Abstract
Understanding the dynamics of the actin filament is essential to a detailed description of their interactions and role in the cell. Previous studies have linked the dynamic properties of actin filaments (F-actin) to three structural elements contributing to a hydrophobic pocket, namely, the hydrophobic loop, the DNase I binding loop, and the C-terminus. Here, we examine how these structural elements are influenced by factors that stabilize or destabilize F-actin, using site-directed spin-labeled (SDSL) electron paramagnetic resonance (EPR), fluorescence, and cross-linking techniques. Specifically, we employ cofilin, an actin destabilizing protein that binds and severs filaments, and phalloidin, a fungal toxin that binds and stabilizes F-actin. We find that cofilin shifts both the DNase I binding loop and the hydrophobic loop away from the C-terminus in F-actin, as demonstrated by weakened spin–spin interactions, and alters the environment of spin probes on residues of these two loops. In contrast, although phalloidin strongly stabilizes F-actin, it causes little or no local change in the environment of the loop residues. This indicates that the stabilizing effect of phalloidin is achieved mainly through constraining structural fluctuations in F-actin and suggests that factors and interactions that control these fluctuations have an important role in the cytoskeleton dynamics.
Actin is an essential protein involved in muscle contraction and many cellular processes. Monomeric actin (G-actin)1 polymerizes into a helical, filamentous (F-actin) form, with more than 150 proteins and binding factors influencing its ability to assemble, bundle, branch, or depolymerize (1, 2). Because actin’s polymerization is incompatible with crystallization, only models of F-actin structure are available. The original and prevalent model of F-actin structure, the “Holmes model”, was constructed by fitting an atomic-resolution G-actin structure to X-ray data obtained from oriented actin fibers (3). There are two distinguishing features of this model. First, there is little change in the body of the actin monomer between the monomeric and polymeric states. Second, the DNase I binding loop of one monomer (residues 38–52) interacts with the C-terminus of the monomer above it in the filament, and with a hydrophobic loop (residues 262–274) on a second monomer, across the actin helix. These cross-monomer interactions were proposed to be important in the establishment of actin filament stability.
Electron microscopy studies and reconstruction of filament images confirm the premise of the Holmes model, that actin polymerization does not involve large conformational changes in the monomer. However, these studies (4) and the more recent analysis of filament structures by Cong et al. (5) revealed that filaments exist in numerous conformational states and are highly dynamic and that subdomain 2 in particular exhibits angular disorder. Other studies have also demonstrated the importance of the hydrophobic loop in filament formation and stability. For example, in yeast actin mutant L180C/L269C/C374A, the loop can be locked to the monomer backbone by disulfide cross-linking, resulting in either filament disruption or inhibition of polymerization, depending on when the cross-linking was achieved (6).
It has been difficult to determine definitively the nature of hydrophobic loop dynamics and its role in filament formation using EM and cross-linking approaches alone. Our subsequent work, using site-directed spin labeling (SDSL) to examine hydrophobic loop dynamics (7), suggested that although the loop existed mainly in a “parked state” along the body of the actin monomer, it could extend part way into the interstrand space along the lines originally suggested by Holmes. Such multiple states of the hydrophobic loop have been revealed also in the structural analysis of actin bundles (5). Structural (8) and computational (9) studies of the DNase I loop have shown that it can exist in either a loop or a helical conformation, but this finding has not been confirmed in another crystallographic investigation (10).
Actin filament stability can be influenced by a change in the state of the bound nucleotide or the interaction of the filament with various actin-binding proteins or drugs. It is likely that these stability changes involve changes in the interaction of the two loops and the C-terminus with one another. This hypothesis is supported by the observation that both the hydrophobic and DNase I loops are dynamic elements in both G-actin (7, 11, 12) and F-actin (7, 12, 13). EM evidence has also shown an angular disorder of subdomain 2 (which contains the DNase I loop) in F-actin (4), and the ability of ATP or ADP-Pi, as opposed to ADP, to stabilize both subdomain 2 and the filament (12, 14).
In this study, we have further investigated the relationship of the dynamics of these structural elements to filament stability using the protein cofilin, an F-actin severer and destabilizer, and the cyclic peptide toxin phalloidin, which stabilizes F-actin. These two factors bind to F-actin in a mutually exclusive manner (15), but the structural basis of their competition has not been elucidated yet. Phalloidin is believed to decrease filament flexibility, or “breathing”, by enhancing interprotomer contacts in the actin helix. Our previous results (7) showed that the level of cross-linking between the wild-type (WT) C374 of one monomer and a substituted cysteine at either residue 265 or 266 in the hydrophobic loop of a second monomer was increased by phalloidin, whereas it was decreased for residues 267 and 269. This result suggested that although phalloidin does not bind near the hydrophobic loop, it can propagate an allosteric change in the filament that alters loop dynamics.
The interaction of cofilin with F-actin has been shown by EM to decrease the electron density of the DNase I loop, implying its induced structural disordering (4). Consistent with this finding is the enhanced subtilisin cleavage between residues 47 and 48 in the DNase I loop in the presence of cofilin (16). EM results (17, 18) and cross-linking data (19) also suggest that changes in another region, in the hydrophobic loop, are important in the alteration of the F-actin structure induced by cofilin.
In spite of this previous work, the dynamic changes in the DNase I loop, the hydrophobic plug, and actin’s C-terminus in connection with changes in actin filament stability are not well-understood. Here, we use a combination of SDSL, fluorescence, and cross-linking techniques to gain insight into the changes in the dynamics of these structural elements caused by the interaction of cofilin and phalloidin with actin.
MATERIALS AND METHODS
Yeast Actin Mutants
Yeast actin mutant strains S265C, S265C/C374A, V266C, V266C/C374A, L267C, L267C/C374A, L269C, L269C/C374A, Q41C, and Q41C/C374S were described previously (20–22).
Protein Purification
Yeast actin was purified by affinity chromatography on a DNaseI column as described by Shvetsov et al. (6). The purified G-actin was then centrifuged at 4000 rpm in Amicon Ultra-4 Ultracel-10k centrifugal filter devices (Millipore, Billerica, MA) to wash and concentrate the actin to a level between 20 and 30 μM in 10 mM MOPS (pH 7.0), 0.2 mM ATP, 1.0 mM DTT, and 0.2 mM CaCl2 or dialyzed overnight into fresh buffer. The purified G-actin was stored on ice, with fresh DTT added daily. For all experiments, and to remove DTT, G-actin was passed through a Sephadex G-50 column equilibrated with 10 mM MOPS (pH 7.0), or HEPES-NaOH (pH 8.0) for cross-linking experiments, 0.2 mM ATP, and 0.2 mM CaCl2. Yeast cofilin was expressed in Escherichia coli BL21(DE3) cells. Cell extracts were applied to an ion exchange column and eluted with a 0 to 500 mM NaCl gradient in 20 mM Tris (pH 7.5) and 1.0 mM DTT. Cofilin eluted at approximately 200 mM salt and was then subjected to gel filtration on a HiLoad 16/60 Superdex 200 gel filtration column (Amersham Biosciences UK Ltd., Buckinghamshire, England) in 20 mM Tris (pH 7.5), 0.2 M NaCl, and 1.0 mM DTT, before being dialyzed into 10 mM Tris (pH 7.5) and 1.0 mM DTT.
Disulfide Cross-Linking Experiments
G-Actin was passed through a Sephadex G-50 column and polymerized at 10 μM for 1 h in the presence or absence of equimolar phalloidin and 3.0 mM MgCl2. CuSO4 (2.0 μM) was added to actin to catalyze disulfide cross-linking at room temperature. Reaction aliquots were taken directly before the addition of CuSO4 (zero time point) and at various time points up to 30 min. Reactions were stopped with excess (2.0 mM) N-ethylmaleimide (NEM), which blocks unreacted cysteine groups. Time point samples were run on SDS–PAGE gels under nonreducing conditions and stained with Coomassie blue for visualization. Gels were scanned, and density values of actin monomer bands were obtained using SigmaGel, version 1.0 (Jandel Scientific, San Rafael, CA). The decay of the actin monomer band intensity with reaction time followed a first-order process. Data were then plotted in SigmaPlot 2000, version 6.0 (SPSS, Inc., Chicago, IL).
Acrylodan Labeling Experiments
G-Actin was passed through a Sephadex G-50 column and polymerized with 2.0 mM MgCl2 for 30 min at 25 °C. After polymerization, cofilin or phalloidin was mixed with actin (1.4:1 molar ratio) and allowed to incubate for 30 min at 25 °C. Prior to experiments, actin samples were diluted to 5.0 μM and then acrylodan (6-acryloyl-2-dimethylaminonaphthalene, Invitrogen, Eugene, OR) was added at a 1:100 acrylodan:actin ratio. Actin labeling was monitored through changes in acrylodan fluorescence, with the excitation wavelength set at 385 nm and the emission monitored at 462 nm. Experiments were conducted using a Photon Technology International (PTI, Lawrenceville, NJ) spectrofluorimeter and FeliX32 Analysis, version 1.1 (PTI).
Electron Microscopy
After being passed through a Sephadex G-50 column, mutant yeast actin was labeled with the MTSL spin probe, as described above, and polymerized with 2.0 mM MgCl2 for 30 min at 25 °C. After polymerization, cofilin or phalloidin was mixed with actin (1.4:1 molar ratio) and allowed to incubate for 30 min at 25 °C. To adsorb actin to carbon-coated holey grids, actin was diluted to 2.5 μM, adsorbed to grids, and negatively stained with 1% uranyl acetate. Grids were observed using a Hitachi H7000 electron microscope at a magnification of 31200×.
Pelleting Assays
Yeast actin was polymerized as described above, at concentrations between 16 and 23 μM. Cofilin or phalloidin was mixed with actin (1.4:1 molar ratio) and allowed to incubate for 30 min at 25 °C. The samples were then centrifuged at 360000g for 30 min, at 18 °C, in a Beckman TLA-100 rotor. The supernatants and pellets were adjusted to the same volumes and analyzed by SDS–PAGE.
Site-Directed Spin Labeling
Yeast single- and double-cysteine substitution mutants of G-actin were passed through Sephadex G-50 columns equilibrated with 10 mM MOPS (pH 7.0), 0.2 mM ATP, and 0.2 mM CaCl2. The mutant G-actin was then incubated for 1 h, at room temperature, in the same buffer containing a 3-fold molar excess (over actin) of a spin labeling reagent {MTSL [(1-oxy-2,2,5,5-tetram-ethylpyrrolinyl-3-methyl)methanethiosulfonate]} that reacts with cysteine residues to generate the side chain, designated R1 (23). For distance measurements, actin was labeled with either pure spin-labeled reagent (MTSL) or a 1:2 (molar ratio) mixture of MTSL with a diamagnetic analogue [(1-methoxy-2,2,5,5-tetramethylpyrrolinyl-3-methyl)methanethiosulfonate], according to the method described in ref 23. Both reagents were kindly provided by K. Hideg (University of Pécs, Pécs, Hungary). Previous work has shown that other nonreactive cysteines present in the actin sequence are not labeled under conditions employed in this work (24). Actin was again passed through Sephadex G-50 columns to remove any unreacted label and was then concentrated to 30 μM using Vivaspin 0.5 mL concentrators (Viva Science, Hannover, Germany) with a 10000 molecular weight cutoff PES membrane. Samples of freshly spin-labeled G-actin were polymerized for at least 20 min at room temperature with 2.0 mM MgCl2. Cofilin or phalloidin was added in excess of actin (1.4:1 molar ratio) and allowed to incubate for 30 min at 25 °C. Spectra of G- and F-actin were recorded at X-band using either a Bruker E580 spectrometer and a cavity (4119HS) or a Varian 109 spectrometer fitted with a loop gap resonator (25). Labeled actin was analyzed in a Suprasil flat cell (volume of 40 μL, Wilmad, Buena, NJ) or a glass capillary (volume of 5.0 μL, VitroCom, Inc., Mountain Lakes, NJ) for each spectrometer, respectively. Each spectrum was collected with a scan time of 30 s, a field scan of 160 G, a modulation amplitude of 1.0 G at 100 kHz, and an incident microwave power of either 10 mW (4119HS) or 2.0 mW (loop gap), as in ref 24. The recorded data are averages of 20–60 scans, are trimmed to 100 G for display, and are normalized to the same number of spins.
Measurement of Interspin Distance
Altenbach et al. (23) showed that the dipolar broadening of EPR spectra can be used to determine interspin distance distributions in doubly spin-labeled proteins at room temperature, as long as the orientation of the interspin vector varies sufficiently slowly. The analysis method used in our previous work (7) has been dramatically improved recently (26) and now uses Tikhonov regularization and nonlinear optimization instead of deconvolution. All results presented here were obtained with the new program created by C. Altenbach using LabVIEW (National Instruments), which is available for download. MTSL or a 1:3 (molar ratio) mixture of MTSL with a diamagnetic analogue was used to obtain the interacting and noninteracting spectra.
Distance distributions are displayed as integrals, meaning that the displayed value for any distance, x, is the fraction of all spins that show a distance closer or equal to x. This has an advantage in that the number of noninteracting spins can be intuitively displayed in the same graph. Here, the background fraction of noninteracting spins due to partial labeling is constant, and changes in the number of noninteracting spins can be directly interpreted as a real change in distance that crosses the distance limit of the method.
RESULTS
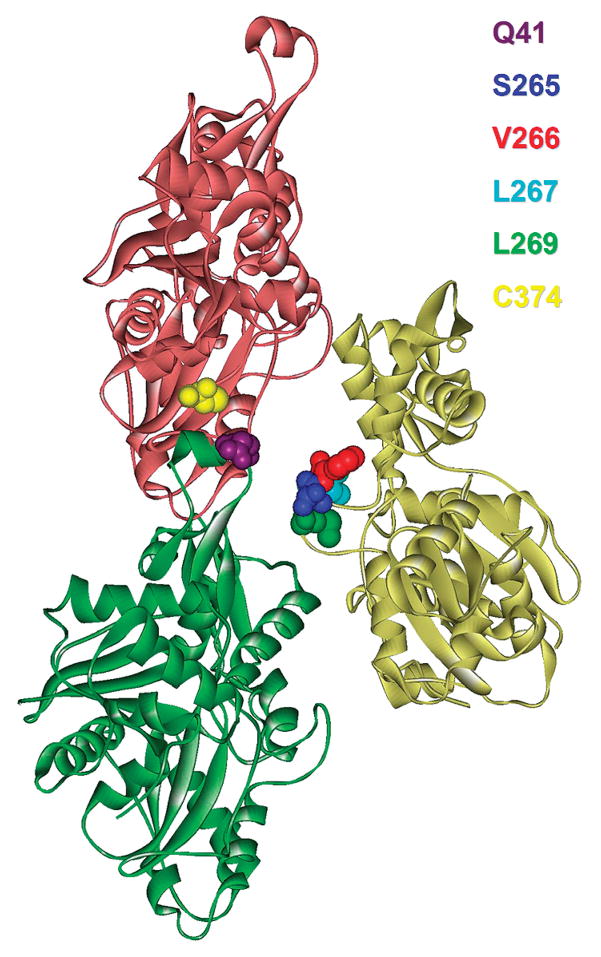
The interaction of three structural elements of actin, the DNase I loop, the hydrophobic loop (between subdomains 3 and 4), and the C-terminus, from three adjoining monomers within the filament, is believed to be important for the stabilization of actin filaments. The relationship between these elements in the actin filament model is shown in Figure 1. The focus of this work was to use SDSL and covalent cross-linking to determine how the filament-destabilizing protein cofilin, and the filament-stabilizing toxin phalloidin, alter these interrelationships. To provide sites for the introduction of the nitroxide spin-label or for disulfide cross-linking, cysteine was substituted for the WT residues at positions 265–267 and 269 in the hydrophobic loop, and at position 41 in the DNase I binding loop (Figure 1). Double mutants were also made in which, in addition to the mutations listed above, the reactive C374 was replaced with alanine in the hydrophobic loop mutants and with serine in the DNase I binding loop mutant.
Figure 1.
Holmes model of the actin filament shown with pertinent residues highlighted. Individual actin protomers are distinguished by their ribbon color. This figure was generated using WebLab ViewerPro, version 4.0.
We first wished to use SDSL to assess the effects of cofilin on the interactions among the C-terminus, the hydrophobic loop, and the DNase I loop. To this end, the single mutant actins were doubly labeled at C374 and the introduced cysteine by the SDSL probe R1. In these experiments, interprobe distances can only be measured if the probes are fewer than 20 Å apart. Within that range, we identified two interspin distance populations with respective means of ~10 and ~14–15 Å for each pair of spin probes. For distances greater than 20 Å, the probes appear as noninteracting. Experiments with G-actin exhibited no interaction between pairs of probes on the same monomer. Evidence of interaction in the actin filament represents intermonomer events either within the same strand (residues 41 and 374) or between strands of the filament (residues 265–267 and 374).
Cofilin Effects
EPR measurements showed no interaction between residue 374 and residue 269 with actin alone or in the presence of either cofilin or phalloidin (Table 1). For each of the remaining hydrophobic loop residues, as shown in Table 1, there was a population of both interacting and noninteracting pairs. Residue 265 showed the greatest degree of interaction, 59%, while 37 and 39% of spin probe populations interacted when residue 374 was paired with residues 266 and 267, respectively.
Table 1.
Interspin Interactions in F-Actina
| mutant | factor | distance (Å) | % interacting |
|---|---|---|---|
| 265R1/374R1 | – | 10.2, 15.6 | 59 |
| cofilin | 8.5, 13.1 | 45 | |
| phalloidin | 10.0, 16.0 | 67 | |
| 266R1/374R1 | – | 8.5, 14.4 | 37 |
| cofilin | 8.5, 13.5 | 29 | |
| phalloidin | 9.0, 15.0 | 49 | |
| 267R1/374R1 | – | 8.7, 15.5 | 39 |
| cofilin | – | 0 | |
| phalloidin | – | 0 | |
| 269R1/374R1 | – | – | 0 |
| cofilin | – | 0 | |
| phalloidin | – | 0 | |
| 41R1/374R1 | – | 8.9, 14.6 | 68 |
| cofilin | 8.4, 14.5 | 41 | |
| phalloidin | 8.9, 14.4 | 70 |
Distances reflect measured distance distribution peak means from doubly R1-labeled F-actin mutants. Each yeast actin mutant had a bimodal distribution of distances. Distance distribution widths may be observed in Figure. Values in the % interacting column reflect measured spin–spin interactions.
As shown in Table 1 and Figures 2 and 3, addition of cofilin caused a substantial change in these C374–hydrophobic loop interactions. In each case, the total percent of interacting pairs decreased substantially, suggestive of changes in the filament structure. In the case of the 267R1/374R1 pair, interaction was eliminated. The average distance of the interaction of 265R1/374R1 changed, moving the residues slightly closer, but the percentage of probes interacting was decreased by 14%. For the 266R1/374R1 pair of spin probes, the range of distances did not change, but the population of interacting spins was decreased by 8%.
Figure 2.
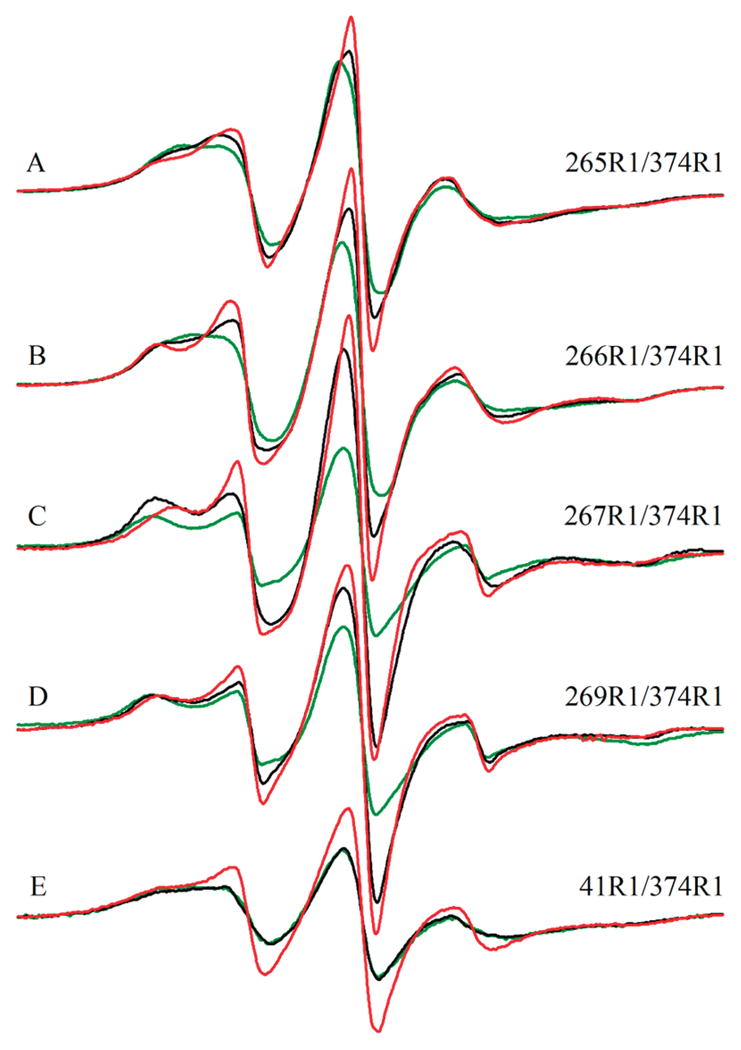
EPR spectra of doubly labeled yeast F-actin mutants alone (black spectra) or in the presence of either cofilin (red spectra) or phalloidin (green spectra): (A) 265R1/374R1, (B) 266R1/374R1, (C) 267R1/374R1, (D) 269R1/374R1, and (E) 41R1/374R1. The displayed field range is 100 G. All spectra are normalized for area.
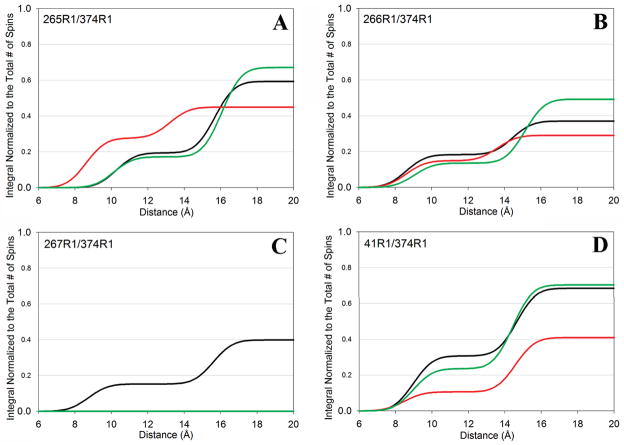
Figure 3.
Integrated distribution of distances for yeast F-actin mutants alone (black lines) or in the presence of cofilin (red lines) or phalloidin (green lines). y axis values represent fractions of the interacting spins. The x-axis depicts distances in angstroms between spin-labels: (A) 265R1/374R1, (B) 266R1/374R1, (C) 267R1/374R1, and (D) 41R1/374R1.
The interactions between residues 41 and 374 (Table 1 and Figures 2 and 3) were similar to those seen by residue 265, though residues 41 and 374 appear closer to each other than residue 374 and the hydrophobic loop residues. The addition of cofilin caused an almost 30% decrease in the percentage of interacting probes, though no change in their distances.
Phalloidin Effects
As shown in Table 1, phalloidin, a filament stabilizer, has a residue-dependent effect on the interprobe interactions. For the 265R1/374R1 pair, the percentage of interacting probes increases from 59 to 67%. A similar effect on interprobe distance is observed for 266R1/374R1. For 267R1/374R1, there is a total loss of interacting probes, indicating a shift of residues away from one another. Thus, phalloidin affects in an opposite way the proximity of N- and C-terminal residues of this loop with respect to C374.
For the 41R1/374R1 interaction (Table 1), the percentage of probes interacting and their distances remain approximately the same, indicating perhaps only small changes in the DNase I loop in the presence of phalloidin. This result implies that phalloidin-induced stabilization of the filament is not caused by a significant narrowing of the distribution of the conformational states of subdomain 2, but perhaps by a decrease in the frequency of the conformational fluctuations of this loop. To assess the effect of phalloidin on the dynamics of the DNase I loop and C-terminus, we determined the ability of C41 and C374 to form a disulfide cross-link. Such cross-linking depends on both distance and orientation parameters. Our results show that phalloidin increases the rate of this cross-linking by ~70% (Figure 4). This result suggests a more “stable” presence of the cross-linking-favoring conformation or orientation of the C41 and C374 sites.
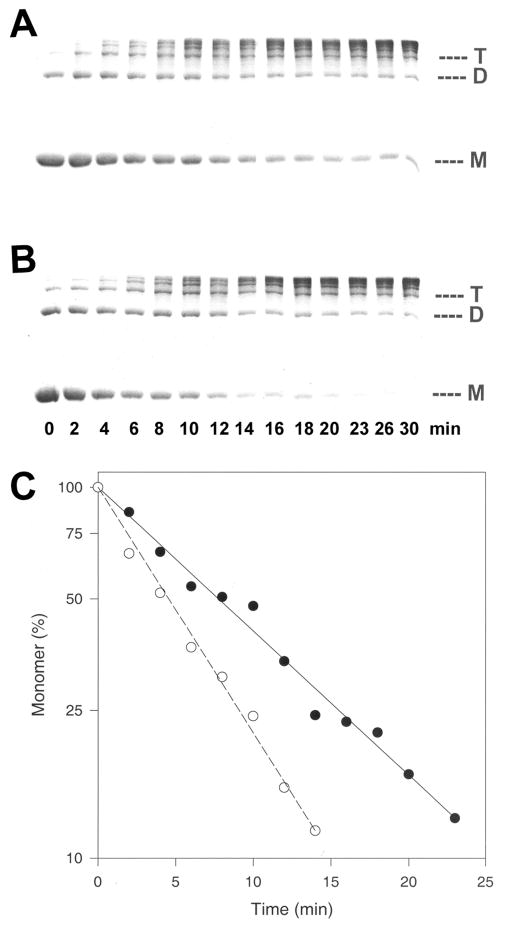
Figure 4.
Disulfide cross-linking experiments with Q41C F-actin in the absence and presence of phalloidin. Time point aliquots were taken and analyzed by SDS–PAGE. (A) SDS–PAGE of Q41C F-actin disulfide cross-linked in the absence of phalloidin. (B) SDS–PAGE of Q41C F-actin disulfide cross-linked in the presence of phalloidin. Symbols M, D, and T correspond to actin monomers, dimers, and trimers, respectively. (C) Semilogarithmic plot of disulfide cross-linking of Q41C F-actin in the absence (●) and presence (○) of phalloidin. The rates of cross-linking were determined using Sigma Plot by fitting the decreasing (with time) percentage of un-cross-linked actin monomers (from panels A and B) to a single-exponential equation. The slopes of the linear plots yielded the first-order rates of disulfide cross-linking of Q41C F-actin in the absence [k = (3.9 ± 0.09) × 10−2 min−1] and in the presence of phalloidin [k = (6.6 ± 0.1) × 10−2 min−1].
Role of the C-Terminal Residue 374 in Distance Changes
The results given above established that cofilin and phalloidin affect the intermonomer interactions between C374 and the two loops under investigation. However, they did not show whether these changes originate from the motion of the 374R1 spin probe at the C-terminus, the loop probes, or perhaps both. To shed light on this question, we examined the EPR spectra of wild-type actin, with a single spin probe at residue 374. Significantly, 374R1 actin exhibited virtually no change in the spectrum of F-actin in the presence of either cofilin or phalloidin (Figures 5A and 6A). Thus, the changes in interspin distances described above are not produced by the motion of the C-terminus to a different environment.
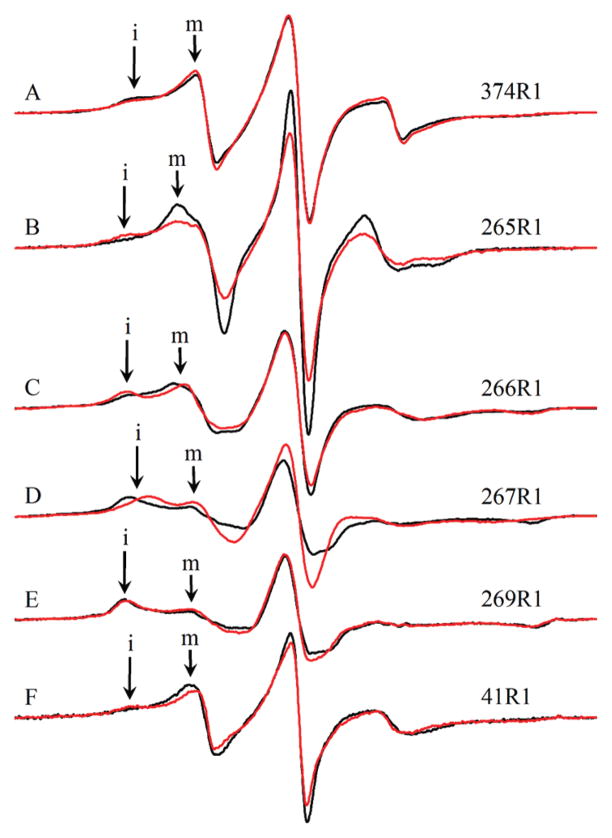
Figure 5.
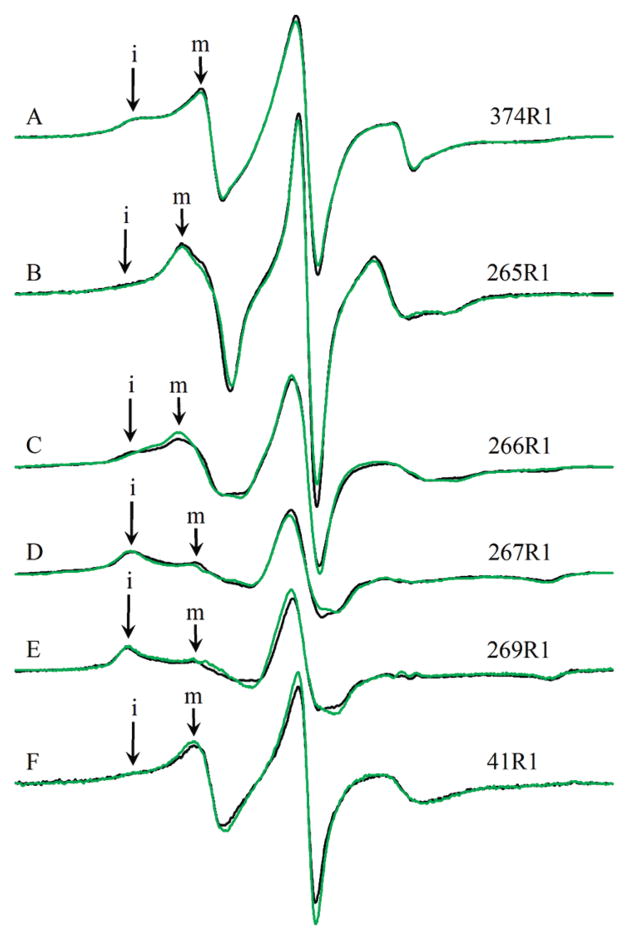
EPR spectra of singly labeled yeast F-actin wild type and actin double mutants in the presence and absence of cofilin: (A) wild-type yeast F-actin (374R1), (B) 265R1, (C) 266R1, (D) 267R1, (E) 269R1, and (F) 41R1. Black spectra are those of F-actin. Red spectra are those of F-actin in the presence of an equimolar amount of yeast cofilin. “i” denotes the immobile component of each spectrum, where labeled. “m” denotes the mobile component of each spectrum, where labeled. The displayed field range is 100 G. All spectra are normalized for area.
Figure 6.
EPR spectra of singly labeled yeast F-actin wild type and actin double mutants in the presence and absence of phalloidin: (A) wild-type yeast actin (374R1), (B) 265R1, (C) 266R1, (D) 267R1, (E) 269R1, and (F) 41R1. Black spectra are those of F-actin. Green spectra are those of F-actin in the presence of an equimolar amount of phalloidin. “i” denotes the immobile component of each spectrum, where labeled. “m” denotes the mobile component of each spectrum, where labeled. The displayed field range is 100 G. All spectra are normalized for area.
Effects of Cofilin and Phalloidin on the Local Environments of the Attached Spin Probes
We next wished to determine the extent to which the changes in interspin distances caused by cofilin and phalloidin were mirrored by changes in the local environment around the individual R1 residues. Changes in the local environment may alter the level of tertiary interaction of R1, which in turn modulates the mobility of the side chain. The mobility is directly encoded by the EPR spectra line shape. The qualitative relation of the line shape to nitroxide mobility has been discussed by Columbus and Hubbell (27), Kusnetsow et al. (28), and Crane et al. (29). The EPR spectra of spin-labeled proteins reflect the motion of the R1 side chain on the nanosecond time scale. The low field region of the spectrum is most sensitive to differences in mobility. Spectral intensity closer to the center line corresponds to less motion-hindered spin-labels, while intensity farther and at lower field corresponds to more hindered spin-labels. Qualitatively, we assign these spin-label states as mobile (m) or immobile (i) and label the regions of the spectra accordingly (Figures 5 and 6). We assessed the effects of cofilin and phalloidin on the mobility of single EPR probes by examining and analyzing changes in their EPR spectra. The 265R1 probe spectrum showed significant changes in the presence of cofilin, indicative of a decreased probe mobility (Figure 5B). A similar but less drastic effect was observed with the probe at residue 266 (Figure 5C). In contrast, the spectrum of 267R1 shows that cofilin actually increases its mobility (Figure 5D). No effect was observed for the probe at 269R1 (Figure 5E). Overall, cofilin increased the mobility of the probe at residue 41 (Figure 5F), although it also seemed to induce the immobilization of a small population of the probed actins.
In contrast to cofilin, phalloidin does not significantly affect the EPR spectra of probes at residue 265, 267, 269, or 41 (Figure 6). It does, however, cause a small increase in the mobility of the probe at residue 266.
Effect of Cofilin and Phalloidin on Acrylodan Reactivity of the Cysteines at Position 374 and Mutated Sites in the Two Loops
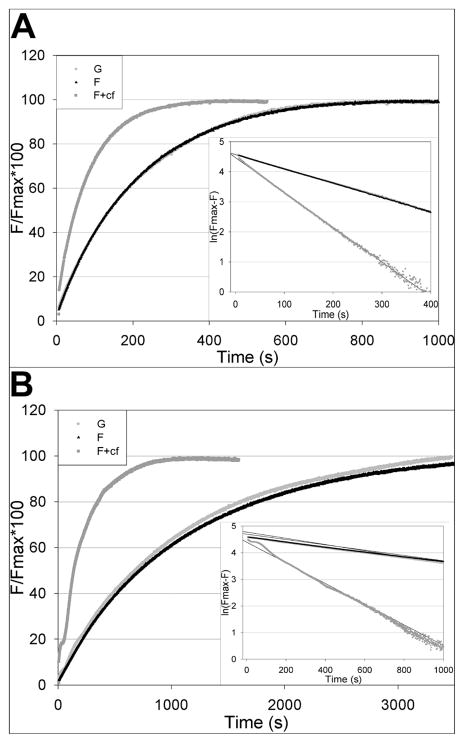
Changes in the conformation of the DNase I and hydrophobic loops brought about by cofilin and phalloidin might alter the reactivity of nucleophilic groups in these loops toward acrylodan, a reactive fluorescent probe. For these experiments, we used the double mutants in which cysteines were present in the mutated sites in both loops but absent from position 374. Thus, each actin had only a single reactive cysteine. Under pseudo-first-order conditions of the reaction, there was no significant difference in the reactivity of G- and F-actin for any of the examined sites. This shows that none of those residues is more shielded from solvent in F-actin than in G-actin. Although cofilin had no effect on the reactivities of the mutant G-actins, it markedly affected the reactivities of cysteines at positions 265 and 41 in F-actin. Figure 7 shows that the reactivities of probes positioned at residues 265 and 41 increased by 3.7- and 2.3-fold, respectively, implying an increase in the degree of solvent exposure of these two residues by cofilin binding. In contrast, phalloidin caused only a small, 36%, decrease in C41 reactivity and an even smaller, 21%, decrease in the reactivity of C265 (data not shown). No change was observed with phalloidin in the reactivity of C374 or other hydrophobic loop residues.
Figure 7.
Plots of acrylodan labeling of yeast actin double mutants in various states. There was no change in the rate of acrylodan labeling of G- or F-actin for the mutants shown. (A) Q41C/C374S actin. The rate of Q41C/C374S F-actin in the presence of cofilin (F+cf) increased approximately 2.3-fold over that of G- or F-actin alone. (B) S265C/C374A actin. The rate of S265C/C374A F-actin in the presence of cofilin increased approximately 3.7-fold over that of G- or F-actin alone. Light gray curves represent data of G-actin. Black curves represent data of F-actin. Dark gray curves represent data of F-actin in the presence of cofilin.
DISCUSSION
The focus of this study was to examine the dynamic aspects of lateral and longitudinal interfaces in F-actin in the presence and absence of filament-stabilizing and -destabilizing factors. To this end, we used phalloidin, cofilin, and yeast actin cysteine mutants of the hydrophobic and DNase I binding loops in experiments involving EPR spectroscopy, fluorescent labeling, and cross-linking methods. SDSL EPR measurements provided information about phalloidin- and cofilin-induced changes in interspin distances between a spin probe at C374 and a second probe at various positions on the two loops. Local spin probe dynamics in these loops was assessed from EPR spectra in double-substitution actin mutants (with C374 mutated to either serine or alanine). Results from cross-linking and acrylodan labeling experiments of the actin mutants supplement the SDSL data and help in the interpretation of local changes at the loop-mediated interprotomer contact regions.
Electron microscopy images showed relatively long and straight actin filaments of the loop mutants in the presence of phalloidin (7). Our SDSL distance data reveal that filament stabilization by phalloidin coincides with an increased level of interactions of spin probes on residues 265 and 266 with a probe on C374 (Table 1 and Figure 3). This increased level of interaction results from a greater fraction of the position 265 and 374 spin probes residing within 20 Å, primarily through an increase in the population of interspin distances in the range of 13–18 Å. This is more pronounced for the position 266 and 374 probes, for which the increase in the level of interaction is clearly dominated by an increase in the population of the 13–18 Å interspin distances. Our disulfide cross-linking results are consistent with the EPR data (7). An increased level of cross-linking of phalloidin-bound filaments of the S265C and V266C mutants, as compared to unbound filaments, supports the SDSL data which show that, on average, these loop residues and residue 374 are close (<20 Å apart) in a greater fraction of actin protomers in F-actin.
A very different picture emerged for residues 267 and 269. SDSL data reveal that spin–spin interactions are abolished in phalloidin-bound filaments between residues 267 and 374, while they are completely absent for the 269–374 spin probe pairs. Moreover, we have previously shown that disulfide cross-linking between residues 267 and 374 and between residues 269 and 374 destroys even the phalloidin-bound filaments (7). We conclude that stable and/or phalloidin-stabilized filaments require that residues 267 and 269 not sample close distances (<20 Å) to C374 in any significant fraction of actin population. This and our previous work (7) support Holmes’ recent model of the F-actin filament (30), in which the hydrophobic loop resides mostly in a parked conformation, and where residues 267 and 269 are oriented away from residue 374 and the opposing actin strand.
Strikingly, phalloidin did not have a strong effect on the interspin distance distribution in Q41C actin filaments, suggesting little change in the population of subdomain 2 conformational states. Thus, the primary filament stabilizing effect of phalloidin is through its impact on interstrand interactions and less, if any, on the DNase I binding loop. Another striking result of this study is the observation that phalloidin has an only marginal impact on the spin probe environment at any of the residues tested here, and it does not affect significantly the labeling of these residues with acrylodan (except for a small inhibition of C41 and C265 labeling). This leads to the conclusion that filament stabilization and its increased stiffness due to phalloidin (31, 32) are caused mainly by the toxin’s direct binding (stapling) of three actin protomers, which results in a decrease in the range and probably frequency of strand separation through conformational fluctuations.
Cofilin, as an actin-destabilizing agent, presents a different situation. Large changes are observed at position 41 in the longitudinal contact region (Table 1 and Figure 3). SDSL experiments reveal an almost 30% decrease in the percent of interacting spins for Q41C cofilin-bound F-actin (Table 1). Though the average distance for interacting spins does not change, the much-lower percentage of interacting spins indicates states in which position 41 is more than 20 Å from C374 in most of the longitudinal pairs of actin in the filament. Disulfide cross-linking experiments with this mutant performed by Bobkov et al. (33) highlight this conclusion, as the cross-linking rates are strongly decreased for Q41C F-actin with bound cofilin. Single-spin SDSL experiments revealed a more mobile residue 41, which may explain the increase in the level of acrylodan labeling by the increased accessibility of position 41 to the reagent. Our data are in agreement with the previous work by Muhlrad et al. (16) in which the level of subtilisin cleavage in the DNase I binding loop increased in F-actin with bound cofilin in comparison to F-actin alone. Moreover, Galkin et al. (4) were not able to fully reconstruct the DNase I binding loop in their EM reconstruction experiments with cofilin-bound F-actin, indicating a disordered or mobile DNase I loop.
Cofilin also has a strong effect on the residues of the hydrophobic loop (Table 1 and Figures 2 and 3). For hydrophobic loop mutants S265C, V266C, and L267C, the percentage of interacting spins was significantly reduced, indicating that a cofilin-induced change in the filament twist moves hydrophobic loop residues from the C-terminus. Cross-linking rates for cofilin-bound F-actin were decreased for S265C, as observed by Bobkov et al. (19), again indicating that C265 does not sample closer distances to C374 as often as in F-actin alone. Furthermore, in contrast to phalloidin, cofilin changed the mobility of single-spin probes for 265R1, 266R1, and 267R1, suggesting that a subpopulation of those residues samples an altered environment in the modified filament structure. These results are consistent with the observations of Cao et al. (34) and support the hypothesis of Paavilainen et al. (35) on the weakening of interactions of the hydrophobic and DNase I binding loops in F-actin, as deduced from the crystal structure of G-actin in complex with the ADF-H homology domain of twinfilin. However, the cofilin-induced shift in interspin distances between probes located at residues 41 and 374 is significantly smaller than the change predicted by the model of Paavilainen et al. (35).
The analysis of cofilin’s effects on both the hydrophobic and DNase I loop environments extends some of our prior observations (19, 33), providing a more detailed view of the changes in filament structure and twist by cofilin. Clearly, the changes caused by cofilin are much bigger than those that are induced by phalloidin, impacting the conformational states of the two loops studied here. This is perhaps not surprising in view of the much stronger structural alteration of F-actin by cofilin than by phalloidin. The extensive perturbation of the interprotomer contact region by cofilin and the propagation of these effects along the filament (36) can provide the mechanistic explanation for the weakening of interprotomer interactions and filament severing. This effect will be especially pronounced in filament segments not occupied by cofilin, i.e., where the allosteric weakening of interprotomer interactions is not compensated by direct cofilin binding and local stabilization through new actin–cofilin–actin binding interfaces (36–38).
In summary, this report provides a residue-detailed understanding of changes in the hydrophobic loop and insight into the changes in the DNase I binding loop and the C-terminus. The complexity of single-spin EPR spectra and interspin distance distributions strengthens the view of multiple conformational states of these three structural elements in actin filaments. The surprisingly small effects of phalloidin on these elements reveal that its main stabilizing effect arises from restricting filament breathing and structure-destabilizing fluctuations. These latter effects must then be the main agonists of cofilin binding to phalloidin-stabilized filaments. It has been proposed before that solution binding of cofilin to F-actin may depend on filament breathing motions (34). Our results add credence to that idea and suggest that cofilin binding to F-actin can be regulated by controlling its structural dynamics. The inhibitory effect of Pi on cofilin binding to F-actin (39) and the different affinities of cofilin for ATP- and ADP-F-actin can be rationalized by the corresponding effects of nucleotides and Pi on filament dynamics.
Table 2.
Effects Induced by Cofilin or Phalloidin on F-Actina
| mutant | cofilin effects
|
phalloidin effects
|
||
|---|---|---|---|---|
| spin mobility | acrylodan labeling | spin mobility | acrylodan labeling | |
| S265C/C374A | ↓ | ↑ | ≈ | (↓) |
| V266C/C374A | ↓ | ≈ | (↑) | ≈ |
| L267C/C374A | (↑) | ≈ | ≈ | ≈ |
| L269C/C374A | ≈ | ≈ | ≈ | ≈ |
| Q41C/C374A | μ | μ | ≈ | (↓) |
The table reflects qualitative effects from either SDSL or acrylodan binding experiments performed on yeast actin double mutants containing only one reactive cysteine. Parentheses denote small changes. Approximately equal to signs denote no change.
Acknowledgments
We thank Dr. Kálmán Hideg for his generous gift of methanethiosulfonate reagents used in SDSL experiments.
Footnotes
This work was supported by grants from U.S. Public Health Service (GM 077190) and the National Science Foundation (MCB 0316269) to E.R., from the U.S. Public Health Service (GM 33689) to P.A.R., from the U.S. Public Health Service (EY 05216) and Jules Stein Professor Endowment to W.L.H., and from the U.S. Public Health Service (Training Grant T32 EY07026) to J.D.S.
Abbreviations: G-actin, globular or monomeric actin; F-actin, filamentous actin; DTT, dithiothreitol; SDSL, site-directed spin labeling; EM, electron microscopy.
References
- 1.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 Update. Nucleic Acids Res. 2004;32:449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dos Remedios CG, Chahbra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin Binding Proteins: Regulation of Cytoskeletal Microfilaments. Physiol Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 3.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic Model of the Actin Filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 4.Galkin VE, Orlova A, VanLoock MS, Shvetsov A, Reisler E, Egelman EH. ADF/Cofilin Use an Intrinsic Mode of F-Actin Instability to Disrupt Actin Filaments. J Cell Biol. 2003;163:1057–1066. doi: 10.1083/jcb.200308144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong Y, Topf M, Sali A, Matsudaira P, Dougherty M, Chiu W, Schmid MF. Crystallographic Conformers of Actin in a Biologically Active Bundle of Filaments. J Mol Biol. 2008;375:331–336. doi: 10.1016/j.jmb.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shvetsov A, Musib R, Phillips M, Rubenstein PA, Reisler E. Locking the Hydrophobic Loop 262–274 to G-Actin Surface by a Disulfide Bridge Prevents Filament Formation. Biochemistry. 2002;41:10787–10793. doi: 10.1021/bi020205f. [DOI] [PubMed] [Google Scholar]
- 7.Scoville D, Stamm JD, Toledo-Warshaviak D, Altenbach C, Phillips M, Shvetsov A, Rubenstein PA, Hubbell WL, Reisler E. Hydrophobic Loop Dynamics and Actin Filament Stability. Biochemistry. 2006;45:13576–13584. doi: 10.1021/bi061229f. [DOI] [PubMed] [Google Scholar]
- 8.Otterbein LR, Graceffa P, Dominguez R. The Crystal Structure of Uncomplexed Actin in the ADP State. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 9.Zheng X, Diraviyam K, Sept D. Nucleotide Effects on the Structure and Dynamics of Actin. Biophys J. 2007;93:1277–1283. doi: 10.1529/biophysj.107.109215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal Structures of Expressed Non-Polymerizable Monomeric Actin in the ADP and ATP States. J Biol Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 11.Tirion MM, ben-Avraham D. Normal Mode Analysis of G-Actin. J Mol Biol. 1993;230:186–195. doi: 10.1006/jmbi.1993.1135. [DOI] [PubMed] [Google Scholar]
- 12.Muhlrad A, Cheung P, Phan BC, Miller C, Reisler E. Dynamic Properties of Actin. Structural Changes Induced by Beryllium Fluoride. J Biol Chem. 1994;269:11852–11858. [PubMed] [Google Scholar]
- 13.Tirion MM, ben-Avraham D, Lorenz M, Holmes KC. Normal Modes and Refinement Parameters for the F-Actin Model. Biophys J. 1995;68:5–12. doi: 10.1016/S0006-3495(95)80156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orlova A, Egelman EH. Structural Basis for the Destabilization of F-Actin by Phosphate Release Following ATP Hydrolysis. J Mol Biol. 1992;227:1043–1053. doi: 10.1016/0022-2836(92)90520-t. [DOI] [PubMed] [Google Scholar]
- 15.Bamburg JR. Proteins of the ADF/Cofilin Family: Essential Regulators of Actin Dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 16.Muhlrad A, Kudryashov D, Peyser YM, Bobkov AA, Almo SC, Reisler E. Cofilin Induced Conformational Changes in F-Actin Expose Subdomain 2 to Proteolysis. J Mol Biol. 2004;342:1559–1567. doi: 10.1016/j.jmb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 17.McGough A, Chiu W. ADF/Cofilin Changes Weakens Lateral Contacts in the Actin Filament. J Mol Biol. 1999;291:513–519. doi: 10.1006/jmbi.1999.2968. [DOI] [PubMed] [Google Scholar]
- 18.Orlova A, Shvetsov A, Galkin VE, Kudryashov DS, Rubenstein PA, Egelman EH, Reisler E. Actin-destabilizing Factors Disrupt Filaments by Means of a Time Reversal of Polymerization. Proc Natl Acad Sci USA. 2004;101:17664–17668. doi: 10.1073/pnas.0407525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bobkov AA, Muhlrad A, Shvetsov A, Benchaar S, Scoville D, Almo SC, Reisler E. Cofilin (ADF) Affects Lateral Contacts in F-Actin. J Mol Biol. 2004;337:93–104. doi: 10.1016/j.jmb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Feng L, Kim E, Lee WL, Miller CJ, Kuang B, Reisler E, Rubenstein A. Fluorescence Probing of Yeast Actin Subdomain 3/4 Hydrophobic Loop 262–274, Actin-Actin and Actin-Myosin Interactions in Actin Filaments. J Biol Chem. 1997;272:16829–16837. doi: 10.1074/jbc.272.27.16829. [DOI] [PubMed] [Google Scholar]
- 21.Musib R, Wang G, Geng L, Rubenstein PA. Effect of Polymerization on the Subdomain 3/4 Loop of Yeast Actin. J Biol Chem. 2002;277:22699–22709. doi: 10.1074/jbc.M203096200. [DOI] [PubMed] [Google Scholar]
- 22.Kim E, Wriggers W, Phillips M, Kokabi K, Rubenstein PA, Reisler E. Cross-Linking Constraints on F-Actin Structure. J Mol Biol. 2000;299:421–429. doi: 10.1006/jmbi.2000.3727. [DOI] [PubMed] [Google Scholar]
- 23.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of Inter-Residue Distances in Spin Labeled Proteins at Physiological Temperatures: Experimental Strategies and Practical Limitations. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 24.Shvetsov A, Stamm JD, Phillips M, Warshaviak D, Altenbach C, Rubenstein PA, Hideg K, Hubbell WL, Reisler E. Conformational Dynamics of Loop 262–274 in G- and F-Actin. Biochemistry. 2006;45:6541–6549. doi: 10.1021/bi052558v. [DOI] [PubMed] [Google Scholar]
- 25.Hubbell WL, Froncisz W, Hyde JS. Continuous and Stopped Flow EPR Spectrometer Based on a Loop Gap Resonator. Rev Sci Instrum. 1987;58:1879–1886. [Google Scholar]
- 26.Altenbach C, Hubbell WL. Improved Data Determination from Dipolar Broadening of EPR Spectra. Biophys J. 2008 (Supplement, Abstract) [Google Scholar]
- 27.Columbus L, Hubbell WL. A New Spin on Protein Dynamics. Trends Biochim Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 28.Kusnetzow AK, Altenbach C, Hubbell WL. Conformational States and Dynamics of Rhodopsin in Micelles and Bilayers. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crane JM, Mao C, Lilly AA, Smith VF, Suo Y, Hubbell WL, Randall LL. Mapping of the Docking of SecA onto the Chaperone SecB by Site-Directed Spin Labeling: Insight into the Mechanism of Ligand Transfer During Protein Export. J Mol Biol. 2005;353:295–307. doi: 10.1016/j.jmb.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR. Electron Cryo-Microscopy Shows how Strong Binding of Myosin to Actin Releases Nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 31.McCullough BR, Blanchoin L, Martiel JL, De La Cruz EM. Cofilin Increases the Bending Flexibility of Actin Filaments: Implications for Severing and Cell Mechanics. J Mol Biol. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isambert H, Venier P, Maggs AC, Fattoum A, Kassab R, Pantaloni D, Carlier MF. Flexibility of Actin Filaments Derived from Thermal Fluctuations. J Biol Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 33.Bobkov A, Muhlrad A, Kokabi K, Vorobiev S, Almo SC, Reisler E. Structural Effects of Cofilin on Longitudinal Contacts in F-Actin. J Mol Biol. 2002;323:739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- 34.Cao W, Goodarzi JP, De La Cruz EM. Energetics and Kinetics of Cooperative Cofilin-Actin Filament Interactions. J Mol Biol. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Paavilainen VO, Oksanen E, Goldman A, Lappalainen P. Structure of the Actin-Depolymerizing Factor Homology Domain in Complex with Actin. J Cell Biol. 2008;182:51–59. doi: 10.1083/jcb.200803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prochniewicz E, Janson N, Thomas DD, De La Cruz EM. Cofilin Increases the Torsional Flexibility and Dynamics of Actin Filaments. J Mol Biol. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 37.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin Filament Severing by Cofilin. J Mol Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrianantoandro E, Pollard TD. Mechanism of Actin Filament Turnover by Severing and Nucleation at Different Concentrations of ADF/Cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Muhlrad A, Pavlov D, Peyser YM, Reisler E. Inorganic Phosphate Regulates the Binding of Cofilin to Actin Filaments. FEBS J. 2006;273:1488–1496. BI801649J. doi: 10.1111/j.1742-4658.2006.05169.x. [DOI] [PubMed] [Google Scholar]