Abstract
Purpose of the review
Epigenetics investigates heritable changes in gene expression occurring without changes in DNA sequence. Several epigenetic mechanisms, including DNA methylation and histone modifications, can change genome function under exogenous influence. We review current evidence indicating that epigenetic alterations mediate effects from exposure to environmental toxicants.
Recent findings
Results from animal models indicate that in-utero or early-life environmental exposures produce effects that can be inherited transgenerationally and are accompanied by epigenetic alterations. The search for human equivalents of the epigenetic mechanisms identified in animal models is under way. Recent investigations have identified a number of environmental toxicants that cause altered methylation of human repetitive elements or genes. Some exposures can alter epigenetic states and the same and/or similar epigenetic alterations can be found in patients with the disease of concern. Based on current evidence, we propose possible models for the interplay between environmental exposures and the human epigenome.
Summary
Several investigations have examined the relation between exposure to environmental chemicals and epigenetics, and identified toxicants that modify epigenetic states. Whether environmental exposures have transgenerational epigenetic effects in humans remains to be elucidated. In spite of the current limitations, available evidence supports the concept that epigenetics holds substantial potential for furthering our understanding of the molecular mechanisms of environmental toxicants, as well as for predicting health-related risks due to conditions of environmental exposure and individual susceptibility.
Keywords: Epigenetics, DNA methylation, Histone modifications, Environmental exposures, Transgenerational effects
Epigenetic Mechanisms
Epigenetics is the study of heritable changes in gene expression that occur without changes in DNA sequence (Wolffe and Guschin, 2000). Epigenetic mechanisms are flexible genomic parameters that can change genome function under exogenous influence, but also provide a mechanism that allows for the stable propagation of gene activity states from one generation of cells to the next. There are at least two kinds of epigenetic information that can be inherited with chromosomes. The first is DNA methylation, the second involves changes in chromatin proteins, usually due to modifications in histone tails.
DNA methylation
DNA methylation is a covalent modification, heritable by somatic cells after cell division. 5-methyl-cytosine (5MeC) represents 2-5% of all cytosines in mammalian genomes and is found primarily on CpG dinucleotides (Lister et al, 2009; Millar et al, 2003; Rossella et al, 2009).
Cytosine methylation of CpG dinucleotides is found in close proximity to critically important cis-elements within promoters and is often associated with a repressed chromatin state and inhibition of transcription (Orphanides and Reinberg, 2002). DNA methylation also plays an important role in the maintenance of genome integrity by transcriptional silencing of repetitive DNA sequences and endogenous transposons (Bestor, 1998; Hedges and Deininger, 2007).
Histone modifications
Histones can be modified by acetylation, methylation, phosphorylation, glycosylation, sumoylation, and ADP ribosylation (Suganuma and Workman, 2008). The most common modifications are acetylation and methylation of lysine residues in the amino terminal of Histone 3 (H3) and Histone 4 (H4). Increased acetylation induces transcription activation, whereas decreased acetylation usually induces transcription repression. Methylation of histones is associated with either repression or activation of transcription, depending on the lysine residue position (Yan and Boyd, 2006).
Environmental Health and Genes: Beyond Gene-Environment Interactions
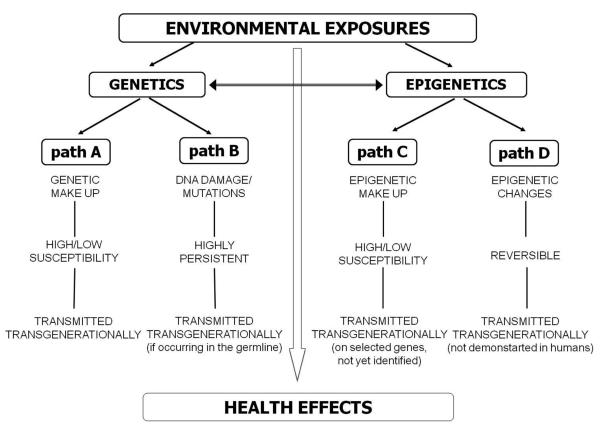
According to the World Health Organization (WHO), more than 13 million deaths annually are due to environmental causes and as much as 24% of disease is caused by exposures that can be averted (Prüss-Üstün, 2006). The list of environmental threats to human health includes a large number of environmental pollutants. For instance, in the third National Report on Human Exposure to Environmental Chemicals by the Center for Disease Control and Prevention, 148 different environmental chemicals that can be detected in blood and urine were found in a sample of the U.S. population. The list of pollutants evaluated included metals, phytoestrogens, polycyclic aromatic hydrocarbons, dioxin-like chemicals, polychlorinated biphenyls, phtalates, and several classes of pesticides (Department of Health and Human Services Centers for Disease Control and Prevention, 2005). In Fig. 1, we propose a categorization of how environmental exposures, with particular interest in environmental toxic chemicals, may interact with genetic and epigenetic mechanisms. In particular, we are interested in contrasting genetic vs. epigenetic mechanisms in their possible interplays with environmental exposures.
Figure 1.
Gene—environment vs epigene—environment interplay: a model of possible genetic and epigenetic paths linking environmental exposures to health effects.
Gene-environment interactions
The interplay between the environment and human genome has been traditionally presented under the framework of gene-environment interactions (figure 1, path A; also indicated as genotype-environment or GxE interaction) (Baccarelli, 2009; Dempfle et al, 2008; Ishibe and Kelsey, 1997; Kraft and Hunter, 2005; London and Romieu, 2009). Under this model, diseases result from interactions between the individual genetic makeup and environmental factors. Geneticists have always held true that the expression of a genetic trait in the phenotype is highly variable, largely depending on the environment to which the individual carrying the trait of concern is subjected. For instance, in patients with phenylketonuria (PKU), which is caused by mutations to a gene coding the liver enzyme phenylalanine hydroxylase (PAH), the amino acid phenylalanine does not get converted into tyrosine and reaches high levels in blood and other tissues (Scriver, 2007). The elevated phenylalanine levels affect brain development leading to mental retardation. However, a low-phenylalanine diet can keep blood phenylalanine low and avoid the severe effects of PKU. The same concept can be approached from the realm of environmental health: some individuals have low risk of developing a disease as a result of an environmental exposure, while others are much more susceptible. For example, individuals that carry genetic polymorphisms that make their cells less capable to respond to oxidative stress, have been found in several investigations to be more susceptible to the cardiovascular and respiratory effects of air pollution, which produces health effects in humans, at least in part, through oxidative stress generation (Baccarelli et al, 2008a; Chahine et al, 2007; Park et al, 2006).
A purely DNA sequence-based approach (naked DNA snapshot) is not sufficient to fully explain the risks of common diseases, which are modulated by other non-genetic or extra-genetic mechanisms. In fact, growing evidence demonstrates that the molecular influences of the environment extend well beyond the interaction with the DNA sequence. Several investigations, as we will discuss in the following sections, have shown that environmental toxicants modify epigenetic states.
Gene-Environment vs Epigene-Environment
In gene-environment interactions, (Fig 1, path A) the genetic polymorphisms that modify the effects of environmental exposures are transmitted transgenerationally according to Mendelian genetics, and the trait determining effect modifications is generally assumed to follow the same genetic model (dominant, co-dominant, recessive) as that of the levels of expression or function of the protein coded by the locus of concern. A second well-established area of interplay (Fig 1, path B) includes the direct effects of environmental exposures on the genome, e.g., DNA damage and/or mutations induced by environmental exposures. In environmental health, the recognition that exposures could produce DNA mutations represented a major landmark for risk assessment and prevention. Consequently, genotoxic agents have been categorized according to their capability to alter DNA sequence and thus increase disease risk (Siemiatycki et al, 2004). Such information has been fundamental to determine environmental risks and shape current regulatory efforts for exposure reduction. In particular potential carcinogenic agents have been carefully tested in in-vitro and in-vivo models of mutagenicity. In human subjects, some of these molecular events may represent early events along the pathways linking carcinogen exposure to cancer. For example, in our own work on the population exposed after the Seveso, Italy accident to high-doses of dioxin (Baccarelli et al, 2008b; Pesatori et al, 2008), a powerful promoting carcinogen in animals, we showed an increased number of t(14;18) translocations detectable in phenotypically normal blood lymphocytes collected from healthy subjects (Baccarelli et al, 2006). This effect may represent an early expansion of lymphocyte clones potentially related with the increased risk of Non-Hodgkin's lymphoma among subjects exposed to high doses of dioxin (Steenland et al, 2004). Environmentally-induced DNA mutations can have a transgenerational effect only if occurring in the germline. For instance, parental exposure to ionizing radiation has been shown to increase the frequency of germline mutations detectable in the next generation (Charles, 2001), and confer a predisposition to cancer (Dubrova et al, 2000).
In principle, the effect-modification model should apply to epigene-environment interactions as well as to gene-environment interactions. Similarly to the effect modifications demonstrated or postulated for genetic polymorphisms (Fig 1, path A), epigenetic differences determining disease risk could make individuals less or more vulnerable to environmental insults (Fig 1, path C). However, to best of our knowledge, a formal concept of epigene-environment interaction has not been yet developed and we are not aware of examples of epigene-environment interactions in environmental health or toxicology studies. In environmental studies, the flexibility of epigenetic states has generated a growing interest in evaluating the direct alterations that environmental exposures may produce on epigenetic states (Fig 1, path D), including changes in DNA methylation and histone modifications. Investigations that evaluated alterations in DNA methylation and histone modifications in response to environmental chemical exposures were reviewed by us in a recent article (Baccarelli and Bollati, 2009). Here, we will discuss the biological basis for potential interplays with epigenetic states that might be activated in the presence of environmental exposures and determine health-related effects. We will also discuss whether available evidence suggests that epigenetics provides biological mechanisms for transgenerational environmental effects.
Epigenetic Reprogramming in the Mammalian Development
In mammals, DNA methylation is essential for embryogenesis, during which methylation patterns change dynamically to adapt embryos to be fit for further differentiation (Reik et al, 2001). Two main waves of genome-wide epigenetic reprogramming characterize mammalian development that take place at the zygote stage and during primordial germ cells formation (Shi and Wu, 2009).
The genome becomes demethylated during preimplantation, to give rise to a totipotent zygote able to generate any cell type. Active DNA demethylation occurring in the paternal genome shortly after fertilization is independent from DNA replication. On the contrary, the maternal genome remains highly methylated and undergoes passive DNA demethylation following embryo development (Mayer et al, 2000; Santos et al, 2002). After the first cell cycle, the maternal allele passively loses methylation through cell divisions up to the blastocyst stage (Shi and Wu, 2009). When implantation occurs, DNA methylation levels are then restored by de novo methylation that triggers cell lineage differentiation (Sassone-Corsi, 2002).
The second reprogramming event occurs also during embryogenesis, but only in the primordial germ cells where DNA methylation patterns are erased at all single copy genes and some repetitive elements (Lees-Murdock and Walsh, 2008).
Similar to the pattern of asymmetric DNA methylation in parental genomes, histone H3K9 trimethylation and dimethylation exhibit asymmetric modifications in the parental pronuclei (Kurdistani et al, 2004; Santos et al, 2005; Valls et al, 2005; Wang et al, 2007; Yoshida et al, 2007).
The Dilemma of Epigenetic Inheritance: can Epigenetic Marks Survive Reprogramming?
The cycles of erasure/reprogramming that take place during embryogenesis raise the question about how and how frequently epigenetic marks are inherited transgenerationally, and whether epigenetic inheritance occurs in humans as well as in animal models (Whitelaw and Whitelaw, 2008). Animal experiments provide us with a few examples suggesting that epigenetic marks that are established during the life of an organism can be passed on to the following generations (Probst et al, 2009). These include epigenetic states of murine genes associated with distinctive phenotypes such as the agouti locus in the viable yellow (Avy/a) mice (determining fur colour variation between yellow and dark brown), the Axin-fused (Axinfu) allele (associated with a kinked tail) (Morgan et al, 1999; Rakyan and Whitelaw, 2003) and the CabpIAP gene, which was identified using a bioinformatic approach and shows sequence homology to the rat CDK5 activator binding protein (Druker et al, 2004).
All these models derive their properties from metastable epialleles, which are alleles that are variably expressed due to epigenetic modifications that are established very early during development (Rakyan et al, 2002). In metastable epialleles, the epigenetic state can be altered and the alteration shows transgenerational inheritance. Metastable alleles are most often determined by the presence of a retrotransposable element (or retrotransposon). In particular, the murine metastable epialleles Avy, AxinFu, and CabpIAP are all associated with contraoriented insertions of an retrotransposable intra-cisternal A particle (IAP) sequence, a family of retrovirus-like genetic elements coding for virus-like particles (Druker et al, 2004; Duhl et al, 1994; Vasicek et al, 1997).
Epigenetic Inheritance and Metastable Alleles in Animal Experiments: The Viable Yellow Agouti Avy Mouse
The viable yellow agouti Avy allele is the most extensively studied murine metastable epiallele. In viable yellow (Avy/a) mice, transcription originating in a IAP retrotransposon inserted upstream of the agouti gene (A) causes ectopic expression of agouti protein, resulting in yellow fur, obesity, diabetes and increased susceptibility to tumours (Morgan et al, 1999). Avy mice display variable expressivity because they are epigenetic mosaics for activity of the retrotransposon: isogenic Avy mice have coats that vary in a continuous spectrum from full yellow, through variegated yellow/agouti, to full agouti (pseudoagouti). The distribution of phenotypes among offspring is related to the phenotype of the dam; when an Avy dam has the agouti phenotype, her offspring is more likely to be agouti. It has been shown that the offspring colour is not the result of a maternally contributed environment, but rather genuinely inherited, and is associated with transmission of a silenced Avy allele through the female germ line (Morganet al, 1999).
Is there enough Evidence for Epigenetic Inheritance in Humans?
Determining whether equivalents of metastable epialleles exist and are frequent in humans poses substantial challenges. There are just a few reports that have been used to suggest inheritance of epigenetic states in humans. For example, in several cases of familial colorectal cancer, the mismatch repair genes MLH1 and MSH2, which usually exhibit low or no methylation, have been found to be silenced due to promoter methylation, and this has been occasionally detected in successive generations (Chan et al, 2006; Hitchins et al, 2007; Suter et al, 2004). However, these were reports of single families and it is has been argued that the promoter methylation identified, even if detected in multiple family members, could be explained by somatic events that occurred after fertilization (Horsthemke, 2007). Whitelaw et al. have recently remarked that, in the light of current evidence, the notion that epigenetic marks can be directly inherited across generations in humans remains contentious (Whitelaw and Whitelaw, 2008).
Environmental Influences on Epigenetic States during early Development in Animal Models
Modifications to the environment during early development can lead to permanent changes in the pattern of epigenetic modifications (Fauque et al, 2007). Pregnant female rats exposed during time of sex determination to the endocrine disruptor vinclozolin have been shown to exhibit in their male offspring transgenerational disease state leading to spermatogenic defects, prostate disease, kidney disease, immune system abnormalities, hypercholesterolemia, and an increased rate of tumor development in the F1-F4 generation offspring (Anway et al, 2005; Anway et al, 2006a; Anwayet al, 2006b; Anway and Skinner, 2006; Chang et al, 2006). Both the F1 generation embryo and F2 generation germ line are directly exposed when an F0 generation pregnant mother is exposed. Therefore, only the F3 generation can provide the first unequivocal signs of transgenerational inheritance. The transgenerational disease states in the vinclozolin F1-F4 generation animals were found to be associated with a transgenerational alteration in the epigenetic programming of the male germ line (Anway et al, 2005).
A second example of epigenetic toxicant is represented by bisphenol A (BPA), a high-production-volume chemical used in the manufacture of polycarbonate plastic. In utero or neonatal exposure to BPA is associated with higher body weight, increased breast and prostate cancer, and altered reproductive function. Dolinoy et al demonstrated that maternal BPA exposure shifted the coat colour distribution of viable yellow agouti (Avy) mouse offspring toward yellow by decreasing CpG (cytosine-guanine dinucleotide) methylation in the IAP sequence upstream of the Agouti gene (Dolinoy et al, 2007a; Waterland, 2009). In addition, CpG methylation was also decreased at the CabpIAP metastable locus. DNA methylation at the Avy locus was similar in tissues from the three germ layers, suggesting that BPA affected epigenetic patterning during early stem cell development.
Moreover, maternal dietary supplementation, with either methyl donors like folic acid or the phytoestrogen genistein, blunted the DNA hypomethylating effect of BPA (Dolinoy et al, 2007a). However, because this investigation was conducted only up to the F2 generation, it did not directly demonstrate inheritance of the epigenetic modifications induced by BPA exposure.
Translating Environmental Epigenetic Effects from Animal Models to Humans
Translating results obtained in mouse models to humans is not straightforward. The agouti model cannot be directly applied to humans, nor is there a human equivalent of kinked tails. One common characteristic of the mouse models described above is that their metastable alleles all include a IAP retrotransposon. IAP sequences are endogenous retrovirus-like mobile elements, present at 1000 copies in the mouse genome. These elements transpose in a replicative manner via an RNA intermediate and its reverse transcription, and their transposition should therefore be tightly controlled by their transcription level (Dupressoir and Heidmann, 1997). In humans, more than one-third of DNA methylation occurs in retrotransposons (Kochanek et al, 1993; Schmid, 1998), which represent a large portion of the human genome (Bernstein et al, 2006). Among these sequences, Alu and LINE-1 retrotrasposons are the most plentiful families representing approximately 30% of the human genome (Babushok and Kazazian, 2007; Grover et al, 2004; Kazazian and Goodier, 2002) and are heavily methylated (Yang et al, 2004). Because of their high representation throughout the genome, Alu and LINE-1 have been proposed as surrogate markers for estimating global DNA methylation level (Weisenberger et al, 2005; Yang et al, 2004), but growing evidence indicates that they could have specific and distinct cellular roles (Wallace et al, 2008). Hypomethylation of repetitive elements favours their activity as retrotransposable sequences and has been suggested to have deleterious effects on cells, initially through insertional mutations (Kazazian, 2004), and later by introducing genome instability through deletions and genomic rearrangements (Gilbert et al, 2002; Ostertag and Kazazian, 2001; Wallace et al, 2008).
In healthy populations, inter-individual methylation variations at Alu or LINE-1 elements from blood DNA have also been associated with risk factors for cancer, neurological and cardiovascular diseases (Baccarelli and Bollati, 2009; Baccarelli et al, 2009; Bollati et al, 2007; Rusiecki et al, 2008). Also the finding of an age-associated decline in repetitive-element methylation in normal tissues of aging individuals suggests a possible role in a variety of common human age-related diseases (Bollati et al, 2009).
Epigenetics and environmental toxicants in humans
Exposure to air pollution, particularly to particulate matter (PM), has been associated with increased morbidity and mortality from cardiorespiratory disease, as well as with lung cancer risk (Baccarelli et al, 2008c; Brook et al, 2004; Peters, 2005; Samet et al, 2000; Vineis and Husgafvel-Pursiainen, 2005). In a human study, we recently demonstrated that promoter methylation of the iNOS (inducible Nitric Oxide Synthase) gene was lower in blood samples of foundry workers with well-characterized exposure to PM with aerodynamic diameter<10 μm (PM10) (Tarantini et al, 2008). iNOS demethylation is expected to increase expression and activity of the iNOS protein, which could in turn contribute to inflammation and oxidative stress generation, which are primary mechanisms linking inhalation of air pollutants to their acute health effects (Alexeeff et al, 2008; Baccarelli et al, 2007; Chahine et al, 2007). In the same study, long-term exposure to PM10 was negatively associated with methylation in both Alu and LINE-1 (Tarantini et al, 2008). In a recent investigation, we showed that exposure to black carbon (BC), a marker of particles from vehicular traffic, was also associated with decreased DNA methylation in LINE-1, measured in 1,097 blood DNA samples from the Normative Aging Study (NAS), an investigation of elderly men in the Boston area. As repetitive element hypomethylation is believed to occur in patients with cancer (Ehrlich, 2002) and cardiovascular disease (Castro et al, 2003), such changes may reproduce epigenetic processes related to disease development and represent mechanisms by which particulate air pollution affects human health (Baccarelli et al, 2008c). Several in-vitro studies have established an association between DNA methylation and environmental metals, which are components of PM, including nickel, cadmium, lead, and particularly arsenic (Bleich et al, 2006; Dolinoy et al, 2007b; McVeigh et al, 2001; Wright and Baccarelli, 2007).
In addition, in an animal study, Yauk et al. showed that sperm DNA of mice exposed to steel plant air was hypermethylated compared to control animals and this change persisted following removal from the environmental exposure (Yauk et al, 2008). This finding calls for further research to determine whether air pollutants produce DNA methylation changes that are transmitted transgenerationally.
In our laboratory, we also investigated whether DNA methylation changes are induced by low-benzene exposure in peripheral blood DNA of gasoline station attendants and traffic police officers. High-level exposure to benzene has been associated with increased risk of acute myelogenous leukemia (AML) (Snyder, 2002). In our study, airborne benzene exposure was associated with a significant reduction in LINE-1 and Alu methylation. Airborne benzene was also associated with hypermethylation in p15 and hypomethylation of the MAGE-1 cancer-antigen gene (Bollati et al, 2007). These findings show that low-level benzene exposure may induce altered DNA methylation reproducing the aberrant epigenetic patterns found in malignant cells, which in most reports have been found to exhibit repetitive element hypomethylation as well as either hyper- or hypomethylation of specific genes, depending on the gene function. Also, benzene-associated demethylation of repetitive elements may help explain the epidemiological data linking benzene exposure with increases risk of multiple myeloma (Costantini et al, 2008; Kirkeleit et al, 2008), which also exhibits reduced methylation in Alu e LINE-1 repetitive elements (Bollati et al, 2007).
Other exposures that are associated with increased risk of hematopoietic malignancies, such as persistent organic pollutants (POPs), have been associated with changes in repetitive element DNA methylation. Rusiecki et al. (Rusiecki et al, 2008) evaluated the relationship between plasma POP concentrations and blood global DNA methylation, estimated in Alu repeated elements, in 70 Greenlandic Inuit, a population presenting some of the highest reported levels of POPs worldwide. In this work, a significant inverse linear relationships was found for DDT, DDE, β-BHC, oxychlordane, α-chlordane, mirex, several PCBs, and sum of all POPs(Rusiecki et al, 2008). Because most of the exposures investigated increase oxidative stress in human tissues, it is possible that production of reactive oxygen species represents a unifying process to account for most of these findings across different chemicals (Baccarelli and Bollati, 2009; Wright and Baccarelli, 2007). Oxidative DNA damage can interfere with the ability of methyltransferases to interact with DNA (Valinluck et al, 2004), thus resulting in a generalized altered methylation of cytosine residues at CpG sites (Turker and Bestor, 1997).
Final Remarks
In the last few years, several investigations have examined the relation between exposure to environmental chemicals and epigenetics, and identified several toxicants that modify epigenetic marks. Most of the studies conducted so far have been focused on DNA methylation, whereas only a few recent investigations have studied the effects of environmental chemicals on histone modifications (Baccarelli and Bollati, 2009). In animal models, environmental effects that might involve epigenetic mechanisms have been shown to be transmitted trans-generationally. Whether transgenerational epigenetic effects of environmental exposures are present in humans, or whether epigenetic inheritance exists at all in humans, remains to be elucidated.
Nonetheless, growing evidence indicates that epigenetics holds substantial potential for developing biological markers to predict which exposures would put exposed subjects at risk and which individuals will be more susceptible to develop disease. In human studies, this will require the use of laboratory methods with enhanced precision, sensitivity, and coverage, so that epigenetic changes can be detected as early as possible and well ahead of disease diagnosis. For several exposures, it has been proven that chemicals can alter epigenetic marks and that the same or similar epigenetic alterations can be found in patients with the disease of concern and/or in diseased tissues. Future prospective investigations are needed to determine whether exposed subjects develop epigenetic alterations over time and, in turn, whether such alterations increase the risk of disease.
References
- Alexeeff SE, Litonjua AA, Wright RO, Baccarelli A, Suh H, Sparrow D, et al. Ozone exposure, antioxidant genes, and lung function in an elderly cohort: VA normative aging study. Occup Environ Med. 2008;65(11):736–742. doi: 10.1136/oem.2007.035253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308(5727):1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006a;147(12):5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Memon MA, Uzumcu M, Skinner MK. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J Androl. 2006b;27(6):868–879. doi: 10.2164/jandrol.106.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147(6 Suppl):S43–49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28(6):527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- Baccarelli A. Breathe deeply into your genes! - Genetic variants and air pollution effects (Editorial) Am J Respir Crit Care Med. 2009;179:431–432. doi: 10.1164/rccm.200812-1808ED. [DOI] [PubMed] [Google Scholar]
- Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21(2):243–251. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Cassano PA, Litonjua A, Park SK, Suh H, Sparrow D, et al. Cardiac autonomic dysfunction: effects from particulate air pollution and protection by dietary methyl nutrients and metabolic polymorphisms. Circulation. 2008a;117(14):1802–1809. doi: 10.1161/CIRCULATIONAHA.107.726067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Giacomini SM, Corbetta C, Landi MT, Bonzini M, Consonni D, et al. Neonatal thyroid function in Seveso 25 years after maternal exposure to dioxin. PLoS Med. 2008b;5(7):e161. doi: 10.1371/journal.pmed.0050161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Hirt C, Pesatori AC, Consonni D, Patterson DG, Jr., Bertazzi PA, et al. t(14;18) translocations in lymphocytes of healthy dioxin-exposed individuals from Seveso, Italy. Carcinogenesis. 2006;27(10):2001–2007. doi: 10.1093/carcin/bgl011. [DOI] [PubMed] [Google Scholar]
- Baccarelli A, Martinelli I, Zanobetti A, Grillo P, Hou LF, Bertazzi PA, et al. Exposure to particulate air pollution and risk of deep vein thrombosis. Arch Intern Med. 2008c;168(9):920–927. doi: 10.1001/archinte.168.9.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179(7):572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Zanobetti A, Martinelli I, Grillo P, Hou L, Lanzani G, et al. Air pollution, smoking, and plasma homocysteine. Environ Health Perspect. 2007;115(2):176–181. doi: 10.1289/ehp.9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bestor TH. The host defence function of genomic methylation patterns. Novartis Found Symp. 1998;214:187–195. doi: 10.1002/9780470515501.ch11. discussion 195-189, 228-132. [DOI] [PubMed] [Google Scholar]
- Bleich S, Lenz B, Ziegenbein M, Beutler S, Frieling H, Kornhuber J, et al. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcohol Clin Exp Res. 2006;30(4):587–591. doi: 10.1111/j.1530-0277.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67(3):876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mechanisms of ageing and development. 2009;130(4):234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook RD, Franklin B, Cascio W, Hong Y, Howard G, Lipsett M, et al. Air pollution and cardiovascular disease: a statement for healthcare professionals from the Expert Panel on Population and Prevention Science of the American Heart Association. Circulation. 2004;109(21):2655–2671. doi: 10.1161/01.CIR.0000128587.30041.C8. [DOI] [PubMed] [Google Scholar]
- Castro R, Rivera I, Ravasco P, Jakobs C, Blom HJ, Camilo ME, et al. 5,10-Methylenetetrahydrofolate reductase 677C-->T and 1298A-->C mutations are genetic determinants of elevated homocysteine. Qjm. 2003;96(4):297–303. doi: 10.1093/qjmed/hcg039. [DOI] [PubMed] [Google Scholar]
- Chahine T, Baccarelli A, Litonjua A, Wright RO, Suh H, Gold DR, et al. Particulate air pollution, oxidative stress genes, and heart rate variability in an elderly cohort. Environ Health Perspect. 2007;115(11):1617–1622. doi: 10.1289/ehp.10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38(10):1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- Chang HS, Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic imprinting of the male germline by endocrine disruptor exposure during gonadal sex determination. Endocrinology. 2006;147(12):5524–5541. doi: 10.1210/en.2006-0987. [DOI] [PubMed] [Google Scholar]
- Charles M. UNSCEAR report 2000: sources and effects of ionizing radiation. United Nations Scientific Comittee on the Effects of Atomic Radiation. J Radiol Prot. 2001;21(1):83–86. doi: 10.1088/0952-4746/21/1/609. [DOI] [PubMed] [Google Scholar]
- Costantini AS, Benvenuti A, Vineis P, Kriebel D, Tumino R, Ramazzotti V, et al. Risk of leukemia and multiple myeloma associated with exposure to benzene and other organic solvents: evidence from the Italian Multicenter Case-control study. Am J Ind Med. 2008;51(11):803–811. doi: 10.1002/ajim.20592. [DOI] [PubMed] [Google Scholar]
- Dempfle A, Scherag A, Hein R, Beckmann L, Chang-Claude J, Schafer H. Gene-environment interactions for complex traits: definitions, methodological requirements and challenges. Eur J Hum Genet. 2008;16(10):1164–1172. doi: 10.1038/ejhg.2008.106. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007a;104(32):13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007b;23(3):297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32(19):5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrova YE, Plumb M, Gutierrez B, Boulton E, Jeffreys AJ. Transgenerational mutation by radiation. Nature. 2000;405(6782):37. doi: 10.1038/35011135. [DOI] [PubMed] [Google Scholar]
- Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8(1):59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- Dupressoir A, Heidmann T. Expression of intracisternal A-particle retrotransposons in primary tumors of oncogene-expressing transgenic mice. Oncogene. 1997;14(24):2951–2958. doi: 10.1038/sj.onc.1201148. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21(35):5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- Fauque P, Jouannet P, Lesaffre C, Ripoche MA, Dandolo L, Vaiman D, et al. Assisted Reproductive Technology affects developmental kinetics, H19 Imprinting Control Region methylation and H19 gene expression in individual mouse embryos. BMC Dev Biol. 2007;7:116. doi: 10.1186/1471-213X-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110(3):315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20(6):813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616(1-2):46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Wong JJ, Suthers G, Suter CM, Martin DI, Hawkins NJ, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356(7):697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- Horsthemke B. Rhythm is not enough. Nat Genet. 2007;39(10):1190–1191. doi: 10.1038/ng1007-1190. [DOI] [PubMed] [Google Scholar]
- Ishibe N, Kelsey KT. Genetic susceptibility to environmental and occupational cancers. Cancer Causes Control. 1997;8(3):504–513. doi: 10.1023/a:1018421724776. [DOI] [PubMed] [Google Scholar]
- Kazazian HH., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303(5664):1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kazazian HH, Jr., Goodier JL. LINE drive. retrotransposition and genome instability. Cell. 2002;110(3):277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- Kirkeleit J, Riise T, Bratveit M, Moen BE. Increased risk of acute myelogenous leukemia and multiple myeloma in a historical cohort of upstream petroleum workers exposed to crude oil. Cancer Causes Control. 2008;19(1):13–23. doi: 10.1007/s10552-007-9065-x. [DOI] [PubMed] [Google Scholar]
- Kochanek S, Renz D, Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. Embo J. 1993;12(3):1141–1151. doi: 10.1002/j.1460-2075.1993.tb05755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Hunter D. Integrating epidemiology and genetic association: the challenge of gene-environment interaction. Philos Trans R Soc Lond B Biol Sci. 2005;360(1460):1609–1616. doi: 10.1098/rstb.2005.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117(6):721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Lees-Murdock DJ, Walsh CP. DNA methylation reprogramming in the germ line. Epigenetics. 2008;3(1):5–13. doi: 10.4161/epi.3.1.5553. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London SJ, Romieu I. Gene by environment interaction in asthma. Annu Rev Public Health. 2009;30:55–80. doi: 10.1146/annurev.publhealth.031308.100151. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- McVeigh GE, Allen PB, Morgan DR, Hanratty CG, Silke B. Nitric oxide modulation of blood vessel tone identified by arterial waveform analysis. Clin Sci (Lond) 2001;100(4):387–393. [PubMed] [Google Scholar]
- Millar D, Holliday R, Grigg G. Five not four: History and significance of the fifth base. In: Beck S, Olek A, editors. The Epigenome, Molecular Hide and Seek. Wiley-VCH Verlag GmbH Co. KGaA; Weinheim: 2003. pp. 3–20. [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23(3):314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108(4):439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr. Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Park SK, O'Neill MS, Wright RO, Hu H, Vokonas PS, Sparrow D, et al. HFE genotype, particulate air pollution, and heart rate variability: a gene-environment interaction. Circulation. 2006;114(25):2798–2805. doi: 10.1161/CIRCULATIONAHA.106.643197. [DOI] [PubMed] [Google Scholar]
- Pesatori AC, Baccarelli A, Consonni D, Lania A, Beck-Peccoz P, Bertazzi PA, et al. Aryl hydrocarbon receptor-interacting protein and pituitary adenomas: a population-based study on subjects exposed to dioxin after the Seveso, Italy, accident. Eur J Endocrinol. 2008;159(6):699–703. doi: 10.1530/EJE-08-0593. [DOI] [PubMed] [Google Scholar]
- Peters A. Particulate matter and heart disease: Evidence from epidemiological studies. Toxicol Appl Pharmacol. 2005;207(2 Suppl):477–482. doi: 10.1016/j.taap.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10(3):192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Prüss-Üstün A. Preventing disease through healthy environments. Towards an estimate of the environmental burden of disease. 2006 [Google Scholar]
- Rakyan V, Whitelaw E. Transgenerational epigenetic inheritance. Curr Biol. 2003;13(1):R6. doi: 10.1016/s0960-9822(02)01377-5. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18(7):348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Rossella F, Polledri E, Bollati V, Baccarelli A, Fustinoni S. Development and validation of a gas chromatography/mass spectrometry method for the assessment of genomic DNA methylation. Rapid Commun Mass Spectrom. 2009;23(17):2637–2646. doi: 10.1002/rcm.4166. [DOI] [PubMed] [Google Scholar]
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116(11):1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samet JM, Dominici F, Curriero FC, Coursac I, Zeger SL. Fine particulate air pollution and mortality in 20 U.S. cities, 1987-1994. N Engl J Med. 2000;343(24):1742–1749. doi: 10.1056/NEJM200012143432401. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280(1):225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296(5576):2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Schmid CW. Does SINE evolution preclude Alu function? Nucleic Acids Res. 1998;26(20):4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriver CR. The PAH gene, phenylketonuria, and a paradigm shift. Hum Mutat. 2007;28(9):831–845. doi: 10.1002/humu.20526. [DOI] [PubMed] [Google Scholar]
- Shi L, Wu J. Epigenetic regulation in mammalian preimplantation embryo development. Reprod Biol Endocrinol. 2009;7:59. doi: 10.1186/1477-7827-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemiatycki J, Richardson L, Straif K, Latreille B, Lakhani R, Campbell S, et al. Listing occupational carcinogens. Environ Health Perspect. 2004;112(15):1447–1459. doi: 10.1289/ehp.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder R. Benzene and leukemia. Crit Rev Toxicol. 2002;32(3):155–210. doi: 10.1080/20024091064219. [DOI] [PubMed] [Google Scholar]
- Steenland K, Bertazzi P, Baccarelli A, Kogevinas M. Dioxin revisited: developments since the 1997 IARC classification of dioxin as a human carcinogen. Environ Health Perspect. 2004;112(13):1265–1268. doi: 10.1289/ehp.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135(4):604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36(5):497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, et al. Effects of Particulate Matter on Genomic DNA Methylation Content and iNOS Promoter Methylation. Environ Health Perspect. 2008 doi: 10.1289/ehp.11898. in press doi:101289/ehp11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turker MS, Bestor TH. Formation of methylation patterns in the mammalian genome. Mutat Res. 1997;386(2):119–130. doi: 10.1016/s1383-5742(96)00048-8. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32(14):4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls E, Sanchez-Molina S, Martinez-Balbas MA. Role of histone modifications in marking and activating genes through mitosis. J Biol Chem. 2005;280(52):42592–42600. doi: 10.1074/jbc.M507407200. [DOI] [PubMed] [Google Scholar]
- Vasicek TJ, Zeng L, Guan XJ, Zhang T, Costantini F, Tilghman SM. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics. 1997;147(2):777–786. doi: 10.1093/genetics/147.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineis P, Husgafvel-Pursiainen K. Air pollution and cancer: biomarker studies in human populations. Carcinogenesis. 2005;26(11):1846–1855. doi: 10.1093/carcin/bgi216. [DOI] [PubMed] [Google Scholar]
- Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419(1-2):75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Kou Z, Zhang Y, Gao S. Dynamic reprogramming of histone acetylation and methylation in the first cell cycle of cloned mouse embryos. Biol Reprod. 2007;77(6):1007–1016. doi: 10.1095/biolreprod.107.063149. [DOI] [PubMed] [Google Scholar]
- Waterland RA. Is epigenetics an important link between early life events and adult disease? Horm Res. 2009;71(Suppl 1):13–16. doi: 10.1159/000178030. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic acids research. 2005;33(21):6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw NC, Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr Opin Genet Dev. 2008;18(3):273–279. doi: 10.1016/j.gde.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Guschin D. Review: chromatin structural features and targets that regulate transcription. J Struct Biol. 2000;129(2-3):102–122. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- Wright RO, Baccarelli A. Metals and neurotoxicology. J Nutr. 2007;137(12):2809–2813. doi: 10.1093/jn/137.12.2809. [DOI] [PubMed] [Google Scholar]
- Yan C, Boyd DD. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol. 2006;26(17):6357–6371. doi: 10.1128/MCB.00311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk C, Polyzos A, Rowan-Carroll A, Somers CM, Godschalk RW, Van Schooten FJ, et al. Germ-line mutations, DNA damage, and global hypermethylation in mice exposed to particulate air pollution in an urban/industrial location. Proc Natl Acad Sci U S A. 2008;105(2):605–610. doi: 10.1073/pnas.0705896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Brahmajosyula M, Shoji S, Amanai M, Perry AC. Epigenetic discrimination by mouse metaphase II oocytes mediates asymmetric chromatin remodeling independently of meiotic exit. Dev Biol. 2007;301(2):464–477. doi: 10.1016/j.ydbio.2006.08.006. [DOI] [PubMed] [Google Scholar]