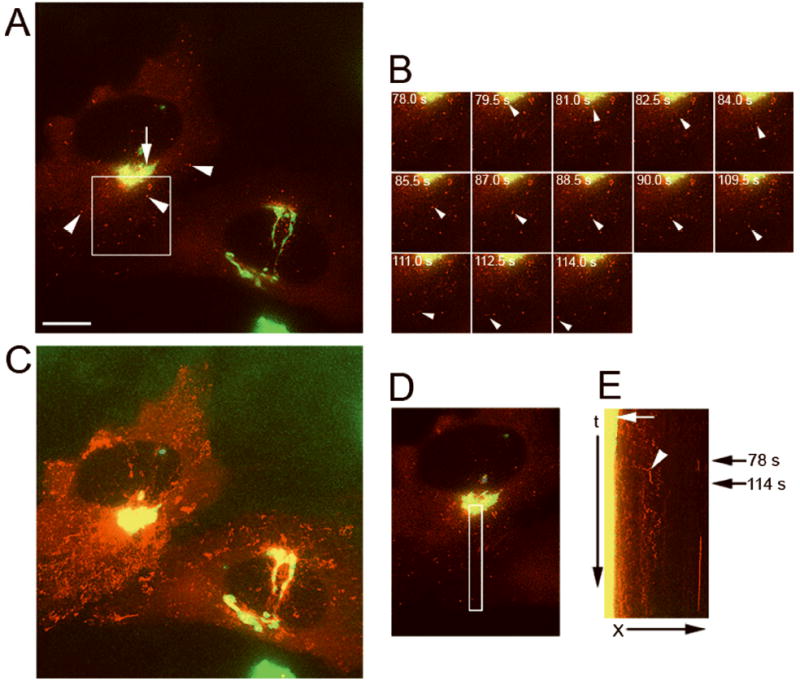
HeLa cells expressing both pGolgi-ECFP and OCRL-1-EYFP were analyzed by fluorescence microscopy. Images at two different emission wavelengths (465 ± 30 nm and 535 ± 30 nm) from the same field were collected every 2 s. to simultaneously visualize pGolgi-ECFP and OCRL-1-EYFP. A) Fluorescence distribution pattern. Red, OCRL-1-EYFP; green, pGolgi-ECFP; yellow, overlapping of OCRL-1-EYFP and pGolgi-ECFP. The arrow shows the colocalization of OCRL-1-EYFP and pGolgi at the Golgi/TGN, arrowheads point to small vesicles present throughout the cytoplasm containing only OCRL-1-EYFP. B) Individual frames of the area boxed in (A). A carrier containing OCRL-1-EYFP breaks up from the TGN and moves towards the cell periphery (white arrowheads). The time in seconds relative to the first image is shown in each frame. C) Maximum pixel projection. OCRL-1-EYFP and pGolgi-ECFP co-localized in the TGN but not in small vesicles present throughout the cytoplasm. D) Area where the events in B (carrier formation and movement to cell periphery) occurred is indicated by a white box. E) Kymograph analysis of the area defined in (D). A lack of movement of TGN compartments results in a vertical trace (yellow, white arrow). Several carriers containing OCRL-1-EYFP are formed in the TGN (red diagonal traces beginning in the yellow vertical trace). The trace that corresponds to carrier budding in (B) is indicated (white arrowhead). Black arrows on the right indicate the time when budding in (B) occurred. X, distance in μm; t, time in seconds. Bar, 10 μm.
Second, the colocalization of Rab31 and OCRL-1 was assessed by time lapse double fluorescence analysis of HeLa cells co-expressing Rab31-EYFP and OCRL-1-ECFP. The results showed that both OCRL-1-ECFP and Rab31-EYFP co-localized in structures of the TGN (r = 0.900 ± 0.044), and in small tubular compartments located throughout the cytoplasm and cell periphery, that are likely to represent endosomes (r = 0.875 ± 0.064) (Figure 5 A). We observed tubular carriers containing both Rab31-EYFP and OCRL-1-ECFP forming from TGN structures (Figure 5 B and C), and they moved to the cell periphery with kinetic and plastic properties similar to those containing only OCRL-1 (see also Video Figure 5).
Figure 5. Rab31 and OCRL-1 colocalize in carriers that bud from the TGN.
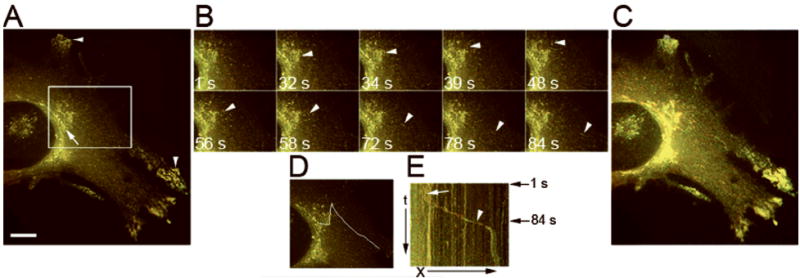
Stably-transfected HeLa cells expressing OCRL-1-ECFP were transiently-transfected with a plasmid encoding Rab31-EYFP. Cells were transferred to recording medium. Formation and the transport of post-TGN carriers were monitored at 32° C by time-lapse fluorescence microscopy, images were collected every 1 s. A) Fluorescence pattern distribution. OCRL-1-ECFP (red), Rab31-EYFP (green) and overlapping of OCRL-1-ECFP and Rab31-EYFP (yellow). Rab31-EYFP and OCRL-1-ECFP co-localize in the TGN (arrow), and in endosomes throughout the cytoplasm (arrowheads). B) Individual frames showing a carrier containing both OCRL-1-ECFP and Rab31-EYFP breaking up from the TGN (white arrowheads). Time in seconds relative to the first image is shown in each frame. C) Maximum pixel projection. OCRL-1-ECFP and Rab31-EYFP co-localized at the TGN and in small vesicles present throughout the cytoplasm. D) Area where the events in B (carrier formation and movement to cell periphery) occurred is indicated by a white lane. E) Kymograph analysis of the area defined in (D). The TGN compartments, vertical yellow trace, is indicated with a white arrow. The carrier formation and its movement, diagonal yellow trace is indicated by the arrowhead. White arrowhead identifies the trace that corresponds to carrier budding shown in (B). Black arrows at the right indicate the time of budding shown in (B). X, distance in μm; t, time in seconds. Bar, 10 μm.
OCRL-1 carriers contain CD-MPR but not CD63 or VSVG
The previous observations suggest that OCRL-1 is involved in the transport of newly synthesized lysosomal hydrolases from TGN to endosomes. Lysosomal hydrolases are transported to endosomes in a complex with MPRs. These complexes dissociate in endosomes, the hydrolases are subsequently delivered to lysosomes and the MPRs recycled to the TGN. To assess the involvement of OCRL-1 in the transport of MPRs, we determined if cationic-dependent MPR (CD-MPR) is recruited into OCRL-1-containing carriers budding from the TGN.
As we and others have demonstrated, the fact that MPRs recycle between TGN and endosomes makes it possible to visualize their transport from TGN to endosomes by fluorescence microscopy analysis of cells expressing MPRs tagged with variants of the green fluorescent protein. Therefore, the presence of OCRL-1 in carriers transporting CD-MPR from the TGN to endosomes was assessed by double fluorescence time-lapse studies of HeLa cells co-expressing OCRL-1-EYFP and CD-MPR-ECFP. HeLa cells stably transfected with plasmid encoding CD-MPR-ECFP were transfected with plasmid encoding OCRL-1-EYFP and, forty eight hours after transfection, the cells were analyzed by double fluorescence microscopy.
As shown in Figure 6 A and E, OCRL-1-EYFP and CD-MPR-ECFP co-localized at the TGN (r = 0.920 ± 0.040) and tubular compartments present all throughout the cytoplasm (0.700 ± 0.075). These studies also showed that CD-MPR-ECFP and OCRL-1-EYFP co-localized in carriers budding from the TGN (Figure 6 B-D, see also Video Figure 6).
Figure 6. OCRL-1 carriers transport CD-MPR from TGN to endosomes.
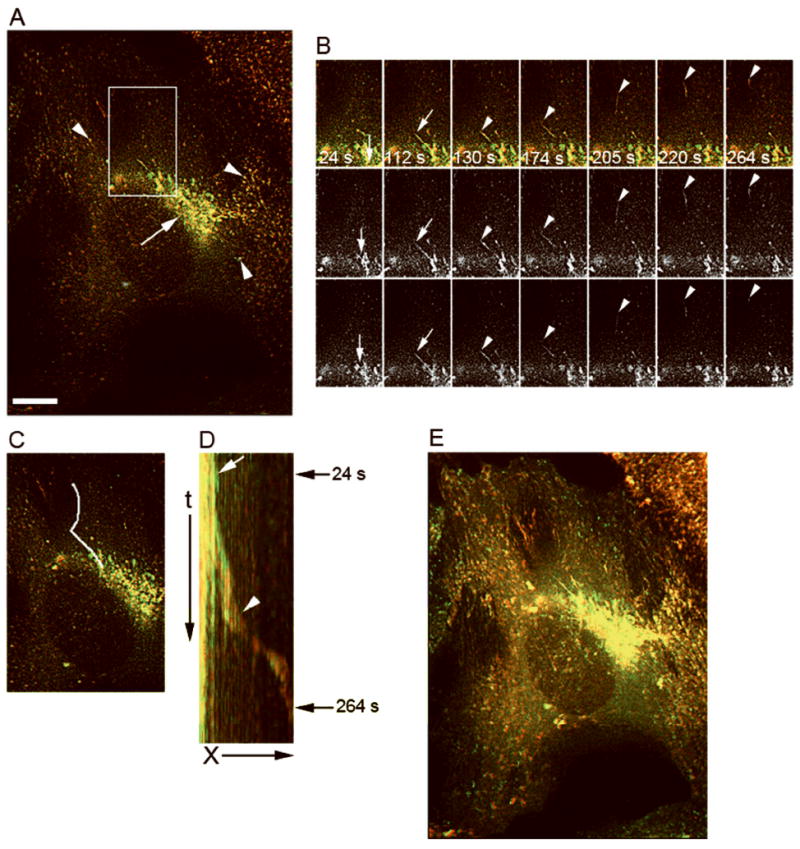
HeLa cells expressing CD-MPR-ECFP and OCRL-1-EYFP were analyzed by fluorescence microscopy. The formation and the transport of post-Golgi carriers were monitored by time-lapse fluorescence microscopy at 32° C. CD-MPR-ECFP and OCRL-1-EYFP were simultaneously visualized, images were collected every 1 s. A) Fluorescence pattern distribution; CD-MPR-ECFP (red), OCRL-1-EYFP (green) and overlapping of CD-MPR-ECFP and OCRL-1-EYFP (yellow). OCRL-1-ECFP and CD-MPR-ECFP colocalized at the TGN (arrow), and in endosomes present throughout the cytoplasm (arrowheads). B) Individual frames showing a TGN tubule that extends (white arrows), and breaks up to form a carrier containing both CD-MPR-ECFP and Rab31-EYFP (white arrowheads). Top panels: merged images; center panels: CD-MPR-ECFP; lower panels: OCRL-1-EYFP. Time relative to the first image is shown in seconds. C) Area where the events showed in B (carrier formation and movement to cell periphery) occurred is indicated by a white line. D) Kymograph analysis of the area defined in (C). A lack of movement of the TGN compartments results in a vertical trace (white arrow). Carrier formation and its movement along the area results in a diagonal trace that begins in the vertical trace (white arrowhead). Arrows at the right indicate the time where the budding showed in (B) occurred. E) Maximum pixel projection. OCRL-1-EYFP and CD-MPR-ECFP co-localized at the TGN and in small vesicles present throughout the cytoplasm X, distance in μm; t, time in seconds. (See animated movie Figure 6). Bar, 10 μm.
These observations indicated that CD-MPR is sorted into OCRL-1-containing carriers. To demonstrate that this sorting is specific, we assessed if newly synthesized CD63 is recruited into OCRL-1 containing carriers. CD63, also known as lysosome integral membrane protein 3, is synthesized in the endoplasmic reticulum and subsequently transported to the Golgi/TGN. CD63 is transported from TGN to endosomes by MPRs independent pathway before being targeted to lysosomes (Rous et al., 2002), and also via the plasma membrane (Janvier and Bonifacino, 2005).
To simultaneously visualize the sorting of CD-MPR and of CD63 to carriers budding from the TGN, Hela cells stably transfected with plasmid encoding OCRL-1-ECFP were transiently transfected with plasmid encoding CD63-EYFP by intranuclear injection. Newly synthesized CD63-EYFP was trapped in the Golgi/TGN by incubating the cells for 3 hours at 20° C in the presence of cycloheximide. The cells were then transferred to recording media at 32 ° C to induce the formation of post-Golgi carriers. Time-lapse photographs visualized the formation of carriers containing OCRL-1-ECFP and of carriers containing CD63-EYFP.
CD63-EYFP and OCRL-1-ECFP partially colocalized in Golgi/TGN compartments (Figure 7 A and H) (r = 0.720 ± 0.074). In contrast, carriers that bud from the TGN contained only a single fluorescent protein. Figure 7 B-D shows the formation of carriers containing only CD63-EYFP, while Figure 7 E-G shows the formation of a carrier containing OCRL-ECFP. Maximum pixel projection analysis (Figure 7 H) confirmed that CD63-EYFP and OCRL-1-ECFP partially colocalized in Golgi/TGN (r= 0.850 ± 0.070) (see also Video Figure 7). In contrast, colocalization is not observed in the small tubular structures located through out the cytoplasm (r = 0.096 ± 0.010).
Figure 7. OCRL-1 and newly synthesized CD63 are sorted to different carriers budding from the TGN.
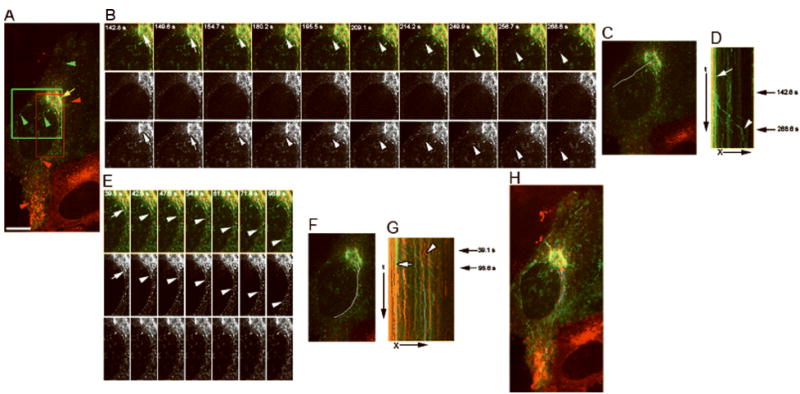
Stably-transfected Hela cells expressing OCRL-1-ECFP were injected intranuclearly with cDNAs encoding CD63-EYFP. Newly synthesized CD63-EYFP was trapped in the TGN and carrier formation visualized by time-lapse double fluorescence microscopy at 32° C. A) Fluorescence distribution pattern. Green, CD63-EYFP; red, OCRL-1-ECFP; yellow overlapping of CD63-EYFP and OCRL-1-ECFP. CD63-EYFP and OCRL-1-ECFP partially co-localized in the Golgi/TGN (arrow), but not in small vesicles present through out the cytoplasm (arrowheads). B) Individual frames of the area marked with green box in (A). Top panels: overlap of images; central panels: OCRL-1-ECFP; lower panel: CD63-EYFP. A TGN tubule containing CD63-EYFP extends (white arrows), and breaks up to form a carrier (white arrowheads). The time relative to the first image is shown in seconds. C) Area where the events showed in B occurred (carrier formation and their movement to cell periphery) is indicated by a white lane. D) Kymograph analysis of the area defined in (C). TGN, vertical traces (red and green lines, white arrow). The green trace that corresponds to the budding of carrier containing CD63-EYFP showed in (B) is indicated by a white arrow head. Black arrows on the right indicate the time where the budding showed in (B) occurred. E) Individual frames of the area marked with red box in (A). A TGN tubule containing CD63-EYFP extend (white arrow), and breaks up to form a carrier (white arrowheads). The time relative to the first image is shown in seconds. F) Area where the events showed in E occurred (carrier formation and its movement to cell periphery) is indicated by a white line. G) Kymograph analysis of the area defined in (F). The green line that corresponds to the TGN compartment from which the carrier was formed is indicated by a white arrow. The green trace that corresponds to the budding of carrier containing CD63-EYFP shown in (E) is indicated by a white arrowhead. Black arrows on the right indicate the time where the budding shown occurred. H) Maximum pixel projection. OCRL-1-EYFP and CD-MPR-ECFP co-localized in the TGN, but not in small vesicles present throughout the cytoplasm. X, distance in μm; t, time in sec. Bar, 10 μm.
To further assess the specificity of sorting, we also studied the recruitment of VSVG to OCRL-1-containing carriers. HeLa cells expressing OCRL-1-ECFP were transfected with plasmid encoding VSVG-Venus (VSVG protein from the ts045 mutant strain of vesicular stomatitis virus was used), and were incubated at 40° C for 10-12 h. to accumulate VSVG-Venus in the ER. Transfected cells were then incubated at 20° C for 3 h., with cycloheximide (100 μg/ml) to trap VSVG-Venus in the TGN. The cells were then transferred to recording media at 32 ° C to induce transport of VSVG from Golgi to PM. VSVG-Venus and OCRL-1-ECFP co-localized at the Golgi/TGN compartments (r = 0.835 ± 0.074) (Figure 8 A and C). In contrast, and similar to the observations in cells expressing CD63 and OCRL-1, carriers budding from the TGN contained only a single fluorescent protein. Figure 8 shows the formation of carriers containing either VSVG-Venus (Figure 8 E-G), or OCRL-1-ECFP (Figure 8 H-J) but not both. These observations were confirmed by time lapse images (maximum pixel projection) that clearly showed no colocalization of VSVG-Venus and OCRL-1-ECFP in either carrier vesicles or endocytic compartments (r = 0.213 ± 0.076) (Figure 8 D, see also Video Figure 8). These results showed that VSVG is not sorted at the TGN into carriers containing OCRL-1.
Figure 8. OCRL-1 and newly synthesized VSVG are sorted to different carriers budding from the TGN.
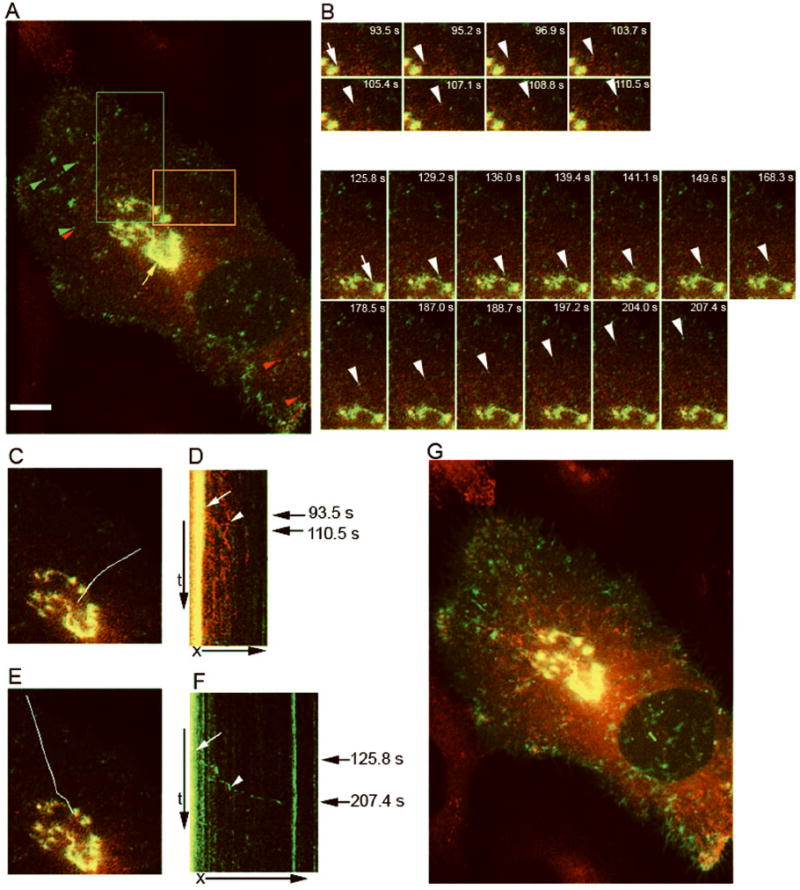
Stably-transfected HeLa cells expressing OCRL-1-ECFP were transfected with plasmid encoding VSVG-Venus, and then incubated at 40° C for 10-12 h to accumulate VSVG-Venus in the ER. Transfected cells were then incubated at 20° C for 3 h, with cycloheximide (100 μg/ml) to trap VSVG-Venus at the TGN. The cells were transferred to recording media at 32 ° C to induce transport of VSVG from Golgi to PM. Time-lapse photographs visualized the formation of carriers containing OCRL-1-ECFP and of carriers containing VSVG-Venus. Images were collected every 1.70 s. A) Fluorescence distribution pattern. Green, VSVG-Venus; red, OCRL-1-ECFP and yellow, overlapping of VSVG-Venus and OCRL-1-ECFP. OCRL-1-ECFP and VSVG-Venus co-localized at the TGN (yellow arrow). Small vesicles present in the cytoplasm only contain VSVG-Venus (green arrowheads) or OCRL-1-ECFP (red arrowheads). B) Top panels, individual frames of the area marked with red box in (A). A TGN tubule containing OCRL-1-ECFP extends (arrow), and breaks up to form a carrier (arrowheads). Bottom panels, individual frames of the area marked with green box in (A). A TGN tubule containing VSVG-Venus extends (arrow), and breaks up to form a carrier (arrowheads). C) Area where the events in B occurred (top panels) is indicated by a white line. D) Kymograph analysis of the area defined in (C). TGN compartments, vertical traces (white arrow). The red trace that corresponds to the budding of carrier containing OCRL-1-ECFP seen in (B) is indicated by a white arrowhead. Arrows at the right indicate the time of budding in (B). E) Area where the events in B occurred (bottom panels) is indicated by a white line. F) Kymograph analysis of the area defined in (E). TGN compartments, vertical traces (white arrow). The green trace that corresponds to the budding of carrier containing VSVG-Venus seen in (B) is indicated by a white arrowhead. Arrows on the right indicate the time of budding in (B). X, distance in μm; t, time in seconds. G) Maximum pixel projection. OCRL-1-ECFP and VSVG-Venus colocalized in the TGN but not in small vesicles present throughout the cytoplasm. Bar, 10 μm.
Depletion of Rab31 caused TGN fragmentation and reduced the amount of OCRL-1 bound to membranes
Our observations raise the possibility that the targeting of OCRL-1 to specific membrane domains depends on Rab31. To test this hypothesis endogenous Rab31 was depleted using siRNA. Treatment of cells with siRNA that target the sequence present in exon 2 (5’-GGAUCACUUUGACCACCACAAC-3’), reduced the expression of Rab31 by 95.00 ± 2.00 % (Figure 9). In contrast, the expression of GAPDH did not change (Figure 9 C and F), indicating that the siRNA effect was specific. Fluorescence microscopy showed that the depletion of Rab31 caused fragmentation of the TGN structure. As shown in cells expressing CD-MPR-ECFP, the TGN breaks up into fragments Figure 9 B). In contrast, the cells treated with non-targeting siRNA showed no alteration of the TGN structure; i.e. CD-MPR-ECFP localized in perinuclear; tubular structures (Figure 9 A). Analysis of cells expressing OCRL-1-ECFP showed that depletion of Rab31 also caused a marked decrease in the amount of OCRL-1-ECFP present in the TGN and small vesicles present in the cell periphery with a parallel increase in the amount of this protein in the cytoplasm (Figure 9 D and E). Similar results were obtained by treating the cells with two other siRNAs that target different segments of the Rab31 sequence [5’-GGAUGCUAAGGAAUACGCU-3 ’ (present in exon 6 ) and 5 ’-GCAGGAUUCAUUUUAUACC-3’ (present in exons 4 and 5)], ruling out an “off-target effect” (results not shown).
Figure 9. Depletion of endogenous Rab31 results in Golgi/TGN fragmentation.
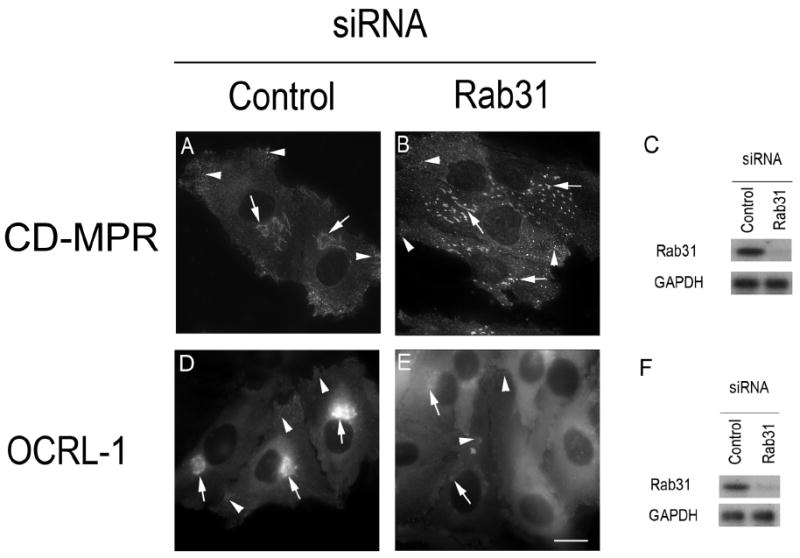
(A and D) HeLa cells were transfected with non targeting siRNA (control), or (B and E) with siRNA targeting a segment of the sequence of Rab31 present in exon 2 ( 5 ’-GGAUCACUUUGACCACCACAAC-3’). After 48 h, the level of Rab31 and GAPDH were determined by immunoblotting (C and F). The effect of Rab31 depletion on the distribution of CD-MPR-ECFP (B) or OCRL-1-ECFP (E) were analyzed by fluorescence microscopy. Arrows indicate the position of Golgi/TGN, while arrowheads indicate the position of small tubular vesicles likely to represent endosomes. Bar, 10 μm.
DISCUSSION
Our previous studies showed that Rab31 is required for the Golgi/TGN organization and for transport of MPRs from TGN to endosomes. Additionally, we showed that Rab31 is involved in the formation of carriers in the TGN(Rodriguez-Gabin et al., 2009). Here, we further extend the knowledge of the mechanism of action of Rab31 by demonstrating its capability to target OCRL-1 to intracellular compartments involved in the transport of MPRs from TGN to endosomes. OCRL-1 is a member of group II inositol polyphosphate 5-phosphatases that regulate the levels of PI (4, 5)P2 and PI(4)P, two signaling molecules involved in the trafficking and Golgi/TGN structure (Vicinanza et al., 2008;Siddhanta et al., 2003). The importance of our results is emphasized by the fact that mutation of OCRL-1 causes the oculocerebrorenal syndrome of Lowe, an X-linked disorder of the nervous system, eyes and kidney (Nussbaum et al., 1997). Pathologic studies of the brain of the OCRL patients showed degenerative demyelination consistent with a primary demyelinating process (Schneider et al., 2001;HABIB R, 1962).
Our results showing that Rab31 interacts with OCRL-1 are in line with the concept that the ability of Rab proteins to regulate membrane transport depends on their capability to interact with a specific set of proteins (Zerial and McBride, 2001). Rab31 interaction with OCRL-1 was first demonstrated using the yeast two-hybrid system. Our data showed that Rab31 interacts with the a isoform of OCRL-1; this isoform is mainly expressed in brain, and as we demonstrated by both reverse transcriptase-PCR studies and immunoblot analysis, it is also expressed in oligodendrocytes. The formation of the Rab31-OCRL-1 complex was confirmed by in vitro GST-Rab31 pull down experiments. The presence of this complex in oligodendrocytes was demonstrated by the fact that OCRL-1 is present in immune complexes generated by treatment of oligodendrocyte lysates with antibodies against Rab31.
Our results indicated that the Rab 31 binding domain is present in the C-terminal region of OCRL-1. OCRL-1 contains three main domains including an inositol polyphosphate 5-phosphatase domain; an ASH (ASPM, SPD2, and Hydin) domain; and a C terminal, catalytically inactive RhoGAP domain. Our yeast two hybrid studies showed that the Rab31 binding domain includes at least two segments of the C-terminal region, one that encompasses residues 540 to 559 and the other that encompass residues 858 to 900. The 540 to 559 segment is located just before the ASH-domain, while the 858 to 900 segment is located after the RhoGAP domain. However, we cannot rule out the involvement in the Rab31 interaction with OCRL-1 of the residues present in the ASH and RhoGAP domains. Similarly to our results, a recent manuscript showed that the whole C-terminal region of OCRL-1 is involved in the binding of this protein with APPL1, a protein that is also a Rab5 effector (Erdmann et al., 2007). In line with previous studies our data showed that Rab5 also interacts with OCRL-1. However, we did not detect interaction of OCRL-1 with Rab1, Rab8 or Rab14 in contrast to the finding of Hyvola et al. (Hyvola et al., 2006). The reason for this difference is not clear.
These results demonstrated that Rab31 interacts with OCRL-1, and confirmed previous findings that Rab5 interacts with OCRL-1. Most importantly, our observations suggest that the involvement of Rab31 in the transport of MPRs from the TGN to endosomes depends, at least in part, on its ability to target OCRL-1 to the cellular compartments located along this transport pathway. Consistent with this hypothesis, we showed that CD-MPR is sorted selectively to OCRL-1 containing carriers, as shown by the findings that pGolgi (a protein that resides in the Golgi) is not recruited to OCRL-1 containing carriers, and that newly synthesized CD63 and VSVG exit the TGN in carriers that do not contain OCRL-1. Indeed, our hypothesis that the role of Rab31 in Golgi/TGN stability depends on a mechanism that involves OCRL-1 is further supported by the fact that Rab31 depletion caused fragmentation of the TGN and reduced the binding of OCRL-1 to TGN and endosomes.
The decreased recruitment of OCRL-1 to the TGN in Rab31-depleted cells raise the possibility that the interaction of Rab 31 with OCRL-1 is essential for the localized production of PI(4)P in the TGN, an event that facilitated the formation of carriers that transport MPRs. This hypothesis is further supported by a large body of evidence. In vivo studies showed that OCRL-1 mainly hydrolyzes PI(4,5)P2 to produce PI(4)P (Zhang et al., 1998). Both lipids are signaling molecules present at the cytosolic surface of the Golgi/TGN membranes where they recruit and/or activate cytosolic proteins that participate in the establishment of the Golgi/TGN structures and in the formation of carriers. Similarly to Rab31 and OCRL-1, PI(4)P localizes mainly in the TGN and it is involved in the transport of MPRs from the TGN to endosomes. Indeed, PI(4)P participated in the recruitment of several proteins required for formation of carriers that transport MPRs including epsinR, the clathrin adaptor protein 1 (AP-1) and GGAs (Brodsky et al., 2001)(Wang et al., 2007). It should be noted that PI(4)P can be generated by the action of phosphatidylinositol 4-kinases (PI4K), PI4KIIα and PI4KIIβ, that are present in the Golgi/TGN (Cockcroft and De Matteis, 2001). PI4KIIα has been shown to participate in the transport of MPRs from TGN to endosomes. However, the fact that the PI(4)P produced by PI4KIIα can be used as precursor to produce PI(4,5)P2 makes it difficult to establish the relative importance of PI4KIIα and OCRL-1 in the TGN to endosomes transport (Godi et al., 1998;Cockcroft and De Matteis, 2001).
PI(4)P is also involved in the traffic of newly synthesized VSVG from TGN to plasma membrane. However, this PI(4)P pool is not likely produced by OCRL-1, since this enzyme is not localized in carriers transporting VSVG, and Rab31 does not participate in the formation of carriers transporting VSVG (Rodriguez-Gabin et al., 2009). It has been shown that the formation of this PI(4)P pool depends of the recruitment of both PI4KIIIβ and PI4KIIα to the TGN (Cockcroft and De Matteis, 2001).
Co-localization of Rab31 and OCRL-1 in carriers transporting MPRs and in endosomes suggests that these proteins are also involved in the targeting and/or fusion of the carriers with endosomes. Additionally, the fact that Rab5 interacts with OCRL-1 suggests that this phosphatase is involved in homotypic fusion of endosomes. Interestingly, depletion of OCRL-1 by siRNA treatment affects the transport from endosomes to TGN (Choudhury et al., 2005).
There are at least 58 different mutations in the OCRL-1 gene described so far in the literature (Monnier et al., 2000). Most of the OCRL patients do not show the presence of detectable transcripts or of detectable phosphatase activity (Lin et al., 1997). Mutation of OCRL-1 causes alterations in CNS white matter including: 1) reduction in quantity, 2) the presence of multiple small round sharp-edged cysts, 3) degenerative demyelination consistent with a primary demyelinating process and 4) gliosis (HABIB R, 1962). These observations highlight the relevance of the Rab31- and OCRL-1-dependent transport in the biology of the oligodendrocyte. Our observations raise the possibility that mutations in OCRL-1 affect Rab31 action, leading to alterations in MPRs transport from TGN to endosomes. This idea is also supported by the finding that OCRL patients present high levels of lysosomal hydrolases in plasma (Ungewickell and Majerus, 1999). Certainly, the MPR transport pathway is used to transport newly synthesized lysosomal hydrolases from the TGN to endosomes. In this pathway, lysosomal hydrolases are transported to endosomes in a complex with cation-dependent and cation-independent MPRs. The complexes dissociate in endosomes, the hydrolases are then delivered to the lysosomes and the MPRs are recycled to the TGN (Griffiths et al., 1988).
The MPR transport pathway is also traveled by proteins that are critical for the oligodendrocyte differentiation and myelin biogenesis, including sortilin, LDL receptor, and transferrin receptor, that are transported from the TGN to endosomes before reaching the cell surface (Bonifacino and Traub, 2003). Evidence implicate this pathway in the transport of certain myelin proteins from TGN to the plasma membrane including L-MAG and MOG (Rodriguez-Gabin et al., 2001). Indeed, alteration of the transport of these proteins could lead to alteration of the myelin biogenesis and stability. It is also possible that mutations in OCRL-1 cause alterations of the endocytic compartments which, in turn, disrupt the formation of myelin. Certainly, endosomes are involved in the biogenesis and turnover of myelin (Larocca and Rodriguez-Gabin, 2002).
In summary, this study demonstrates that OCRL-1 is a novel binding partner of Rab31. Our observations also indicated that Rab31 and OCRL-1 are involved in the transport of MPRs from TGN to endosomes. This knowledge, not only contributes to the understanding of how this transport pathway is regulated, but it also provides essential information for explaining the brain pathology observed in OCRL patients.
Supplementary Material
Acknowledgments
We want to dedicate this manuscript to the memory of Dr. Eduardo Soto; friend, mentor and colleague; to express our gratitude for his influence on and fostering of our careers.
We thank Dr. Maurice Rapport and Dr. Teresa DiLorenzo for editing and commenting on the article. We thank Dr. Robert Nussbaum for the gift of the mouse monoclonal antibodies against OCRL-1. We thank Dr. Erik Snapp for the gift of the VSVG-Venus expression plasmid. We thank Mr. Michael Cammer, Director of light Microcopy and Image Analysis, for his valuable help in processing of the cell images. This work was supported by NIH grant RO1 NS 47500-01 to J.N.L., S. Q. was supported by NIH training grant NS 07098, G. A. was supported by a grant from the Multiple Sclerosis Society of Canada
References
- 1.Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. see comments. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong J. How do Rab proteins function in membrane traffic? Int J Biochem Cell Biol. 2000;32:303–307. doi: 10.1016/s1357-2725(99)00112-0. [DOI] [PubMed] [Google Scholar]
- 3.Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, Kay GF, Whitelaw E. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 5.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 6.Bouverat BP, Krueger WH, Coetzee T, Bansal R, Pfeiffer SE. Expression of rab GTP-binding proteins during oligodendrocyte differentiation in culture. J Neurosci Res. 2000;59:446–453. doi: 10.1002/(SICI)1097-4547(20000201)59:3<446::AID-JNR20>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- 8.Burcelin R, Rodriguez-Gabin AG, Charron MJ, Almazan G, Larocca JN. Molecular analysis of the monomeric GTP-binding proteins of oligodendrocytes. Brain Res Mol Brain Res. 1997;50:9–15. doi: 10.1016/s0169-328x(97)00151-4. [DOI] [PubMed] [Google Scholar]
- 9.Choudhury R, Diao A, Zhang F, Eisenberg E, Saint-Pol A, Williams C, Konstantakopoulos A, Lucocq J, Johannes L, Rabouille C, Greene LE, Lowe M. Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell. 2005;16:3467–3479. doi: 10.1091/mbc.E05-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cockcroft S, De Matteis MA. Inositol lipids as spatial regulators of membrane traffic. J Membr Biol. 2001;180:187–194. doi: 10.1007/s002320010069. [DOI] [PubMed] [Google Scholar]
- 11.De Matteis M, Godi A, Corda D. Phosphoinositides and the golgi complex. Curr Opin Cell Biol. 2002;14:434–447. doi: 10.1016/s0955-0674(02)00357-5. [DOI] [PubMed] [Google Scholar]
- 12.Dressman MA, Olivos-Glander IM, Nussbaum RL, Suchy SF. Ocrl1, a PtdIns(4,5)P(2) 5-phosphatase, is localized to the trans-Golgi network of fibroblasts and epithelial cells. J Histochem Cytochem. 2000;48:179–190. doi: 10.1177/002215540004800203. [DOI] [PubMed] [Google Scholar]
- 13.Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S, Paradise S, Modregger J, Biemesderfer D, Toomre D, De Camilli P. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell. 2007;13:377–390. doi: 10.1016/j.devcel.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faucherre A, Desbois P, Satre V, Lunardi J, Dorseuil O, Gacon G. Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum Mol Genet. 2003;12:2449–2456. doi: 10.1093/hmg/ddg250. [DOI] [PubMed] [Google Scholar]
- 15.Godi A, Santone I, Pertile P, Devarajan P, Stabach PR, Morrow JS, Di Tullio G, Polishchuk R, Petrucci TC, Luini A, De Matteis MA. ADP ribosylation factor regulates spectrin binding to the Golgi complex. Proc Natl Acad Sci U S A. 1998;95:8607–8612. doi: 10.1073/pnas.95.15.8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths G, Hoflack B, Simons K, Mellman I, Kornfeld S. The mannose 6-phosphate receptor and the biogenesis of lysosomes. Cell. 1988;52:329–341. doi: 10.1016/s0092-8674(88)80026-6. [DOI] [PubMed] [Google Scholar]
- 17.HABIB R, BARGETON E, BRISSAUD HE, RAYNAUD J, LE BALL JC. CONSTATATIONS ANATOMIQUES CHEZ UN ENFANT ATTEINT D’UN SYNDROME DE LOWE. Archives Francaises de Pediatrie. 1962:945–960. [PubMed] [Google Scholar]
- 18.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. see comments. [DOI] [PubMed] [Google Scholar]
- 19.Huber LA, Madison DL, Simons K, Pfeiffer SE. Myelin membrane biogenesis by oligodendrocytes. Developmental regulation of low molecular weight GTP-binding proteins. FEBS Lett. 1994;347:273–278. doi: 10.1016/0014-5793(94)00562-1. [DOI] [PubMed] [Google Scholar]
- 20.Hyvola N, Diao A, McKenzie E, Skippen A, Cockcroft S, Lowe M. Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J. 2006;25:3750–3761. doi: 10.1038/sj.emboj.7601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janvier K, Bonifacino JS. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol Biol Cell. 2005;16:4231–4242. doi: 10.1091/mbc.E05-03-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 23.Larocca JN, Almazan G. Acetylcholine agonists stimulate mitogen-activated protein kinase in oligodendrocyte progenitors by muscarinic receptors. J Neurosci Res. 1997;50:743–754. doi: 10.1002/(SICI)1097-4547(19971201)50:5<743::AID-JNR11>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Larocca JN, Rodriguez-Gabin AG. Myelin biogenesis: vesicle transport in oligodendrocytes. Neurochem Res. 2002;27:1313–1329. doi: 10.1023/a:1021667515030. [DOI] [PubMed] [Google Scholar]
- 25.Lin T, Orrison BM, Leahey AM, Suchy SF, Bernard DJ, Lewis RA, Nussbaum RL. Spectrum of mutations in the OCRL1 gene in the Lowe oculocerebrorenal syndrome. Am J Hum Genet. 1997;60:1384–1388. doi: 10.1086/515471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madison DL, Kruger WH, Kim T, Pfeiffer SE. Differential expression of rab3 isoforms in oligodendrocytes and astrocytes. J Neurosci Res. 1996;45:258–268. doi: 10.1002/(SICI)1097-4547(19960801)45:3<258::AID-JNR7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 27.Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103(Pt 3):857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 28.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monnier N, Satre V, Lerouge E, Berthoin F, Lunardi J. OCRL1 mutation analysis in French Lowe syndrome patients: implications for molecular diagnosis strategy and genetic counseling. Hum Mutat. 2000;16:157–165. doi: 10.1002/1098-1004(200008)16:2<157::AID-HUMU8>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Morell P. Myelin formation, structure, and biochemistry. In: Siegel B, editor. Basic Neuroschemistry: Molecular, Cellular, and Medical Aspects. New York: Raven Press; 1994. pp. 147–195. [Google Scholar]
- 31.Mullins C, Bonifacino JS. The molecular machinery for lysosome biogenesis. Bioessays. 2001;23:333–343. doi: 10.1002/bies.1048. [DOI] [PubMed] [Google Scholar]
- 32.Nussbaum RL, Orrison BM, Janne PA, Charnas L, Chinault AC. Physical mapping and genomic structure of the Lowe syndrome gene OCRL1. Hum Genet. 1997;99:145–150. doi: 10.1007/s004390050329. [DOI] [PubMed] [Google Scholar]
- 33.Pfeffer S. A model for Rab GTPase localization. Biochem Soc Trans. 2005;33:627–630. doi: 10.1042/BST0330627. [DOI] [PubMed] [Google Scholar]
- 34.Raine CS. In: Morphology of Myelin and Myelination. second. Morell P, editor. New York and London: Plenum Press. Myelin. Morell, P.; 1984. pp. 1–41. [Google Scholar]
- 35.Rodriguez-Gabin AG, Almazan G, Larocca JN. Vesicle transport in oligodendrocytes: probable role of Rab40c protein. J Neurosci Res. 2004;76:758–770. doi: 10.1002/jnr.20121. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Gabin AG, Cammer M, Almazan G, Charron M, Larocca JN. Role of rRAB22b, an oligodendrocyte protein, in regulation of transport of vesicles from trans Golgi to endocytic compartments. J Neurosci Res. 2001;66:1149–1160. doi: 10.1002/jnr.1253. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Gabin AG, Yin X, Si Q, Larocca JN. Transport of mannose-6-phosphate receptors from the trans-Golgi network to endosomes requires Rab31. Exp Cell Res. 2009;315:2215–2230. doi: 10.1016/j.yexcr.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouille Y, Rohn W, Hoflack B. Targeting of lysosomal proteins. Semin Cell Dev Biol. 2000;11:165–171. doi: 10.1006/scdb.2000.0168. [DOI] [PubMed] [Google Scholar]
- 39.Rous BA, Reaves BJ, Ihrke G, Briggs JA, Gray SR, Stephens DJ, Banting G, Luzio JP. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol Biol Cell. 2002;13:1071–1082. doi: 10.1091/mbc.01-08-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider JF, Boltshauser E, Neuhaus TJ, Rauscher C, Martin E. MRI and proton spectroscopy in Lowe syndrome. Neuropediatrics. 2001;32:45–48. doi: 10.1055/s-2001-12221. [DOI] [PubMed] [Google Scholar]
- 41.Siddhanta A, Radulescu A, Stankewich MC, Morrow JS, Shields D. Fragmentation of the Golgi apparatus. A role for beta III spectrin and synthesis of phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2003;278:1957–1965. doi: 10.1074/jbc.M209137200. [DOI] [PubMed] [Google Scholar]
- 42.Ungewickell AJ, Majerus PW. Increased levels of plasma lysosomal enzymes in patients with Lowe syndrome. Proc Natl Acad Sci U S A. 1999;96:13342–13344. doi: 10.1073/pnas.96.23.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vicinanza M, D’Angelo G, Di Campli A, De Matteis MA. Function and dysfunction of the PI system in membrane trafficking. EMBO J. 2008;27:2457–2470. doi: 10.1038/emboj.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Sun HQ, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 46.Zerial M, Stenmark H. Rab GTPases in vesicular transport. Curr Opin Cell Biol. 1993;5:613–620. doi: 10.1016/0955-0674(93)90130-i. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Hartz PA, Philip E, Racusen LC, Majerus PW. Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1998;273:1574–1582. doi: 10.1074/jbc.273.3.1574. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Jefferson AB, Auethavekiat V, Majerus PW. The protein deficient in Lowe syndrome is a phosphatidylinositol-4,5-bisphosphate 5-phosphatase. Proc Natl Acad Sci U S A. 1995;92:4853–4856. doi: 10.1073/pnas.92.11.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


