Abstract
The α-hydroxytroplone, manicol (5,7-dihydroxy-2-isopropenyl-9-methyl-1,2,3,4-tetrahydro-benzocyclohepten-6-one) potently and specifically inhibits ribonuclease H (RNase H) activity of human immunodeficiency virus reverse transcriptase (HIV RT) in vitro. However, manicol was ineffective in reducing virus replication in culture. Ongoing efforts to improve the potency and specificity over the lead compound led us to synthesize 14 manicol derivatives that retain the divalent metal-chelating α-hydroxytropolone pharmacophore. These efforts were augmented by a high resolution structure of p66/p51 HIV-1 RT containing the nonnucleoside reverse transcriptase inhibitor (NNRTI), TMC278 and manicol in the DNA polymerase and RNase H active sites, respectively. We demonstrate here that several modified α-hydroxytropolones exhibit antiviral activity at non-cytotoxic concentrations. Inclusion of RNase H active site mutants indicated that manicol analogs can occupy an additional site in or around the DNA polymerase catalytic center. Collectively, our studies will promote future structure-based design of improved α-hydroxytropolones to complement the NRTI and NNRTI currently in clinical use.
Introduction
Synthesis of double-stranded, integration-competent DNA in retroviruses proceeds through an RNA/DNA hybrid intermediate, whose (+) RNA genome must be removed to facilitate second, or (+)-strand DNA synthesis1. Hybrid hydrolysis is mediated by the C-terminal RNase H domain of the virus-coded reverse transcriptase (RT). RT of human immunodeficiency virus (HIV), the etiological agent of acquired immunodeficiency syndrome (AIDS), is a p66/p51 heterodimer of asymmetrically-organized subunits processed from the 165 kDa gag/pol polyprotein precursor2, 3. Abrogating RNase H function was demonstrated almost two decades ago to inhibit enzyme activity in vitro and virus replication in culture4, 5, thereby demonstrating that RNase H antagonists might be combined with nucleoside and nonnucleoside RT inhibitors currently in clinical use as components of highly active antiretroviral therapy (HAART). Since these early reports, however, there been a paucity of data on clinical trials with small molecule RNase H inhibitors, possibly reflecting their toxicity, lack of selectivity, or poor cellular penetration.
Development of HIV RNase H inhibitors has recently been encouraged by documentation of several structurally-dissimilar classes of antagonists derived from natural products or via chemical synthesis. Examples of the former include the α-hydroxytropolones6, 7, dimeric lactones8, madurahydroxylactones9, and 1,3,4,5-tetragalloylapiitol10 (Figure 1), while synthetic entities include N-hydroxyimides, diketo acids, pyrimidinol carboxylates11, vinylogous ureas12, 13, thiocarbamates and triazoles14. While many of these compounds target the RNase H active site, our recent work suggests that vinylogous ureas represent a class of allosteric inhibitors that influence RNase H active site geometry by binding to the p51 thumb subdomain13. In addition to this expanding collection of small molecules, X-ray crystallography has provided high resolution information on enzyme:inhibitor complexes with either the isolated RNase H domain or the intact p66/p51 heterodimer11, 15–18, which should aid future structure-based efforts.
Figure 1.
Chemical structures of HIV-1 RNase H inhibitors derived from natural products.
Although potently and selectively inhibiting RNase H activity of HIV-1 and HIV-2 RT, the natural products, β-thujaplicinol (2,7-dihydroxy-4-isopropyl-cyclohepta-2,4,6-triene) and manicol (5,7-dihydroxy-2-isopropenyl-9-methyl-1,2,3,4-tetrahydro-benzocyclohepten-6-one) were ineffective in reducing virus replication in culture, most likely due to toxicity caused by inhibition of cellular enzymes19. In order to overcome the toxicity issue and improve potency and selectivity, our efforts have been focused on derivatization of these inhibitors. Our previous structure-activity relationship studies showed that three oxygen ligands of inhibitors are critical while substitutions elsewhere on the heptatriene ring were less deleterious6. Indeed, epoxidation of the alkene moiety of manicol did not interfere with the biological activity (IC50 = 0.45 µM). This terminal alkene was therefore chosen as a target site for modification because of its versatile chemistry. The synthesis, in vitro evaluation and antiviral activity of 14 novel manicol derivatives are the subject of this communication. In addition to providing the first report of α-hydroxytropolones with antiviral activity against HIV-1, we present here the high resolution crystal structure of p66/p51 HIV-1 RT containing the NNRTI, 18 (TMC278)20 in the DNA polymerase domain and manicol complexed with divalent metal at the RNase H active site. Interestingly, the bound structure of manicol differs in conformation from that of the bulk manicol structure reported by Polonsky et al.21. The high resolution cocrystal structure, together with our revised manicol structure, will allow us to develop future generations of RNase H inhibitors with greater selectivity, as well as examine the effect of simultaneous application of DNA polymerase and RNase H inhibitors on HIV replication.
Results
Structure of HIV-1 RT Containing Manicol and the NNRTI, 18
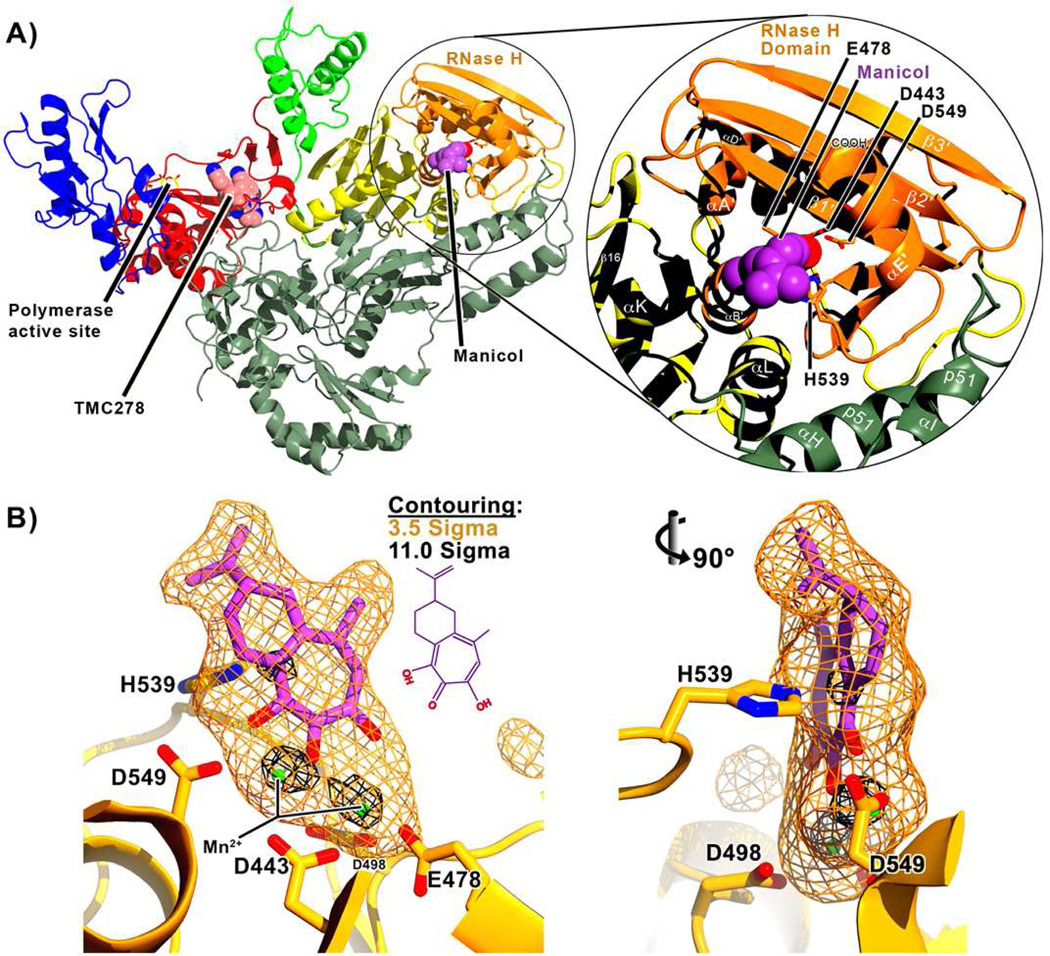
Although a high resolution structure for HIV-1 RT containing β-thujaplicinol was recently solved15, the related natural product, manicol is more readily amenable to modification, since it posseses a double bond in its side chain. In order to guide synthesis of future α-hydroxytropolones, we therefore solved the structure of p66/p51 HIV-1 RT containing 18 in the DNA polymerase domain and manicol at the RNase H active site. The structure, refined at 2.7 Å resolution (Table 1), revealed unambiguous electron density maps for manicol binding (Figure 2). Two strong Mn2+ peaks in the electron density map within the RNase H active site and correspond to the divalent cation positions "A" and "B" (following the convention for a two-cation mechanism of hydrolysis22, 23). The carbonyl oxygen and both hydroxyls of the tropolone ring coordinate the divalent cations in a manner similar to that previously observed in the RT/β-thujaplicinol structure15.
Table 1.
Data collection and refinement statistics for p66/p51 HIV-1 RT/18/manicol cocrystal.
| (a) Unit Cell Parameters (Space Group C2) | |
| a (Å): | 163.9 |
| b (Å): | 73.0 |
| c (Å): | 108.4 |
| β (Å): | 101.1 |
| (b) Data Collection | |
| Resolution Range: | 45 – 2.7Å |
| Rsym(%): | 6.3 |
| Average I/σ: | 16.1 |
| Completeness (%): | 99.5 |
| Unique Reflections/Multiplicity: | 34841/6.2 |
| (c) Refinement | |
| Sigma Cutoff: | 0.0 |
| Resolution Range Used (Å): | 45 – 2.7 |
| Completeness of Used Reflections (%): | 95.8 |
| R-factor/Rfree(%): | 23.2/25.1 |
| Cross-Validated Coordinate Error (Å): 24 | 0.36 |
| No. of Protein/Solvent Atoms: | 7907/96 |
| No. of Ligand/Cation Atoms: | 54/2 |
| Average B-Factors (Å2): | |
| Protein/Solvent: | 76.7/60.3 |
| Ligands/Cations: | 77.5/68.0 |
| RMS Bond Lengths (Å)/Angles (°):53 | 0.010/1.27 |
| (d) Ramachandran Regions | |
| Most Favored: | 95.0 |
| Additional Allowed: | 4.2% |
| Generous or Disallowed: | 0% |
Figure 2.
Structure of HIV-1 RT containing the NNRTI, 18 and manicol (a) A ribbon diagram of the structure of RT in complex with both 18 (pink) and manicol (violet). RT is a heterodimer comprising p66 (in color) and p51 (gray) subunits. The subdomains of p66 are shown in color as indicated. Right inset: A close-up view of the RNase H domain and nearby secondary structural elements. Helices and beta-sheets are labeled according to reference53. Side chains for the RNase H active site carboxylates (D443, E478, H539, and D549) are shown. (b) Manicol at the RNase H active site. A simulated annealing Fo-Fc electron density omit map is shown in which manicol and two Mn2+ cations have been omitted from the phasing. The map is contoured both at the 3.5 sigma (orange) and 11.0 sigma (black) levels. The manicol molecule modeled into this density is shown with its carbon atoms in violet. Two different vantage points are shown rotated 90 degrees from each other about the vertical axis. The inset (center top) gives the chemical structure of manicol.
Unlike β-thujaplicinol, manicol forms extensive contacts with the imidazole ring of His539 (Figure 3(a)), including contacts with interatomic distances of under 3 Å. Since tropolone ring systems can have aromatic character24, the His539-manicol interaction is an example of an aromatic-aromatic interaction. As reviewed in References 25–27, interactions between the imidazole side chain of histidine and another aromatic ring has been documented in a number of configurations. These include parallel stacking of the aromatic rings and edge-on-face orientations, in which a histidine C/N-H serves as a hydrogen-bonding donor. While in many cases the histidine C/N-H points toward the center of the other aromatic ring, there are examples in which the imidazole ring edge is offset from the center of the other ring, as seen in the His539-manicol interaction. Conventionally, to be considered a hydrogen-bonding interaction, the angle between C/N-H to the center of the aromatic ring should be greater than 120°, and the distance between the C/N and the ring should be less than 3.5 Å. Both criteria are satisfied in the His539-manicol interaction
Figure 3.
(a) Interaction distances (dashed lines, Å) between manicol (magenta carbon atoms), two Mn2+ cations (green), and RT. The same vantage points are shown as in Figure 1 (b). Divalent cations are labeled A and B following the convention for a two-cation mechanism of hydrolysis. Numbering for the manicol carbon atoms is shown (black, left image). Most of the close contacts that manicol forms with RT are with amino acid residue His539. (b) Comparison of manicol conformations. The manicol conformation we observed (yellow carbon atoms) is superimposed on a conformation reported from a small molecule crystallization study of manicol21 (magenta carbon atoms). The carbon numbering used in the deposited PDB coordinates is shown. We find that the alicyclic carbons pucker in the opposite direction than previously reported.
At 2.7 Å resolution, we cannot completely rule out an alternative possibility. Tropolone rings are believed to behave as electron-deficient aromatic rings (“tropylium”), as discussed in reference 15. If manicol forms a tropylium ion, then the tropolone ring carries a positive charge, while the three oxygens may share a resonant negative charge that contributes to the coordination of the two observed Mn2+ cations at the RNase H active site. These circumstances raise the question of whether His539 may serve as a Lewis base, in which one of the lone pair electrons of nitrogen atom serves as an electron donor to the delocalized positive charge of the tropolone ring and stabilizes the tropylium ion, analogous to anion-π interactions that have recently been reported both in small organic molecule and biological structures28, 29. In this case, we suspect that proximity of His539 to the manicol ring system would produce excessive electronegative repulsion if not mediated by a hydrogen-bonding proton.
The electron density clearly indicates that the alicyclic carbon atoms of manicol pucker in the direction of the His539 side-chain, with the 2-isopropenyl substituent occupying the equatorial position on the C10 carbon. This observation is in contrast to a previous small molecule crystal structure study of Polonsky et al.21 which reported that the 2-isopropenyl substituent occupied the axial orientation (Figure 3(b)). In both our results and the Polonsky et al. study, the chiral center at the C10 carbon has an S configuration. In the RT/β-thujaplicinol structure15, one hydroxyl group of the tropolone ring came within hydrogen bonding distance of the side-chain carboxylates of the catalytically-essential residues Glu478 and Asp498. Significantly, manicol pivots away from these residues and loses these interactions in favor of contacts with His539 and a 2.4 Å contact between one of the tropolone hydroxyls and the side-chain carboxylate of Asp549. Manicol does, however, retain hydrophobic interactions with Glu478 and Asp498 (contact distances ranging from 3.4 Å to 4.0 Å).
Other structures of either the p66/p51 RT heterodimer or the isolated 15 kDa HIV RNaseH domain have recently been published in which an RNase H inhibitor has been demonstrated to occupy the RNase H active site by coordinating two active site Mn2+ cations. These include β-thujaplicinol15, a pyrimidinol carboxylic acid derivative11, and several naphthyridinone derivatives18. Su et al. observed that most of these compounds position their metal-coordinating moieties in approximately the same plane with the metal ions18. It was therefore suggested that it might be helpful to design RNase H inhibitors on the assumption that the metal-chelating moiety would lie in the same plane with the metals. However, one of the naphthyridinone derivatives, MK3, was not co-planar with the Mn2+ cations in these studies. Likewise, when comparing manicol to β-thujaplicinol, we observe that, while the tropolone ring of the latter is essentially co-planar with the metal cations, the manicol counterpart is not. The full significance of this observation is not clear but may be related to the favorable interactions that manicol can form with the His539 side-chain if the tropolone ring moves out of the plane of the metal cations. Another possible contributing factor could be the molecular orbital alignment of the outer shell electrons about the central tropolone oxygen as it chelates both Mn2+ cations. If that oxygen behaves predominantly like a carbonyl, in which the outer shell electrons form sp2 hybrid orbitals and a π-orbital, then the lone pair electrons that coordinate the Mn2+ cations might be expected to favor a trigonal planar arrangement. Conversely, if the oxygen carries a formal negative charge, in which the outer shell electrons predominantly form sp3 hybrid orbitals, then a roughly tetrahedral (non-planar) geometry might be favored for the lone pair electrons that coordinate the cations.
Manicol Derivatization
The synthesis of manicol analogs is depicted in Scheme 1. Manicol epoxide 16 was synthesized according to the reported procedure21. In vitro studies indicated that manicol epoxide retained its efficacy as an RNase H inhibitor. Opening of epoxide 16 with a variety of amines catalyzed by stoichiometric LiClO4 afforded analogues 1–5. Addition of selected thiols required Et3N or NaH and resulted in sulfides 6–8. Sulfides 6 and 7 were oxidized with m-CPBA to either sulfoxide 10 or sulfones 9 and 11 by adjusting the reaction temperature. Starting from diacetyl-protected manicol intermediate 15, dihydroxylation of the olefin functionality using OsO4/NMO followed by the deprotection of the acetyl groups gave analogue 12. Alternatively, in the presence of OsO4 and NaIO4, the in situ dihydroxylation/oxidative cleavage of 15 furnished ketone 17, which could be reduced to alcohol 13 with NaBH4 or converted to amine 14 via reductive amination. It should be noted that all of the tested analogues (1–14) were obtained as a mixture of stereoisomers.
Scheme 1.
Syntheses of manicol derivatives 1 – 14.
In Vitro Inhibition of RNase H Activity
Using a previously reported high throughput, fluorescence-based RNase H assay30, Table 2 provides the IC50 values for compounds 1 – 14. Compound 9 was slightly more potent than manicol (IC50 0.24 µM vs 0.6 µM, respectively), while a 3-4-fold decrease in activity was observed for compound 2 (IC50 1.9 µM). All remaining compounds fell within this range. Since the high throughput RNase H assay examines non-specific, “polymerase-independent” RNase H activity defined by the spatial separation of the DNA polymerase and RNase H active site of HIV-1 RT31, we examined whether α-hydroxytropolones altered cleavage specificity on a more biologically-relevant substrate, namely the polypurine tract (PPT) primer, which must be processed from the RNA/DNA replication intermediate to initiate (+) strand DNA synthesis1. Inhibition of RNase H activity on this model PPT-containing RNA/DNA hybrid is illustrated in Figure 4(a), and quantification of cleavage data in Figure 4(b). In this experiment, compounds 1 – 14 were assayed at a final concentration of 20 µM.
Table 2.
Inhibition of HIV-1 RNase H enzymatic activity and the cytopathic effect of α-hydroxytropolones on HIV-1RF virus replication on CEM-SS cells.
| Inhibitor | In vitro | Viral Replication | |
|---|---|---|---|
| IC50 (µM) | EC50 (µM) | CC50 (µM) | |
| β-Thujaplicinol | 0.21a | n.p.b | 2.3 |
| Manicol | 0.60a | n.p. | 13.6 |
| 1 | 0.82 ± 0.08 | 10.2 | > 50 |
| 2 | 1.9 ± 0.4 | 42.1 | > 50 |
| 3 | 0.50 ± 0.07 | < 50%c | > 50 |
| 4 | 1.2 ± 0.1 | n.p. | > 50 |
| 5 | 1.2 ± 0.2 | 7.4 | 31.7 |
| 6 | 0.38 ± 0.07 | < 50% | 25.8 |
| 7 | 0.93 ± 0.10 | 11.5 | 17.8 |
| 8 | 1.3 ± 0.2 | 4.2 | 17.4 |
| 9 | 0.24 ± 0.02 | < 50% | 10.4 |
| 10 | 1.3 ± 0.2 | 14.5 | > 50 |
| 11 | 0.51 ± 0.10 | 21.2 | > 50 |
| 12 | 1.9 ± 0.2 | n.p. | > 50 |
| 13 | 0.68 ± 0.03 | 6.9 | 26.1 |
| 14 | 0.96 ± 0.07 | 10.6 | 16.7 |
Reference 14.
n.p. = no protective effect on cytopathicity of virus.
<50% = protective effect less than 50% of control, i.e. weak activity.
Figure 4.
α-Hydroxytropolone inhibition of RNase H-mediated release of the HIV-1 PPT primer from (+) RNA. Panel (a) Upper, schematic model of RNA/DNA substrate, indicating the RNase H cleavage site at the PPT/U3 junction (<>). The PPT is indicated by the shaded box. Lower, PPT cleavage assay. Lane C1, no enzyme; Lane C2, no inhibitor; Lane B, β-thujaplicinol; Lane M, manicol. The PPT/U3 cleavage product is indicated. (b), Quantification of PPT cleavage data. All inhibitors were evaluated at 20 µM.
In all instances, primary RNase H-mediated hydrolysis occurred at the 5’-pG<>pA-3’ PPT/U3 junction, with additional cleavage at the immediately adjacent 5’-pG-pG-3’ sequences. Thus, while differing levels of inhibition were observed with compounds 1 – 14, none altered cleavage specificity in the PPT assay. In keeping with the data of Table 2, compound 9 was almost 100% effective in inhibiting PPT/U3 cleavage, while compounds 2, 4, 10, and 14 were poorly active. Interestingly, some inhibitors, e.g. compounds 3 and 11, were ineffective in inhibiting PPT/U3 cleavage while more active on the non-specific RNA/DNA hybrid. In contrast, compound 5 was more active in blocking PPT/U3 cleavage than inhibiting polymerase-independent cleavage on the non-specific substrate. Data of Table 2 and Figure 3 thus illustrate potential benefits of including model systems that mimic some of the more complex RNase H-mediated events in HIV replication as screening tools.
Inhibition of DNA Polymerase Activity by Manicol Analogs
Although our crystallographic data indicated a single manicol binding site in the RNase H domain, Didierjean et al.7 have demonstrated that dihydroxytropolones can inhibit both the DNA polymerase and RNase H functions of HIV-1 RT. In addition, Su et al., recently presented crystallographic evidence that a naphthyridinone-based inhibitor, indentified through their RNase H screening efforts, bound close to the NNRTI binding site of the DNA polymerase domain in a metal-independent manner18. Such “action from a distance” is not unexpected, since similar observations have been reported with both NNRTIs32 and a dihydroxybenzoyl-naphthyl hydrazone-based inhibitor16. In order to examine the specificity of our manicol analogs, we therefore determined their effect on the DNA polymerase activity of wild type p66/p51 HIV-1 RT and RNase H-deficient derivatives, p66EQ/p51 and p66DA/p51, which harbor mutations in one of the catalytically-essential amino acids contacted by manicol (E478 and D549, respectively, Figure 3), that would be predicted to interfere with α-hydroxytropolone binding. Inhibition of DNA polymerase activity of RNase H-deficient enzymes would be diagnostic of a second binding mode within, or proximal to, the DNA polymerase active site, possibly resulting from more extensive modification of manicol.
The results of our analysis are presented in Figure 5. In general, manicol analogs had minimal effect on DNA polymerase activity of RNase H-deficient p66EQ/p51 and p66DA/p51 RT, suggesting specificity for the RNase H active site. However, two exceptions were noted, namely compounds 5 and 10, which significantly and consistently inhibited DNA-dependent DNA polymerase activity of both mutants. Although crystallographic evidence is presently unavailable, compounds 5 and 10 thus appear capable of occupying a second site on HIV-1 RT.
Figure 5.
Influence of RNase H inhibitors on the DNA-dependent DNA polymerase activity of HIV-1 RT. Panel (a), wild type enzyme; Panel (b), RNase H mutant p66E478Q/p51; Panel (c), RNase H mutant p66D549A/p51. Lane notation is as in the legend to Figure 3. All inhibitors were tested at 20 µM. P, unextended primer; P+10, fully-extended primer.
Antiviral Activity of Manicol Analogs
The parent compounds β-thujaplicinol and manicol showed no protection against the cytopathic effect of HIV, with CC50 values of 2.3 µM and 13.6 µM (Table 2), consistent with previous reports from our laboratory. Compounds 4 and 12 also showed no protection against virus cytopathicity but were not cytotoxic up to a concentration of 50 µM. Compounds 3, 6, and 9 showed slight protection against cytopathicity. However in each case the protection did not reach 50% of control cell growth, and these compounds were therefore judged inactive.
Compounds 1, 2, 5, 7, 8, 10, 11, 13, and 14 all showed protection against the cytopathicity of the virus, with EC50 values ranging from 4.2 µM (compound 8) to 42.1 µM (compound 2). In vitro therapeutic indices (CC50/EC50) were quite modest, ranging from 1.2 to 4.9 (compound 1). It was encouraging, however, that the cytotoxicity of all of the active compounds was reduced compared to their manicol precursor.
Modeling Compounds 14 and 9 into the RNase H Active Site
To gain insight into how manicol modification affects inhibitor binding, we performed energy minimization experiments on model RNase H-inhibitor complexes in which compound 14 and 9 were substituted for manicol. These derivatives were selected for modeling to determine whether their distinctive amine or sulfide constituents, respectively, would be predicted to form hydrogen bonds, or other stabilizing interactions, with RNase H residues – particularly His539. Models were constructed using coordinates from the RT-manicol cocrystal structure reported herein as described in the Experimental section and were subjected to two rounds of energy minimization.
As illustrated in Figure 6 [A], the amino group of compound 14 is several angstroms from the His539 side chain and oriented toward solvent rather than the imidazole ring. This may reflect electrostatic repulsion between charged or partially charged nitrogen atoms within the two groups, or because interaction between the phenyl substituent of compound 14 and the His539 imidazole ring is more favorable. Modeling of compound 9 (Figure 6 [B]) suggests that both sulfonyl oxygens are located within hydrogen bonding distance of His539 Nδ. Moreover, since the partial charges of the sulfonyl oxygens and the δ-nitrogen are predicted to be opposed, electrostatic interaction between these two groups may serve to stabilize the enzyme-inhibitor complex. In the cases of these two compounds, different chemical moieties appear to mediate their interaction with His539. Nevertheless, both models of Figure 6 suggest that IC50 differences among manicol derivatives most likely reflect changes in their interaction with the side chain of this conserved residue, and that stabilizing this interaction is the key to improving these inhibitors.
Figure 6.
Modeling of compound 14 [A] and 9 [B] into the RNase H active site of HIV-1 RT. For convenience, only the RNase H active site is illustrated. The Mn2+ cations are represented as as dark gray spheres. Atoms of residue H539 and compounds 14 and 9 are shown as spheres sized to reflect their van der Waals radii, and colored as follows: carbon (H539) = cyan; carbon (compound 14 and 9) = purple; nitrogen = blue; oxygen = red; sulfur = yellow; hydrogen = white.
Discussion
Successful combination antiretroviral therapy will require continued efforts to develop a variety of small molecule antagonists that interact synergistically at different sites of their target molecule. For HIV-1 RT, this has been exemplified by NRTI/NNRTI combinations targeting the DNA polymerase active site and a hydrophobic pocket at the base of the p66 thumb, respectively. With regard to RNase H function, Su et al.18, have recently published the structure of HIV-1 RT containing nevirapine in the NNRTI binding pocket and a naphthyridinone-containing inhibitor at the RNase H active site. In the present study, we present a complementary high-resolution structure of RT containing the second generation diarylpyrimidine derivative 18 in the NNRTI binding site and manicol at the RNase H active site. Moreover, we show that manicol derivatives retain in vitro potency and also inhibit HIV-1 replication, albeit with low therapeutic indices. Since the structure of RNase H active site is relatively flat and open, allowing a large substrate to be accommodated, an important consideration is to design inhibitors that interact with active site residues and coordinate divalent metal ions in order to improve potency and selectivity. His539 is clearly an attractive target based on its important role in catalysis and the fact that it can interact with other aromatic residues as well as be hydrogen bonded with positive or negative charged molecules. Our co-crystal structure shows how manicol could achieve favorable interactions with His539, yet maintain its ability to coordinate divalent metal ions. Modeling studies of Figure 6 also indicate that a number of additional interactions with His539 could be achieved by introducing sulfonyl or aromatic groups into manicol. In addition, adding bulky substituents to manicol might be beneficial by reducing cellular toxicity via decreasing binding affinity for other cellular metalloenzymes, which may explain enhanced antiviral activity in cell culture we observed here. Thus, while clinical evaluation of α-hydroxytropolones is premature, our work provides an important platform for structure-based drug design, as well as in vivo studies to better characterize their mode of action.
Despite our structural and biochemical studies, it remains to be established that RNase H is the in vivo target of our novel α-hydroxytropolones. Inhibition of RNase H activity predicts that virus replication would be interrupted at the step of (−) strand DNA transfer, where nascent DNA relocates to the 3’ end of the viral genome via homology between the 5’ and 3’ repeat (R) regions of the (+) viral genome33. Successful strand transfer requires RNase H-mediated cleavage of the RNA/DNA replication intermediate to allow the newly-synthesized (−) strand DNA to dissociate from the donor and access the acceptor template. Accumulation of (−) strong-stop DNA would thus indicate loss in RNase H function. An alternative strategy would be the isolation of RNase H inhibitor-resistant mutants and localization of the inactivating lesion to the RNase H domain. Although the therapeutic indices determined here are low, they should be sufficient to allow such in vivo manipulations.
Another issue that can be addressed is the “dynamic copy choice” model of Nikolenko et al.34, who have proposed that perturbing the balance between the DNA polymerase and RNase H activities may promote dissociation of the replication complex and interrupt virus replication. By introducing mutations into the RNase H domain which inhibit hydrolysis, these authors demonstrated increased NRTI resistance, inferring that increased residency at the DNA polymerase catalytic center provides more time for excision of the chain terminating NRTI. One implication of this model would be that RNase H inhibitors could not be used in combination with AZT in a clinical setting. Rather than employ mutant enzymes that may have subtle effects on both catalytic activities, AZT sensitivity can now be assessed in vivo in the presence of an RNase H inhibitor. In the event that an NRTI/RNase H inhibitor combination proves deleterious, studies of Shaw-Reid et al.35 and Budihas et al.6 have shown that NNRTIs are synergistic with both diketo acid- or α-hydroxytropolone-based RNase H inhibitors, suggesting that regimens including NNRTI and RNase H inhibitors might have therapeutic benefits.
Finally, in a manner analogous to the NRTI/NNRTI combinations, the possibility of allosteric and active site RNase H inhibitors is worth considering. Recently, we reported a structure-activity relationship of several vinylogous ureas that inhibit HIV-1 RNase H activity in the sub- to low micromolar range13. Protein footprinting12, molecular modeling13 and in vitro site-directed mutagenesis (Chung, S. et al., unpublished results) collectively implicate the p51 thumb as the vinylogous urea binding site, suggesting they function by altering RT subunit or subdomain geometry. As mentioned earlier, our crystallographic analysis indicates that manicol makes multiple contacts with the highly conserved His539 of the flexible “His loop”. At the same time, modeling studies of Chung et al.13 indicate that the vinylogous urea binding site is also in the vicinity of His539. Conceivably, a bidentate inhibitor comprising these two structural classes may combine the properties of both to provide improved selectivity.
Experimental Section
General All air or moisture sensitive reactions were performed under positive pressure of nitrogen with oven-dried glassware. Anhydrous solvents such as tetrahydrofuran (THF), toluene, dichloromethane, N,N-dimethylformamide (DMF), acetonitrile, methanol and triethylamine were purchased from Sigma-Aldrich. Purifications of the compounds were performed on Waters UPLC or Biotage SP systems. Samples were analyzed for purity on an Agilent 1200 series LC/MS equipped with a Zorbax™ Eclipse XDB-C18 reverse phase (5 micron, 4.6×150 mm) column having a flow rate of 0.8 ml/min. The mobile phase was a mixture of acetonitrile and H2O each containing 0.05% trifluoroacetic acid. Gradient of 4% to 100% acetonitrile (0.05% TFA) over 7 minutes with flow rate of 0.8 ml/min using Luna C18 3 micron 3x75 mm column. All of the analogues for assay have purity greater than 95%. High resolution mass spectrometry was recorded on Agilent 6210 Time-of-Flight LC/MS system. Note: all of the final analogues are the mixture of diastereomers which are inseparable on preparative HPLC. Purified oligonucleotides for the fluorescence-based RNase H were purchased from TriLink Biotechnologies (San Diego, CA). His-p66/His-p51 HIV-1 RT, derived from the HIV-1HXB2 isolate36, was expressed and purified as previously reported. This enzyme is identical in sequence with respect to the residues discussed in modeling studies.
5,7-Dihydroxy-2-(1-hydroxy-1-methyl-2-piperidin-1-yl-ethyl)-9-methyl-1,2,3,4-tetrahydro benzocyclohepten-6-one (1). To a solution of manicol epoxide (16) (7 mg, 0.027 mmol) in acetonitrile (0.5 ml) was added piperidine (18.2 mg, 0.213 mmol, 8.0 eq.) and lithium perchlorate (5.7 mg, 0.053 mmol, 2 eq.). The mixture was refluxed for 1 h. After cooling to room temperature, another 1.5 ml acetonitrile was added and the mixture was directly subject to preparative HPLC purification to give the desired product 1 as a brownish solid (4 mg, 54%). 1H NMR (400 MHz, DMSO-d6) δ 8.83-8.58 (br.s., 1H), 7.37 (s, 1H), 5.75 (s, 1H), 5.55-5.23 (br.s., 1H), 3.62-3.40 (m, 2H), 3.29-3.17 (m, 2H), 3.13-2.96 (m, 3H), 2.92-2.67 (m, 2H), 2.59-2.50 (m, 3H), 2.41 (s, 3H), 2.05-1.85 (m, 1H), 1.85-1.68 (m, 4H), 1.67-1.57 (m, 1H), 1.52-1.37 (m, 1H), 1.29 and 1.25 (s, 3H); LC/MS: Retention time: 3.449 min; HRMS: m/z (M+H+) = 348.2176 (Calculated for C20H30NO4S = 348.2175).
5,7-Dihydroxy-2-(1-hydroxy-2-imadazol-1-yl-1-methyl-ethyl)-9-methyl-1,2,3,4-tetrahydro benzocyclohepten-6-one (2) was prepared in the same manner as 1 except by using imidazole as a nucleophile. 1H NMR (400 MHz, DMSO-d6) δ 9.03 and 9.01 (s, 1H), 7.72-7.64 (m, 2H), 7.38 and 7.36 (s, 1H), 5.25-5.03 (br.s., 1H), 4.36-4.21 (m, 2H), 3.80-3.30 (br.s., 2H), 3.20-3.00 (m, 1H), 3.00-2.60 (m, 2H), 2.44 and 2.40 (s, 3H), 2.62-2.47 (m, 1H), 2.25-1.92 (m, 1H), 1.75-1.48 (m, 1H), 1.45-1.25 (m, 1H), 1.01 and 1.00 (s, 3H) ;LC/MS: Retention time: 3.337 min; HRMS: m/z (M+H+) = 331.1651 (Calculated for C18H23N2O4 = 331.1658).
2-(2-Diethylamino-1-hydroxy-1-methyl-ethyl)-5,7-dihydroxy-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (3) was prepared in the same manner as 1 except by using diethylamine as a nucleophile. 1H NMR (400 MHz, DMSO-d6) δ 8.56-8.33 (br.s., 2H), 7.38 and 7.37 (s, 1H), 5.50-5.28 (br.s., 1H), 3.95-3.35 (m, 4H), 3.34-3.10 (m, 4H), 3.10-2.90 (m, 1H), 2.87-2.55 (m, 1H), 2.54 and 2.52 (s, 3H), 2.25-1.85 (m, 1H), 1.75-1.58 (m, 1H), 1.35-1.15 (m, 10H); LC/MS: Retention time: 3.427 min; HRMS: m/z (M+H+) = 336.2171 (Calculated for C19H30NO4 = 336.2175).
2-[2-(2-Fluoro-benzylamino)-1-hydroxy-1-methyl-ethyl]-5,7-dihydroxy-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (4) was prepared in the same manner as 1 except by using 2-fluoroaniline as a nucleophile. 1H NMR (400 MHz, DMSO-d6) δ 8.90-8.60 (br.s., 2H), 7.70-7.61 (m, 1H), 7.54-7.45 (m, 1H), 7.36 and 7.35 (s, 1H), 7.34-7.35 (m, 2H), 5.48-5.15 (br.s., 1H), 4.25 (s, 2H), 3.20-2.90 (m, 3H), 2.90-2.61 (m, 2H), 2.38 and 2.34 (s, 3H), 2.50-2.35 (m, 2H), 2.15-1.92 (m, 1H), 1.80-1.65 (m, 1H), 1.33-1.13 (m, 1H), 1.20 and 1.18 (s, 3H); LC/MS: Retention time: 3.844 min; HRMS: m/z (M+H+) = 388.1914 (Calculated for C22H27FNO4 = 388.1924).
5,7-Dihydroxy-2-(1-hydroxy-1-methyl-2-phenylamino-ethyl)-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (5) was prepared in the same manner as 1 except by using aniline as a nucleophile. 1H NMR (400 MHz, CDCl3) δ 7.50 and 7.49 (s, 1H), 7.34-7.25 (m, 2H), 7.05-6.94 (m, 3H), 3.43-3.17 (m, 2H), 3.04-2.82 (m, 2H), 2.64-2.33 (m, 2H), 2.48 and 2.46 (s, 3H), 2.23-2.03 (m, 1H), 2.02-1.84 (m, 2H), 1.53-1.39 (m, 1H), 1.38 and 1.31 (s, 3H). LC/MS: Retention time min: 4.715 min; HRMS: m/z (M+H+) = 356.1864 (Calculated for C21H26NO4 = 356.1862).
2-(2-Ethylsulfanyl-1-hydroxy-1-methyl-ethyl)-5,7-dihydroxy-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (6) was prepared in the same manner as 5 except by using ethanethiol as a nucleophile. 1H NMR (400 MHz, CDCl3) δ 7.49 and 7.48 (s, 1H), 3.28-3.24 and 3.24-3.19 (m, 1H), 3.03-2.78 (m, 2H), 2.76-2.64 (m, 2H), 2.60-2.50 (m, 1H), 2.48 and 2.47 (s, 3H), 2.16-2.02 (m, 2H), 1.83-1.71 (m, 1H), 1.70-1.59 (m, 1H), 1.56-1.25 (m, 4H), 1.40 and 1.38 (s, 3H); LC/MS: Retention time: 5.529 min; HRMS: m/z (M+H+) = 325.1466 (Calculated for C17H25O4S = 325.1474).
5,7-Dihydroxy-2-(1-hydroxy-1-methyl-2-phenylsulfanyl-ethyl)-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (7). To a solution of 16 (15 mg, 0.057 mmol) in THF (1 ml) was added thiophenol (0.20 ml, 1.94 mmol, 34 eq.) and Et3N (0.08 ml, 0.57 mmol, 10 eq.) and the solution was refluxed overnight. After cooling to room temperature, the mixture was directly purified by HPLC to give the desired product 7 as a light yellow solid (10 mg, 47%). 1H NMR (400 MHz, CDCl3) δ 7.50 and 7.49 (s, 1H), 7.46-7.39 (m, 2H), 7.33-7.18 (m, 3H), 3.37-3.10 (m, 3H), 3.05-2.91 (m, 1H), 2.91-2.76 (m, 1H), 2.67-2.50 (m, 2H), 2.48 and 2.42 (s, 3H), 1.98-1.85 (m, 1H), 1.49-1.33 (m, 1H), 1.32 and 1.26 (s, 3H); LC/MS: Retention time: 6.101 min; HRMS: m/z (M+H+) = 373.1472 (Calculated for C21H25O4S = 373.1474).
2-(2-Benzylsulfanyl-1-hydroxy-1-methyl-ethyl)-5,7-dihydroxy-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (8). To a solution of BnSH (26 mg, 0.21 mmol) in MeOH (1 ml) was added NaH (4.6 mg, 0.19 mmol) and the mixture was stirred for 10 min. 16 (5 mg, 0.019 mmol) was added and the mixture was stirred overnight at room temperature and directly purified by preparative HPLC to give the desired product 8 (3 mg, 58%). 1H NMR (400 MHz, DMSO-d6) δ 7.35 and 7.34 (s, 1H), 7.33-7.28 (m, 4H), 7.25-7.20 (m, 1H), 3.79-3.76 (m, 2H), 3.06-2.94 (m, 1H), 2.82-2.60 (m, 5H), 2.48-2.36 (m, 1H), 2.34 and 2.33 (s, 3H), 1.30-1.16 (m, 2H), 1.15 and 1.13 (s, 3H); Retention time: 6.157 min; HRMS: m/z (M+H+) = 387.1620 (Calculated for C22H27O4S = 387.1630).
2-(2-Ethanesulfonyl-1-hydroxy-1-methyl-ethyl)-5,7-dihydroxy-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (9) was prepared through oxidation of corresponding sulfide 6 with m-CPBA in CH2Cl2 at room temperature. 1H NMR (400 MHz, DMSO-d6) δ 7.36 and 7.35 (s, 1H), 3.42-3.26 (m, 2H), 3.11-2.98 (m, 1H), 2.89-2.67 (m, 2H), 2.60-2.45 (m, 4H), 2.40 and 2.39 (s, 3H), 2.02-1.79 (m, 1H), 1.77-1.65 (m, 1H), 1.45-1.12 (m, 3H), 1.07 and 1.05 (s, 3H); LC/MS: Retention time: 4.323; HRMS: m/z (M+H+) = 357.1370 (Calculated for C17H25O6S = 357.1372).
2-(2-Benzylsulfanyl-1-hydroxy-1-methyl-ethyl)-5,7-dihydroxy-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (10) was prepared by oxidation of corresponding sulfide with m-CPBA at −78°C, while 2-(2-benzenesulfinyl-1-hydroxy-1-methyl-ethyl)-5,7-dihydroxy-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (11) was prepared by oxidation using m-CPBA at room temperature. 10: 1H NMR (400 MHz, DMSO-d6) δ 7.72-7.66 (m, 2H), 7.63-7.52 (m, 3H), 7.37 and 7.36 (s, 1H), 3.23-2.80 (m, 4H), 2.62-2.50 (m, 2H), 2.42 and 2.41 (s, 3H), 2.15-1.90 (m, 3H), 1.40-1.15 (m, 4H); LC/MS: Retention time: 4.767 min; HRMS: m/z (M+H+) = 389.1422 (Calculated for C21H25O5S = 389.1423). 11: 1H NMR (400 MHz, DMSO-d6) δ 10.61 (s, 1H), 9.98 (s, 1H), 7.96-7.87 (m, 2H), 7.74-7.66 (m, 1H), 7.75-7.58 (m, 1H), 7.45-7.38 (m, 2H), 4.10-3.50 (m, 3H), 2.64-2.54 (m, 2H), 2.53 and 2.52 (s, 3H), 2.34-2.18 (m, 2H), 2.17-1.94 (m, 1H), 1.94-1.76 (m, 1H), 1.43-1.28 (m, 1H), 1.24 and 1.23 (s, 3H). LC/MS Retention time: 5.123 min; HRMS: m/z (M+H+) = 405.1354 (Calculated for C21H25O6S = 405.1360).
2-(1,2-Dihydroxy-1-methyl-ethyl)-5,7-dihydroxy-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (12). To a solution of diacetate 15 (5 mg, 0.014 mmol) in acetone/H2O (0.19 ml/0.02 ml) was added 2.5wt% tert-BuOH OsO4 solution (7.7 µl, 0.757 µmol) and N-methylmorpholine oxide (3.4 mg, 0.029 mmol) and the mixture was stirred for 3 h. EtOAc (10 ml) was added and the solution was washed with 10% aqueous Na2SO3 solution and brine. The organic layer was dried over MgSO4. After the removal of EtOAc, the crude product was redissolved in MeOH (2 ml) and the refluxed for 5 h. The solution was concentrated to 2 ml and directly subject to preparative HPLC purification to give the desired diol 12 (3 mg, 71%). 1H NMR (400 MHz, DMSO-d6) δ 7.36 and 7.35 (s, 1H), 3.42-3.27 (m, 2H), 3.11-2.97 (m, 1H), 2.89-2.65 (m, 2H), 2.58-2.44 (m, 1H), 2.39 and 2.38 (s, 3H), 2.00-1.81 (m, 1H), 1.76-1.64 (m, 1H), 1.36-1.20 (m, 2H), 1.06 and 1.05 (s, 3H); LC/MS: Retention time: 3.795 min; HRMS: m/z (M+H+) = 281.1386 (Calculated for C15H21O5 = 281.1389).
5,7-Dihydroxy-2-(1-hydroxy-ethyl)-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (13). To a solution of 15 (22 mg, 0.07 mmol) in tert-BuOH/H2O (1.8 ml/0.36 ml) was added 2.5wt% tert-BuOH OsO4 solution (0.42 ml, 0.042 mmol), NaIO4 (123 mg, 0.58 mmol) and NaHCO3 (81 mg, 0.96 mmol) and the mixture was stirred 2 h at room temperature. 10% Na2SO3 (1.5 ml) was added and the mixture was stirred for 0.5 h and extracted with CH2Cl2 (3x10 ml) and the combined organic layers were washed with brine and dried over Na2SO4. After the removal of organic solvent, the residue was purified by column chromatography (EtOAc/Hexane 1/1) to give the desired ketone 17 (20 mg, 90%). 17 was then dissolved in MeOH (2 ml) and NaBH4 (6 mg, 0.16 mmol) was added and stirred at room temperature. for 0.5 h. After the complete reduction of the ketone monitored by LC/MS, the mixture was further refluxed for 5 h and directly purified by preparative HPLC to give 13. 1H NMR (400 MHz, CDCl3) δ 7.51 and 7.50 (s, 1H), 3.84-3.77 and 3.76-3.63 (m, 1H), 3.29-3.18 (m, 1H), 3.05-2.94 (m, 2H), 2.84 and 2.79 (d, J= 3.9 Hz, 1H), 2.70-2.51 (m, 1H), 2.51 and 2.48 (s, 3H), 2.16-2.06 and 2.03-1.93 (m, 1H), 1.78-1.66 (m, 1H), 1.49-1.34 (m, 1H), 1.31 (d, J= 6.3 Hz, 3H); LC/MS: Retention time: 4.315 min; HRMS: m/z (M+H+) = 251.1280 (Calculated for C14H19O4 = 251.1283).
2-(1-Benzylamino-ethyl)-5,7-Dihydroxy-9-methyl-1,2,3,4-tetrahydrobenzocyclohepten-6-one (14). To a solution of 17 (5 mg, 0.015 mmol) in 1,2-dichloroethane (0.1 ml) was added benzylamine (1.6 µl, 0.016 mmol) and sodium triacetoxyborohydride (4.5 mg, 0.021 mmol) and the mixture was stirred overnight at room temperature. LC/MS indicated the formation of de-acetylated product 14. The mixture was directly purified by preparative HPLC to give desired product 14 (3 mg, 59%). 1H NMR (400 MHz, DMSO-d6) δ 9.03-8.85 (br.s., 1H), 8.73-8.56 (br.s., 1H), 7.63-7.51 (m, 2H), 7.51-7.40 (m, 3H), 7.37 (s, 1H), 4.37-4.14 (m, 2H), 3.45-3.27 (m, 1H), 3.20-3.00 (m, 1H), 2.97-2.63 (m, 2H), 2.58-2.44 (m, 2H), 2.39 and 2.37 (s, 3H), 2.25-2.05 (m, 1H), 2.03-1.93 and 1.92-1.83 (m, 1H), 1.42-1.28 (m, 4H). LC/MS: Retention time: 3.913 min; HRMS: m/z (M+H+) = 340.1908 (Calculated for C21H26NO3 = 340.1913).
HIV-1 RT Expression and Purification for Biochemical Analysis. (His)6-tagged p66/p51 HIV-1HXB2 RT and the RNase H-deficient mutants were expressed from recombinant Escherichia coli and purified by a combination of immobilized metal affinity and ion exchange chromatography as previously described37. Purified enzymes were stored at −20°C in a buffer of 50 mM Tris/HCl, pH 7.0, 25 mM NaCl, 1 mM EDTA, 1 mM dithiothereitol and 50% (v/v) glycerol.
RNase H Inhibitor Analysis. IC50 values were determined as previously reported6, using an 18-nucleotide 3’-fluourescein-labeled RNA annealed to a complementary 18-nucleotide 5’-dabsyl-labeled DNA. Cleavage of the HIV-1 polypurine tract (PPT) primer was performed with a 29 nt Cy5-labeled RNA (5’-Cy5-UUU UAA AAG AAA AGG GGG G*AC UGG AAG GG-3’, where *represents the PPT 3’ terminus) hybridized to a 40 nt DNA (5’-ATT AGC CCT TCC AGT CCC CCC TTT TCT TTT AAA AAG TGG C-3’). The reaction was initiated by adding 1 µL of 100 mM MgCl2 to 9 µL of mixture containing 4 ng enzyme, 200 nM substrate, 20 µM α-hydroxytropolones in 50 mM Tris, pH 8.0, 80 mM KCl, 2 mM DTT, and 10% DMSO at 37°C and quenched with 10 µL of a gel-loading buffer after 10 min. Hydrolysis products were fractionated by denaturing polyacrylamide gel electrophoresis and visualized by fluorescent imaging (Typhoon Trio+, GE Healthcare).
DNA Polymerase Assay. DNA-dependent DNA synthesis was measured on a fluorescently-labeled duplex DNA prepared by annealing a 33-nt template, 5’-CAC TGC TCA AGA AGT TCC AAT CCT AAA TAC ATA -3’, to the 5’-Cy5 -labeled primer 5’-ATG TAT GGG TAT GTA TTT AGG-3’. Polymerization was initiated by adding 1 µL of 2 mM dNTPs to 9 µL of mixture containing 4 ng enzyme, 200 nM substrate, 20 µM α-hydroxytropolones in 10 mM Tris, pH 7.8, 80 mM KCl, 1 mM DTT, 10 mM MgCl2, and 10% DMSO at 37°C. DNA synthesis was quenched with 10 µL of a gel-loading buffer after 10 min and reaction products were analyzed by denaturing polyacrylamide gel electrophoresis and fluorescent imaging.
HIV-1 Cytopathicity assay. This assay was conducted as previously reported38. Samples were dissolved in DMSO at 10 mM and diluted to a final high concentration of 50 µM in the 96-well assay plate, with 2-fold dilutions made to a low concentration of 0.78 µM. All samples were tested in duplicate. The HIV-1 virus strain RF was used to infect CEM-SS cells. Compound cytotoxicity was measured in the same assay plate using uninfected cells. Regression analysis was used to estimate the effective concentration (EC50) as well as the cytotoxic concentration (CC50).
Expression and Purification of HIV-1 RT for structural studies. An HIV-1 RT variant designated RT52A was used for X-ray diffraction studies. In this variant, which was optimized for crystallization of RT with nucleoside or non-nucleoside RT inhibitors (NRTIs and NNRTIs, respectively), the p66 subunit was truncated at residue 555. The p51 subunit contained a HRV14 3C protease cleavable N-terminal hexahistidine tag and was truncated at residue 428. Both subunits contained the mutation C280S. The p66 subunit also contained the mutations K172A and K173A39. RT52A was expressed and purified as previously described39. Briefly, 1 mM IPTG was used to induce BL21-CodonPlus®-RIL (Stratagene) containing a plasmid encoding both subunits of RT69A at an OD600 of 0.9 and the culture was incubated for three hours at 37°C. The cells were pelleted and lysed by sonication. Protein was purified by Ni-NTA according to manufacturers’ recommendations (Qiagen) with the following modifications: each buffer contained 600 mM NaCl, no lysozyme was added, and a 1.2 M NaCl wash was added. Eluted protein was incubated with HRV14 3C protease overnight at 4°C. A Mono Q purification step was performed as described40. The protein was concentrated to 20 mg/ml in "RT storage buffer" (10 mM Tris pH 8.0, 75 mM NaCl) and stored at −80°C.
Crystallization and Data Collection. HIV-1 RT52A was co-crystallized with manicol and 18 at 4°C by vapor diffusion in microseeded hanging drops containing 1.2 µl each of 20 mg/ml protein (in a solution of 9.2 mM Tris pH 8.0, 68.7 mM NaCl, 3.6 mM manganese sulfate, 0.7 mM tris(2-carboxyethyl) phosphine (TCEP), 0.27% (w/v) β-ocytl glucopyranoside, 7% (v/v) DMSO, 0.9 mM manicol, and 0.7 mM 18, pre-incubated for 30 minutes on ice) and a reservoir solution containing 50 mM HEPES pH 7.5, 100 mM ammonium sulfate, 15 mM manganese sulfate, 10 mM spermine, 5 mM TCEP, and 11% (w/w) PEG 8000. The chosen crystal was soaked for 120 seconds in a solution containing 50 mM HEPES pH 7.5, 50 mM NaCl, 100 mM ammonium sulfate, 15 mM manganese sulfate, 10 mM spermine, 15% (w/w) PEG 8000, 5% (w/w) PEG 400, 10% (v/v) DMSO, 11% (v/v) ethylene glycol, 6.5% (w/v) trimethylamine-N-oxide (TMAO), 0.69 mM manicol, and 0.34 mM 18. The crystal was subsequently flash-cooled and stored in liquid N2. X-ray data were collected at 100K and a wavelength of 1.1 Å at the National Synchrotron Light Source at Brookhaven National Laboratories, Beamline X25. The data were processed using the HKL-DENZO/SCALEPACK software suite41, 42.
Structure Determination and Refinement. Phases for the diffraction data were determined by molecular replacement with the CCP4 program PHASER43, using an RT/18 structure (PDB accession number 2ZD1)44 as an initial search model. Stepwise model building and refinement were conducted using the "O" graphics package45, the Coot graphics package46, and CNS47 with a bulk solvent correction (Table 1). Water molecules were built in both manually in Coot and using the program Refmac/ARP/wARP in the CCP4 software suite48–52. The geometry of the inhibitor and refinement of the RNH active site were improved by energy minimization using the Impact and PrimeX facilities of the Schrödinger software package (Schrödinger, LLC).
Molecular Modeling. A model of the HIV-1 RNase H domain in complex with an inhibitor and two Mn2+ cations was constructed from the HIV-1 RT-manicol co-crystal structure using Discovery Studio 2.0 (Accelrys, San Diego, CA). To create the starting structure for energy minimization, RT was truncated to include only the RNase H domain, the inhibitors were constructed directly from manicol using various “build” functions, and the CHARMm forcefield was applied to the entire structure via the “Simulation” tool. The modified structure was then subjected to two rounds of energy minimization (500 iterations each). Inhibitor atoms outside of the tropolone and cyclohexane rings, as well as the entire “His-loop” and side chains of α-helix E’, were permitted full flexibility according to the dictates of the minimization algorithm. The positions of other atoms were held fixed in order to minimize perturbation of the overall complex. Minimized structures were saved as PDB files and imported into PyMOL (Delano Scientific, LLC, San Francisco, CA) to generate the final image.
Acknowledgements
SC, JR, JB, JW and SLG are supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health. JJ and CT were supported by the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research and the Intramural Research Program of the National Human Genome Research Institute. DMH and EA were supported by NIH MERIT Award AI 27690.
Abbreviations Used
- RNase
Ribonuclease
- HIV
Human immunodeficiency virus
- RT
Reverse transcriptase
- NNRTI
nonnucleoside RT inhibitor
- NRTI
Nucleoside RT inhibitor
- AIDS
Acquired immunodeficiency syndrome
- HAART
Highly active antiretroviral therapy
- m-CPBA
meta-Chloroperbenzoic acid
- NMO
N-Methyl morpholine oxide
- PPT
Polypurine tract
- AZT
Azidothymidine
- DMSO
Dimethyl sulfoxide
- THF
Tetrahydrofuran
- DMF
N,N-Dimethyl formamide
References
- 1.Champoux JJ. Roles of Ribonuclease H in Reverse Transcription. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1993. pp. 103–118. [Google Scholar]
- 2.Di Marzo Veronese F, Copeland TD, DeVico AL, Rahman R, Oroszlan S, Gallo RC, Sarngadharan MG. Characterization of Highly Immunogenic p66/p51 as the Reverse Transcriptase of HTLV-III/LAV. Science (Washington, D. C., 1883-) 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- 3.Mous J, Heimer EP, Le Grice SF. Processing protease and reverse transcriptase from human immunodeficiency virus type I polyprotein in Escherichia coli. J Virol. 1988;62:1433–1436. doi: 10.1128/jvi.62.4.1433-1436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schatz O, Cromme FV, Gruninger-Leitch F, Le Grice SF. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989;257:311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- 5.Tisdale M, Schulze T, Larder BA, Moelling K. Mutations within the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase abolish virus infectivity. J Gen Virol. 1991;72(Pt 1):59–66. doi: 10.1099/0022-1317-72-1-59. [DOI] [PubMed] [Google Scholar]
- 6.Budihas SR, Gorshkova I, Gaidamakov S, Wamiru A, Bona MK, Parniak MA, Crouch RJ, McMahon JB, Beutler JA, Le Grice SF. Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 2005;33:1249–1256. doi: 10.1093/nar/gki268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didierjean J, Isel C, Querre F, Mouscadet JF, Aubertin AM, Valnot JY, Piettre SR, Marquet R. Inhibition of human immunodeficiency virus type 1 reverse transcriptase, RNase H, and integrase activities by hydroxytropolones. Antimicrob Agents Chemother. 2005;49:4884–4894. doi: 10.1128/AAC.49.12.4884-4894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dat NT, Bae K, Wamiru A, McMahon JB, Le Grice SF, Bona M, Beutler JA, Kim YH. A dimeric lactone from Ardisia japonica with inhibitory activity for HIV-1 and HIV-2 ribonuclease H. J Nat Prod. 2007;70:839–841. doi: 10.1021/np060359m. [DOI] [PubMed] [Google Scholar]
- 9.Marchand C, Beutler JA, Wamiru A, Budihas S, Mollmann U, Heinisch L, Mellors JW, Le Grice SF, Pommier Y. Madurahydroxylactone derivatives as dual inhibitors of human immunodeficiency virus type 1 integrase and RNase H. Antimicrob Agents Chemother. 2008;52:361–364. doi: 10.1128/AAC.00883-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takada K, Bermingham A, O'Keefe BR, Wamiru A, Beutler JA, Le Grice SF, Lloyd J, Gustafson KR, McMahon JB. An HIV RNase H inhibitory 1,3,4,5-tetragalloylapiitol from the African plant Hylodendron gabunensis. J Nat Prod. 2007;70:1647–1649. doi: 10.1021/np0702279. [DOI] [PubMed] [Google Scholar]
- 11.Kirschberg TA, Balakrishnan M, Squires NH, Barnes T, Brendza KM, Chen X, Eisenberg EJ, Jin W, Kutty N, Leavitt S, Liclican A, Liu Q, Liu X, Mak J, Perry JK, Wang M, Watkins WJ, Lansdon EB. RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information. J Med Chem. 2009;52:5781–5784. doi: 10.1021/jm900597q. [DOI] [PubMed] [Google Scholar]
- 12.Wendeler M, Lee HF, Bermingham A, Miller JT, Chertov O, Bona MK, Baichoo NS, Ehteshami M, Beutler J, O'Keefe BR, Gotte M, Kvaratskhelia M, Le Grice S. Vinylogous ureas as a novel class of inhibitors of reverse transcriptase-associated ribonuclease H activity. ACS Chem Biol. 2008;3:635–644. doi: 10.1021/cb8001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung S, Wendeler M, Rausch JW, Beilhartz G, Gotte M, O'Keefe BR, Bermingham A, Beutler JA, Liu SX, Zhuang XW, Le Grice SFJ. Structure-Activity Analysis of Vinylogous Urea Inhibitors of Human Immunodeficiency Virus-Encoded Ribonuclease H. Antimicrobial Agents and Chemotherapy. 2010;54:3913–3921. doi: 10.1128/AAC.00434-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Grandi M, Olson M, Prashad AS, Bebernitz G, Luckay A, Mullen S, Hu YB, Krishnamurthy G, Pitts K, O'Connell J. Small molecule inhibitors of HIV RT Ribonuclease H. Bioorganic & Medicinal Chemistry Letters. 2010;20:398–402. doi: 10.1016/j.bmcl.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 15.Himmel DM, Maegley KA, Pauly TA, Bauman JD, Das K, Dharia C, Clark AD, Jr, Ryan K, Hickey MJ, Love RA, Hughes SH, Bergqvist S, Arnold E. Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site. Structure. 2009;17:1625–1635. doi: 10.1016/j.str.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himmel DM, Sarafianos SG, Dharmasena S, Hossain MM, McCoy-Simandle K, Ilina T, Clark AD, Jr, Knight JL, Julias JG, Clark PK, Krogh-Jespersen K, Levy RM, Hughes SH, Parniak MA, Arnold E. HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. ACS Chem Biol. 2006;1:702–712. doi: 10.1021/cb600303y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klumpp K, Mirzadegan T. Recent progress in the design of small molecule inhibitors of HIV RNase H. Curr Pharm Des. 2006;12:1909–1922. doi: 10.2174/138161206776873653. [DOI] [PubMed] [Google Scholar]
- 18.Su HP, Yan YW, Prasad GS, Smith RF, Daniels CL, Abeywickrema PD, Reid JC, Loughran HM, Kornienko M, Sharma S, Grobler JA, Xu B, Sardana V, Allison TJ, Williams PD, Darke PL, Hazuda DJ, Munshi S. Structural Basis for the Inhibition of RNase H Activity of HIV-1 Reverse Transcriptase by RNase H Active Site-Directed Inhibitors. Journal of Virology. 2010;84:7625–7633. doi: 10.1128/JVI.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa Y, Tayama K. Mechanism of mitochondrial dysfunction and cytotoxicity induced by tropolones in isolated rat hepatocytes. Chemico-Biological Interactions. 1998;116:45–60. doi: 10.1016/s0009-2797(98)00078-7. [DOI] [PubMed] [Google Scholar]
- 20.Boone LR. Next-generation HIV-1 non-nucleoside reverse transcriptase inhibitors. Current Opinion in Investigational Drugs. 2006;7:128–135. [PubMed] [Google Scholar]
- 21.Polonsky J, Beloeil JC, Prange T, Pascard C, Jacquemin H, Donnelly DMX, Kenny PTM. Manicol - a Sesquiterpenoid Hydroxytropolone from Dulacia-Guianensis - a Revised Structure (X-Ray-Analysis) Tetrahedron. 1983;39:2647–2655. [Google Scholar]
- 22.Yang W, Hendrickson WA, Crouch RJ, Satow Y. Structure of Ribonuclease-H Phased at 2-a Resolution by Mad Analysis of the Selenomethionyl Protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 23.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luzzati V. Traitement Statistique Des Erreurs Dans La Determination Des Structures Cristallines. Acta Crystallographica. 1952;5:802–810. [Google Scholar]
- 25.Baker EN. Hydrogen bonding in biological marcomolecules. In: Rossman MG, Arnold E, editors. International Tables for Crystallography, Volume F: Crystallography of Biological Macromolecule. Boston: Kluwer Academic Publishers; 2001. pp. 546–552. [Google Scholar]
- 26.Bhattacharyya R, Saha RP, Samanta U, Chakrabarti P. Geometry of interaction of the histidine ring with other planar and basic residues. Journal of Proteome Research. 2003;2:255–263. doi: 10.1021/pr025584d. [DOI] [PubMed] [Google Scholar]
- 27.Chakrabarti P, Bhattacharyya R. Geometry of nonbonded interactions involving planar groups in proteins. Progress in Biophysics & Molecular Biology. 2007;95:83–137. doi: 10.1016/j.pbiomolbio.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Schottel BL, Chifotides HT, Dunbar KR. Anion-pi interactions. Chemical Society Reviews. 2008;37:68–83. doi: 10.1039/b614208g. [DOI] [PubMed] [Google Scholar]
- 29.Estarellas C, Frontera A, Quinonero D, Deya PM. Relevant Anion-pi Interactions in Biological Systems: The Case of Urate Oxidase. Angewandte Chemie-International Edition. 2011;50:415–418. doi: 10.1002/anie.201005635. [DOI] [PubMed] [Google Scholar]
- 30.Parniak MA, Min KL, Budihas SR, Le Grice SFJ, Beutler JA. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Analytical Biochemistry. 2003;322:33–39. doi: 10.1016/j.ab.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Gopalakrishnan V, Peliska JA, Benkovic SJ. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hang JQ, Li Y, Yang Y, Cammack N, Mirzadegan T, Klumpp K. Substrate-dependent inhibition or stimulation of HIV RNase H activity by non-nucleoside reverse transcriptase inhibitors (NNRTIs) Biochem Biophys Res Commun. 2007;352:341–350. doi: 10.1016/j.bbrc.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 33.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 34.Nikolenko GN, Palmer S, Maldarelli F, Mellors JW, Coffin JM, Pathak VK. Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: Balance between RNase H activity and nucleotide excision. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2093–2098. doi: 10.1073/pnas.0409823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw-Reid CA, Munshi V, Graham P, Wolfe A, Witmer M, Danzeisen R, Olsen DB, Carroll SS, Embrey M, Wai JS, Miller MD, Cole JL, Hazuda DJ. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J Biol Chem. 2003;278:2777–2780. doi: 10.1074/jbc.C200621200. [DOI] [PubMed] [Google Scholar]
- 36.Ratner L, Fisher A, Jagodzinski LL, Mitsuya H, Liou RS, Gallo RC, Wongstaal F. Complete Nucleotide-Sequences of Functional Clones of the Aids Virus. Aids Research and Human Retroviruses. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 37.Le Grice SF, Gruninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem. 1990;187:307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- 38.Weislow OS, Kiser R, Fine DL, Bader J, Shoemaker RH, Boyd MR. New soluble-formazan assay for HIV-1 cytopathic effects: application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J Natl Cancer Inst. 1989;81:577–586. doi: 10.1093/jnci/81.8.577. [DOI] [PubMed] [Google Scholar]
- 39.Bauman JD, Das K, Ho WC, Baweja M, Himmel DM, Clark AD, Jr, Oren DA, Boyer PL, Hughes SH, Shatkin AJ, Arnold E. Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design. Nucleic Acids Res. 2008;36:5083–5092. doi: 10.1093/nar/gkn464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark AD, Jr, Jacobo-Molina A, Clark P, Hughes SH, Arnold E. Crystallization of human immunodeficiency virus type 1 reverse transcriptase with and without nucleic acid substrates, inhibitors, and an antibody Fab fragment. Methods Enzymol. 1995;262:171–185. doi: 10.1016/0076-6879(95)62017-6. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. DENZO and SCALEPACK. In: Rossman MG, Arnold E, editors. International Tables for Crystallography Volume F: Crystallography of Biological Macromolecules. First ed. Boston: Kluwer Academic Publishers; 2001. pp. 226–235. [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. In: Charles W, Carter J, Sweet RM, editors. Methods in Enzymology: Marcomolecular Crystallography Part A. First ed. Vol. 276. New York: Academic Press, Inc; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 43.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 44.Das K, Bauman JD, Clark AD, Jr, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci U S A. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 49.Lamzin VS, Perrakis A, Wilson KS. The ARP/wARP Suite for Automated Construction and Refinement of Protein Models. In: Rossman MG, Arnold E, editors. International Tables for Crystallography Volume F: Crystallography of Biological Macromolecules. First ed. Boston: Kluwer Academic Publishers; 2001. pp. 720–722. [Google Scholar]
- 50.Perrakis A, Sixma TK, Wilson KS, Lamzin VS. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 51.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 52.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 53.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Hizi A, Hughes SH, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]