Abstract
“Genomic medicine” refers to the diagnosis, optimized management, and treatment of disease—as well as screening, counseling, and disease gene identification—in the context of information provided by an individual patient’s personal genome. Genomic medicine, to some extent synonymous with “personalized medicine,” has been made possible by recent advances in genome technologies. Genomic medicine represents a new approach to health care and disease management that attempts to optimize the care of a patient based upon information gleaned from his or her personal genome sequence. In this review, we describe recent progress in genomic medicine as it relates to neurological disease. Many neurological disorders either segregate as Mendelian phenotypes or occur sporadically in association with a new mutation in a single gene. Heritability also contributes to other neurological conditions that appear to exhibit more complex genetics. In addition to discussing current knowledge in this field, we offer suggestions for maximizing the utility of genomic information in clinical practice as the field of genomic medicine unfolds.
Introduction
We have entered the personal genomic era. Complete diploid genomes of multiple individuals have been sequenced (Schuster et al. 2010; Bentley et al. 2008; Wang et al. 2008; Ahn et al. 2009; Wheeler et al. 2008), in some cases offering diagnostic (Lupski et al. 2010) or even therapeutic (Worthey et al. 2010) insights. Genomic methods, including array comparative genomic hybridization (aCGH) and single-nucleotide polymorphism microarrays (“SNP-chips”) have already proven their clinical utility by providing information about the content of individual patients’ genomes; whole-genome sequencing (WGS) and its more streamlined counterpart, whole-exome sequencing (WES), appear to be on the verge of doing so. Although these genomic methodologies differ in many ways from “traditional” medical tests (e.g. complete blood counts, electroencephalography, or magnetic resonance imaging) and even from single-locus genetic tests (i.e. DNA sequencing of ‘candidate genes’ or enzyme assays for diseases considered during a differential diagnosis), they have similarly established their place among the tools available to physicians to anticipate, diagnose, and effectively manage illness in individual patients.
The neurological diseases represent a vast category of illness encompassing diseases of varying severity, age of onset, clinical presentation, genetic influences, and therapeutic responses. Many have a specific genetic basis (OMIM; http://www.ncbi.nlm.nih.gov/omim); whether this refers to an established cause-and-effect relationship between genotype at a single locus and phenotype (as in single-gene Mendelian disorders caused by highly penetrant variants) or to a genotype predisposing to illness (with possible modification by the environment or by other features of the genome) varies among different neurological diseases. The substantial number and variety of neurological conditions for which genomic variation plays a role in the susceptibility, course, and/or treatability of disease makes this an exciting class of disorders to consider in the context of genomic medicine.
We provide a working definition of genomic medicine, explain relevant genomic technologies and the types of genome variation they detect, and then review recent achievements in the field of genomic medicine as they pertain to selected neurological conditions. Conditions that have been reviewed recently (or concurrently in this special issue of Human Genetics) in a genomic context and those beyond the scope of this short review, for example, nervous system cancer and tumor syndromes, stroke, deafness, dementia, Parkinson disease, and autism, will not be covered. Rather, we will focus on a few neurological conditions most illustrative of the principles and possibilities of genomic medicine. We will conclude the article by offering suggestions for strategically maximizing the value of personal genome information in the practice of genomic medicine, particularly as this discipline pertains to neurological disease.
What is genomic medicine?
The term “genetic medicine” entered the scientific vocabulary almost four decades ago (Osmundsen 1973). This concept, made more relevant to clinical practice by the advent of disease gene mapping (Botstein et al. 1980; Gusella et al. 1983), gene identification by positional cloning and other methods (Royer-Pokora et al. 1986; Riordan et al. 1989; Wallace et al. 1990), and clinical mutation testing (Nowak 1994), referred—and still refers—to medicine in the context of an individual’s genotype at a single locus. Related terms like “pharmacogenetics” (the impact of genotype on pharmacology) had already been in use for some time (Evans and Clarke 1961).
The term “genomic medicine” arose in the late 1990s (Sikorski and Peters 1997; Beaudet 1999). In his 1998 Presidential Address to the American Society of Human Genetics, Dr. Arthur Beaudet described it as “the routine use of genotypic analysis, usually in the form of DNA testing, to enhance the quality of medical care” (Beaudet 1999). Since that time, the initial phases of the Human Genome Project (HGP) have been completed (International Human Genome Sequencing Consortium 2001, 2004), which allows a modified definition of genomic medicine to be constructed: The screening, diagnosis, counseling, and/or treatment of individual patients that is enhanced by knowledge of their genomes (and, in certain situations, their cancer genome(s) and/or the genomes of their microbial flora; Auffray et al. 2011). By this definition, genomic medicine contrasts with genetic medicine in that multiple loci are considered, a broadening of our thinking to incorporate genome-wide (as opposed to locus-specific) information (Table 1). Importantly, the genome is finite and contains all of the genetic information required for the existence of an individual living organism. The sum total of genome variation, including both simple nucleotide variation (SNV) and copy number variation (CNV), specifies much or most of the biological individuality of a person. For the purpose of this review, genomic medicine will also refer to disease gene identification, both in inherited and sporadic disease, using genomic technologies.
Table 1.
The scope of genetic and genomic medicine
| Genetic medicine (locus-specific) | Genomic medicine (genomic-scale assays) | |
|---|---|---|
| Molecular diagnosis | Hypothesis-driven, based on clinical suspicion (one or a few genes tested) |
Diagnosis by genomic techniques: aCGH, SNP arrays, exome or whole-genome sequencing, karyotype |
| Risk assessment | Carrier testing of relatives of affected individuals | “Personal genomic panels” (DTC or physician- referred) |
| Pharmacology | Pharmacogenetics | Pharmacogenomics |
| Treatment | Considers the patient’s genotype at a single locus and pharmacogenetic data; genetic tumor typing |
Incorporates modifier loci and pharmacogenomic data into management decisions; genomic tumor typing |
| Disease gene/susceptibility locus identification |
Hypothesis-driven gene sequencing, positional cloning; Mendelian inheritance patterns preferred |
aCGH, exome or whole-genome sequencing, GWAS, linkage mapping; complex inheritance patterns acceptable; discovery of modifier loci |
| Genocentric? | Usually | Sometimes |
aCGH array comparative genomic hybridization, SNP single-nucleotide polymorphism, DTC direct-to-consumer, genocentric interpretation of variation focuses on genes and the genetic code
Recent technological advances
Disease gene discovery is entering the realm of the clinic. Although the research laboratory remains paramount in this pursuit, clinical diagnostic laboratories have in recent years established many novel gene–phenotype connections (Samuels et al. 2008; Vissers et al. 2010b). As some familiarity with the genomic technologies used in gene discovery is necessary to understand their capabilities and limitations, a brief description of each follows. When appropriate, citations are chosen from studies of neurological disease.
Microarrays
Microarrays consist of small pieces of the human genome arrayed on a glass slide and enable simultaneous genome-wide interrogation of multiple loci. The more sequences placed on the array, the higher the resolution of genomic interrogation, analogous to the better ‘picture’ resolved by increasing the pixel count of a digital camera, CT/MRI machine, etc. Microarrays were originally designed to measure relative mRNA expression (expression arrays; Schena et al. 1995), then subsequently co-opted to measure relative genomic content (aCGH; Pinkel et al. 1998), invaluable for detecting genomic rearrangements (deletions and duplications) genome wide. It became apparent by the 1990s that many disease phenotypes could result from structural changes of the human genome; these afflictions were referred to as genomic disorders (Lupski 1998, 2009). Clinically, aCGH has been used to diagnose patients with genomic disorders originating from both large (Cheung et al. 2005) and small (Boone et al. 2010) genomic copy number changes. In research and clinical settings, aCGH has been used to discover new disease loci by associating specific CNVs with clinical phenotypes. Such disease-associated CNVs can include genomic intervals encompassing many genes (Potocki et al. 2007, 2000), affect individual genes (Walsh et al. 2008), or even occur in non-coding regions of the genome (Smyk et al. 2007).
Another type of microarray is the SNP (single-nucleotide polymorphism) array, which enables genotyping of >105 SNPs throughout the genome. It is used clinically to detect absence of heterozygosity (AOH) owing to uniparental disomy or consanguinity, and even allows an estimation of the degree of relatedness of parents of consanguineous offspring (Altug-Teber et al. 2005; Schaaf et al. 2011). AOH detection enables homozygosity mapping, which can aid in disease gene identification in recessive conditions (Gundesli et al. 2010). SNP arrays are also the technology used in modern linkage mapping (a technique to map and identify disease loci responsible for Mendelian disorders) (Sas et al. 2010) and genome-wide association studies (GWAS; Grant and Hakonarson 2008).
GWAS
The goal of GWAS is to identify either risk genotypes (i.e. those found more commonly in a cohort of unrelated patients with a disease) or protective genotypes (found more commonly in unaffected control individuals) at one or more SNPs throughout the genome. As multiple loci may be identified in a single GWAS, this technique is amenable to the study of diseases with an oligo- or polygenic origin (i.e. an individual’s genotypes at a few or several loci can each contribute to disease risk) and of genetically heterogeneous single-gene disorders (i.e. a mutation in any one of several genes results in the same clinical phenotype). Susceptibility to many common diseases is thought to have these types of genetic underpinnings; referring to this and to the large number of patients (difficult to enroll in studies of rare diseases) and the relatively large contribution to disease heritability that a given haplotype must make to be identified, GWAS is said to be ideal for the study of “common diseases [caused by] common variants” (i.e. investigating the CDCV hypothesis).
An unanticipated finding has been the repeated observation that even well-executed GWAS may only account for a small fraction of the heritability of many diseases (Bogardus 2009). The “rare variants, common disease” (RVCD) hypothesis suggests that multitudinous, rare variants, potentially at many different loci (i.e. constituting genetic heterogeneity), may contribute to the remaining unexplained heritability. For rare variants that are ‘non-ancient,’ or even de novo in the affected individual, the haplotype in which they exist will be uninformative to GWAS. This hypothesis has played out in the case of Parkinson disease, where high-throughput sequencing of a single gene (GBA) may explain more heritability of disease burden than GWAS in certain populations (Sidransky et al. 2009; Tsuji 2010).
Resequencing
Once a single haploid reference human genome had been completed (International Human Genome Sequencing Consortium 2001), genome resequencing could begin. In resequencing (referred to below as “sequencing”), newly derived sequences are mapped to the reference genome, which allows the sum total of variation in a personal genome to be identified by virtue of comparison to the reference and specific variations to be correlated to specific phenotypes. Only with the invention of massively parallel sequencing (MPS) using next-generation sequencing technology (methods that are faster and cheaper than Sanger dideoxynucleotide sequencing, the method used to complete the reference human genome) (Metzker 2010) has routine genomic sequencing become feasible. Two general approaches to genome sequencing exist: (1) whole-genome sequencing (WGS) and (2) whole-exome sequencing (WES), in which exonic sequence (~1–2% of the genome) is enriched via a ‘capture’ step, allowing preferential sequencing of exons to reduce time and cost (Okou et al. 2007; Hodges et al.2007; Albert et al. 2007). Two drawbacks of exome sequencing are that mutations in non-exonic sequence or poorly captured exons may be missed and that very little, if any, CNV data are provided.
Linkage mapping in Mendelian disease may eventually be replaced by sequencing, especially as SNPs are usually not themselves the disease-causing or predisposing variants. Sequencing has not yet been used successfully to replace GWAS in cases of multi-gene disease.
Genomic panels
Diseases displaying genetic heterogeneity arise from mutations in any of several genes. Thus, an efficient way to test clinically for mutations is for a physician to order a panel test, in which multiple genes are sequenced. This type of genomic panel can be diagnostic, whereas others are predictive.
Predictive panel tests fall into several categories: carrier tests, disease susceptibility tests, and pharmacogenomic tests. A further distinction is that some are direct-to-consumer genetic tests (DTC-GTs) while others are ordered by physicians. Predictive tests generally use SNP arrays to assess an individual’s genome for both known disease-causing mutations and disease-associated SNPs. In the latter case, the strength of the disease association varies widely among SNPs (United States National Human Genome Research Institute; http://www.genome.gov/gwastudies/). In the case of one currently offered DTC-GT panel, 12 of 24 (50%) carrier status tests involve neurological/muscular conditions (e.g. familial dysautonomia and limb-girdle muscular dystrophy), 10 of 18 (56%) pharmacogenetic tests involve neurological/psychiatric conditions (e.g. response to interferon-beta therapy and naltrexone treatment response), and 27 of 93 (29%) disease risk tests involve neurological/psychiatric conditions (e.g. restless leg syndrome and stroke; only five of the 27 conditions (19%) are “conditions and traits for which there are genetic associations supported by multiple, large, peer-reviewed studies”) (https://www.23andme.com/health/all/). Legal and ethical issues of DTC-GT are discussed in an accompanying article in this issue of Human Genetics (Caulfield 2011).
Internet resources
The large number of known genetic conditions (even among the neurological disorders), genes involved in them, variants reported in these genes, and the incredible rate with which discoveries in these areas are reported have made online sources the ideal repository for this information. Table 2 constitutes a brief list of internet resources valuable for the practice of genomic medicine. These and other online resources are discussed by De Sevo (2010), Waggoner and Pagon (2009), and Mangan et al. (2009).
Table 2.
Internet resources for genomic medicine
| Website | URL | Use |
|---|---|---|
| Online Mendelian inheritance in man (OMIM) |
http://www.ncbi.nlm.nih.gov/omim | Standardized descriptions of all known Mendelian disorders and many genes |
| University of California, Santa Cruz (UCSC) Genome Browser |
http://www.genome.ucsc.edu/ | Browsable reference genome sequences for multiple organisms and copious annotation |
| Ensembl Genome Browser | http://www.ensembl.org/index.html | Browsable reference genome sequences for multiple organisms and copious annotation |
| GeneReviews | http://www.ncbi.nlm.nih.gov/sites/GeneTests/review?db=GeneTests | Freely available, expert-authored and reviewed descriptions of conditions with a genetic basis |
| GeneTests | http://www.ncbi.nlm.nih.gov/sites/GeneTests/?db=GeneTests | Lists of clinical genetic tests (and testing laboratories) for various conditions |
| Human Gene Mutation Database (HGMD) |
http://www.hgmd.cf.ac.uk/ | A gene-centric database of all disease-causing mutations |
| Some data require a paid subscription to access | ||
| Database of Genotype and Phenotype (dbGaP) |
http://www.ncbi.nlm.nih.gov/gap | Repository of genotypic information linked to phenotypic data. Two data ‘levels’ (controlled and publically available) |
| Database of Genomic Variants | http://projects.tcag.ca/variation/ | A catalog of benign structural genomic variation |
| dbVar (Database of Genomic Structural Variation) |
http://www.ncbi.nlm.nih.gov/dbvar/ | Downloadable data from studies of structural variation |
| DECIPHER | http://decipher.sanger.ac.uk/ | A catalog of pathogenic structural genomic variation |
| dbSNP | http://www.ncbi.nlm.nih.gov/projects/SNP/ | A database of nucleotide polymorphisms discovered in various populations |
| United States National Institutes of Health Catalog of Published Genome-Wide Association Studies |
http://www.genome.gov/gwastudies/ | Compendium of published GWAS |
Mutation types
“Classic” examples of mutations causing human disease are often point mutations (e.g. HbS in sickle cell anemia; Ingram 1959). Although illustrative of the genetic code, this type of mutation does not portray the entire picture with respect to genomic variation and the genomic basis of disease. Indeed, visible structural mutations (aneuploidy, translocation), genomic/genic/intragenic/non-coding copy-number variation, inversions, microsatellite expansion, indels, mutations in non-coding RNAs, regulatory sequences, and other non-coding regions of the genome, and epimutations may lead to neurological (and other) diseases. A brief, illustrative list of neurological conditions resulting from various mutation types constitutes Table 3. Furthermore, a given disorder may be caused by any of several mutation types (Fig. 1). At the present time, a dichotomy exists between genomic methods that detect SNVs (detectable with SNP arrays and sequencing) and those that detect CNVs (best detected using aCGH). It is anticipated that whole-genome sequencing may eventually rival aCGH at efficiently detecting CNVs (Mills et al. 2011).
Table 3.
Mutations of various types cause neurological disease
| Mutation type | Mutation subtype(s) | Example(s)a |
|---|---|---|
| Structural variation | ||
| Chromosomal abnormalities visible by conventional karyotype |
Aneuploidy | Down syndrome (MIM 190685) |
| Translocation | X-linked mental retardation 58 (MIM 300210; Zemni et al. 2000) |
|
| Copy-number variation (CNV) | Genomic deletion | Hereditary neuropathy with liability to pressure palsies (MIM 162500) |
| Genomic duplication | Charcot–Marie–Tooth disease, type 1A (MIM 118220) |
|
| Whole gene deletion | Autosomal recessive early onset Parkinson disease 6 (MIM 605909; PINK1 deletion; Marongiu et al. 2007) |
|
| Whole gene duplication | Early-onset Alzheimer disease with cerebral amyloid angiopathy (MIM 104300; APP duplication; Rovelet-Lecrux et al. 2006; Delabar et al. 1987) |
|
| Intragenic Deletion | Duchenne muscular dystrophy (MIM 310200; deletions in DMD) |
|
| Intragenic duplication | Autosomal dominant dopa-responsive dystonia (MIM 128230; duplication in GCH1; Ling et al. 2011) |
|
| Non-coding | X-linked spastic paraplegia type 2 (MIM 312920; duplication 136 kilobases downstream of PLP1; Lee et al. 2006) |
|
| Inversion | – | Duchenne muscular dystrophy (MIM 310200) and other features (inv(X)(p21.2q22.2); Saito-Ohara et al. 2002) |
| Microsatellite expansion | Triplet repeat (polyglutamine) | Huntington disease (MIM 143100; expansion in HTT) |
| Triplet repeat (polyalanine) | Congenital central hypoventilation syndrome (MIM 209880; expansion in PHOXB2) |
|
| Triplet repeat (other) | Fragile X mental retardation syndrome (MIM 300624; CGG repeat expansion in FMR1 5′ UTR) |
|
| Myotonic dystrophy 1 (MIM 160900; CTG repeat expansion in DMPK 3′UTR) |
||
| SNV | ||
| Insertions/deletions (indels) | Frameshift, splice site, addition or deletion of amino acid(s) |
Many neurological disorders with a genetic basis |
| Point mutation | Missense, nonsense, frameshift, splice site |
Many neurological disorders with a genetic basis |
| Epigenetic | – | Prader-Willi syndrome (MIM 176270; maternal heterodisomy of non-mutated chromosome 15; Nicholls et al. 1989) |
| Mutations in small RNAs | – | Autosomal dominant deafness 50 (MIM 613074; mutations in miR-96; Mencía et al. 2009) |
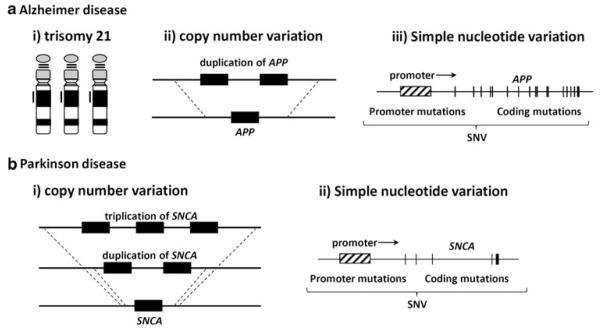
Just as multiple diseases may result from a given mutation type, multiple mutation types may cause a given disease (see Fig. 1). SNV simple nucleotide variation
Fig. 1.
Multiple mutation types can lead to the same phenotype. Two examples are shown in which multiple mutation types can increase gene dosage, leading to a neurological phenotype. a Increased expression of APP, owing to (i) trisomy 21, (ii) duplication of APP, or (iii) mutations in regulatory or coding regions of the gene can result in Alzheimer disease. b Increased expression of SNCA, owing to (i) duplication or triplication of SNCA or (ii) mutations in regulatory or coding regions of the gene can result in Parkinson disease. SNV simple nucleotide variation [reproduced, with permission, from Shiga et al. (2008)]
Applicability to neurological conditions
Intellectual disability
Intellectual disability or ID (previously referred to as mental retardation) can be caused either by environmental insults or by genetic lesions and may exist in isolation (nonsyndromic ID) or as part of a complex of symptoms (syndromic ID). A large number of genes has been implicated in ID (Leiden Open Variation Database Mental Retardation Database; http://www.LOVD.nl/MR). The number of genes reported to be associated with X-linked intellectual disability (XLID), the most common mode of inheritance in ID, is climbing towards 100 (Chiurazzi et al. 2008); a figure of ~10% of X-chromosome genes is given by Tarpey et al. (2009). Molecular diagnostic tests for XLID have become quite sophisticated (GeneTests; http://www.ncbi.nlm.nih.gov/sites/GeneTests/?db=GeneTests). For example, next-generation sequencing of the exons and flanking intronic sequence of 92 genes implicated in XLID has just been introduced by one diagnostic laboratory (http://www.ggc.org/diagnostics/next-gen-sequencing.htm). Yet, despite the large number of known XLID genes and tests to detect mutations in them, substantial missing heritability exists in this condition. As such, XLID has been the subject of several recent large-scale gene- and mutation-finding efforts.
Tarpey et al. (2009) sequenced the coding exons of most X-chromosome genes in 208 ID families where pedigrees were consistent with X-linked inheritance and previous mutation detection efforts had failed. Mutations were detected in both known ID-related genes and newly proposed ones, including SYP and ZNF711. As a family based approach was used, only mutations which segregated with disease in males and obligate carrier females were considered candidates. This approach, although powerful, was only informative for a minority (35/208; 17%) of families. This may be on account of disease-causing mutations in non-captured sequence (including non-annotated exons), synonymous mutations being overlooked (intentionally) as part of the computational mutation-finding algorithm, or CNVs that were not detected by exome sequencing, among other reasons.
Another family based approach was that of Whibley et al. (2010), who performed high-density X-chromosome aCGH in 251 families with evidence of XLID, most of which were mutation-negative families from the study by Tarpey et al. (2009). CNVs deemed likely to be pathogenic, which ranged from 2 kb to 11 Mb in size, were found in approximately 10% of families. These CNVs interrupted or encompassed both known XLID genes and new candidates (PTCHD1, WDR13, FAAH2, and GSPT2). Complementing this study of X-linked ID CNVs, the search for autosomal CNVs associated with ID has also been fruitful (reviewed in Vissers et al. 2010b).
The aforementioned XLID studies were performed on families in which segregation of the mutations with disease in males and carrier females strengthened evidence for causation. Furthermore, the inheritance pattern (X-linked) limited the genomic scope of capture sequencing and aCGH. Vissers et al. (2010a), contrastingly, performed whole-exome sequencing on 10 pre-screened subjects with sporadic mental retardation. In each case, both parents were available for testing, which allowed the search for potentially causative mutations to be narrowed to include only de novo variants. Assuming autosomal dominant causality, the authors identified a heterozygous, de novo, non-synonymous mutation in each of nine different genes, six of which were suggested to explain sporadic ID in as many patients. Two of these were in known ID genes, and four were in genes that made ‘biological sense,’ though no functional experiments were performed. Of the remaining three patients, one had inherited a mutation in a known XLID gene from a carrier mother in whom the mutation occurred de novo. These data suggest that new mutations can contribute significantly to ID (reviewed by Lupski 2010). Whole-genome sequencing in ID is likely imminent, though issues of data analysis, replication, appropriate use of controls, and functional studies will remain important to consider.
Much progress has been made finding genes responsible for Mendelian syndromes involving ID. The gene for Kabuki syndrome (MIM 147920), MLL2, was recently identified by whole-exome sequencing of a cohort of Kabuki syndrome patients and confirmed by subsequent MLL2 sequencing in multiple other affected individuals (Ng et al. 2010). Similarly, a gene for Schinzel–Giedion midface retraction syndrome (MIM 269150), SETBP1, was discovered by Hoischen et al. (2010) by exome sequencing.
Part of the excitement of genomic medicine is the potential to understand how genotypes at more than one locus might modify a given phenotype. Girirajan et al. (2010) identified a rare, recurrent genomic deletion at 16p12.1 associated with ID/developmental delay that, in combination with other CNVs, has been proposed to cause disease in a “two-hit” fashion. Alone, the CNV was found to predispose to neuropsychiatric disease (for example in transmitting parents). Interestingly, probands with this CNV were more likely to have an additional large CNV in their genome than were controls, and those with two CNVs had symptoms more severe than those with only the 16p12.1 deletion. Biological reasons for this phenomenon can be hypothesized, although these will have to be tested by experimentation in the future. Furthermore, other explanations for the observed data will have to be ruled out. It is likely that many other two-hit pairs of loci exist, and that three-hit, four-hit, and higher order models, as proposed for digenic (Kajiwara et al. 1994) and triallelic (Katsanis et al. 2001) inheritance, may eventually more accurately explain the risk of ID/developmental delay and other phenotypes.
Pain
Although not always thought of as a disease itself, pain is certainly a phenomenon of neurological origin and importance. Genome-wide studies of pain are limited in number, though a handful of single-gene pain or pain insensitivity syndromes have provided insights. Paroxysmal extreme pain disorder (PEXPD; MIM 167400) consists of episodic and intense rectal, mandibular, and/or ocular pain. Fertleman et al. (2006) mapped this condition to chromosome 2q using linkage analysis in a single, large pedigree. They subsequently identified causative heterozygous missense mutations in SCN9A, a gene in this region that encodes the alpha subunit of the Na(v)1.7 voltage-gated sodium channel, in multiple PEXPD families and individuals. Intriguingly, heterozygous gain of function mutations in this gene have also been shown to cause autosomal-dominant primary erythermalgia (MIM 133020), a disorder of vascular congestion/dilation and burning pain of the distal lower extremities that is precipitated by heat and other factors (Drenth and Michiels 1992). Functional in vitro analysis demonstrated that PEXPD-causing and primary erythermalgia-causing mutations have differing effects on cellular electrophysiology (Fertleman et al. 2006). Thus, these are allelic channelopathies that arise from separable ion conduction phenomena. Furthermore, carbamazepine, which effectively treats PEXPD but not primary erythermalgia, rectifies the electrophysiological abnormalities caused by PEXPD mutations, but not those of primary erythermalgia mutations. This example is instructive as to how genotype may dictate not only phenotype, but also pharmacologic response.
Very interestingly, homozygous loss of function mutations in SCN9A can cause a third disorder linked to this locus, autosomal recessive congenital indifference to pain (MIM 243000). Cox et al. (2006) mapped this condition in three families to 2q and identified homozygous nonsense mutations in the gene. They demonstrated that Na(v)1.7 function was nonexistent in cells with these mutations and suggested that SCN9A and the channel it encodes are attractive targets for drug development to treat pain.
Two recent studies have identified genetic risk factors for neuropathic pain by combining human and model organism genomic approaches. Costigan et al. (2010) performed genome-wide expression analysis in dorsal root ganglia of rat models of neuropathic pain following nerve injury. The expression of KCNS1, a potassium channel alpha subunit, was found to be decreased following nerve injury. The authors then tested for sequence differences at this locus in patient cohorts, and a risk allele for neuropathic pain was identified. Nissenbaum et al. (2010) mapped a mouse quantitative trait locus (QTL) for pain to a ~4 Mb region of the mouse genome. Subsequent expression and informatic analyses identified CACNG2 as a candidate pain susceptibility gene in this region, further implicated by experiments in CACNG2 hypomorphic mice. The authors then described CACNG2 polymorphisms that segregated with chronic neuropathic pain in post-surgical human subjects. These studies, each unique in methodology, provide examples of how model organism genomics may inform human genomics, identifying susceptibility loci that could aid drug development and/or risk stratification in patients.
Epilepsy
Over 20 genes have been implicated in the monogenic epilepsies (Rees 2010). Most of these constitute channelopathies or result from some other manner of dysfunctional modulation of neurotransmission. Many display autosomal dominant inheritance, suggesting that pathogenicity may derive from haploinsufficiency or altered stoichiometry of channel subunits, from a dominant-negative effect, or from gain of function. Mutations in putative epilepsy genes may be tested electrophysiologically in vivo and in vitro, providing evidence for or against their implication in the disease. Despite remarkable progress in family-based mapping and identification of epilepsy disease genes, over half of epilepsy patients remain genetically undiagnosable (Rees 2010). Unfortunately, both a recent case–control approach examining candidate genes (Cavalleri et al. 2007) and GWAS (Kasperavičiūte et al. 2010) failed to find additional susceptibility loci. Equally unfortunate for patients is that a sophisticated and effective genome-based algorithm for treatment optimization, ideal for a genetically heterogeneous condition for which multiple treatment options exist, is not yet a reality (Anderson 2008).
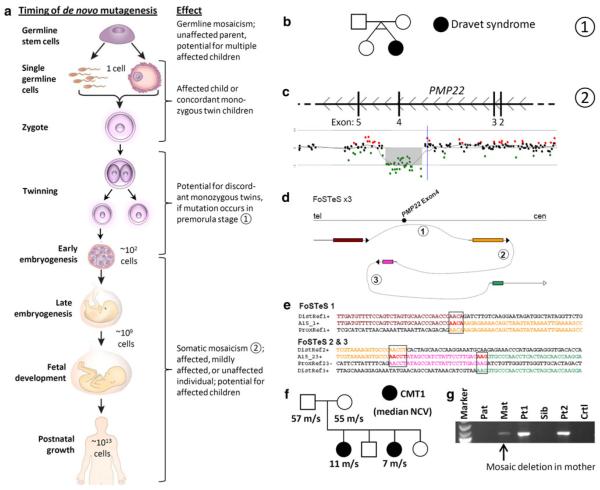
Multiple mutations have been described in the SCN1A gene, which cause a handful of related epilepsy phenotypes including Dravet syndrome (also known as severe myoclonic epilepsy of infancy [SMEI]; MIM 607208) and generalized epilepsy with febrile seizures plus (GEFS+; MIM 604233) (Human Gene Mutation Database; http://www.hgmd.cf.ac.uk/ac/index.php). Individuals with somatic or germline mosaicism for this gene have been described (Gennaro et al. 2006). Interestingly, and expectedly, a mosaic, transmitting parent may have mild or nonexistent symptoms, whereas that individual’s child, who is expected to have the mutation in all of his or her (brain) cells, is fully affected. Furthermore, Vadlamudi et al. (2010) recently described a set of monozygotic twins discordant for Dravet syndrome in which the affected individual had a mutation in SCN1A and the unaffected twin did not. These and similar studies indicate that the concept of mosaicism is important in epilepsy and support the idea that mutations can arise at any stage of development (Fig. 2) (Lupski 2010). In addition, they suggest something more profound: that perhaps many patients have epilepsies that result from mutations during their own development, such that the brain contains cells with a mutation—potentially even in a known epilepsy gene—that is not present in ‘testable tissues’ (most commonly blood or potentially fibroblasts) (Lindhout 2008). These patients would evade a genetic diagnosis. In addition, perhaps in some focal epilepsies only part of the brain carries a mutation, or perhaps some types of epilepsies can only arise when two genetically distinct populations of cells co-exist in a single individual’s brain. As Lindhout (2008) suggests, these hypotheses could be tested by searching for somatic mutations in postmortem or surgically resected brain samples from patients with epilepsy (and other neurological disorders!). One can imagine an initiative akin to The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/abouttcga) or the Catalogue of Somatic Mutations in Cancer (COSMIC, http://www.sanger.ac.uk/genetics/CGP/cosmic/) in which genome-wide methods like aCGH or genome/exome sequencing are used, although a simple ‘candidate’ gene approach focusing on known disease genes may be sufficient as an initial, proof of principal experiment.
Fig. 2.
The timing of de novo mutagenesis. a New mutations may occur at any time during development. The effect on the individual and his or her offspring is dependent upon mutation timing and the cell type in which the mutation occurs [adapted from (Lupski 2010)]. b Mutations occurring during very early embryogenesis may result in discordant monozygotic twins [see also (Vadlamudi et al. 2010)]. c–g A complex intragenic deletion in PMP22 illustrates that mitotic mutational processes are a mechanism of de novo mutagenesis and mosaicism [adapted from (Zhang et al. 2009)]. c Array CGH detects a deletion of exon 4 of PMP22. d, e Deletion breakpoint sequencing identifies complexity and microhomology, hallmarks of DNA replication-based (i.e. mitotic) CNV-generating mechanisms [e.g. fork stalling and template switching (FoSTeS) and microhomology-mediated break-induced replication (MMBIR)]. f, g Two siblings with the PMP22 exon 4 deletion show diminished nerve conduction velocity (NCV; meters/second (m/s)), consistent with Charcot–Marie–Tooth disease, type 1 (CMT1). Their mother is mosaic (in blood) for the deletion as detected by breakpoint PCR, but does not have symptoms of CMT1
Multiple sclerosis
Multiple sclerosis (MS) is a complex disease that affects multiple central nervous system substructures and is characterized by inflammation, demyelination, and neuronal pathology (Hauser and Oksenberg 2006). Although epidemiologic studies indicate that an individual’s environment contributes heavily to disease susceptibility (Ebers 2008), a modest level of heritability is indicated by familial aggregation (Oksenberg and Baranzini 2010). Several susceptibility alleles or haplotypes have been found by GWAS and candidate-gene approaches, the highest risk of which correspond to HLA genes on chromosome 6p21–p23 (Svejgaard 2008; Oksenberg and Baranzini 2010). De Jager et al. (2009) extracted additional statistical power from several GWAS by pooling their results in a meta-analysis. Risk alleles in three new non-HLA genes were identified, all of which were replicated in follow-up studies on independent control and MS-afflicted individuals.
None of the GWAS-identified risk genotypes are completely penetrant; as well, no families transmitting the MS phenotype with a clear inheritance pattern have been described. Despite these indications of the genetic complexity of MS, Baranzini et al. (2010) took a whole genome, whole epigenome, and whole transcriptome sequencing approach to determine whether detectable changes might explain disease in a small number of individuals. Specifically, these authors sequenced the entire genomes of one pair of MS-discordant monozygotic twins, and the epigenomes and mRNA transcriptomes of CD4+ T cells from three pairs of MS-discordant monozygotic twins. No informative differences were revealed in any of the three arms of the study, including informative indels, SNPs (even confirmed MS susceptibility SNPs), HLA haplotypes, CNVs, methylation differences, or mRNA expression changes.
The question has arisen whether clinical genetic testing might have some utility to estimate MS risk, diagnose the disease, or offer prognostic information. In one recent analysis, Sawcer et al. (2010) argued that doing so at this time would offer few, if any, patients any medically actionable information. Future MS genomic studies are needed to assemble a usable, multi-locus model of MS susceptibility and progression. Population-scale whole-genome sequencing and elegant computational analyses are likely to be the necessary technologies in this pursuit.
Two recent studies (Ramagopalan et al. 2009, 2010) provided exciting biological evidence to support the idea that the genetic and environmental influences on MS susceptibility may interact (Ebers 2008). Latitude is the environmental variable most predictive of MS risk (Lauer 1997), an effect proposed to be mediated by vitamin D via differences in sunlight exposure (Ebers 2008). Ramagopalan et al. described a vitamin D responsive element in the MS risk-associated locus HLA-DRB1 (Ramagopalan et al. 2009), then subsequently identified vitamin D receptor-binding sites genome-wide via chromatin immunoprecipitation followed by sequencing (ChIP-seq), providing experimental evidence for binding sites that overlap with several susceptibility loci for MS and other autoimmune diseases (Ramagopalan et al. 2010).
Studies that probe the interface of the environment and the genome may prove to be particularly useful in that an individual’s environment could potentially be modified; thus, such gene × environment studies might suggest options for disease prevention, modification, or cure. An instructive example of this involves dietary modification of another neurological disorder—ataxia. Familial isolated deficiency of vitamin E (VED; MIM 277460) is a recessive disorder caused by mutations in the gene encoding α-tocopherol transfer protein, TTPA (Ouahchi et al. 1995). Deficiency of this protein prohibits α-tocopherols, members of the vitamin E family of molecules, from being incorporated into plasma VLDL (Traber et al. 1990) and results in vitamin E deficiency and resultant neurological symptoms—most strikingly a Friedreich-like ataxia (Harding et al. 1985). Vitamin E supplementation slows progression of the disease or may even lead to some improvement of neurological symptoms (Gabsi et al. 2001; Mariotti et al. 2004). One report of presymptomatic treatment prevented symptom onset in the younger siblings of an affected patient for the 5-year duration of the study (Amiel et al. 1995).
Charcot–Marie–Tooth disease
Charcot–Marie–Tooth disease (CMT) is the most common inherited neurological disease (Szigeti and Lupski 2009). Patients with CMT experience progressive deterioration of the peripheral nerves with secondary muscle wasting and weakness in a distal distribution (i.e. distal symmetric polyneuropathy, or DSP) (England et al. 2009a, b). The disease is extremely heterogeneous both clinically and molecularly. Clinical subtypes include Charcot–Marie–Tooth disease, type 1 (CMT1), Charcot–Marie–Tooth disease, type 2 (CMT2), Dejerine–Sottas neuropathy (DSN; MIM 145900), congenital hypomyelinating neuropathy (CHN; MIM 605253), and Roussy–Levy syndrome (RLS; MIM 180800) (Jani-Acsadi et al. 2008). Electrophysiological studies enable a distinction between the two major classes of the disease: demyelinating, with symmetrically slowed nerve conduction velocity (NCV); and axonal, which is associated with normal NCV but reduced muscle action potentials.
Thus far, over 40 different genetic loci have been linked to CMT; for approximately 30 of these loci, specific genes have been identified (Bird 2011). These genes encode proteins involved in myelination, axonal transport, Schwann cell differentiation, signal transduction, mitochondrial function, protein translation, and single-stranded DNA break repair (Szigeti and Lupski 2009). CMT displays autosomal dominant, autosomal recessive, or X-linked transmission, predominately depending on the locus/gene involved; intriguingly, however, for mutations in some loci either dominant or recessive inheritance may be observed depending on the specific mutation (De Jonghe et al. 1997; Keller and Chance 1999; Nelis et al. 1999). The most prevalent form of Charcot–Marie–Tooth disease, CMT1A (MIM 118200), is caused in the vast majority of cases by copy-number gain of the PMP22 gene and a gene dosage effect (Lupski et al. 1991, 1992; Raeymaekers et al. 1991). This condition was the first genomic disorder described (Lupski 1998, 2009). The CMT1A 1.4 Mb genomic duplication in 17p12 results from unequal crossing over of homologous chromosomes at repeated sequences that flank the duplicated region (Pentao et al. 1992). The reciprocal deletion leads to hereditary neuropathy with liability to pressure palsies (HNPP; MIM 162500) that manifests with a recurrent nerve dysfunction secondary to nerve compression (Chance et al. 1993, 1994). Point mutations in PMP22 can lead to autosomal-dominant or autosomal-recessive forms of CMT (Roa et al. 1993; Shy et al. 2006).
Despite the significant advancements in CMT research, there are still multiple unidentified CMT genes and likely many genetic factors affecting the phenotypic expressivity that remain to be discovered. Progress in these areas has the potential to accelerate in the coming years owing to recent technological advances. For example, the application of whole-genome sequencing in a patient with recessive CMT recently enabled identification of two different causative alleles in the SH3TC2 gene and documented the first successful application of this powerful technique for the identification of disease-causing mutations (Lupski et al. 2010). In addition, as family members of the affected individual were available for testing, it was demonstrated that carriers for either mutation exhibited milder, distinct, dominant neuropathic symptoms. These included an electrophysiologically characterized axonal neuropathy segregating with a missense, potentially gain of function variant and susceptibility to carpal tunnel syndrome (CTS) segregating with a nonsense allele; the latter variant demonstrated, as has been shown by Young et al. (1997) and Potocki et al. (1999) for PMP22, that haploinsufficiency of a CMT gene can result in genetic susceptibility to the common complex trait of CTS.
Spinal muscular atrophy
Spinal muscular atrophy (SMA) is one of the most common autosomal-recessive nervous system disorders, and is characterized by degeneration of the alpha motor neurons in the spinal cord and brain stem nuclei leading to progressive muscle weakness (Wang et al. 2007; Lunn and Wang 2008). Patients with SMA present with a broad clinical spectrum ranging from death in early infancy to normal adult life with only mild weakness. SMA is classically divided into four types (SMA I–IV) based on the age of onset and highest function achieved. All types of SMA are caused by disruption of the SMN1 gene that is localized to chromosome 5q13 (Melki et al. 1990; Lefebvre et al. 1995). This gene-harboring region is unstable and subject to intrachromosomal rearrangements including gene duplications, conversions, and deletions. This fact delayed the cloning of the disease-causing gene (Burghes 1997; McLean et al. 1994).
There are two paralogous (telomeric and centromeric) elements of ~500 kb in the SMA region, with SMN1 localized to the telomeric copy and the SMN2 gene localized to the centromeric copy. Increased SMN2 number and reduction or absence of SMN1, resulting from gene conversion from SMN1 to SMN2, occasionally occurs on account of the genomic architecture at this locus (Burghes 1997). SMN1 is a fully active gene that is translated into full-length protein (Lefebvre et al. 1995). SMN2 differs from this gene by only five nucleotides; one of these changes causes abnormal splicing and skipping of exon 7 in 90% of SMN2 mRNA (Lorson et al. 1999). However, 10% of SMN2 mRNA retains exon 7, yielding fully active protein (Lefebvre et al. 1995). Quantitative analyses of SMN2 copy number in SMA patients has shown that higher copy number correlates with milder phenotype (Mailman et al. 2002). This provides an interesting example of how genomic architecture may play a role not only in generating disease-causing mutations, but also in modulating disease phenotype.
Another mechanism explaining clinical heterogeneity of SMA involves the existence of modifiers in trans. Recently, Plastin 3 was identified as a potential modifier of the clinical course of SMA through transcriptome-wide differential expression analysis (Oprea et al. 2008). The work of Oprea et al. (2008) suggests that Plastin 3 may have an effect on axonogenesis that is altered in individuals lacking SMN expression and acts as a protective modifier of SMA.
Duchenne muscular dystrophy
Duchenne muscular dystrophy (DMD; MIM 310200), Becker muscular dystrophy (MIM 300376), and X-linked dilated cardiomyopathy (MIM 302045) are allelic disorders caused by mutations of the dystrophin (DMD) gene. The most severe form, DMD, starts early in life and is characterized by progressive muscle weakness (Darras et al. 2008). The dystrophin gene is localized to Xp21 and encodes a large glycoprotein. The gene spans 2.4 Mb and consists of 79 exons (Koenig et al. 1987). Deletions of one or more exons are prevalent and observed in up to 65% of patients whereas duplications of gene fragments are documented in 5–15% of subjects with DMD (Den Dunnen et al. 1989), suggesting that point mutations and potentially other structural variants such as inversions account for the remaining portion of cases of the disease. In dystrophanopathies, the severity of the phenotype varies with the level of expression of dystrophin and whether or not the translational reading frame has been disrupted by mutation (Aartsma-Rus et al. 2006). Gene rearrangements in dystrophin are clustered in the 5′ and central part of the gene (Den Dunnen et al. 1989). To date, few benign CNVs have been identified in the DMD gene; one study showed the presence of benign CNVs in 6/341 subjects, all of which were localized to intronic sequences (del Gaudio et al. 2008). Interestingly, there are not many low copy repeats (LCRs) or other repetitive sequences that might promote genome instability, rendering the region prone to gene rearrangements within the DMD gene; indeed, only 3/26 breakpoints found by del Gaudio et al. localized in proximity to repetitive sequences.
A search for genetic factors able to modify the clinical course of DMD resulted in the identification of the SPP1 (osteopontin) gene as a potential genetic modifier of the disease (Pegoraro et al. 2011). A polymorphism localized to the SPP1 gene promoter was shown to change promoter activity, resulting in lower SPP1 mRNA production. The effect of SPP1 genotype on DMD progression was relatively substantial—similar to the effect of the pharmacologic use of steroids (about 1 year difference in the time to loss of ambulation).
Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS), known also as Lou Gehrig’s disease, is a progressive degenerative disease of both lower and upper motor neurons (Wijesekera and Leigh 2009). Patients with ALS present with focal muscle weakness, bulbar symptoms, muscle fasciculations, and neuropsychiatric deficits. It has been estimated that familial forms of ALS account for 5–10% of cases. Mutations in superoxide dismutase 1 (SOD1) (Rosen et al. 1993) are the most frequent genetic culprit in familial ALS (FALS), being found in 20% of patients (Andersen 2006). Other genes commonly involved in FALS include FUS (also known as TLS; mutated in ALS6 [MIM 608030]) (Kwiatkowski et al. 2009; Vance et al. 2009) and TARDBP (mutated in ALS10 [MIM 612069]) (Yokoseki et al. 2008; Sreedharan et al. 2008; Kabashi et al. 2008); however, mutations in these genes are only found in up to 8% of subjects with ALS (Valdmanis et al. 2009). Collectively, mutations in known genes explain disease in only 25–30% of familial ALS cases (Tsuji 2010).
Two FALS-associated genes provide interesting examples of how different mutations at a single locus may result in multiple disease phenotypes. Missense mutations in senataxin (SETX) have been reported in patients with juvenile amyotrophic lateral sclerosis 4 (ALS4; MIM 602433) (Chen et al. 2004). SETX mutations are also the cause of autosomal recessive spinocerebellar ataxia-1 (SCAR1; MIM 606002) (Moreira et al. 2004), also known as ataxia-ocular apraxia-2 (AOA2). Similarly, heterozygous mutations of FIG4, known to cause an autosomal recessive form of Charcot–Marie–Tooth disease (CMT4J; MIM 611228) when homozygous or compound heterozygous mutations occur (Chow et al. 2007), have recently been reported in patients with ALS11 (MIM 612577) and adult-onset primary lateral sclerosis (PLSA1; MIM 611637) (Chow et al. 2009).
Recent studies argue against CNVs playing a large role in the pathogenesis of ALS (Blauw et al. 2008, 2010), with one exception (Broom et al. 2008). GWA studies have identified a few genes that may play a role in the pathogenesis of ALS (Valdmanis et al. 2009), for example DPP6 in different populations of European ancestry (van Es et al. 2008), however additional investigation is needed to confirm these observed associations. One recent hypothesis proposes a role for retroelements in pathogenesis of ALS, as it was found that the activity of reverse transcriptase in plasma of subjects with ALS was higher than in controls, however the meaning and significance of this finding is currently obscure (Mougeot et al. 2009).
Conclusion
Even before the initial phases of the HGP were complete, the inception of the “postgenomic era” was claimed (Scangos 1997). Although a scientific achievement of Herculean importance, the HGP provided only a single haploid genome sequence representing an individual who was healthy at the time the DNA sample was obtained. Countless questions concerning genomic variation among populations with differing ancestry, among persons afflicted with heritable disease, and even among healthy individuals within a family remained unanswered. The continuing insights provided by the International HapMap Project (International HapMap3 Consortium 2010), 1000 Genomes Project (The 1000 Genomes Project Consortium 2010), and other genomic studies large and small—and the new questions that arise from this work—remind us that we are most certainly in the genomic era, and that, as personal genomes are probed in various ways, personal genomics is becoming a reality.
Two current challenges that must be overcome are an incomplete reference genome (The 1000 Genomes Project Consortium 2010) and incomplete or incorrectly annotated mutation databases. Particularly in the case of predictive genomic testing, it will be of the utmost importance for clinicians to thoughtfully consider the disease-causing potential of rare or novel mutations in view of published evidence. Prudent questions should include: did a study or studies reporting a mutation as disease-causing fail to find it in a statistically significant number of appropriately selected controls? Might there exist protective variants that reduce penetrance in certain ethnic groups or individuals? Have healthy family members been tested? Has mosaicism been considered? Has the patient’s mutation been confirmed by a second methodology? Wheeler et al. (2008), in sequencing the genome of an apparently healthy adult, found multiple risk alleles and purportedly disease-causing variants, and Lupski et al. (2010), in a patient with Charcot–Marie–Tooth disease, identified a purportedly pathogenic variant documented in a mutation database as being associated with a “persistent vegetative state;” these studies indicate that the above questions will need to be answered even in the case of screening and carrier testing.
Physicians must consider the clinical utility of a genomic test before it is ordered. This is no different from ordering traditional medical tests. Unfortunately, in any genomic panel, the utility of each individual result will depend on the locus tested and the variant detected. Therefore, clinicians should become familiar with each component of a panel test and decide, in consultation with the patient, whether ordering it would be of greater potential benefit than harm. Given the vast amount of genomic variation that exists among healthy individuals (The 1000 Genomes Project Consortium 2010; Conrad et al. 2010; International HapMap3 Consortium 2010), it is likely that no result of a genomic test will be completely ‘normal’ (i.e. completely free of carrier mutations, risk alleles, variants of unknown significance, or disease-causing mutations with reduced or age-dependent penetrance). Findings such as these may be unsettling to patients and should be considered before any genomic test is utilized.
We have attempted to provide evidence for both the promise and limitations of whole-genome resequencing and its potential for genomic medicine. This technology will soon become commonplace in both research studies and clinical practice. Even the most cheaply and easily obtainable sequence data requires proper analysis—a current and future challenge (Mardis 2010). For example, one current difficulty for sequencing-based mutation detection is the identification of genomic inversions and variants within or consisting of repeated sequences. Another such hurdle to overcome is assigning significance to mutations found in non-coding regions of the genome (i.e. establishing the genomic code). Yet another is making sense of genomic contributions to complex traits: might multiple common mutations with minor effects interact to manifest a phenotype? Or might oligogenic inheritance in the individual with one or a few variants of major effects—but tremendous genetic heterogeneity in the population of patients—be at play? And what of gene–environment interactions? Both innovative bioinformaticists and computational biologists will be required to meet these and similar challenges.
The neurogenomics community—composed of clinicians, scientists, informaticists, and even patients and healthy volunteers—should be proud of the accomplishments of the past and excited about the prospects of the future to improve human quality of life and explain some of the mysteries of the human nervous system and brain—our most unique and defining organ.
Acknowledgments
P.M.B. is a fellow of the Baylor College of Medicine Medical Scientist Training Program (T32GM007330-34). This work was supported in part by a National Eye Institute (NEI) Training Program Grant (T32EY007102) (P.M.B) from the United States National Institutes of Health (NIH), and by a National Institute of Neurological Disorders and Stroke (NINDS) Grant (R01NS058529) (J.R.L.) from the NIH. J.R.L. is a paid consultant for Athena Diagnostics and Ion Torrent Systems and is a co-inventor on multiple United States and European patents related to molecular diagnostics. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis offered in the Medical Genetics Laboratory.
Contributor Information
Philip M. Boone, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Room 604B, Houston, TX 77030, USA
Wojciech Wiszniewski, Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Room 604B, Houston, TX 77030, USA.
James R. Lupski, Department of Pediatrics, Baylor College of Medicine, Houston, TX 77030, USA; Texas Children’s Hospital, Houston, TX 77030, USA
References
- Aartsma-Rus A, Van Deutekom JCT, Fokkema IF, Van Ommen G-JB, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34(2):135–144. doi: 10.1002/mus.20586. doi:10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- Ahn S-M, Kim T-H, Lee S, Kim D, Ghang H, Kim DS, Kim BC, Kim SY, Kim WY, Kim C, Park D, Lee YS, Kim S, Reja R, Jho S, Kim CG, Cha JY, Kim KH, Lee B, Bhak J, Kim SJ. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19(9):1622–1629. doi: 10.1101/gr.092197.109. doi:10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, Weinstock GM, Gibbs RA. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4(11):903–905. doi: 10.1038/nmeth1111. doi:10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Altug-Teber Ö , Dufke A, Poths S, Mau-Holzmann UA, Bastepe M, Colleaux L, Cormier-Daire V, Eggermann T, Gillessen-Kaesbach G, Bonin M, Riess O. A rapid microarray based whole genome analysis for detection of uniparental disomy. Hum Mutat. 2005;26(2):153–159. doi: 10.1002/humu.20198. doi:10.1002/humu.20198. [DOI] [PubMed] [Google Scholar]
- Amiel J, Maziere JC, Beucler I, Koenig M, Reutenauer L, Loux N, Bonnefont D, Feo C, Landrieu P. Familial isolated vitamin E deficiency. Extensive study of a large family with a 5-year therapeutic follow-up. J Inherit Metab Dis. 1995;18(3):333–340. doi: 10.1007/BF00710425. [DOI] [PubMed] [Google Scholar]
- Andersen PM. Amyotrophic lateral sclerosis associated with mutations in the CuZn superoxide dismutase gene. Curr Neurol Neurosci Rep. 2006;6(1):37–46. doi: 10.1007/s11910-996-0008-9. [DOI] [PubMed] [Google Scholar]
- Anderson GD. Pharmacokinetic, pharmacodynamic, and pharmacogenetic targeted therapy of antiepileptic drugs. Ther Drug Monit. 2008;30(2):173–180. doi: 10.1097/FTD.0b013e318167d11b. doi:10.1097/FTD.0b013e318167d11b. [DOI] [PubMed] [Google Scholar]
- Auffray C, Caulfield T, Khoury MJ, Lupski JR, Schwab M, Veenstra T. Genome Medicine: past present and future. Genome Med. 2011;3(1):6. doi: 10.1186/gm220. doi:10.1186/gm220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, Zhang L, Farmer AD, Bell CJ, Kim RW, May GD, Woodward JE, Caillier SJ, McElroy JP, Gomez R, Pando MJ, Clendenen LE, Ganusova EE, Schilkey FD, Ramaraj T, Khan OA, Huntley JJ, Luo S, Kwok P-Y, Wu TD, Schroth GP, Oksenberg JR, Hauser SL, Kingsmore SF. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464(7293):1351–1356. doi: 10.1038/nature08990. doi:10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudet AL. ASHG presidential address. Making genomic medicine a reality. Am J Hum Genet. 1999;64(1):1–13. doi: 10.1086/302217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Cheetham RK, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IMJ, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DMD, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara MCE, Chang S, Cooley RN, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fajardo KV Fuentes, Furey W Scott, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Jones TA Huw, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ng B Ling, Novo SM, O’Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, Pinkard DC, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Rodriguez A Chiva, Roe PM, Rogers J, Bacigalupo MC Rogert, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Sohna JE Sohna, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, vande Vondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. doi:10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird TD. Charcot–Marie–Tooth Hereditary Neuropathy Overview. In: Pagon RA, Bird TD, Dolan CR, et al., editors. GeneReviews. University of Washington; Seattle (WA): 2011. [Internet], 2010/03/20 edn. NBK1358 [bookaccession] [PubMed] [Google Scholar]
- Blauw HM, Veldink JH, van Es MA, van Vught PW, Saris CGJ, van der Zwaag B, Franke L, Burbach JPH, Wokke JH, Ophoff RA, van den Berg LH. Copy-number variation in sporadic amyotrophic lateral sclerosis: a genome-wide screen. Lancet Neurol. 2008;7(4):319–326. doi: 10.1016/S1474-4422(08)70048-6. doi:10.1016/S1474-4422(08)70048-6. [DOI] [PubMed] [Google Scholar]
- Blauw HM, Al-Chalabi A, Andersen PM, van Vught PWJ, Diekstra FP, van Es MA, Saris CGJ, Groen EJN, van Rheenen W, Koppers M, van’t Slot R, Strengman E, Estrada K, Rivadeneira F, Hofman A, Uitterlinden AG, Kiemeney LA, Vermeulen SHM, Birve A, Waibel S, Meyer T, Cronin S, McLaughlin RL, Hardiman O, Sapp PC, Tobin MD, Wain LV, Tomik B, Slowik A, Lemmens R, Rujescu D, Schulte C, Gasser T, Brown RH, Jr, Landers JE, Robberecht W, Ludolph AC, Ophoff RA, Veldink JH, Van den Berg LH. A large genome scan for rare CNVs in amyotrophic lateral sclerosis. Hum Mol Genet. 2010;19(20):4091–4099. doi: 10.1093/hmg/ddq323. doi:10.1093/hmg/ddq323. [DOI] [PubMed] [Google Scholar]
- Bogardus C. Missing heritability and GWAS utility. Obesity (Silver Spring) 2009;17(2):209–210. doi: 10.1038/oby.2008.613. doi:10.1038/oby.2008.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Bacino CA, Shaw CA, Eng PA, Hixson PM, Pursley AN, Kang SH-L, Yang Y, Wiszniewska J, Nowakowska BA, del Gaudio D, Xia Z, Simpson-Patel G, Immken LL, Gibson JB, Tsai AC-H, Bowers JA, Reimschisel TE, Schaaf CP, Potocki L, Scaglia F, Gambin T, Sykulski M, Bartnik M, Derwinska K, Wisniowiecka-Kowalnik B, Lalani SR, Probst FJ, Bi W, Beaudet AL, Patel A, Lupski JR, Cheung SW, Stankiewicz P. Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat. 2010;31(12):1326–1342. doi: 10.1002/humu.21360. doi:10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- Broom WJ, Greenway M, Sadri-Vakili G, Russ C, Auwarter KE, Glajch KE, Dupre N, Swingler RJ, Purcell S, Hayward C, Sapp PC, McKenna-Yasek D, Valdmanis PN, Bouchard JP, Meininger V, Hosler BA, Glass JD, Polack M, Rouleau GA, Cha JH, Hardiman O, Brown RH., Jr 50 bp deletion in the promoter for superoxide dismutase 1 (SOD1) reduces SOD1 expression in vitro and may correlate with increased age of onset of sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2008;9(4):229–237. doi: 10.1080/17482960802103107. doi:10.1080/17482960802103107. [DOI] [PubMed] [Google Scholar]
- Burghes AHM. When is a deletion not a deletion? When it is converted. Am J Hum Genet. 1997;61(1):9–15. doi: 10.1086/513913. doi:10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulfield T. Direct-to-consumer testing: if consumers are not anxious, why are policymakers? Hum Genet. 2011 doi: 10.1007/s00439-011-0987-8. doi:10.1007/s00439-011-0987-8. [DOI] [PubMed] [Google Scholar]
- Cavalleri GL, Weale ME, Shianna KV, Singh R, Lynch JM, Grinton B, Szoeke C, Murphy K, Kinirons P, O’Rourke D, Ge D, Depondt C, Claeys KG, Pandolfo M, Gumbs C, Walley N, McNamara J, Mulley JC, Linney KN, Sheffield LJ, Radtke RA, Tate SK, Chissoe SL, Gibson RA, Hosford D, Stanton A, Graves TD, Hanna MG, Eriksson K, Kantanen A-M, Kalviainen R, O’Brien TJ, Sander JW, Duncan JS, Scheffer IE, Berkovic SF, Wood NW, Doherty CP, Delanty N, Sisodiya SM, Goldstein DB. Multicentre search for genetic susceptibility loci in sporadic epilepsy syndrome and seizure types: a case–control study. Lancet Neurol. 2007;6(11):970–980. doi: 10.1016/S1474-4422(07)70247-8. doi:10.1016/S1474-4422(07)70247-8. [DOI] [PubMed] [Google Scholar]
- Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, Swanson PD, Odelberg SJ, Disteche CM, Bird TD. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72(1):143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR. Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet. 1994;3(2):223–228. doi: 10.1093/hmg/3.2.223. [DOI] [PubMed] [Google Scholar]
- Chen Y-Z, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, Nicholson GA, Auer-Grumbach M, Wagner K, De Jonghe P, Griffin JW, Fischbeck KH, Timmerman V, Cornblath DR, Chance PF. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74(6):1128–1135. doi: 10.1086/421054. doi:10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Yu W, Li J, Ou Z, Patel A, Yatsenko SA, Cooper ML, Furman P, Stankiewicz P, Lupski JR, Chinault AC, Beaudet AL. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7(6):422–432. doi: 10.1097/01.gim.0000170992.63691.32. doi:00125817-200507000-00008. [DOI] [PubMed] [Google Scholar]
- Chiurazzi P, Schwartz CE, Gecz J, Neri G. XLMR genes: update 2007. Eur J Hum Genet. 2008;16(4):422–434. doi: 10.1038/sj.ejhg.5201994. doi:10.1038/sj.ejhg.5201994. [DOI] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, Lupski JR, Weisman LS, Meisler MH. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448(7149):68–72. doi: 10.1038/nature05876. doi:10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, Figlewicz D, Brown RH, Meisler MH. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84(1):85–88. doi: 10.1016/j.ajhg.2008.12.010. doi:10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AWC, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Wellcome Trust Case Consortium. Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464(7289):704–712. doi: 10.1038/nature08516. doi:10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, Wu T, Kiselycznyk C, Poddar M, Lu Y, Diatchenko L, Smith S, Cobos EJ, Zaykin D, Allchorne A, Shen PH, Nikolajsen L, Karppinen J, Männikkö M, Kelempisioti A, Goldman D, Maixner W, Geschwind DH, Max MB, Seltzer Z, Woolf CJ. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 2010;133(9):2519–2527. doi: 10.1093/brain/awq195. doi:10.1093/brain/awq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, Karbani G, Jafri H, Mannan J, Raashid Y, Al-Gazali L, Hamamy H, Valente EM, Gorman S, Williams R, McHale DP, Wood JN, Gribble FM, Woods CG. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444(7121):894–898. doi: 10.1038/nature05413. doi:10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darras BT, Korf BR, Urion DK. Dystrophinopathies. In: Pagon RA, Bird TD, Dolan CR, et al., editors. GeneReviews. University of Washington; Seattle (WA): 2008. [Internet], 2010/03/20 edn. NBK1119 [bookaccession] [PubMed] [Google Scholar]
- De Jager PL, Jia X, Wang J, de Bakker PIW, Ottoboni L, Aggarwal NT, Piccio L, Raychaudhuri S, Tran D, Aubin C, Briskin R, Romano S, International MS Genetics Consortium. Baranzini SE, McCauley JL, Pericak-Vance MA, Haines JL, Gibson RA, Naeglin Y, Uitdehaag B, Matthews PM, Kappos L, Polman C, McArdle WL, Strachan DP, Evans D, Cross AH, Daly MJ, Compston A, Sawcer SJ, Weiner HL, Hauser SL, Hafler DA, Oksenberg JR. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41(7):776–782. doi: 10.1038/ng.401. doi:10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonghe P, Timmerman V, Nelis E, Martin JJ, Van Broeckhoven C. Charcot–Marie–Tooth disease and related peripheral neuropathies. J Peripher Nerv Syst. 1997;2(4):370–387. [PubMed] [Google Scholar]
- De Sevo MR. Genetics and genomics resources for nurses. J Nurs Educ. 2010;49(8):470–474. doi: 10.3928/01484834-20100524-01. doi:10.3928/01484834-20100524-01. [DOI] [PubMed] [Google Scholar]
- del Gaudio D, Yang Y, Boggs BA, Schmitt ES, Lee JA, Sahoo T, Pham HT, Wiszniewska J, Chinault AC, Beaudet AL, Eng CM. Molecular diagnosis of Duchenne/Becker muscular dystrophy: enhanced detection of dystrophin gene rearrangements by oligonucleotide array-comparative genomic hybridization. Hum Mutat. 2008;29(9):1100–1107. doi: 10.1002/humu.20841. doi:10.1002/humu.20841. [DOI] [PubMed] [Google Scholar]
- Delabar J-M, Goldgaber D, Lamour Y, Nicole A, Huret J-L, de Grouchy J, Brown P, Gajdusek DC, Sinet P-M. β amyloid gene duplication in Alzheimer’s disease and karyotypically normal Down syndrome. Science. 1987;235:1390–1392. doi: 10.1126/science.2950593. [DOI] [PubMed] [Google Scholar]
- Den Dunnen JT, Grootscholten PM, Bakker E, Blonden LAJ, Ginjaar HB, Wapenaar MC, van Paassen HMB, van Broeckhoven C, Pearson PL, van Ommen GJB. Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet. 1989;45(6):835–847. [PMC free article] [PubMed] [Google Scholar]
- Drenth JPH, Michiels JJ. Clinical characteristics and pathophysiology of erythromelalgia and erythermalgia. Am J Med. 1992;93(1):111–114. doi: 10.1016/0002-9343(92)90696-9. doi:0002-9343(92)90696-9. [DOI] [PubMed] [Google Scholar]
- Ebers GC. Environmental factors and multiple sclerosis. Lancet Neurol. 2008;7(3):268–277. doi: 10.1016/S1474-4422(08)70042-5. doi:10.1016/S1474-4422(08)70042-5. [DOI] [PubMed] [Google Scholar]
- England JD, Gronseth GS, Franklin G, Carter GT, Kinsella LJ, Cohen JA, Asbury AK, Szigeti K, Lupski JR, Latov N, Lewis RA, Low PA, Fisher MA, Herrmann DN, Howard JF, Jr, Lauria G, Miller RG, Polydefkis M, Sumner AJ. Practice Parameter: evaluation of distal symmetric polyneuropathy: role of autonomic testing, nerve biopsy, and skin biopsy (an evidence-based review). Report of the American Academy of Neurology, American Association of Neuromuscular and Electrodiagnostic Medicine, and American Academy of Physical Medicine and Rehabilitation. Neurology. 2009a;72(2):177–184. doi: 10.1212/01.wnl.0000336345.70511.0f. doi:10.1212/01.wnl.0000336345.70511.0f. [DOI] [PubMed] [Google Scholar]
- England JD, Gronseth GS, Franklin G, Carter GT, Kinsella LJ, Cohen JA, Asbury AK, Szigeti K, Lupski JR, Latov N, Lewis RA, Low PA, Fisher MA, Herrmann DN, Howard JF, Jr, Lauria G, Miller RG, Polydefkis M, Sumner AJ. Practice Parameter: evaluation of distal symmetric polyneuropathy: role of laboratory and genetic testing (an evidence-based review). Report of the American Academy of Neurology, American Association of Neuromuscular and Electrodiagnostic Medicine, and American Academy of Physical Medicine and Rehabilitation. Neurology. 2009b;72(2):185–192. doi: 10.1212/01.wnl.0000336370.51010.a1. doi:10.1212/01.wnl.0000336370.51010.a1. [DOI] [PubMed] [Google Scholar]
- Evans DAP, Clarke CA. Pharmacogenetics. Br Med Bull. 1961;17:234–240. doi: 10.1093/oxfordjournals.bmb.a069915. [DOI] [PubMed] [Google Scholar]
- Fertleman CR, Baker MD, Parker KA, Moffatt S, Elmslie FV, Abrahamsen B, Ostman J, Klugbauer N, Wood JN, Gardiner RM, Rees M. SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron. 2006;52(5):767–774. doi: 10.1016/j.neuron.2006.10.006. doi:10.1016/j.neuron.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Gabsi S, Gouider-Khouja N, Belal S, Fki M, Kefi M, Turki I, Ben Hamida M, Kayden H, Mebazaa R, Hentati F. Effect of vitamin E supplementation in patients with ataxia with vitamin E deficiency. Eur J Neurol. 2001;8(5):477–481. doi: 10.1046/j.1468-1331.2001.00273.x. doi:ene273. [DOI] [PubMed] [Google Scholar]
- Gennaro E, Santorelli FM, Bertini E, Buti D, Gaggero R, Gobbi G, Lini M, Granata T, Freri E, Parmeggiani A, Striano P, Veggiotti P, Cardinali S, Bricarelli FD, Minetti C, Zara F. Somatic and germline mosaicisms in Severe Myoclonic Epilepsy of Infancy. Biochem Biophys Res Commun. 2006;341(2):489–493. doi: 10.1016/j.bbrc.2005.12.209. doi:10.1016/j.bbrc.2005.12.209. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W, McDonald MT, Hoo JJ, French BN, Hudson C, Johnson JP, Ozmore JR, Moeschler JB, Surti U, Escobar LF, El-Khechen D, Gorski JL, Kussmann J, Salbert B, Lacassie Y, Biser A, McDonald-McGinn DM, Zackai EH, Deardorff MA, Shaikh TH, Haan E, Friend KL, Fichera M, Romano C, Gécz J, DeLisi LE, Sebat J, King M-C, Shaffer LG, Eichler EE. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42(3):203–209. doi: 10.1038/ng.534. doi:10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SFA, Hakonarson H. Microarray technology and applications in the arena of genome-wide association. Clin Chem. 2008;54(7):1116–1124. doi: 10.1373/clinchem.2008.105395. doi:10.1373/clinchem.2008.105395. [DOI] [PubMed] [Google Scholar]
- Gundesli H, Talim B, Korkusuz P, Balci-Hayta B, Cirak S, Akarsu NA, Topaloglu H, Dincer P. Mutation in exon 1f of PLEC, leading to disruption of plectin isoform 1f, causes autosomal-recessive limb-girdle muscular dystrophy. Am J Hum Genet. 2010;87(6):834–841. doi: 10.1016/j.ajhg.2010.10.017. doi:10.1016/j.ajhg.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella JF, Wexler NS, Conneally PM, Naylor SL, Anderson MA, Tanzi RE, Watkins PC, Ottina K, Wallace MR, Sakaguchi AY, Young AB, Shoulson I, Bonilla E, Martin JB. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306(5940):234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- Harding AE, Matthews S, Jones S, Ellis CJ, Booth IW, Muller DP. Spinocerebellar degeneration associated with a selective defect of vitamin E absorption. N Engl J Med. 1985;313(1):32–35. doi: 10.1056/NEJM198507043130107. doi:10.1056/NEJM198507043130107. [DOI] [PubMed] [Google Scholar]
- Hauser SL, Oksenberg JR. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron. 2006;52(1):61–76. doi: 10.1016/j.neuron.2006.09.011. doi:10.1016/j.neuron.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, McCombie WR. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39(12):1522–1527. doi: 10.1038/ng.2007.42. doi:10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Hoischen A, van Bon BWM, Gilissen C, Arts P, van Lier B, Steehouwer M, de Vries P, de Reuver R, Wieskamp N, Mortier G, Devriendt K, Amorim MZ, Revencu N, Kidd A, Barbosa M, Turner A, Smith J, Oley C, Henderson A, Hayes IM, Thompson EM, Brunner HG, de Vries BBA, Veltman JA. De novo mutations of SETBP1 cause Schinzel–Giedion syndrome. Nat Genet. 2010;42(6):483–485. doi: 10.1038/ng.581. doi:10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- Ingram VM. Abnormal human haemoglobins. III. The chemical difference between normal and sickle cell haemoglobins. Biochim Biophys Acta. 1959;36:402–411. doi: 10.1016/0006-3002(59)90183-0. [DOI] [PubMed] [Google Scholar]
- International HapMap3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. doi:10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. doi:10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. doi:10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jani-Acsadi A, Krajewski K, Shy ME. Charcot–Marie–Tooth neuropathies: diagnosis and management. Semin Neurol. 2008;28(2):185–194. doi: 10.1055/s-2008-1062264. doi:10.1055/s-2008-1062264. [DOI] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard J-P, Lacomblez L, Pochigaeva K, Salachas F, Pradat P-F, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40(5):572–574. doi: 10.1038/ng.132. doi:10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264(5165):1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- Kasperavičiūte D, Catarino CB, Heinzen EL, Depondt C, Cavalleri GL, Caboclo LO, Tate SK, Jamnadas-Khoda J, Chinthapalli K, Clayton LMS, Shianna KV, Radtke RA, Mikati MA, Gallentine WB, Husain AM, Alhusaini S, Leppert D, Middleton LT, Gibson RA, Johnson MR, Matthews PM, Hosford D, Heuser K, Amos L, Ortega M, Zumsteg D, Wieser H-G, Steinhoff BJ, Krämer G, Hansen J, Dorn T, Kantanen A-M, Gjerstad L, Peuralinna T, Hernandez DG, Eriksson KJ, Kälviäinen RK, Doherty CP, Wood NW, Pandolfo M, Duncan JS, Sander JW, Delanty N, Goldstein DB, Sisodiya SM. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain. 2010;133(7):2136–2147. doi: 10.1093/brain/awq130. doi:10.1093/brain/awq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293(5538):2256–2259. doi: 10.1126/science.1063525. doi:10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- Keller MP, Chance PF. Inherited peripheral neuropathy. Semin Neurol. 1999;19(4):353–362. doi: 10.1055/s-2008-1040850. doi:10.1055/s-2008-1040850. [DOI] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50(3):509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. doi:10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lauer K. Ecologic studies of multiple sclerosis. Neurology. 1997;49(2 Suppl 2):S18–S26. doi: 10.1212/wnl.49.2_suppl_2.s18. [DOI] [PubMed] [Google Scholar]
- Lee JA, Madrid RE, Sperle K, Ritterson CM, Hobson GM, Garbern J, Lupski JR, Inoue K. Spastic paraplegia type 2 associated with axonal neuropathy and apparent PLP1 position effect. Ann Neurol. 2006;59:398–403. doi: 10.1002/ana.20732. doi:10.1002/ana.20732. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Le Paslier D, Frézal J, Cohen D, Weissenbach J, Munnich A, Melki J. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lindhout D. Somatic mosaicism as a basic epileptogenic mechanism? Brain. 2008;131(4):900–901. doi: 10.1093/brain/awn056. doi:10.1093/brain/awn056. [DOI] [PubMed] [Google Scholar]
- Ling H, Polke JM, Sweeney MG, Haworth A, Sandford CA, Heales SJR, Wood NW, Davis MB, Lees AJ. An intragenic duplication in guanosine triphosphate cyclohydrolase-1 gene in a dopa-responsive dystonia family. Mov Disord. 2011 doi: 10.1002/mds.23593. doi:10.1002/mds.23593. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn MR, Wang CH. Spinal muscular atrophy. Lancet. 2008;371(9630):2120–2133. doi: 10.1016/S0140-6736(08)60921-6. doi:10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14(10):417–422. doi: 10.1016/s0168-9525(98)01555-8. doi:S0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders ten years on. Genome Med. 2009;1(4):42. doi: 10.1186/gm42. doi:10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. New mutations and intellectual function. Nat Genet. 2010;42(12):1036–1038. doi: 10.1038/ng1210-1036. doi:10.1038/ng1210-1036. [DOI] [PubMed] [Google Scholar]
- Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, Chakravarti A, Patel PI. DNA duplication associated with Charcot–Marie–Tooth disease type 1A. Cell. 1991;66(2):219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Wise CA, Kuwano A, Pentao L, Parke JT, Glaze DG, Ledbetter DH, Greenberg F, Patel PI. Gene dosage is a mechanism for Charcot–Marie–Tooth disease type 1A. Nat Genet. 1992;1(1):29–33. doi: 10.1038/ng0492-29. doi:10.1038/ng0492-29. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Deiros D Rio, Chen DCY, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N Engl J Med. 2010;362(13):1181–1191. doi: 10.1056/NEJMoa0908094. doi:10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Heinz JW, Papp AC, Snyder PJ, Sedra MS, Wirth B, Burghes AHM, Prior TW. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med. 2002;4(1):20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- Mangan ME, Williams JM, Kuhn RM, Lathe WC., 3rd The UCSC genome browser: what every molecular biologist should know. Curr Protoc Mol Biol. 2009 doi: 10.1002/0471142727.mb1909s88. Chap 19:Unit19 19. doi:10.1002/0471142727.mb1909s88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. The $1,000 genome, the $100,000 analysis? Genome Med. 2010;2(11):84. doi: 10.1186/gm205. doi:10.1186/gm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti C, Gellera C, Rimoldi M, Mineri R, Uziel G, Zorzi G, Pareyson D, Piccolo G, Gambi D, Piacentini S, Squitieri F, Capra R, Castellotti B, Di Donato S. Ataxia with isolated vitamin E deficiency: neurological phenotype, clinical follow-up and novel mutations in TTPA gene in Italian families. Neurol Sci. 2004;25(3):130–137. doi: 10.1007/s10072-004-0246-z. doi:10.1007/s10072-004-0246-z. [DOI] [PubMed] [Google Scholar]
- McLean MD, Roy N, MacKenzie AE, Salih M, Burghes AHM, Simard L, Korneluk RG, Ikeda J-E, Surh L. Two 5q13 simple tandem repeat loci are in linkage disequilibrium with type 1 spinal muscular atrophy. Hum Mol Genet. 1994;3(11):1951–1956. doi: 10.1093/hmg/3.11.1951. [DOI] [PubMed] [Google Scholar]
- Melki J, Abdelhak S, Sheth P, Bachelot MF, Burlet P, Marcadet A, Aicardi J, Barois A, Carriere JP, Fardeau M, Fontan D, Ponsot G, Billette T, Angelini C, Barbosa C, Ferriere G, Lanzi G, Ottolini A, Babron MC, Cohen D, Hanauer A, Clerget-Darpoux F, Lathrop M, Munnich A, Frezal J. Gene for chronic proximal spinal muscular atrophies maps to chromosome 5q. Nature. 1990;344(6268):767–768. doi: 10.1038/344767a0. doi:10.1038/344767a0. [DOI] [PubMed] [Google Scholar]
- Mencía Á , Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MÁ. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. doi:10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. doi:10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, Chinwalla A, Conrad DF, Fu Y, Grubert F, Hajirasouliha I, Hormozdiari F, Iakoucheva LM, Iqbal Z, Kang S, Kidd JM, Konkel MK, Korn J, Khurana E, Kural D, Lam HYK, Leng J, Li R, Li Y, Lin C-Y, Luo R, Mu XJ, Nemesh J, Peckham HE, Rausch T, Scally A, Shi X, Stromberg MP, Stütz AM, Urban AE, Walker JA, Wu J, Zhang Y, Zhang ZD, Batzer MA, Ding L, Marth GT, McVean G, Sebat J, Snyder M, Wang J, Ye K, Eichler EE, Gerstein MB, Hurles ME, Lee C, McCarroll SA, Korbel JO, 1000 Genomes Project Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332):59–65. doi: 10.1038/nature09708. doi:10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira M-C, Klur S, Watanabe M, Németh AH, Le Ber I, Moniz J-C, Tranchant C, Aubourg P, Tazir M, Schöls L, Pandolfo M, Schulz JB, Pouget J, Calvas P, Shizuka-Ikeda M, Shoji M, Tanaka M, Izatt L, Shaw CE, M’Zahem A, Dunne E, Bomont P, Benhassine T, Bouslam N, Stevanin G, Brice A, Guimarães J, Mendonça P, Barbot C, Coutinho P, Sequeiros J, Dürr A, Warter J-M, Koenig M. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36(3):225–227. doi: 10.1038/ng1303. doi:10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- Mougeot J-L, Richardson-Milazi S, Brooks BR. Whole-genome association studies of sporadic amyotrophic lateral sclerosis: are retroelements involved? Trends Mol Med. 2009;15(4):148–158. doi: 10.1016/j.molmed.2009.02.005. doi:10.1016/j.molmed.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Nelis E, Haites N, Van Broeckhoven C. Mutations in the peripheral myelin genes and associated genes in inherited peripheral neuropathies. Hum Mutat. 1999;13(1):11–28. doi: 10.1002/(SICI)1098-1004(1999)13:1<11::AID-HUMU2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42(9):790–793. doi: 10.1038/ng.646. doi:10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Knoll JHM, Butler MG, Karam S, Lalande M. Genetic imprinting suggested by maternal heterodisomy in non-deletion Prader-Willi syndrome. Nature. 1989;342:281–285. doi: 10.1038/342281a0. doi:10.1038/342281a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissenbaum J, Devor M, Seltzer Z, Gebauer M, Michaelis M, Tal M, Dorfman R, Abitbul-Yarkoni M, Lu Y, Elahipanah T, del Canho S, Minert A, Fried K, Persson A-K, Shpigler H, Shabo E, Yakir B, Pisanté A, Darvasi A. Susceptibility to chronic pain following nerve injury is genetically affected by CACNG2. Genome Res. 2010;20(9):1180–1190. doi: 10.1101/gr.104976.110. doi:10.1101/gr.104976.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak R. Genetic testing set for takeoff. Science. 1994;265(5171):464–467. doi: 10.1126/science.8036486. [DOI] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4(11):907–909. doi: 10.1038/nmeth1109. doi:10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Oksenberg JR, Baranzini SE. Multiple sclerosis genetics—is the glass half full, or half empty? Nat Rev Neurol. 2010;6(8):429–437. doi: 10.1038/nrneurol.2010.91. doi:10.1038/nrneurol.2010.91. [DOI] [PubMed] [Google Scholar]
- Oprea GE, Kröber S, McWhorter ML, Rossoll W, Müller S, Krawczak M, Bassell GJ, Beattie CE, Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320(5875):524–527. doi: 10.1126/science.1155085. doi:10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmundsen JA. We are all mutants—preventive genetic medicine: a growing clinical field troubled by a confusion of ethicists. Med Dimens. 1973;2(2):5+–7+. [PubMed] [Google Scholar]
- Ouahchi K, Arita M, Kayden H, Hentati F, Ben Hamida M, Sokol R, Arai H, Inoue K, Mandel J-L, Koenig M. Ataxia with isolated vitamin E deficiency is caused by mutations in the alpha-tocopherol transfer protein. Nat Genet. 1995;9(2):141–145. doi: 10.1038/ng0295-141. doi:10.1038/ng0295-141. [DOI] [PubMed] [Google Scholar]
- Pegoraro E, Hoffman EP, Piva L, Gavassini BF, Cagnin S, Ermani M, Bello L, Soraru G, Pacchioni B, Bonifati MD, Lanfranchi G, Angelini C, Kesari A, Lee I, Gordish-Dressman H, Devaney JM, McDonald CM. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76(3):219–226. doi: 10.1212/WNL.0b013e318207afeb. doi:10.1212/WNL.0b013e318207afeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR. Charcot–Marie–Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet. 1992;2(4):292–300. doi: 10.1038/ng1292-292. doi:10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo W-L, Chen C, Zhai Y, Dairkee SH, Ljung B-M, Gray JW, Albertson DG. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20(2):207–211. doi: 10.1038/2524. doi:10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Koeuth T, Killian J, Iannaccone ST, Shapira SK, Kashork CD, Spikes AS, Shaffer LG, Lupski JR. DNA rearrangements on both homologues of chromosome 17 in a mildly delayed individual with a family history of autosomal dominant carpal tunnel syndrome. Am J Hum Genet. 1999;64(2):471–478. doi: 10.1086/302240. doi:10.1086/302240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, Shaffer LG, Lupski JR. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith–Magenis microdeletion. Nat Genet. 2000;24(1):84–87. doi: 10.1038/71743. doi:10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, Friedman EM, Glaze D, Krull K, Lee JA, Lewis RA, Mendoza-Londono R, Robbins-Furman P, Shaw C, Shi X, Weissenberger G, Withers M, Yatsenko SA, Zackai EH, Stankiewicz P, Lupski JR. Characterization of Potocki–Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am J Hum Genet. 2007;80(4):633–649. doi: 10.1086/512864. doi:10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeymaekers P, Timmerman V, Nelis E, De Jonghe P, Hoogendijk JE, Baas F, Barker DF, Martin JJ, De Visser M, Bolhuis PA, The HMSN Collaborative Research Group Duplication in chromosome 17p11.2 in Charcot–Marie–Tooth neuropathy type 1a (CMT 1a) Neuromuscul Disord. 1991;1(2):93–97. doi: 10.1016/0960-8966(91)90055-w. [DOI] [PubMed] [Google Scholar]
- Ramagopalan SV, Maugeri NJ, Handunnetthi L, Lincoln MR, Orton S-M, Dyment DA, DeLuca GC, Herrera BM, Chao MJ, Sadovnick AD, Ebers GC, Knight JC. Expression of the multiple sclerosis-associated MHC class II Allele HLA-DRB1*1501 is regulated by vitamin D. PLoS Genet. 2009;5(2):e1000369. doi: 10.1371/journal.pgen.1000369. doi:10.1371/journal.pgen.1000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, Handunnetthi L, Handel AE, Disanto G, Orton S-M, Watson CT, Morahan JM, Giovannoni G, Ponting CP, Ebers GC, Knight JC. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20(10):1352–1360. doi: 10.1101/gr.107920.110. doi:10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees MI. The genetics of epilepsy—the past, the present and future. Seizure. 2010;19(10):680–683. doi: 10.1016/j.seizure.2010.10.029. doi:10.1016/j.seizure.2010.10.029. [DOI] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J-L, Drumm ML, Iannuzzi MC, Collins FS, Tsui L-C. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245(4922):1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Roa BB, Dyck PJ, Marks HG, Chance PF, Lupski JR. Dejerine–Sottas syndrome associated with point mutation in the peripheral myelin protein 22 (PMP22) gene. Nat Genet. 1993;5(3):269–273. doi: 10.1038/ng1193-269. doi:10.1038/ng1193-269. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng H-X, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung W-Y, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak-Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH., Jr Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. doi:10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. doi:10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- Royer-Pokora B, Kunkel LM, Monaco AP, Goff SC, Newburger PE, Baehner RL, Cole FS, Curnutte JT, Orkin SH. Cloning the gene for an inherited human disorder—chronic granulomatous disease—on the basis of its chromosomal location. Nature. 1986;322(6074):32–38. doi: 10.1038/322032a0. doi:10.1038/322032a0. [DOI] [PubMed] [Google Scholar]
- Saito-Ohara F, Fukuda Y, Ito M, Agarwala KL, Hayashi M, Matsuo M, Imoto I, Yamakawa K, Nakamura Y, Inazawa J. The Xq22 inversion breakpoint interrupted a novel Ras-like GTPase gene in a patient with Duchenne muscular dystrophy and profound mental retardation. Am J Hum Genet. 2002;71:637–645. doi: 10.1086/342208. doi:10.1086/342208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels ME, Orr A, Guernsey DL, Dooley K, Riddell C, Hodgkinson K, Ludman M, Pullman D. Is gene discovery research or diagnosis? Genet Med. 2008;10(6):385–390. doi: 10.1097/GIM.0b013e3181770172. doi:10.1097/GIM.0b013e3181770172. [DOI] [PubMed] [Google Scholar]
- Sas AMG, Di Fonzo A, Bakker SLM, Simons EJ, Oostra BA, Maat-Kievit AJ, Boon AJW, Bonifati V. Autosomal dominant restless legs syndrome maps to chromosome 20p13 (RLS-5) in a Dutch kindred. Mov Disord. 2010;25(11):1715–1722. doi: 10.1002/mds.23248. doi:10.1002/mds.23248. [DOI] [PubMed] [Google Scholar]
- Sawcer S, Ban M, Wason J, Dudbridge F. What role for genetics in the prediction of multiple sclerosis? Ann Neurol. 2010;67(1):3–10. doi: 10.1002/ana.21911. doi:10.1002/ana.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scangos G. Drug discovery in the postgenomic era. Nat Biotechnol. 1997;15(12):1220–1221. doi: 10.1038/nbt1197-1220. doi:10.1038/nbt1197-1220. [DOI] [PubMed] [Google Scholar]
- Schaaf CP, Scott DA, Wiszniewska J, Beaudet AL. Identification of incestuous parental relationships by SNP-based DNA microarrays. Lancet. 2011;377(9765):555–556. doi: 10.1016/S0140-6736(11)60201-8. doi:10.1016/S0140-6736(11)60201-8. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, Alkan C, Kidd JM, Sun Y, Drautz DI, Bouffard P, Muzny DM, Reid JG, Nazareth LV, Wang Q, Burhans R, Riemer C, Wittekindt NE, Moorjani P, Tindall EA, Danko CG, Teo WS, Buboltz AM, Zhang Z, Ma Q, Oosthuysen A, Steenkamp AW, Oostuisen H, Venter P, Gajewski J, Zhang Y, Pugh BF, Makova KD, Nekrutenko A, Mardis ER, Patterson N, Pringle TH, Chiaromonte F, Mullikin JC, Eichler EE, Hardison RC, Gibbs RA, Harkins TT, Hayes VM. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463(7283):943–947. doi: 10.1038/nature08795. doi:10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga K, Inoue K, Lupski JR. Mendelian, nonMendelian, and multigenic inheritance and complex traits. In: Rosenberg RN, DiMauro S, Paulson HL, Ptacek L, Nestler EJ, editors. The molecular and genetic basis of neurologic and psychiatric disease. 4th edn Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2008. p. 31. [Google Scholar]
- Shy ME, Scavina MT, Clark A, Krajewski KM, Li J, Kamholz J, Kolodny E, Szigeti K, Fischer RA, Saifi GM, Scherer SS, Lupski JR. T118M PMP22 mutation causes partial loss of function and HNPP-like neuropathy. Ann Neurol. 2006;59(2):358–364. doi: 10.1002/ana.20777. doi:10.1002/ana.20777. [DOI] [PubMed] [Google Scholar]
- Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, Chen C-M, Clark LN, Condroyer C, De Marco EV, Dürr A, Eblan MJ, Fahn S, Farrer MJ, Fung H-C, Gan-Or Z, Gasser T, Gershoni-Baruch R, Giladi N, Griffith A, Gurevich T, Januario C, Kropp P, Lang AE, Lee-Chen G-J, Lesage S, Marder K, Mata IF, Mirelman A, Mitsui J, Mizuta I, Nicoletti G, Oliveira C, Ottman R, Orr-Urtreger A, Pereira LV, Quattrone A, Rogaeva E, Rolfs A, Rosenbaum H, Rozenberg R, Samii A, Samaddar T, Schulte C, Sharma M, Singleton A, Spitz M, Tan E-K, Tayebi N, Toda T, Troiano AR, Tsuji S, Wittstock M, Wolfsberg TG, Wu Y-R, Zabetian CP, Zhao Y, Ziegler SG. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009;361(17):1651–1661. doi: 10.1056/NEJMoa0901281. doi:10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R, Peters R. Genomic medicine. Internet resources for medical genetics. JAMA. 1997;278(15):1212–1213. doi: 10.1001/jama.278.15.1212b. [DOI] [PubMed] [Google Scholar]
- Smyk M, Berg JS, Pursley A, Curtis FK, Fernandez BA, Bien-Willner GA, Lupski JR, Cheung SW, Stankiewicz P. Male-to-female sex reversal associated with an 250 kb deletion upstream of NR0B1 (DAX1) Hum Genet. 2007;122(1):63–70. doi: 10.1007/s00439-007-0373-8. doi:10.1007/s00439-007-0373-8. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. doi: 10.1126/science.1154584. doi:10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejgaard A. The immunogenetics of multiple sclerosis. Immunogenetics. 2008;60(6):275–286. doi: 10.1007/s00251-008-0295-1. doi:10.1007/s00251-008-0295-1. [DOI] [PubMed] [Google Scholar]
- Szigeti K, Lupski JR. Charcot–Marie–Tooth disease. Eur J Hum Genet. 2009;17(6):703–710. doi: 10.1038/ejhg.2009.31. doi:10.1038/ejhg.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O’Meara S, Latimer C, Dicks E, Menzies A, Stephens P, Blow M, Greenman C, Xue Y, Tyler-Smith C, Thompson D, Gray K, Andrews J, Barthorpe S, Buck G, Cole J, Dunmore R, Jones D, Maddison M, Mironenko T, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Teague J, Butler A, Jenkinson A, Jia M, Richardson D, Shepherd R, Wooster R, Tejada MI, Martinez F, Carvill G, Goliath R, de Brouwer APM, van Bokhoven H, Van Esch H, Chelly J, Raynaud M, Ropers H-H, Abidi FE, Srivastava AK, Cox J, Luo Y, Mallya U, Moon J, Parnau J, Mohammed S, Tolmie JL, Shoubridge C, Corbett M, Gardner A, Haan E, Rujirabanjerd S, Shaw M, Vandeleur L, Fullston T, Easton DF, Boyle J, Partington M, Hackett A, Field M, Skinner C, Stevenson RE, Bobrow M, Turner G, Schwartz CE, Gecz J, Raymond FL, Futreal PA, Stratton MR. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41(5):535–543. doi: 10.1038/ng.367. doi:10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. doi:10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traber MG, Sokol RJ, Burton GW, Ingold KU, Papas AM, Huffaker JE, Kayden HJ. Impaired ability of patients with familial isolated vitamin E deficiency to incorporate alpha-tocopherol into lipoproteins secreted by the liver. J Clin Invest. 1990;85(2):397–407. doi: 10.1172/JCI114452. doi:10.1172/JCI114452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji S. Genetics of neurodegenerative diseases: insights from high-throughput resequencing. Hum Mol Genet. 2010;19(R1):R65–R70. doi: 10.1093/hmg/ddq162. doi:10.1093/hmg/ddq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi L, Dibbens LM, Lawrence KM, Iona X, McMahon JM, Murrell W, Mackay-Sim A, Scheffer IE, Berkovic SF. Timing of de novo mutagenesis—a twin study of sodium-channel mutations. N Engl J Med. 2010;363(14):1335–1340. doi: 10.1056/NEJMoa0910752. doi:10.1056/NEJMoa0910752. [DOI] [PubMed] [Google Scholar]
- Valdmanis PN, Daoud H, Dion PA, Rouleau GA. Recent advances in the genetics of amyotrophic lateral sclerosis. Curr Neurol Neurosci Rep. 2009;9(3):198–205. doi: 10.1007/s11910-009-0030-9. [DOI] [PubMed] [Google Scholar]
- van Es MA, van Vught PWJ, Blauw HM, Franke L, Saris CGJ, Van Den Bosch L, de Jong SW, de Jong V, Baas F, van’t Slot R, Lemmens R, Schelhaas HJ, Birve A, Sleegers K, Van Broeckhoven C, Schymick JC, Traynor BJ, Wokke JHJ, Wijmenga C, Robberecht W, Andersen PM, Veldink JH, Ophoff RA, van den Berg LH. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2008;40(1):29–31. doi: 10.1038/ng.2007.52. doi:10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo J-M, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. doi:10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LELM, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BWM, Hoischen A, de Vries BBA, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nat Genet. 2010a;42(12):1109–1112. doi: 10.1038/ng.712. doi:10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Vissers LELM, de Vries BBA, Veltman JA. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J Med Genet. 2010b;47(5):289–297. doi: 10.1136/jmg.2009.072942. doi:10.1136/jmg.2009.072942. [DOI] [PubMed] [Google Scholar]
- Waggoner DJ, Pagon RA. Internet resources in medical genetics. Curr Protoc Hum Genet. 2009 doi: 10.1002/0471142905.hg0912s62. Chap 9, Unit 9, 12. doi:10.1002/0471142905.hg0912s62. [DOI] [PubMed] [Google Scholar]
- Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, Fountain JW, Brereton A, Nicholson J, Mitchell AL, Brownstein BH, Collins FS. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249(4965):181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King M-C, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539–543. doi: 10.1126/science.1155174. doi:10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wang CH, Finkel RS, Bertini ES, Schroth M, Simonds A, Wong B, Aloysius A, Morrison L, Main M, Crawford TO, Trela A. Consensus statement for standard of care in spinal muscular atrophy. J Child Neurol. 2007;22(8):1027–1049. doi: 10.1177/0883073807305788. doi:10.1177/0883073807305788. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, Guo Y, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z, Zheng H, Hellmann I, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, Qin J, Ma L, Li G, Yang Z, Zhang G, Yang B, Yu C, Liang F, Li W, Li S, Li D, Ni P, Ruan J, Li Q, Zhu H, Liu D, Lu Z, Li N, Guo G, Zhang J, Ye J, Fang L, Hao Q, Chen Q, Liang Y, Su Y, San A, Ping C, Yang S, Chen F, Li L, Zhou K, Zheng H, Ren Y, Yang L, Gao Y, Yang G, Li Z, Feng X, Kristiansen K, Wong GK-S, Nielsen R, Durbin R, Bolund L, Zhang X, Li S, Yang H, Wang J. The diploid genome sequence of an Asian individual. Nature. 2008;456(7218):60–65. doi: 10.1038/nature07484. doi:10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen Y-J, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song X, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–876. doi: 10.1038/nature06884. doi:10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Whibley AC, Plagnol V, Tarpey PS, Abidi F, Fullston T, Choma MK, Boucher CA, Shepherd L, Willatt L, Parkin G, Smith R, Futreal PA, Shaw M, Boyle J, Licata A, Skinner C, Stevenson RE, Turner G, Field M, Hackett A, Schwartz CE, Gecz J, Stratton MR, Raymond FL. Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am J Hum Genet. 2010;87(2):173–188. doi: 10.1016/j.ajhg.2010.06.017. doi:10.1016/j.ajhg.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesekera LC, Leigh PN. Amyotrophic lateral sclerosis. Orphanet J Rare Dis. 2009;4:3. doi: 10.1186/1750-1172-4-3. doi:10.1186/1750-1172-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, Basehore MJ, Broeckel U, Tomita-Mitchell A, Arca MJ, Casper JT, Margolis DA, Bick DP, Hessner MJ, Routes JM, Verbsky JW, Jacob HJ, Dimmock DP. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2010 doi: 10.1097/GIM.0b013e3182088158. doi:10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- Yokoseki A, Shiga A, Tan C-F, Tagawa A, Kaneko H, Koyama A, Eguchi H, Tsujino A, Ikeuchi T, Kakita A, Okamoto K, Nishizawa M, Takahashi H, Onodera O. TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann Neurol. 2008;63(4):538–542. doi: 10.1002/ana.21392. doi:10.1002/ana.21392. [DOI] [PubMed] [Google Scholar]
- Young P, Wiebusch H, Stögbauer F, Ringelstein B, Assmann G, Funke H. A novel frameshift mutation in PMP22 accounts for hereditary neuropathy with liability to pressure palsies. Neurology. 1997;48(2):450–452. doi: 10.1212/wnl.48.2.450. [DOI] [PubMed] [Google Scholar]
- Zemni R, Bienvenu T, Vinet MC, Sefiani A, Carrié A, Billuart P, McDonell N, Couvert P, Francis F, Chafey P, Fauchereau F, Friocourt G, des Portes V, Cardona A, Frints S, Meindl A, Brandau O, Ronce N, Moraine C, van Bokhoven H, Ropers HH, Sudbrak R, Kahn A, Fryns JP, Beldjord C, Chelly J. A new gene involved in X-linked mental retardation identified by analysis of an X;2 balanced translocation. Nat Genet. 2000;24:167–170. doi: 10.1038/72829. doi:10.1038/72829. [DOI] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41(7):849–853. doi: 10.1038/ng.399. doi:10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]