Abstract
Proteoglycomics is a systematic study of structure, expression, and function of proteoglycans, a posttranslationally modified subset of a proteome. Although relying on the established technologies of proteomics and glycomics, proteoglycomics research requires unique approaches for elucidating structure–function relationships of both proteoglycan components, glycosaminoglycan chain, and core protein. This review discusses our current understanding of structure and function of proteoglycans, major players in the development, normal physiology, and disease. A brief outline of the proteoglycomic sample preparation and analysis is provided along with examples of several recent proteoglycomic studies. Unique challenges in the characterization of glycosaminoglycan component of proteoglycans are discussed, with emphasis on the many analytical tools used and the types of information they provide.
Introduction
Proteoglycans (PGs) are a diverse group of glycoconjugates constituted by various core proteins posttranslationally modified with linear, anionic polysaccharide, glycosaminoglycans (GAGs) consisting of repeating disaccharides. Ubiquitously found throughout the extracellular matrix (ECM), PGs are also found on virtually all cell surfaces and in basement membranes of different tissues. Through their interactions with proteins, PGs mediate many biological processes including cell–cell and cell–matrix interactions, growth factor sequestration, chemokine and cytokine activation, microbial recognition, tissue morphogenesis during embryonic development, and cell migration and proliferation (Capila and Linhardt, 2002; Cattaruzza and Perris, 2006; Esko and Selleck, 2002; Garner et al., 2008; Kreuger et al., 2006).
Proteoglycomics combines the proteomics of the protein core and the glycomics of the GAG chains and can serve to identify new PGs, catalog subsets of the proteome that show the level, type, and structure of GAG substitution, quantify PGs, investigate structure–function relationships, and study the impact of PGs in development or disease (Raman et al., 2005, 2006; Timmer et al., 2007; Tissot et al., 2009). Characterization of PGs is a challenging task because of the tremendous structural diversity of these bioconjugates arising from multiple isoforms of their core proteins and variations in glycoforms of their GAG components. Decoding the fine structures of GAGs from PGs is important because GAGs play numerous biological roles in cancer, the coagulation cascade, and microbial pathogenesis.
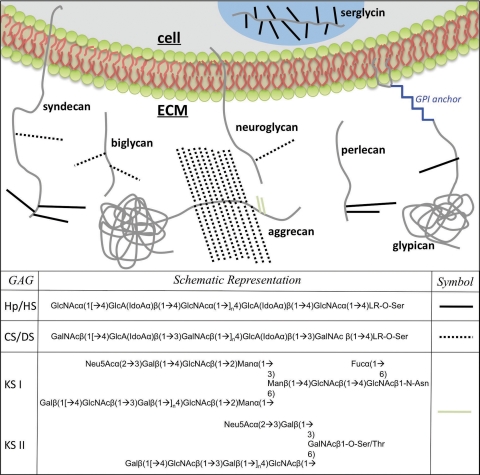
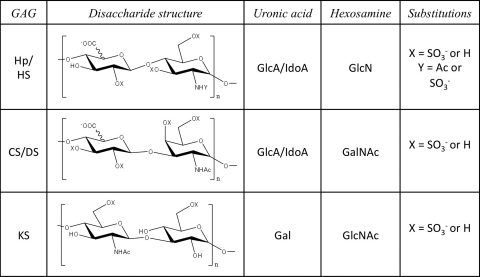
Most linear GAG chains are formed from repeating disaccharide units of hexosamine and hexuronic acid with keratan sulfate, comprised of hexosamine and galactose, being the exception. Even without their protein core, GAGs are highly complex and diverse structures due to the large number of possible combinations of chain lengths, saccharide sequences and compositions, sulfo group substitution, and domain arrangements and distributions. The different types of GAG chains, defined by their repeating disaccharide units, including heparin/heparan sulfate (Hp/HS), chondroitin/dermatan sulfate (CS/DS), and keratan sulfate (KS) (Figs. 1 and 2), lead to the organization of PGs. Main classifications for PGs include HSPGs, CSPGs, KSPGs, hyalectins, and SLRPs (Table 1).
FIG. 1.
A diagram of various proteoglycans in a cell and the ECM is shown. The detailed structure of the LR is defined in the text.
FIG. 2.
Possible disaccharide structures of the various glycosaminoglycans.
Table 1.
Properties of Common Proteoglycans (Esko et al., 2008)
| PG class | Proteoglycan | Number/type of GAG chains | Core protein size (kDa) |
|---|---|---|---|
| HSPG | Serglycin | 10–15 Hp/CS | 10–19 |
| Glypicans 1–6 | 1–3 HS | ∼60 | |
| Betaglycan | 1 HS, 1CS | 110 | |
| Syndecans 1–6 | 1–3 CS, 1–2 HS | 31–45 | |
| KSPG/SLRP | Lumican | KS I | 37 |
| Keratocan | KS I | 37 | |
| SLRP | Decorin | 1 DS/CS | 36 |
| Biglycan | 1–2 DS/CS | 38 | |
| Hyalectin | Aggrecan | ∼100 CS, KS II | 208–220 |
| Neurocan | 1–2 CS | 145 | |
| Brevican | 0–4 CS | 96 | |
| Versican | 12–15 CS | 265 | |
| CSPG | Bikunin | 1 CS | 18 |
| Neuroglycan C | 0–1 CS, part-time PG | ∼60 | |
| Leprecan | 1–2 CS | 82 | |
| Phosphacan | 2–5 CS | 175 | |
| Thrombomodulin | 1 CS | 58 |
Adding to complexity, PG protein core expression levels and GAGylation vary by cell type, as do other posttranslational modifications (PTMs) of the protein core, such as phosphorylation, N-terminal methylation, sulfation, and N-linked or O-linked glycosylation. Lipid attachment, including N-myristoylation, prenylation, and palmitoylation, as well as glycolipid addition, is also possible. PTM of the protein core can affect the function of PGs. For example, modifications in the versican and decorin core proteins are linked to colon adenocarcinoma (Theocharis, 2002), and acylation of an HSPG with myristate and palmitate occurs in carcinoma cells (Iozzo et al., 1990). The PTM of a lumican core protein interacting with melanoma cells has been shown to affect integrin distribution in cell membranes (Brezillon et al., 2009). PTMs and selective metabolic modifications (Fux et al., 2009; Nawroth et al., 2007) can clearly alter or fine tune PG function. Glypican-1, for example, is S-nitrosylated on its protein core to allow processing for polyamine uptake (Fransson, 2003). However, when large GAG chains are attached, they often dominate the physical, chemical, and biological properties of PGs.
PG core proteins vary in size from 10 to 400 kDa. Core protein isoforms exist for glypicans and syndecans (Table 1). For example, glypican 1 is an HSPG and a member of the glypican family, each member of which contains a conserved 14-cysteine residue region. Syndecans 1–4 form a family of HS/CS PGs each containing a single-pass, highly conserved transmembrane domain and two conserved cytoplasmic domains (Bass et al., 2009; Bulow and Hobert, 2006).
A tetrasacharide linkage sequence, GlcAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser, is conserved in the HS, CS, and DS GAG chains of PGs. HS/Hp GAG and CS/DS GAG biosynthesis of the linkage region requires the sequential action of Xyl transferase, Gal transferase β4GalT7, Gal transferase β3GalT6, GlcA transferase I, as well as still undefined sulfotransferases and kinases. Although CS/DS relies on GalNAcT1, HS/Hp instead relies on the enzymes EXTL3 and α4 GlcNAc transferase I for addition of the first hexosamine residue. Once this residue is in place, chain elongation of CS/DS proceeds through the consecutive action of β3GlcA transferase, β4GalNAc transferase, GlcA C5-epimerase, 4-O-sulfotransferases, 6-O-sulfotransferases, and 2-O-sulfotransferase. HS/Hp biosynthesis instead requires EXT1/EXT2, β4 GlcA transferase/α4 GlcNAc transferase, followed by GlcNAc N-deacetylase/N-sulfotransferases, then GlcA C5-epimerase, 2-O-sulfotransferase, 6-O-sulfotransferase, and 3-O-sulfotransferases (Carlsson et al., 2008; Esko and Selleck, 2002; Sugahara and Kitagawa, 2002).
Part-time PGs can be found with or without attached GAG chain or where GAG is replaced by a truncated oligosaccharide. A well-known part-time PG, neuroglycan C (NGC), is a unique transmembrane CSPG found only in the central nervous system (CNS). NGC is thought to play a role in retina and cerebellum development and is present in its non-PG form in undeveloped brain, but its mature form is decorated with CS (Aono et al., 2004; Oohira et al., 2004). In addition to six possible CS attachment sites, among which only one is occupied, NGC core protein contains other PTMs such as phosphorylation, O- and N-glycosylation (Sims and Reinberg, 2008).
HSPGs, the most prevalent PGs in animal cells, mediate embryonic development through binding to fibroblast growth factors and their receptors, controling homeostasis, DNA replication, cell growth, lipid metabolism, and membrane permeability (Dreyfuss et al., 2009; Lin, 2004; Nybakken and Perrimon, 2002; Perrimon and Bernfield, 2000; Schlessinger et al., 2000; Schwartz and Domowicz, 2004; Yamada et al., 2009; Zhang et al., 2007). The major disaccharide-repeating unit of HS is formed by N-acetylglucosamine and glucuronic acid. Syndecans 1–4, glypicans 1–6, betaglycan, perlecan, agrin, serglycin, and collagen type XVIII are all members of HSPG family, with core protein sizes ranging from 10–400 kDa, and with 1–15 HS chains. Syndecan, betaglycan, and serglycin can be glycosylated with both HS and CS chains. Syndecan 1 has five GAG attachment sites, which can be differentially occupied by both HS and CS chains. There are six different glypicans bound to cell surfaces by glycosylphosphatidylinositol (GPI) anchors. Glypicans are expressed during development and linked specifically to fibroblast morphogenesis (Fransson, 2003).
The most abundant CSPGs include aggrecan, versican, neurocan, and brevican. CS chains are formed from repeating N-acetylgalactosamine and glucuronic acid disaccharides, while the DS chains contain some (1–100% of total uronic acid) iduronic acid (Sugahara et al., 2003). Aggrecan, the principal cartilage PG is the largest CSPG with an average of 100 CS chains and a protein core of 208–220 kD (Gargiulo et al., 2009). Aggrecan belongs to the family of small leucine rich proteoglycans (SLRPs), which also includes versican, decorin, biglycan, leprecan, phosphacan, thrombomodulin, CD44, NG2, and serglycin (Kinsella et al., 2004; Ruhland et al., 2007; Schaefer and Iozzo, 2008; Wadhwa et al., 2004). DSPGs have been linked to cardiovascular disease, tumorigenesis, wound repair, fibrosis, and infections (Trowbridge and Gallo, 2002).
KSPGs are classified as either corneal type (KSI, N-glycosidic linkage to asparagine residue) or skeletal type (KSII, O-glycosidic linkage to serine of threonine) depending on the GAG attachment to the core protein. Numerous KSI PGs are also SLRPs that regulate collagen fibril assembly. KSPG is absent in the cornea of patients with macular corneal dystrophy leading to blindness (Akama et al., 2000; Bulow and Hobert, 2006).
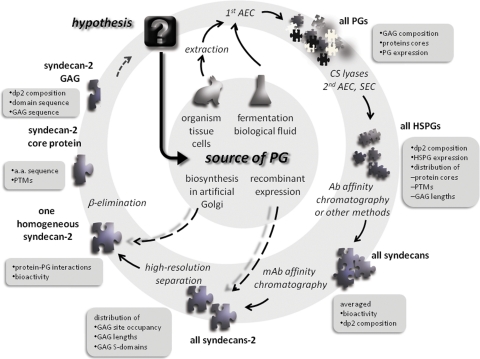
The application of proteoglycomics for the systematic characterization of PG expression and structure holds a great potential for learning physiological roles of these glycoconjugates and determining their impact on disease pathology and developmental biology. The aim of this review is to survey current strategies employed in proteoglycomics and discuss future technological and scientific challenges presented by this area of “omics” studies. Recent developments in the isolation and characterization of PGs and their structural components from cultured cells and tissue samples and the types of information gained are discussed (Fig. 3).
FIG. 3.
Solving the proteoglycomic puzzle: An example of proteoglycomic workflow. The outer circle shows possible analytical tools and is linked to the types of information obtained at each step (boxes).
PG Isolation for Analysis
The GAG component imparts unique physicochemical properties to PGs setting them apart from the rest of the posttranslationally glycosylated proteome. The large hydrodynamic radius and high net negative charge of linear GAG chains make the isolation of PGs a fairly straightforward task. PGs from defatted, minced, powdered, or homogenized tissues or cells can be extracted with a buffered solution of guanidinium chloride containing a detergent and/or protease inhibitors. Large, heavily glycosylated PGs, such as perlecan or aggrecan with molecular densities comparable to those of nucleic acids, can be further purified from the extracts by cesium chloride density gradient centrifugation (Iozzo, 2001). Anion-exchange chromatography (AEC) separates PGs based on the high negative charge of their GAG component and is usually performed using a buffered solution of urea containing a detergent, and/or sodium chloride, which disrupt interactions between PGs and other species in the sample and improve selectivity and efficiency of the PG binding to ion-exchanger. PGs that are present in biological fluids or secreted into the fermentation media can be enriched by directly applying a PG-containing sample diluted with an appropriate buffer to an AEC column (Iozzo, 2001).
Guanidinium chloride extraction followed by AEC in a urea buffer is a classical protocol; however, it is suboptimal for the extraction of small tissue samples in high throughput. Turnbull and coworkers developed a more rapid and efficient method for the PG extraction from cells and tissues with a phenol/guanidine reagent, TRIzol®, and demonstrated its utility for the isolation of HSPGs for the HS disaccharide analysis (Guimond et al., 2009). PGs partition exclusively into the aqueous phase of TRIzol®/chloroform and can be extracted in <1 h. The extract can be applied directly to the anion-exchange medium, which eliminates the buffer exchange step to urea, thus minimizing sample loss. These workers reported quantitative recovery of HSPGs from submilligram amounts of tissues and from several thousand cells in sufficient amounts for the HS structural profiling and bioassay.
Characterization of PG Structural Components
Crude PGs can be further fractionated into their main structural components, core protein and GAG with the linkage region (LR) oligosaccharide, to reduce sample complexity. An established procedure for the GAG release from the core protein is base-catalyzed β-elimination under reducing conditions, which generates a xylitol residue at the GAG reducing end. Prior to the β-elimination, protein can be degraded using a nonspecific protease such as pronase, or actinase to generate peptidoglycans (pGs) and peptides. Proteins, peptides, and neutral glycans can be separated from GAGs and pGs by reverse-phase (RP) chromatography, AEC, or polyacrylamide gel electrophoresis (PAGE). Nucleic acids can be removed from the crude pG or GAG fraction by degradation with endonuclease followed by size-exclusion chromatography (SEC) or dialysis.
Isolated GAGs can be further purified and/or characterized using highly specific lyases (Gu et al., 1993; Jandik et al., 1994). Enzymatic depolymerization is arguably the most important tool in the process of GAG structural characterization. Several CS/DS and HS/Hp lyases, well characterized in terms of their substrate specificity and kinetic parameters, are routinely used for complete depolymerization of GAGs into the building block disaccharides or partial depolymerization to oligosaccharides. Current bioengineering efforts are directed toward expanding the enzymology toolbox to express GAG degrading enzymes with tailored specificities (Hyun et al., 2009a, 2009b; Prabhakar et al., 2005, 2009 Reitinger et al., 2009). In systematic studies such as proteoglycomics, disaccharide analysis is the first step in assessing changes in the GAG compositional profile. Disaccharide composition can be determined using a combination of on-line separation methods such as capillary electrophoresis (Amon et al., 2008; Volpi et al., 2008; Zaia, 2009), ion-pairing RP liquid chromatography (LC) (Doneanu et al., 2009; Korir et al., 2008; Zhang et al., 2009b), hydrophilic interaction LC (HILIC) (Zaia, 2009) in conjunction with MS or UV absorbance detection. Introducing a fluorophore by chemical derivatization of disaccharides greatly enhances their detection sensitivity (Deakin and Lyon, 2008; Hitchcock et al., 2008a; Skidmore et al., 2009) and is a basis for such detection methods as laser-induced fluorescence (Hitchcock et al., 2008b; Korir and Larive, 2009; Skidmore et al., 2009) and fluorophore-assisted carbohydrate electrophoresis (Volpi and Maccari, 2006). Oligosaccharides obtained by partial enzymatic or chemical depolymerization under controlled conditions represent larger pieces of GAG structural information, and their characterization is part of the domain sequencing process. The sample amount requirements for oligosaccharide sequence determination are usually higher than those for disaccharide analysis due to the multidimensional character of their purification. Nuclear magnetic resonance (NMR) spectroscopy is an invaluable method of GAG characterization providing unambiguous structural information for fractionated or purified oligosaccharides (Korir and Larive, 2007, 2009). However, its limited utility in complex mixture analysis precludes NMR spectroscopy from becoming a routine technique for GAG sequencing. (For detailed discussion of current methods for GAG structural characterization, see review by Joseph Zaia in this issue.)
In studies involving PG core protein sequencing or identification, GAGs can be released from their Ser attachment site using β-elimination followed by Michael addition (BEMAD) of DTT or biotin pentylamine (BPA) (Wells et al., 2002). The covalent attachment of DTT or BPA to the dehydroserine residue, formed upon the elimination of GAG, stabilizes the peptide toward degradation, introduces a mass tag that could be isotopically labeled for comparative proteomics, and provides a thiol (DTT) or avidin (BPA) affinity handle at the GAG attachment site. The BEMAD procedure is applicable to both types of O-glycosylation and has been used for mapping O-GlcNAc modifications in the rat brain proteins enriched by anti-O-GlcNAc immunoaffinity chromatography (Wells et al., 2002) and recently, O-GlcNAc PTMs in contractile proteins of rat skeletal muscle (Hedou et al., 2009). Esko and coworkers (2008) used the BEMAD of DTT tagging approach to identify 9 novel CSPGs in Caenorhabditis elegans (Olson et al., 2006).
GAG-protein LR tetrasaccharide (GlcAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser) is a subject of many studies owing to the role it is believed to play in the initiation of GAG chain biosynthesis. Modifications of the LR tetrasaccharide, for example, 2-O-phosphorylation of Xyl and 4-O-sulfonation and 6-O-sulfonation of Gal residues have been correlated with changes in number and type of GAG chains present in PGs (De Waard et al., 1992; Izumikawa et al., 2008; Koike et al., 2009; Matsuno et al., 2007; Nadanaka et al., 1999; Sakaguchi et al., 2001; Ueno et al., 2001; Yamada et al., 2002). The LR oligosaccharides can be purified from crude PG fractions by nonspecific proteolysis, followed by isolation of pGs and their treatment with GAG lysases. SEC can separate the peptide–oligosaccharide conjugate from disaccharides, and the oligosaccharide can be released by β-elimination. Kakehi and coworkers have developed an apparatus for rapid release of O-glycans, AutoGlycoCutter, which decreases the β-elimination reaction time from several hours to several minutes and generates reducing oligosaccharides that can be derivatized with fluorescent tags for downstream sensitive analysis (Matsuno et al., 2007).
MS Analysis and Bioinformatics
Because of the significant differences in ionization characteristics between GAG and protein, these components of PGs are characterized by mass spectrometry (MS) individually and the resulting data are handled separately. MS analysis of intact PGs is a virtually impossible task because of their GAG's low ionization efficiency, size heterogeneity, thermal lability, and propensity to undergo extensive H/Na exchange (Chi et al., 2008; Enghild et al., 1999). When the protein is an object of study, GAGs are exhaustively digested and separated from the protein fraction. The proteins are then separated by PAGE, and the resulting bands are subjected to the in-gel digestion followed by MS and/or MS2 analysis. The LR stub can be removed by β-elimination prior to the MS analysis with or without the tagging and enrichment of GAG attachment sites. In a so-called shotgun approach, intact PGs can be subjected to electrophoresis on a short SDS-PAGE gel followed by in-gel digestion (Talusan et al., 2005). Alternatively, a PG mixture can be proteolyzed and the resulting pGs separated from unmodified peptides by anion-exchange chromatography and/or size-exclusion chromatography for the shotgun LC-MS2 proteomics analysis (Olson et al., 2006). Using bioinformatics tools, the peptides can be identified by matching their MS2 spectra against a database of theoretical MS2 spectra for every possible peptide in the protein database (Armengaud, 2009; Olson et al., 2006).
MS analysis of intact GAGs released from PGs remains incredibly difficult; however, domain sequencing of HS and CS/DS GAGs is a widely applied strategy for GAG characterization (Zaia, 2009). Currently, there are no bioinformatics tools designed specifically for depositing and retrieving GAG structural information, but the glycobioinformatics community is active, and with a constant influx of GAG structural information, the development of bioinformatics tool for managing this information is inevitable (Lutteke et al., 2006; Tissot et al., 2008, 2009; Von Der Lieth et al., 2006). Protein databases such as for example Swiss-Prot contain information on putative GAGylation sites as well as known GAG modifications of proteins as annotations to protein sequence.
MS analysis of GAG oligosaccharides generates complex mass spectra in which single species may appear as a series of peaks due to the H/cation exchange and sulfo group loss. Manual interpretation of MS and MS2 data sets generated in the process of GAG oligosaccharide analysis is a daunting task that may take many hours or even days. Recently, a publicly available glycomics software tool, GlycoWorkbench has been developed by the EUROCarbDB initiative to assist the manual interpretation of MS and MS2 data in high-throughput projects (Ceroni et al., 2008; Tissot et al., 2008). GlycoWorkbench expedites MS and MS2 peak annotation, enables rapid structure assembly using several symbolic notations, and uses sequence-encoding formats compatible with the existing structure databases.
Spatiotemporal profiling
Discovery proteoglycomics
Discovery proteoglycomics explores an ensemble of PGs present in an organism, tissue, or cell type, develops analytical strategies, and provides initial data for further, in-depth studies. Recent examples of exploratory studies from our laboratory include profiling zebrafish HS and CS at different stages of development (Zhang et al., 2009a), characterization of mosquito HS and CS (Sinnis et al., 2007) and HS from various murine tissues (Warda et al., 2006) including murine embryonic stem cells (Nairn et al., 2007), and characterization of human liver HS (Vongchan et al., 2005). The focus of these experiments was the GAG component of PGs. Isolated and purified GAGs were enzymatically depolymerized into constituent disaccharides, and the disaccharide compositional analyses were carried out using RP ion-pair (IP) LC-MS (Zhang et al., 2009b) and HPLC with postcolumn fluorescence detection (Sinnis et al., 2007; Vongchan et al., 2005; Zhang et al., 2009b).
Comparative proteoglycomics
Perhaps, the most important question addressed by “omics” studies in general, and proteoglycomics in particular, is how the PG structure and expression levels vary as a function of environmental or physiological conditions. The goals of comparative proteoglycomic experiments are to identify diagnostic markers, therapeutic targets, and improve our understanding of biological pathways mediated by PGs. Due to the limited space, only five recent examples of comparative studies are discussed here, each employing a different analytical strategy for quantifying the expression of PGs or their GAG components in biological systems under different conditions.
Zaia and coworkers profiled structural changes in CS released from healthy and osteoarthritic cartilage and connective tissues from subjects of different age (Hitchcock et al., 2008b). The study required microgram amounts of CSPGs and afforded compositional analysis of LR tetrasaccharides, nonreducing end oligosaccharides, and internal disaccharides using isotopically labeled anthranilic acid (d0-AA and d4-AA) derivatization and capillary hydrophilic interaction chromatography (HILIC)-MS and MS2 analysis. Quantification of various glycoforms was based on comparing MS signal intensities of d0-AA-derivatized internal standards and d4-AA-derivatized tissue-derived disaccharides. The CS chain lengths were estimated from the ratio of total ion abundances (TIA) of saturated reducing-end oligosaccharides to TIA of unsaturated internal oligosaccharides. Adult human cartilage CS was found to have shorter chains than CS in juvenile bovine cartilage; also, the ratio of N-acetylgalactosamine 6-O-sulfo groups to 4-O-sulfo groups increased with age. The highlights of this study include the streamlined MS-compatible isolation of CS, high sensitivity of detection, and simultaneous MS characterization of the LR, internal chain, and NRE oligosaccharide composition enabled by the HILIC chromatographic microseparation (Hitchcock et al., 2008a).
Diagnostic value of cell-surface HSPG, glypican-3 (GPC3) as a marker for hepatocellular carcinoma (HCC) was assessed using immunohistochemistry by Wang and coworkers (2008). A total of 221 liver specimens were immunostained using anti-GPC3 mAb, and GPC3 was detected in 84 of the 111 HCC specimens, whereas none of the non-HCC lesions showed detectable GPC3. Immunofluorescence microscopy imaging permits visualization of PG expression and spatial distribution in cells and tissues, and currently a number of well-characterized monoclonal antibodies (mAbs) are available commercially that react specifically with PG-related epitopes. Two IgM type mAbs, 10E4 and 3G10, first characterized by David et al. (1992) have been used in several studies for the HSPG immunodetection (David et al., 1992; Thompson et al., 2007; Van Den Born et al., 2005; Yokoyama et al., 1999). An epitope recognized by mAb 10E4 is present in native HS and contains at least one N-sulfo group, whereas mAb 3G10 does not react with intact HS but recognizes HS or HSPGs treated with heparinase III (E.C.4.2.2.8), which generates an unsaturated uronic acid residue attached to an N-acetylglucosamine through a β 1–4 linkage (David et al., 1992). The saccharide structures recognized by anti-HS mAbs HepSS1, JM13, JM403, and 10E4 are described by Van Den Born et al. (Van Den Born et al., 2005).
A number of anti-HS, domain-specific mAbs generated using phage display and biopanning technology have been characterized followed the pioneering work of Van Kuppevelt (Van Kuppevelt et al., 1998). Phage display mAbs bind GAG epitopes with higher affinity and specificity than the IgM type mAbs generated by immunization and have been used in spatial profiling of HS structural domains in kidney (Dennissen et al., 2002; Van Kuppevelt et al., 1998), uterine myometrium (Van Kuppevelt et al., 1998), developing mouse intercostal muscle (Jenniskens et al., 2002), developing rat lung (Thompson et al., 2007), as well as in characterization of eight anticoagulant heparinoids (Wijnhoven et al., 2008).
Antibodies against epitopes specific to other GAG types have been generated, characterized, and used for immunofluorescent detection of PGs in rat brain (Suzuki et al., 2008), mosquito midgut (Dinglasan et al., 2007), ovarian adenocarcinomas (Ten Dam et al., 2007), and human skin (Sorrell et al., 1999) to name just a few. In addition, mAbs raised against neo-epitopes generated by PG proteolysis have been extensively used for studying the distribution and catabolism of connective tissue PGs in normal and arthritic cartilage (Caterson et al., 2000; Fosang et al., 1995; Karsdal et al., 2008; Smith et al., 2009; Sumer et al., 2007).
Lauer et al. (2007) employed fluorophore-assisted carbohydrate electrophoresis (FACE) to examine the changes in the kidney HS disaccharide composition at different time points from the onset of streptozotocin-induced diabetes in rats. Structural analysis of the glomerular and glomerular basement membrane (GBM) HS in healthy and diabetic rat kidneys demonstrated that insulin-dependent diabetes mellitus is marked by a significant decrease in the HS N-sulfo groups. As control animals aged from 3 days to 8 weeks, the amount of N-acetylated disaccharides from the GBM HSPGs decreased while the amount of N-sulfo group containing disaccharides increased. Within the same age group, the diabetic rat GBM contained HS with a lower amount of N-sulfo groups compared to control animals. Thus, both the time of sacrifice and the selection of the specific cell type for HS compositional profiling were important experimental parameters in identifying the disease marker (Lauer et al., 2007).
Stone and coworkers used MS-based proteomics approach to assess the extracellular PG composition in preatherosclerotic lesions from the internal carotid artery and internal thoracic artery, two sites in human vasculature with different propensities for developing atherosclerosis (Talusan et al., 2005). Proteoglycans extracted from the carefully dissected tissues were purified using AEC and subjected to electrophoresis in short SDS-PAGE gel. The portion of the gel containing PGs was subjected to in-gel trypsinolysis, and the resulting peptides were separated by nanoscale RP-HPLC with ESI MS and MS2 detection. Identification and relative quantification of the PG core proteins was achieved using MS signal of their tryptic peptides. The preatherosclerotic intimal extracts were complex mixtures of PGs containing versican, perlecan, biglycan, lumican, decorin, aggrecan, and the two SLRPs fibromodulin and prolargin/PRELP (proline arginine-rich end leucine-rich repeat protein). There was a statistically significant enhancement in the MS signal intensities of lumican-derived peptides in the atherosclerosis-prone internal carotid artery compared to atherosclerosis-resistant internal thoracic artery. The proteomics data was consistent with the results of immunohistochemical staining using mAbs against fibromodulin, biglycan, and lumican, which showed no significant difference in chemiluminescence signals for biglycan and fibromodulin, but a marked increase in the signal for the carotid artery lumican (Talusan et al., 2005).
An interdisciplinary approach combined the GAG disaccharide analysis with the assessment of the levels of transcripts encoding for proteins involved in GAG biosynthesis to examine the role of GAGs in murine embryonic stem cell (ESC) differentiation (Nairn et al., 2007). The GAG disaccharide analysis, performed using RP-IP-HPLC with postcolumn fluorescence detection showed that as ESC differentiated to embryoid bodies (EB) and extraembryonic endodermal cells (EXE), their overall GAG content increased and the sulfation patterns changed. The relative levels of gene expression for the PG core proteins, transporter proteins, and biosynthetic enzymes involved in GAG precursor biosynthesis and catabolism, GAG LR synthesis, and chain extension and modification were determined by quantitative real-time polymerase chain reaction (qRT-PCR) for ESC, EB, and E × E. The CS/DSPG and HSPG core protein mRNA levels increased >10-fold in 39% cases, remained unchanged (<10-fold change) in 56% cases, and decreased >10-fold in 3% cases. Consistent with the increased amounts of 2-O- and 4,6-O-sulfo disaccharides determined by GAG disaccharide analysis, mRNA levels were increased for CS/DS GlcA 2-O-sulfotransferase and GlcNAc 4,6-O-sulfotransferase in EB and EXE cells compared to ESC, demonstrating the utility of this approach in predicting glycome composition and structure from the transcript levels of biosynthetic enzymes.
Future Directions
The “omics” approach to the PG characterization and elucidation of their structure–function relationships is a technology driven, high-throughput analysis of dynamic changes in this subset of the posttranslationally glycosylated proteome. Advances in analytical technology leading to a more sensitive detection and automated data handling make it possible to study such complex and chemically unique class of bioconjugates as PGs.
The emerging field of proteoglycomics has many uncharted territories. Perhaps, the major breakthrough in proteoglycomics will be the development of an amplification mechanism for GAG analysis. Such a development would improve both analytical sensitivity and selectivity. Single-cell PG analysis today may seem like science fiction, but not long ago, the same could be said about the structural analysis of an intact GAG chain by MSn.
A general trend toward miniaturization (Comelli et al., 2006; De Paz and Seeberger, 2008; Liu et al., 2009; Miller and Wheeler, 2009) will also influence proteoglycomics research. Our laboratory is developing a microfluidic device that mimics Golgi organelle where GAG biosynthesis takes place (Martin et al., 2009). Application of this approach in proteoglycomics would allow studying how different enzyme combinations affect the fine structure of GAG–protein interactions between the resulting PG with defined structure and other participants of a biological pathway. As our understanding of protein glycosylation grows, the bioinformatics applications in the field of glycobiology will improve, and with that, patterns of dynamic changes in this PTM will emerge.
Imaging the glycome in vivo is an emerging strategy, which permits visualizing the dynamics of the cell-surface glycan expression, internalization, and trafficking in real time (Baskin et al., 2007; Laughlin et al., 2008; Laughlin and Bertozzi, 2009). In one approach, azido-functionalized sugars (peracetylated N-azidoacetylgalactosamine, N-azidogalactosamine, or N-azidomannose) serve as a covalent attachment site for imaging probes and are installed into glycans using the cell's metabolic machinery (Baskin et al., 2007). Fluorescent probes conjugated to a difluorinated cyclooctyne, which reacts rapidly with azides under physiological conditions, can be introduced at various time points of the organism development using the so-called copper-free click chemistry (Baskin et al., 2007). The click chemistry-based in vivo imaging has been used solely for visualizing cell-surface glycans, due to the impermeability of cell membrane to the fluorescent probe. In principle, azido-sugar-nucleotides can be metabolically introduced into GAGs; thus, it would be interesting to see this strategy implemented in the proteoglycomics studies.
Advances in technology offer additional promise in the field of proteoglycomics. Recombinant technology and metabolic engineering might be used to prepare large quantities of pure PGs. Novel bioinformatics for the statistical treatment of data and improved computational analysis might allow an improved understanding of domain and sequence patterns in nontemplate driven GAG biosynthesis. Advances in synthetic technologies should lead to stable isotopic labeling by metabolic incorporation facilitating structural studies by MS and NMR or the metabolic incorporation of reporter groups. Separation technology such as polyacrylamide gel and capillary electrophoresis with improved methods of detection such as laser-induced fluorescence might afford single homogeneous GAG or PG species for sequence analysis. MS and MS/MS of a single GAG chain and new sugar-specific fragmentation methods offer the potential of automated high throughput sequencing. Miniaturization using microfluidics, microarray technology offers signal amplification, and improved sensitivity potentially leading to single cell analysis. Improvements in atomic force microscopy and X-ray crystallography might one day afford primary structure and higher order structural information on PGs.
Acknowledgments
The authors thank the NIH in the forms of Grants GM38060, GM090257, HL62244, HL094463, RR023764, as well as the NY State Stem Cell Foundation N08G-264, for generously supporting this research.
Author Disclosure Statement
No competing financial interests exist.
References
- Akama T.O. Nishida K. Nakayama J. Watanabe H. Ozaki K. Nakamura T., et al. Macular corneal dystrophy type I and type II are caused by distinct mutations in a new sulphotransferase gene. Nat Genet. 2000;26:237–241. doi: 10.1038/79987. [DOI] [PubMed] [Google Scholar]
- Amon S. Zamfir A.D. Rizzi A. Glycosylation analysis of glycoproteins and proteoglycans using capillary electrophoresis-mass spectrometry strategies. Electrophoresis. 2008;29:2485–2507. doi: 10.1002/elps.200800105. [DOI] [PubMed] [Google Scholar]
- Aono S. Tokita Y. Shuo T. Yamauchi S. Matsui F. Nakanishi K., et al. Glycosylation site for chondroitin sulfate on the neural part-time proteoglycan, neuroglycan C. J Biol Chem. 2004;279:46536–46541. doi: 10.1074/jbc.M403263200. [DOI] [PubMed] [Google Scholar]
- Armengaud J. A perfect genome annotation is within reach with the proteomics and genomics alliance. Curr Opin Microbiol. 2009;12:292–300. doi: 10.1016/j.mib.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Baskin J.M. Prescher J.A. Laughlin S.T. Agard N.J. Chang P.V. Miller I.A., et al. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass M.D. Morgan M.R. Humphries M.J. Syndecans shed their reputation as inert molecules. Sci Signal. 2009;2:pe18. doi: 10.1126/scisignal.264pe18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezillon S. Radwanska A. Zeltz C. Malkowski A. Ploton D. Bobichon H., et al. Lumican core protein inhibits melanoma cell migration via alterations of focal adhesion complexes. Cancer Lett. 2009;283:92–100. doi: 10.1016/j.canlet.2009.03.032. [DOI] [PubMed] [Google Scholar]
- Bulow H.E. Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- Capila I. Linhardt R.J. Heparin–protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Carlsson P. Presto J. Spillmann D. Lindahl U. Kjellen L. Heparin/heparan sulfate biosynthesis: processive formation of N-sulfated domains. J Biol Chem. 2008;283:20008–20014. doi: 10.1074/jbc.M801652200. [DOI] [PubMed] [Google Scholar]
- Caterson B. Flannery C.R. Hughes C.E. Little C.B. Mechanisms involved in cartilage proteoglycan catabolism. Matrix Biol. 2000;19:333–344. doi: 10.1016/s0945-053x(00)00078-0. [DOI] [PubMed] [Google Scholar]
- Cattaruzza S. Perris R. Approaching the proteoglycome: molecular interactions of proteoglycans and their functional output. Macromol Biosci. 2006;6:667–680. doi: 10.1002/mabi.200600100. [DOI] [PubMed] [Google Scholar]
- Ceroni A. Maass K. Geyer H. Geyer R. Dell A. Haslam S.M. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 2008;7:1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- Chi L. Wolff J.J. Laremore T.N. Restaino O.F. Xie J. Schiraldi C., et al. Structural analysis of bikunin glycosaminoglycan. J Am Chem Soc. 2008;130:2617–2625. doi: 10.1021/ja0778500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comelli E.M. Head S.R. Gilmartin T. Whisenant T. Haslam S.M. North S.J., et al. A focused microarray approach to functional glycomics: transcriptional regulation of the glycome. Glycobiology. 2006;16:117–131. doi: 10.1093/glycob/cwj048. [DOI] [PubMed] [Google Scholar]
- David G. Bai X.M. van der Schueren B. Cassiman J.J. van den Berghe H. Developmental changes in heparan sulfate expression: in situ detection with mAbs. J. Cell Biol. 1992;119:961–975. doi: 10.1083/jcb.119.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin J.A. Lyon M. A simplified and sensitive fluorescent method for disaccharide analysis of both heparan sulfate and chondroitin/dermatan sulfates from biological samples. Glycobiology. 2008;18:483–491. doi: 10.1093/glycob/cwn028. [DOI] [PubMed] [Google Scholar]
- Dennissen M.A.B.A. Jenniskens G.J. Pieffers M. Versteeg E.M.M. Petitou M. Veerkamp J.H., et al. Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J Biol Chem. 2002;277:10982–10986. doi: 10.1074/jbc.M104852200. [DOI] [PubMed] [Google Scholar]
- De Paz J.L. Seeberger P.H. Deciphering the glycosaminoglycan code with the help of microarrays. Mol Biosyst. 2008;4:707–711. doi: 10.1039/b802217h. [DOI] [PubMed] [Google Scholar]
- De Waard P. Vliegenthart J.F. Harada T. Sugahara K. Structural studies on sulfated oligosaccharides derived from the carbohydrate-protein linkage region of chondroitin 6-sulfate proteoglycans of shark cartilage. II. Seven compounds containing 2 or 3 sulfate residues. J Biol Chem. 1992;267:6036–6043. [PubMed] [Google Scholar]
- Dinglasan R.R. Alaganan A. Ghosh A.K. Saito A. van Kuppevelt T.H. Jacobs-Lorena M. Plasmodium falciparum ookinetes require mosquito midgut chondroitin sulfate proteoglycans for cell invasion. Proc Natl Acad Sciences USA. 2007;104:15882–15887. doi: 10.1073/pnas.0706340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doneanu C.E. Chen W. Gebler J.C. Analysis of oligosaccharides derived from heparin by ion-pair reversed-phase chromatography/mass spectrometry. Anal Chem. 2009;81:3485–3499. doi: 10.1021/ac802770r. [DOI] [PubMed] [Google Scholar]
- Dreyfuss J.L. Regatieri C.V. Jarrouge T.R. Cavalheiro R.P. Sampaio L.O. Nader H.B. Heparan sulfate proteoglycans: structure, protein interactions and cell signaling. An Acad Bras Cienc. 2009;81:409–429. doi: 10.1590/s0001-37652009000300007. [DOI] [PubMed] [Google Scholar]
- Enghild J.J. Thogersen I.B. Cheng F. Fransson L.A. Roepstorff P. Rahbek-Nielsen H. Organization of the inter-alpha-inhibitor heavy chains on the chondroitin sulfate originating from Ser(10) of bikunin: posttranslational modification of IalphaI-derived bikunin. Biochemistry. 1999;38:11804–11813. doi: 10.1021/bi9908540. [DOI] [PubMed] [Google Scholar]
- Esko J.D. Selleck S.B. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Esko J.D. Kimata K. Lindahl U. Essentials of Glycobiology. 2nd. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2008. Proteoglycans and sulfated glycoaminoglycans. [PubMed] [Google Scholar]
- Fosang A.J. Last K. Gardiner P. Jackson D.C. Brown L. Development of a cleavage-site-specific monoclonal antibody for detecting metalloproteinase-derived aggrecan fragments: detection of fragments in human synovial fluids. Biochem J. 1995;310(Pt 1):337–343. doi: 10.1042/bj3100337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson L.A. Glypicans. Int J Biochem Cell Biol. 2003;35:125–129. doi: 10.1016/s1357-2725(02)00095-x. [DOI] [PubMed] [Google Scholar]
- Fux L. Feibish N. Cohen-Kaplan V. Gingis-Velitski S. Feld S. Geffen C., et al. Structure–function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 2009;69:1758–1767. doi: 10.1158/0008-5472.CAN-08-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargiulo V. Lanzetta R. Parrilli M. de Castro C. Structural analysis of chondroitin sulfate from Scyliorhinus canicula: a useful source of this polysaccharide. Glycobiology. 2009;19:1485–1491. doi: 10.1093/glycob/cwp123. [DOI] [PubMed] [Google Scholar]
- Garner O.B. Yamaguchi Y. Esko J.D. Videm V. Small changes in lymphocyte development and activation in mice through tissue-specific alteration of heparan sulphate. Immunology. 2008;125:420–429. doi: 10.1111/j.1365-2567.2008.02856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu K. Liu J. Pervin A. Linhardt R.J. Comparison of the activity of two chondroitin AC lyases on dermatan sulfate. Carbohydr Res. 1993;244:369–377. doi: 10.1016/0008-6215(83)85014-9. [DOI] [PubMed] [Google Scholar]
- Guimond S.E. Puvirajesinghe T.M. Skidmore M.A. Kalus I. Dierks T. Yates E.A., et al. Rapid purification and high sensitivity analysis of heparan sulfate from cells and tissues: toward glycomics profiling. J Biol Chem. 2009;284:25714–25722. doi: 10.1074/jbc.M109.032755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedou J. Bastide B. Page A. Michalski J.C. Morelle W. Mapping of O-linked beta-N-acetylglucosamine modification sites in key contractile proteins of rat skeletal muscle. Proteomics. 2009;9:2139–2148. doi: 10.1002/pmic.200800617. [DOI] [PubMed] [Google Scholar]
- Hitchcock A.M. Bowman M.J. Staples G.O. Zaia J. Improved workup for glycosaminoglycan disaccharide analysis using CE with LIF detection. Electrophoresis. 2008a;29:4538–4548. doi: 10.1002/elps.200800335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock A.M. Yates K.E. Costello C.E. Zaia J. Comparative glycomics of connective tissue glycosaminoglycans. Proteomics. 2008b;8:1384–1397. doi: 10.1002/pmic.200700787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y.J. Lee J.H. Kim D.H. Cloning, overexpression, and characterization of recombinant heparinase III from Bacteroides stercoris HJ-15. Appl Microbiol Biotechnol. 2009a Nov 12; doi: 10.1007/s00253-009-2327-7. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Hyun Y.J. Lee K.S. Kim D.H. Cloning, expression and characterization of acharan sulfate-degrading heparin lyase II from Bacteroides stercoris HJ-15. J Appl Microbiol. 2009b;108:226–235. doi: 10.1111/j.1365-2672.2009.04418.x. [DOI] [PubMed] [Google Scholar]
- Iozzo R.V. Proteoglycan Protocols. Humana Press; Totowa, NJ: 2001. [Google Scholar]
- Iozzo R.V. Kovalszky I. Hacobian N. Schick P.K. Ellingson J.S. Dodge G.R. Fatty acylation of heparan sulfate proteoglycan from human colon carcinoma cells. J Biol Chem. 1990;265:19980–19989. [PubMed] [Google Scholar]
- Izumikawa T. Koike T. Shiozawa S. Sugahara K. Tamura J.-I. Kitagawa H. Identification of chondroitin sulfate glucuronyltransferase as chondroitin synthase-3 involved in chondroitin polymerization: chondroitin polymerization is achieved by multiple enzyme complexes consisting of chondroitin synthase family members. J Biol Chem. 2008;283:11396–11406. doi: 10.1074/jbc.M707549200. [DOI] [PubMed] [Google Scholar]
- Jandik K.A. Gu K. Linhardt R.J. Action pattern of polysaccharide lyases on glycosaminoglycans. Glycobiology. 1994;4:289–296. doi: 10.1093/glycob/4.3.289. [DOI] [PubMed] [Google Scholar]
- Jenniskens G.J. Hafmans T. Veerkamp J.H. van Kuppevelt T.H. Spatiotemporal distribution of heparan sulfate epitopes during myogenesis and synaptogenesis: a study in developing mouse intercostal muscle. Dev Dynam. 2002;225:70–79. doi: 10.1002/dvdy.10138. [DOI] [PubMed] [Google Scholar]
- Karsdal M.A. Madsen S.H. Christiansen C. Henriksen K. Fosang A.J. Sondergaard B.C. Cartilage degradation is fully reversible in the presence of aggrecanase but not matrix metalloproteinase activity. Arthritis Res Ther. 2008;10:R63. doi: 10.1186/ar2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella M.G. Bressler S.L. Wight T.N. The regulated synthesis of versican, decorin, and biglycan: extracellular matrix proteoglycans that influence cellular phenotype. Crit Rev Eukaryot Gene Expr. 2004;14:203–234. doi: 10.1615/critreveukaryotgeneexpr.v14.i3.40. [DOI] [PubMed] [Google Scholar]
- Koike T. Izumikawa T. Tamura J. Kitagawa H. FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan-protein linkage region. Biochem J. 2009;421:157–162. doi: 10.1042/BJ20090474. [DOI] [PubMed] [Google Scholar]
- Korir A.K. Larive C.K. On-line NMR detection of microgram quantities of heparin-derived oligosaccharides and their structure elucidation by microcoil NMR. Anal Bioanal Chem. 2007;388:1707–1716. doi: 10.1007/s00216-007-1400-2. [DOI] [PubMed] [Google Scholar]
- Korir A.K. Larive C.K. Advances in the separation, sensitive detection, and characterization of heparin and heparan sulfate. Anal Bioanal Chem. 2009;393:155–169. doi: 10.1007/s00216-008-2412-2. [DOI] [PubMed] [Google Scholar]
- Korir A.K. Limtiaco J.F. Gutierrez S.M. Larive C.K. Ultraperformance ion-pair liquid chromatography coupled to electrospray time-of-flight mass spectrometry for compositional profiling and quantification of heparin and heparan sulfate. Anal Chem. 2008;80:1297–1306. doi: 10.1021/ac702235u. [DOI] [PubMed] [Google Scholar]
- Kreuger J. Spillmann D. Li J.-P. Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer M.E. Hascall V.C. Wang A. Heparan sulfate analysis from diabetic rat glomeruli. J Biol Chem. 2007;282:843–852. doi: 10.1074/jbc.M608823200. [DOI] [PubMed] [Google Scholar]
- Laughlin S.T. Bertozzi C.R. Imaging the glycome. Proc Natl Acad Sci USA. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin S.T. Baskin J.M. Amacher S.L. Bertozzi C.R. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- Liu Y. Palma A.S. Feizi T. Carbohydrate microarrays: key developments in glycobiology. Biol Chem. 2009;390:647–656. doi: 10.1515/BC.2009.071. [DOI] [PubMed] [Google Scholar]
- Lutteke T. Bohne-Lang A. Loss A. Goetz T. Frank M. von der Lieth C.-W. Glycosciences.de: an Internet portal to support glycomics and glycobiology research. Glycobiology. 2006;16:71R–81R. doi: 10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]
- Martin J.G. Gupta M. Xu Y. Akella S. Liu J. Dordick J.S., et al. Toward an artificial Golgi: redesigning the biological activities of heparan sulfate on a digital microfluidic chip. J Am Chem Soc. 2009;131:11041–11048. doi: 10.1021/ja903038d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno Y.-K. Yamada K. Tanabe A. Kinoshita M. Maruyama S.-Z. Osaka Y.-S., et al. Development of an apparatus for rapid release of oligosaccharides at the glycosaminoglycan-protein linkage region in chondroitin sulfate-type proteoglycans. Anal Biochem. 2007;362:245–257. doi: 10.1016/j.ab.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Miller E.M. Wheeler A.R. Digital bioanalysis. Anal Bioanal Chem. 2009;393:419–426. doi: 10.1007/s00216-008-2397-x. [DOI] [PubMed] [Google Scholar]
- Nadanaka S. Kitagawa H. Goto F. Tamura J. Neumann K.W. Ogawa T., et al. Involvement of the core protein in the first beta-N-acetylgalactosamine transfer to the glycosaminoglycan–protein linkage-region tetrasaccharide and in the subsequent polymerization: the critical determining step for chondroitin sulphate biosynthesis. Biochem J. 1999;340:353–357. [PMC free article] [PubMed] [Google Scholar]
- Nairn A.V. Kinoshita-Toyoda A. Toyoda H. Xie J. Harris K. Dalton S., et al. Glycomics of proteoglycan biosynthesis in murine embryonic stem cell differentiation. J Proteome Res. 2007;6:4374–4387. doi: 10.1021/pr070446f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroth R. van Zante A. Cervantes S. McManus M. Hebrok M. Rosen S.D. Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS One. 2007;2:e392. doi: 10.1371/journal.pone.0000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken K. Perrimon N. Heparan sulfate proteoglycan modulation of developmental signaling in Drosophila. Biochim Biophys Acta. 2002;1573:280–291. doi: 10.1016/s0304-4165(02)00395-1. [DOI] [PubMed] [Google Scholar]
- Olson S.K. Bishop J.R. Yates J.R. Oegema K. Esko J.D. Identification of novel chondroitin proteoglycans in Caenorhabditis elegans: embryonic cell division depends on CPG-1 and CPG-2. J Cell Biol. 2006;173:985–994. doi: 10.1083/jcb.200603003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oohira A. Shuo T. Tokita Y. Nakanishi K. Aono S. Neuroglycan C, a brain-specific part-time proteoglycan, with a particular multidomain structure. Glycoconj J. 2004;21:53–57. doi: 10.1023/B:GLYC.0000043748.90896.83. [DOI] [PubMed] [Google Scholar]
- Perrimon N. Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- Prabhakar V. Capila I. Bosques C.J. Pojasek K. Sasisekharan R. Chondroitinase ABC I from Proteus vulgaris: cloning, recombinant expression and active site identification. Biochem J. 2005;386:103–112. doi: 10.1042/BJ20041222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar V. Capila I. Soundararajan V. Raman R. Sasisekharan R. Recombinant expression, purification, and biochemical characterization of chondroitinase ABC II from Proteus vulgaris. J Biol Chem. 2009;284:974–982. doi: 10.1074/jbc.M806630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman R. Raguram S. Venkataraman G. Paulson J.C. Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat Methods. 2005;2:817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- Raman R. Venkataraman M. Ramakrishnan S. Lang W. Raguram S. Sasisekharan R. Advancing glycomics: implementation strategies at the consortium for functional glycomics. Glycobiology. 2006;16:82R–90R. doi: 10.1093/glycob/cwj080. [DOI] [PubMed] [Google Scholar]
- Reitinger S. Mullegger J. Greiderer B. Nielsen J.E. Lepperdinger G. Designed human serum hyaluronidase 1 variant, HYAL1DeltaL, exhibits activity up to pH 5.9. J Biol Chem. 2009;284:19173–19177. doi: 10.1074/jbc.C109.004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhland C. Schonherr E. Robenek H. Hansen U. Iozzo R.V. Bruckner P., et al. The glycosaminoglycan chain of decorin plays an important role in collagen fibril formation at the early stages of fibrillogenesis. FEBS J. 2007;274:4246–4255. doi: 10.1111/j.1742-4658.2007.05951.x. [DOI] [PubMed] [Google Scholar]
- Sakaguchi H. Watanabe M. Ueoka C. Sugiyama E. Taketomi T. Yamada S., et al. Isolation of reducing oligosaccharide chains from the chondroitin/dermatan sulfate–protein linkage region and preparation of analytical probes by fluorescent labeling with 2-aminobenzamide. J Biochem. 2001;129:107–118. doi: 10.1093/oxfordjournals.jbchem.a002820. [DOI] [PubMed] [Google Scholar]
- Schaefer L. Iozzo R.V. Biological functions of the small leucine-rich proteoglycans: from genetics to signal transduction. J Biol Chem. 2008;283:21305–21309. doi: 10.1074/jbc.R800020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J. Plotnikov A.N. Ibrahimi O.A. Eliseenkova A.V. Yeh B.K. Yayon A., et al. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- Schwartz N.B. Domowicz M. Proteoglycans in brain development. Glycoconj J. 2004;21:329–341. doi: 10.1023/B:GLYC.0000046278.34016.36. [DOI] [PubMed] [Google Scholar]
- Sims R.J., III Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- Sinnis P. Coppi A. Toida T. Toyoda H. Kinoshita-Toyoda A. Xie J., et al. Mosquito heparan sulfate and its potential role in malaria infection and transmission. J Biol Chem. 2007;282:25376–25384. doi: 10.1074/jbc.M704698200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skidmore M. Atrih A. Yates E. Turnbull J.E. Labelling Heparan Sulphate Saccharides with Chromophore, Fluorescence and Mass Tags for HPLC and MS Separations. Humana Press; Clifton, NJ: 2009. [DOI] [PubMed] [Google Scholar]
- Smith S. Whitelock J. Iozzo R. Little C. Melrose J. Topographical variation in the distributions of versican, aggrecan and perlecan in the foetal human spine reflects their diverse functional roles in spinal development. Histochem Cell Biol. 2009 Aug 11; doi: 10.1007/s00418-009-0623-z. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Sorrell J.M. Carrino D. Baber M. Asselineau D. Caplan A. A monoclonal antibody which recognizes a glycosaminoglycan epitope in both dermatan sulfate and chondroitin sulfate proteoglycans of human skin. Histochem J. 1999;31:549–558. doi: 10.1023/a:1003896124595. [DOI] [PubMed] [Google Scholar]
- Sugahara K. Kitagawa H. Heparin and heparan sulfate biosynthesis. IUBMB Life. 2002;54:163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- Sugahara K. Mikami T. Uyama T. Mizuguchi S. Nomura K. Kitagawa H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr Opin Struct Biol. 2003;13:612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Sumer E.U. Sondergaard B.C. Rousseau J.C. Delmas P.D. Fosang A.J. Karsdal M.A., et al. MMP and non-MMP-mediated release of aggrecan and its fragments from articular cartilage: a comparative study of three different aggrecan and glycosaminoglycan assays. Osteoarthritis Cartilage. 2007;15:212–221. doi: 10.1016/j.joca.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Suzuki K. Yamamoto K. Kariya Y. Maeda H. Ishimaru T. Miyaura S., et al. Generation and characterization of a series of monoclonal antibodies that specifically recognize [HexA(±2S)-GlcNAc]n epitopes in heparan sulfate. Glycoconj J. 2008;25:703–712. doi: 10.1007/s10719-008-9130-z. [DOI] [PubMed] [Google Scholar]
- Talusan P. Bedri S. Yang S. Kattapuram T. Silva N. Roughley P.J., et al. Analysis of intimal proteoglycans in atherosclerosis-prone and atherosclerosis-resistant human arteries by mass spectrometry. Mol Cell Proteomics. 2005;4:1350–1357. doi: 10.1074/mcp.M500088-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Dam G.B. van de Westerlo E.M.A. Purushothaman A. Stan R.V. Bulten J. Sweep F.C.G.J., et al. Antibody GD3G7 selected against embryonic glycosaminoglycans defines chondroitin sulfate-E domains highly up-regulated in ovarian cancer and involved in vascular endothelial growth factor binding. Am J Pathol. 2007;171:1324–1333. doi: 10.2353/ajpath.2007.070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theocharis A.D. Human colon adenocarcinoma is associated with specific post-translational modifications of versican and decorin. Biochim Biophys Acta. 2002;1588:165–172. doi: 10.1016/s0925-4439(02)00161-8. [DOI] [PubMed] [Google Scholar]
- Thompson S. Connell M. Fernig D. Ten Dam G. van Kuppevelt T. Turnbull J., et al. Novel ‘phage display antibodies identify distinct heparan sulfate domains in developing mammalian lung. Pediatr Surg Int. 2007;23:411–417. doi: 10.1007/s00383-006-1864-8. [DOI] [PubMed] [Google Scholar]
- Timmer M.S. Stocker B.L. Seeberger P.H. Probing glycomics. Curr Opin Chem Biol. 2007;11:59–65. doi: 10.1016/j.cbpa.2006.11.040. [DOI] [PubMed] [Google Scholar]
- Tissot B. Ceroni A. Powell A.K. Morris H.R. Yates E.A. Turnbull J.E., et al. Software tool for the structural determination of glycosaminoglycans by mass spectrometry. Anal Chem. 2008;80:9204–9212. doi: 10.1021/ac8013753. [DOI] [PubMed] [Google Scholar]
- Tissot B. North S.J. Ceroni A. Pang P.C. Panico M. Rosati F., et al. Glycoproteomics: past, present and future. FEBS Lett. 2009;583:1728–1735. doi: 10.1016/j.febslet.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge J.M. Gallo R.L. Dermatan sulfate: new functions from an old glycosaminoglycan. Glycobiology. 2002;12:117R–125R. doi: 10.1093/glycob/cwf066. [DOI] [PubMed] [Google Scholar]
- Ueno M. Yamada S. Zako M. Bernfield M. Sugahara K. Structural characterization of heparan sulfate and chondroitin sulfate of syndecan-1 purified from normal murine mammary gland epithelial cells. Common phosphorylation of xylose and differential sulfation of galactose in the protein linkage region tetrasaccharide sequence. J Biol Chem. 2001;276:29134–29140. doi: 10.1074/jbc.M102089200. [DOI] [PubMed] [Google Scholar]
- Van Den Born J. Salmivirta K. Henttinen T. Östman N. Ishimaru T. Miyaura S., et al. Novel Heparan Sulfate Structures Revealed by Monoclonal Antibodies. J Biol Chem. 2005;280:20516–20523. doi: 10.1074/jbc.M502065200. [DOI] [PubMed] [Google Scholar]
- Van Kuppevelt T.H. Dennissen M.A.B.A. van Venrooij W.J. Hoet R.M.A. Veerkamp J.H. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. J Biol Chem. 1998;273:12960–12966. doi: 10.1074/jbc.273.21.12960. [DOI] [PubMed] [Google Scholar]
- Volpi N. Maccari F. Electrophoretic approaches to the analysis of complex polysaccharides. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;834:1–13. doi: 10.1016/j.jchromb.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Volpi N. Maccari F. Linhardt R.J. Capillary electrophoresis of complex natural polysaccharides. Electrophoresis. 2008;29:3095–3106. doi: 10.1002/elps.200800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Der Lieth C.-W. Lütteke T. Frank M. The role of informatics in glycobiology research with special emphasis on automatic interpretation of MS spectra. Biochim Biophys Acta (BBA) 2006;1760:568–577. doi: 10.1016/j.bbagen.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Vongchan P. Warda M. Toyoda H. Toida T. Marks R.M. Linhardt R.J. Structural characterization of human liver heparan sulfate. Biochim Biophys Acta. 2005;1721:1–8. doi: 10.1016/j.bbagen.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Wadhwa S. Embree M.C. Bi Y. Young M.F. Regulation, regulatory activities, and function of biglycan. Crit Rev Eukaryot Gene Expr. 2004;14:301–315. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.50. [DOI] [PubMed] [Google Scholar]
- Wang H.L. Anatelli F. Zhai Q.J. Adley B. Chuang S.T. Yang X.J. Glypican-3 as a useful diagnostic marker that distinguishes hepatocellular carcinoma from benign hepatocellular mass lesions. Arch Pathol Lab Med. 2008;132:1723–1728. doi: 10.5858/132.11.1723. [DOI] [PubMed] [Google Scholar]
- Warda M. Toida T. Zhang F. Sun P. Munoz E. Xie J., et al. Isolation and characterization of heparan sulfate from various murine tissues. Glycoconj J. 2006;23:555–563. doi: 10.1007/s10719-006-7668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L. Vosseller K. Cole R.N. Cronshaw J.M. Matunis M.J. Hart G.W. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- Wijnhoven T. van de Westerlo E. Smits N. Lensen J. Rops A. van der Vlag J., et al. Characterization of anticoagulant heparinoids by immunoprofiling. Glycoconjugate J. 2008;25:177–185. doi: 10.1007/s10719-007-9070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S. Okada Y. Ueno M. Iwata S. Deepa S.S. Nishimura S., et al. Determination of the glycosaminoglycan-protein linkage region oligosaccharide structures of proteoglycans from Drosophila melanogaster and Caenorhabditis elegans. J Biol Chem. 2002;277:31877–31886. doi: 10.1074/jbc.M205078200. [DOI] [PubMed] [Google Scholar]
- Yamada S. Onishi M. Fujinawa R. Tadokoro Y. Okabayashi K. Asashima M., et al. Structural and functional changes of sulfated glycosaminoglycans in Xenopus laevis during embryogenesis. Glycobiology. 2009;19:488–498. doi: 10.1093/glycob/cwp005. [DOI] [PubMed] [Google Scholar]
- Yokoyama H. Sato K. Okudaira M. Morita C. Takahashi C. Suzuki D., et al. Serum and urinary concentrations of heparan sulfate in patients with diabetic nephropathy. Kidney Int. 1999;56:650–658. doi: 10.1046/j.1523-1755.1999.00591.x. [DOI] [PubMed] [Google Scholar]
- Zaia J. On-line separations combined with MS for analysis of glycosaminoglycans. Mass Spectrom Rev. 2009;28:254–272. doi: 10.1002/mas.20200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. McLellan J.S. Ayala A.M. Leahy D.J. Linhardt R.J. Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry. 2007;46:3933–3941. doi: 10.1021/bi6025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. Zhang Z. Thistle R. McKeen L. Hosoyama S. Toida T., et al. Structural characterization of glycosaminoglycans from zebrafish in different ages. Glycoconj J. 2009a;26:211–218. doi: 10.1007/s10719-008-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. Xie J. Liu H. Liu J. Linhardt R.J. Quantification of heparan sulfate disaccharides using ion-pairing reversed-phase microflow high-performance liquid chromatography with electrospray ionization trap mass spectrometry. Anal Chem. 2009b;81:4349–4355. doi: 10.1021/ac9001707. [DOI] [PMC free article] [PubMed] [Google Scholar]