Abstract
Genetic regulation of mammalian heart size is poorly understood. Hippo signaling represents an organ-size control pathway in Drosophila, where it also inhibits cell proliferation and promotes apoptosis in imaginal discs. To determine whether Hippo signaling controls mammalian heart size, we inactivated Hippo pathway components in the developing mouse heart. Hippo-deficient embryos had overgrown hearts with elevated cardiomyocyte proliferation. Gene expression profiling and chromatin immunoprecipitation revealed that Hippo signaling negatively regulates a subset of Wnt target genes. Genetic interaction studies indicated that β-catenin heterozygosity suppressed the Hippo cardiomyocyte overgrowth phenotype. Furthermore, the Hippo effector Yap interacts with β-catenin on Sox2 and Snai2 genes. These data uncover a nuclear interaction between Hippo and Wnt signaling that restricts cardiomyocyte proliferation and controls heart size.
In mammals, organ-extrinsic influences such as nutritional status, circulating growth factors, and hormones have a large impact on organ size control (1). Several lines of evidence indicate that there are also organ-intrinsic mechanisms to modulate organ size (1, 2). Insight into genetic pathways regulating organ size has come from Drosophila, where two main organ-size control pathways are bone morphogenetic protein (Bmp) and Hippo signaling pathways (3, 4). Whether these growth control mechanisms are broadly conserved in mammalian organs remains unclear.
Organ size is important in cardiac development, because the heart must be large enough to generate a physiological cardiac output but not so large as to block cardiac outflow, as in obstructive cardiomyopathies. The mammalian core Hippo signaling components include Ste20 family kinases Mst1 and Mst2, which are homologous to Drosophila Hippo. Mst kinases form an active complex with WW repeat scaffolding protein Salvador (Salv), also called WW45, that phosphorylates large tumor suppressor homolog (Lats) kinase. Mammals have two Lats genes, Lats1 and Lats2, which are homologous to Drosophila Warts. Lats kinases complex with Mob to phosphorylate Yap and Taz, two related transcriptional coactivators. Upon phosphorylation, Yap and Taz, the most downstream Hippo signaling components, are excluded from the nucleus and are transcriptionally inactive.
To determine whether the Hippo signaling pathway functions in the determination of mammalian heart size, we inactivated the single mammalian Salv ortholog in the mouse heart by using a Salv conditional null allele and the Nkx2.5 cre allele that directs cardiac cre activity (5–7). Yap phosphorylation at Ser127 (pYAP) was examined in Nkx2.5cre:Salvf/f (Salv CKO) mutants via Western blot and immunofluorescence (IF) (fig. S1). IF of embryonic day 9.5 (E9.5) sagittal sections revealed pYap signal in both second heart field (SHF) progenitors and outflow tract and ventricular cardiomyocytes (fig. S1, A to D). Salv CKO hearts had reduced pYAP but no change in total Yap, revealing that Hippo signaling is active in the developing heart and that Salv deletion reduces cardiac Hippo activity (fig. S1, E and F).
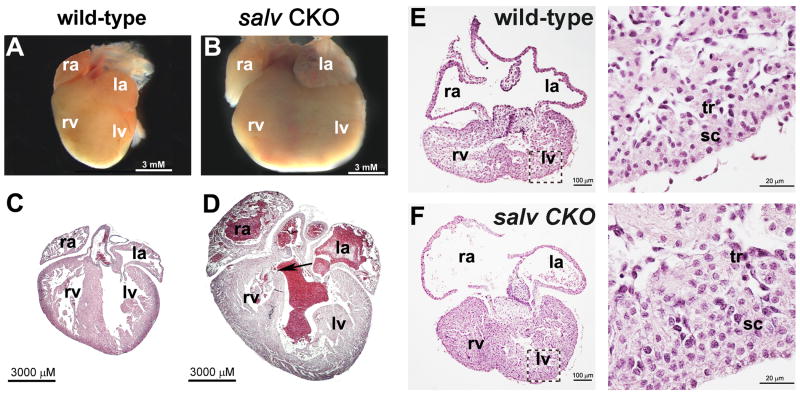
Salv CKO mutants survived development, but most mutants expired postnatally with obvious heart enlargement or cardiomegaly (Fig. 1, A and B, and table S1). Histological examination of Salv CKO mutant hearts revealed that, although organ size is affected, arterioventricular connections and arrangement of chambers and valves were unaffected (Fig. 1, C and D), consistent with preserved patterning observed in Hippo mutant imaginal discs (4). Hearts of some Salv CKO mutant animals had a ventricular septal defect (VSD), indicating that Hippo signaling regulates ventricular septation (Fig. 1D). Because VSD can cause heart failure, we confined further analysis of Salv CKO mutants to stages before ventricular septation is completed, E14.5 and earlier stages.
Fig. 1.
Salvador mutant cardiomegaly. (A to D) Control [(A) and (C)] and Salv CKO [(B) and (D)] P2 neonate hearts. ra indicates right atrium; la, left atrium; rv, right ventricle; lv, left ventricle. Hearts in (A) and (B) were sectioned and stained with hematoxylin and eosin (H&E), as shown in (C) and (D). Arrow, ventricular septum defect. (E and F) H&E-stained control (E) and Salv CKO (F) hearts. High magnification of (E) and (F) are shown in the right-hand images; subcompact, sc; trabecular, tr myocardium. Control genotype is Nkx2.5cre; Salvf/+.
Salv CKO mutant hearts had expansion of trabecular and subcompact ventricular myocardial layers, thickened ventricular walls, and enlarged ventricular chambers without a change in myocardial cell size (Fig. 1, E and F, and fig. S2). Lats2 and Mst1/2 CKO E11.5 mutant hearts had similar myocardial expansion phenotypes (fig. S3).
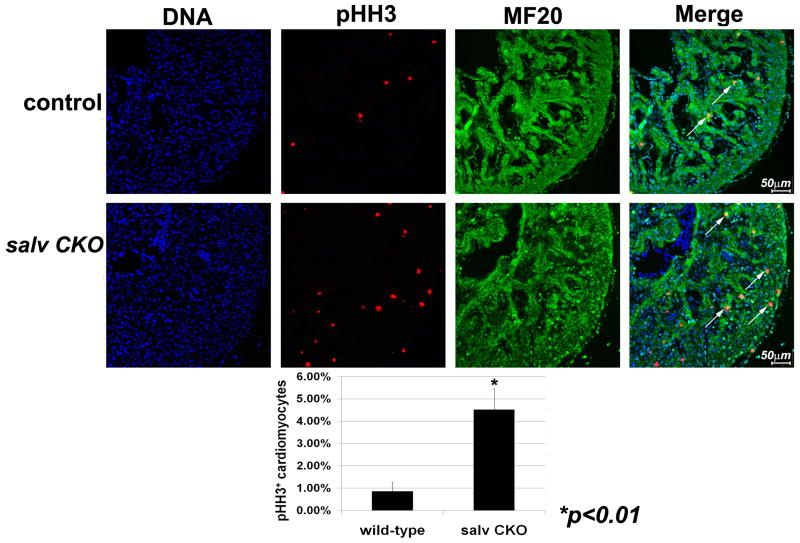
We investigated cardiomyocyte proliferation by double immunostaining with phosphorylated Ser10 histone H3 (pHH3) antibodies to detect mitotic cells and sarcomeric myosin (α-MF20) (Fig. 2, top and middle). Less than 1% of control ventricular cardiomyocytes were pHH3-positive, whereas about 4.5% of Salv CKO mutant cardiomyocytes were pHH3-positive, indicating excessive cardiac proliferation in cardiomyocytes (Fig. 2, bottom). Cardiomyocyte proliferation was elevated in both left and right Salv CKO ventricles. Cell counting indicated elevated ventricular cardiomyocte number in Salv CKO mutants but no changes in ventricular smooth muscle (α-SMA) or cardiac fibroblasts (α-DDR2) (fig. S4).
Fig. 2.
Cardiomyocyte proliferation in Salvador mutant ventricles. E12.5 control (top) and Salv CKO (middle) coronal sections of left ventricles: stain with TO-PRO-3, blue; α-pHH3, red; and α-MF20, green. Arrows, pHH3/MF20-positive cells. (Bottom) pHH3-positive cardiomyocytes quantification. Control genotype is Nkx2.5cre; Salvf/+.
SHF cardiac progenitors that contribute to right ventricle had normal cellular proliferation levels in Salv CKO, Lats2 CKO, and Mst1/2 CKO mutants, indicating that Salv CKO mutant cardiomegaly was unlikely to be due to elevated cardiac progenitor number (fig. S5).
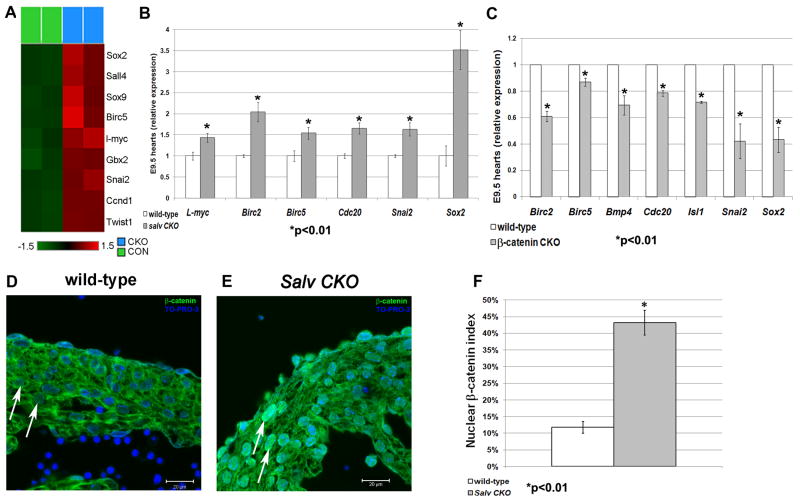
Microarray revealed that known canonical Wnt target genes were up-regulated in Salv CKO embryos (Fig. 3A). This Wnt-Hippo signature included Sox2, which has been implicated in cardiac repair and cell reprogramming (8, 9); Snai2/Slug, a tumorigenesis factor having roles in epithelial-mesenchymal transition and cell growth (10); and the anti-apoptosis gene Birc5/Survivin (11).
Fig. 3.
Salvador deletion potentiates canonical Wnt signaling. Heat map (A) and qRT-PCR validation (B) showing relative transcript levels of Wnt/β-catenin target genes. Values were determined as a mean of three samples ± SD with glyceraldehyde-3-phosphate dehydrogenase control. (C) qRT-PCR of Wnt/β-catenin target genes in β-catenin CKO hearts. (D and E) IF images with quantification (F) of E12.5 heart sections: stain with TO-PRO-3, blue, and α-β-catenin, green. Nuclear β-catenin identified by β-catenin/TO-PRO-3 signal overlap (arrows). Control genotype is Nkx2.5cre; Salvf/+.
Reverse transcription quantitative polymerase chain reaction (qRT-PCR) validated up-regulated expression of Sox2 and Snai2, as well as cdc20, l-Myc, Birc2, and Birc5, in Salv CKO embryonic hearts (Fig. 3B). With the exception of l-Myc, reduced expression was observed in Mef2ccre; β-cateninf/f mutants (Fig. 3C). Other β-catenin–regulated genes, such as Fgf10 and Isl1, were unchanged in Salv CKO hearts, indicating incomplete overlap between Yap targets and β-catenin targets (fig. S6). In situ hybridization revealed Sox2 and Snai2 up-regulation in Salv CKO E12.5 hearts (fig. S6).
E12.5 hearts were immunostained with β-catenin–specific antibodies to assay the nuclear β-catenin index, a readout for cells receiving a Wnt signal. Whereas β-catenin localized to plasma membrane junctions and cytosol of control myocardium, Salv CKO mutants had a fourfold increase in nuclear β-catenin staining (Fig. 3, D to F). These findings indicate that canonical Wnt signaling in cardiomyocytes is derepressed upon Salv deletion and support the notion that Hippo signaling inhibits Wnt/β-catenin to regulate heart size.
To obtain genetic evidence that Hippo signaling negatively regulates Wnt/ β-catenin–dependent cardiac growth, we crossed Salv CKO mice to a β-catenin conditional null allele to generate Nkx2.5cre; Salvf/f;βcatf/+ (Salv/βcatf/+ CKO) embryos that are Hippo-deficient with reduced β-catenin dosage.
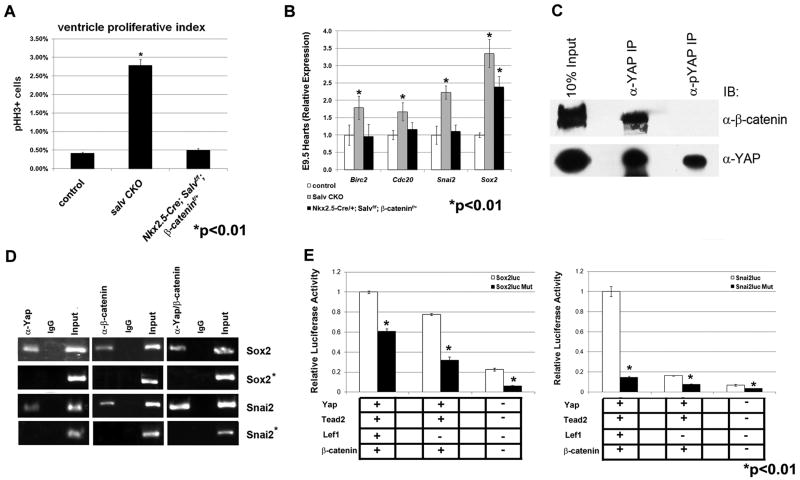
Myocardial thickness of Salv/βcatf/+ CKO hearts was significantly reduced by comparison to Salv CKO (figs. S7, B and C, and S2). Ventricular proliferation rates, as well as subcompact and trabecular myocardial thickness, in Salv/βcatf/+ CKO; mutants resemble those of control (Fig. 4A and figs. S2 and S7, A and C). qRT-PCR indicated that Sox2, Snai2, Birc2, and Cdc20 expression levels reverted to control or were lower in hearts with reduced β-catenin dosage (Fig. 4B). Suppression of the Hippo myocardial overgrowth phenotype by reduced β-catenin indicates that Wnt signaling is required for up-regulated cardiomyocyte proliferation and cardiomegaly in Hippo mutants.
Fig. 4.
Yap interacts with β-catenin. (A) Quantification of pHH3 indices in control, salv CKO, and Nkx2.5cre; Salvf/f; β-catenin F/+ E12.5 hearts. (B) qRT-PCR with indicated genes. (C) Immunoprecipitation/Western with indicated antibodies. IB, immunoblot. (D) ChIP and sequential ChIP with indicated antibodies. Control ChIP sites proximal to Sox2 and Snai2 loci were tested (asterisks). Refer to fig. S7D for ChIP assay design. IgG, immunoglobulin G. (E) Luciferase reporter assays with co-transfected factors.
To investigate whether Yap and β-catenin are in the same molecular complex, we immunoprecipitated protein extracts from E14.5 hearts with Yap and pYAP-specific antibodies (Fig. 4C). Western blotting with antibodies against β-catenin revealed that β-catenin forms a complex only with nuclear, nonphosphorylated Yap, indicating that the Yap/β-catenin molecular interaction is nuclear (Fig. 4C).
Because Yap associates with DNA-binding Tead/Tef transcription factors whereas β-catenin binds to Lef/Tcf factors (12, 13), we examined surrounding genomic sequence of genes within the Wnt-Hippo expression signature for Tead/Tef and Lef/Tcf binding sites. For Sox2 and Snai2, candidate Yap/Tead binding sites were identified both upstream and downstream of open reading frames (fig. S7D). Conserved Tcf/Lef binding elements (CTTTG) were in close proximity to Sox2 and Snai2 downstream candidate Yap/Tead sites (fig. S7D).
Chromatin immunoprecipitation (ChIP) revealed that Yap and β-catenin were recruited to Sox2 and Snai2 downstream regions but not upstream sites (Fig. 4D). Sequential ChIP revealed that Yap and β-catenin concurrently occupied Sox2 and Snai2, suggesting that Yap and β-catenin are contained within a common regulatory complex on Sox2 and Snai2 (Fig. 4D).
Transfection experiments indicated that Yap and β-catenin transfection along with their DNA binding co-factors induced luciferase expression from both Sox2 and Snai2 reporter plasmids. Luciferase reporter activity was significantly reduced by preventing Yap or β-catenin recruitment to the reporter either individually or concurrently (Fig. 4E). These findings support the model that Yap and β-catenin recruitment to Sox2 and Snai2 chromatin through their respective DNA binding partners potentiates transcriptional activity.
The Yap and β-catenin interaction on genes such as Snai2 and Sox2 uncovers a nuclear mechanism for antagonistic control of cardiomyocyte growth by Hippo and canonical Wnt signaling. Our model is supported by genetic suppression of Hippo-enlarged hearts by reduced β-catenin. Hippo signaling inhibits a pro-growth Wnt/β-catenin-Yap interaction in differentiating cardiomyocytes as the heart transitions from rapid progenitor cell growth to more measured growth in the maturing heart.
Although our pYap data indicate that there is some Hippo activity in E9.5 SHF cardiac progenitors, cell proliferation in Hippo mutant SHF was unchanged from control. This may reflect Salv-independent Hippo activity and perhaps overlapping functions with other Hippo pathway kinases (i.e., Lats1). Our data support the previous observation that Hippo signaling was low in E10.5 myocardium (14). Although Salv CKO cardiomyocyte cell size was unchanged, this question requires further study under stressed conditions (15).
Hippo regulates growth and progenitor genes like Sox2, Snai2, Ccdn1, Cdc20, and l-Myc in cardiomyocytes. In livers overexpressing Yap and Hippo loss-of-function mutants, expression of c-Myc and Ccdn1 is up-regulated, suggesting shared mechanisms between liver and heart (4). Although apoptosis inhibitors Birc2 and Birc5 were up-regulated in Salv CKO mutant hearts, apoptosis was unchanged.
Important recent work uncovered a repressive cytoplasmic interaction between pTaz and Dvl in kidney (16). Our findings, uncovering a nuclear interaction between Wnt and Hippo, suggest a two-tiered mechanism by which Hippo negatively modulates Wnt signaling in multiple contexts (fig. S8).
Wnt signaling has distinct functions in the two cardiac progenitor fields (13). Nonetheless, we find no proliferation difference between first heart field–derived left ventricle and SHF-derived right ventricle in Salv CKO hearts, indicating shared organ size-control mechanisms for the two myocardial lineages. Also Wnt signaling is low in myocardium even though Wnt ligands are expressed (17). Our findings suggest that, in Hippo-low cardiac progenitors, progrowth genes regulated by Hippo and Wnt are actively transcribed. In cardiomyocytes, Hippo signaling restricts Yap from the nucleus, resulting in diminution of Hippo-Wnt–regulated genes.
Supplementary Material
Acknowledgments
We thank J. Epstein, S. Sasakura, T. Gridley, M. Sudol, F. Long, and B. Amendt for reagents. Supported by NIH grants (J.F.M. and R.L.J.), T32 DE15355-04 (M.B.C.), R01HD052785 and R01HD060579 (R.L.J.), AHA09PRE2150024 (J.W.), and AHA10POST4140029 (T.H.). Microarray data (accession number GSE27259) can be accessed at the Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/geo/).
Footnotes
Supporting Online Material: www.sciencemag.org/cgi/content/full/332/6028/458/DC1, Materials and Methods, Figs. S1 to S9, Tables S1 and S2, References
References and Notes
- 1.Conlon I, Raff M. Cell. 1999;96:235. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 2.Stanger BZ, Tanaka AJ, Melton DA. Nature. 2007;445:886. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 3.Affolter M, Basler K. Nat Rev Genet. 2007;8:663. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 4.Pan D. Dev Cell. 2010;19:491. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu L, et al. Proc Natl Acad Sci USA. 2010;107:1437. [Google Scholar]
- 6.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Genesis. 2001;31:176. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 7.Materials and methods are available as supporting material on Science Online.
- 8.Koyanagi M, et al. Circ Res. 2010;106:1290. doi: 10.1161/CIRCRESAHA.109.206045. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Cell. 2006;126:663. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Cano A, et al. Nat Cell Biol. 2000;2:76. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 11.Islam A, et al. Oncogene. 2000;19:617. doi: 10.1038/sj.onc.1203358. [DOI] [PubMed] [Google Scholar]
- 12.Wu S, Liu Y, Zheng Y, Dong J, Pan D. Dev Cell. 2008;14:388. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Tzahor E. Dev Cell. 2007;13:10. doi: 10.1016/j.devcel.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Ota M, Sasaki H. Development. 2008;135:4059. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 15.Matsui Y, et al. Circ Res. 2008;103:1309. doi: 10.1161/CIRCRESAHA.108.180042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varelas X, et al. Dev Cell. 2010;18:579. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Cohen ED, Tian Y, Morrisey EE. Development. 2008;135:789. doi: 10.1242/dev.016865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.