Abstract
Oxidative stress and protein carbonylation is implicated in aging and various diseases such as neurodegenerative disorders, diabetes, and cancer. Therefore, the accurate identification and quantification of protein carbonylation may lead to the discovery of new biomarkers. We have developed a new method that combines avidin affinity selection of carbonylated proteins with iTRAQ labeling and LC fractionation of intact proteins. This simple LC-based workflow is an effective technique to reduce sample complexity, minimize technical variation, and enable simultaneous quantification of four samples. This method was used to determine protein oxidation in an iron accumulating mutant of Saccharomyces cerevisiae exposed to oxidative stress. Overall, 31 proteins were identified with 99% peptide confidence, and of those, 27 proteins were quantified. Most of the identified proteins were associated with energy metabolism (32.3%), and cellular defense, transport, and folding (38.7%), suggesting a drop in energy production and reducing power of the cells due to the damage of glycolytic enzymes and decrease in activity of enzymes involved in protein protection and regeneration. In addition, the oxidation sites of seven proteins were identified and their estimated position also indicated a potential impact on the enzymatic activities. Predicted 3D structures of peroxiredoxin (TSA1) and thioredoxin II (TRX2) revealed close proximity of all oxidized amino acid residues to the protein active sites.
Introduction
Oxidative stress (OS) is defined as an imbalance between processes producing reactive oxygen species (ROS) and antioxidant cascades, which removes and prevents the formation of ROS. Superoxide anions (O2•−), hydroxyl radicals (OH·) and hydrogen peroxide in the presence of transition metal ions are well-known ROS that cause cellular damage (Jamieson, 1998; Sayre et al., 2008; Sies, 1997) through chemical modifications of specific biomolecules, particularly lipids and proteins. Direct protein oxidation results in the formation of carbonyl groups (aldehydes and ketones) on specific amino acids such as Arg (glutamic semialdehyde), Lys (2-aminoadipic semialdehyde), Pro (glutamic semialdehyde), and Thr (2-amino-3-ketobutyric acid) (Levine and Stadtman, 2001), leading to irreversible and irreparable protein damage (Nystrom, 2005). Protein carbonylation is associated with age-related disorders and diseases such as Parkinson's (Berg et al., 2001), Alzheimer's (Aliev et al., 2002), diabetes (Maritim et al., 2003) and cancer (Akman, 2003; Sayre et al., 2008; Sies, 1997). In addition, specific sets of proteins appear to be prone to carbonylation in starvation, aging, or disease states (Cabiscol et al., 2000; Levine, 2002; Tamarit et al., 1998). Therefore, the accurate identification and quantification of protein carbonylation is of great interest, as it may lead to the discovery of new predictive and diagnostic biomarkers. Traditional techniques for determination of carbonylated proteins include derivatization with 2,4-dinitrophenylhydrazine (DNP) (England and Cotter, 2004; Korolainen et al., 2002; Kristensen et al., 2004; Talent et al., 1998) or with biotin–hydrazide (Yoo and Regnier, 2004) followed by gel-based protein separation and affinity staining by avidin–FITC. Recently, various methods based on the enrichment of biotin–hydrazide labeled proteins with avidin or streptavidin affinity chromatography, and their identification using nanoLC-MS/MS have been published (Mirzaei and Regnier, 2005; Soreghan et al., 2003; Thomas et al., 2005). Although promising, the methods only provide semiquantitative data due to the ion suppression effect that occurs during LC-ESI-MS analysis. To overcome this challenge and increase the accuracy and reproducibility of LC-ESI-MS-based techniques, numerous stable-isotope labeling strategies have been developed, most of which target the primary amines generated by tryptic digestion of the proteins. To eliminate a potential inconsistency in proteolysis, several approaches of intact protein labeling have been reported. For example, isotope-coded protein labeling (ICPL) is a strategy based on coding lysine side chains and α-amino groups where primary amine coding is achieved using d0/d4 isotopomers of N-hydroxyl succinimide activated nicotinic acid (Schmidt et al., 2005). The ICPL includes protein labeling and 1D- or 2D-gel separation followed by trypsin digestion and LC-MS/MS analysis (Schmidt et al., 2005). This method was evaluated and validated using an Escherichia coli proteome spiked with five standard proteins. The standard proteins were quantified with an average SD of 8.3% and average error of 4.6%. Unfortunately, although this labeling reagent showed high accuracy and reproducibility, the quantification is achieved through the first dimension of MS/MS (scan mode), and it may prove to be challenging for complex samples. In recent years, commercially available iTRAQ reagents have become widely used in proteomics research. These reagents are global amine reactive tags that label the N-terminal and lysine residues of peptides (Regnier and Julka, 2006; Ross et al., 2004). The reproducibility, variation, and accuracy of multiplexed protein quantification through peptide derivatization has been evaluated (Boehm et al., 2007; Chong et al., 2006; Gan et al., 2007; Regnier and Julka, 2006; Ross et al., 2004) and many quantification studies have been performed with the proteins obtained from various cell types (Aggarwal et al., 2004; Borner et al., 2006; Hardt et al., 2005; Lund et al., 2007; Reinders et al., 2006; Unwin et al., 2006). In 2007, Wiese et al. showed the first intact protein labeling strategy using iTRAQ (Wiese et al., 2007). The labeled proteins were separated by SDS-gel electrophoresis followed by in gel digestion and peptide MS/MS analysis. The dynamic range of the method was tested with a differentially labeled glucose oxidase in concentration ratios ranging from 1:10 to 10:1, showing an average SD of 12%. In addition, two protein samples containing five standard proteins at different ratios were differentially labeled with iTRAQ-115 and iTRAQ-117, respectively, and their SD observed in MALDI-MS/MS was about 10%.
In this study, we have identified and quantified the dynamic changes in protein carbonyl content using a unique iron accumulating Δyfh1 yeast mutant exposed to oxidative stress. Protein YFH1 is a homologue to the human mitochondrial protein frataxin (encoded by the chromosomal FRDA gene) that is responsible for an autosomal recessive neurodegenerative disorder, Friedreich's ataxia (FA), affecting about 1 in 50,000 people (Campuzano et al., 1996). Homology between yeast and human frataxin suggests that both proteins carry out fundamental cell functions conserved across different species (Huynen et al., 2001). Previous yeast studies demonstrated that the deletion of YFH1 caused an accumulation of iron in mitochondria (Adamec et al., 2000; Babcock et al., 1997; Foury and Cazzalini, 1997; Radisky et al., 1999), which is an essential element in energy metabolism. Above a certain threshold, however, iron represents a potentially dangerous constituent in cell function as it reacts with oxygen and hydrogen peroxide to produce high levels of ROS that may irreversibly damage cells (Jeong and David, 2006). Because the pathways of disease progression are still not completely understood, it is important to quantify the target proteins affected by OS in different stages to explain the relationship between OS and disease progression. Carbonylated proteins were identified and quantified using a newly developed LC-based labeling method. Oxidized proteins were modified with biotin hydrazide in an LC-based approach, and isolated by affinity chromatography using immobilized avidin. The proteins were then labeled with iTRAQ reagents, separated on RPLC, digested with Glu-C, and analyzed by MS/MS. Although gel-based methods have demonstrated promising results, in-solution digestion is generally less time-consuming and has better protein coverage than in-gel digestion (Roe and Griffin, 2006). This simple LC-based workflow with iTRAQ protein labeling is an effective methodology to remove potential inhibitors of digestion and reduce sample complexity, thus enabling simultaneous quantification of four samples.
Materials and Methods
Ultralink immobilized monomeric avidin, EZ-Link biotin hydrazides, D-biotin, biotinylated bovine serum albumin (BSA), bond-Breaker™ TCEP Solution (500 mM stock), and Slide-A-Lyzer (MWCO = 10,000) for dialysis were purchased from Pierce (Rockford, IL). Standard chemicals, BSA, protease inhibitor cocktail, and IGEPAL CA-630 were purchased from Sigma (St. Louis, MO). The iTRAQTM reagents multiplex kit and proteomic grade trypsin (TPCK treated) were obtained from Applied Biosystems Inc. (ABI, Foster City, CA). Endoproteinase Glu-C enzyme was purchased from Roche Diagnostics Co. (Indianapolis, IN).
Growth conditions and stress induction
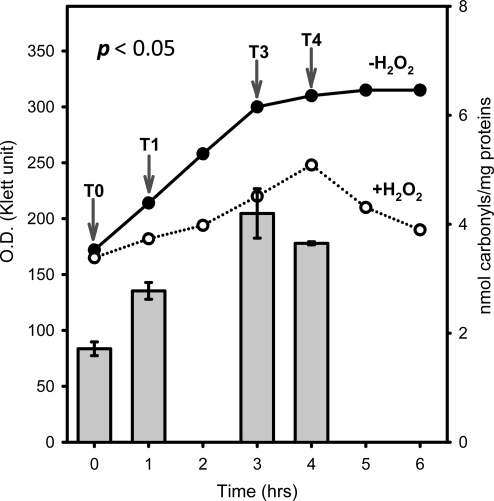
An overnight culture of frataxin homologue deletion strain Saccharomyces cerevisiae 10546A (Δyfh1 FY MATa ura3-52 HIS3 leu2Δ1 LYS2 trp1Δ63 YDL120w (4,531)::kanMX4) (EUROSCARF, Germany) was inoculated in 400-mL YEPD media (1% yeast extract, 2% peptone, 2% glucose) at an OD ∼50 KU (Klett Unit) and cells were grown at 30°C to OD of ∼160 KU. At this time, 1 mL of 8 M H2O2 was added (20 mM final concentration in media), and the culture was shaken for an additional 4 h at 30°C. For analysis, aliquots of cell culture were collected just before H2O2 addition (T0), and 1 (T1), 3 (T3), and 4 (T4) h following exposure (Kim et al., 2010).
Biotinylation and enrichment of oxidized proteins
The modified method described by Kim et al. (2010) was used to isolate oxidized proteins. Cells from the 400-mL culture were harvested into two 250-mL Beckman centrifuge tubes by centrifugation (3,000 × g) for 10 min at 4°C, washed twice in cold water, and the combined pellet immediately resuspended into 50 mL conical tubes with equal volumes of BH Lysis Buffer (5 mM biotin hydrazide; 4.5 mM NaH2PO4; 49.4 mM Na2HPO4; 48.4 mM NaCl, 5 mM KCl; 20% glycerol: 1% 2-mercaptoethanol, 0.3% IGEPAL CA-630; protease inhibitor cocktail; pH 7.4). Cells were disrupted by vortexing with glass beads (425–600 μm; acid washed; amount equal to pellet weight) for 1 min followed by 1 min rest on ice and repeated five times. The cellular debris was removed by centrifugation at 20,000 × g for 30 min at 4°C. The supernatant was mixed with an equal volume of R Buffer (30 mM sodium cyanoborohydride; 4.5 mM NaH2PO4; 49.4 mM Na2HPO4; 48.4 mM NaCl, 5 mM KCl; pH 7.4) and incubated for 40 min on ice. To remove excess biotin hydrazide and detergent, the sample was dialyzed against PBS buffer (0.1 M sodium phosphate, 0.15 M NaCl, pH 7.4) followed by determination of the protein concentration with Bio-Rad protein assay kit (standard curve was generated using BSA). To isolate biotinylated proteins, a stainless steel column (4.6 × 100 mm) was packed with Ultralinked immobilized monomeric avidin at 100 psi and binding capacity was evaluated using biotinylated BSA. The column was equilibrated with 17-mL PBS buffer followed by sample injection (30 mL with protein concentration at 2 mg/mL) and washing with 17 mL of PBS buffer to remove any unbound proteins. Biotinylated proteins were then eluted with 17 mL of elution buffer (2 mM D-biotin in 50 mM PBS). At the end of each run the column was regenerated with 17 mL of 0.1 M glycine (pH 2.8) and reequilibrated with 17 mL PBS buffer. Protein elution was monitored at A280 during the entire run. To remove the excess D-biotin, fractions containing the biotinylated proteins were pooled (10 mL), dialyzed against 10 mM HEPES buffer (pH 8.5), and stored at −80°C.
Labeling of a model protein in protein level
Labeling of standard protein
BSA was dissolved at a concentration of 1 mg/mL in 6 M Urea and 0.5 M HEPES buffer (pH 8.5). Aliquots (20 μL) were reduced with 2 μL of 0.5 M TCEP at 37°C for 1 h and alkylated with 2 μL 0.5 M iodoacetamide by incubating the mixtures at room temperature in the dark for 30 min. The aliquots were then diluted with 16 μL of water and labeled for 2 h with corresponding iTRAQ reagents (114, 115, 116, or 117) previously diluted in 50 μL ethanol. Reactions were stopped by adding 2 μL of 10 mM lysine to each tube and further incubation for 20 min at room temperature. Differentially labeled BSA samples were then combined into a single tube as needed.
Labeling of biotinylated proteins
Dialyzed samples obtained from affinity chromatography were lyophilized and resuspended in 100 μL 6 M Urea (final concentration of the proteins ∼5 mg/mL). Samples were reduced with 10 μL of 0.5 M TCEP at 37°C for 1 h, alkylated with 10 μL 0.5 M iodoacetamide at room temperature for 30 min, and diluted with 80 μL of 6 M Urea. Before labeling reaction, iTRAQ reagents were diluted with 50 μL ethanol, and five tubes of the same iTRAQ were combined. Samples corresponding to T0, T1, T3, and T4 were then derivatized by iTRAQ reagent 114, 115, 116, and 117, respectively, for 2 h at room temperature. Reactions were stopped with 10 μL of 10 mM lysine followed by incubation for 20 min. Samples were then combined into a single tube, diluted 10 × with dilution buffer (8/3 M urea in 150 mM HEPES) to maintain ethanol concentration under 10% and further separated by RPLC.
Reversed-phase chromatography of labeled proteins
Chromatographic separation of the iTRAQ labeled proteins was carried out on the Agilent 1100 HPLC system and POROS R1 column (4.6 × 100 mm; 10-μm particle) (Applied Biosystems, Framingham, MA) using a linear gradient between solvent A (0.1% TFA in deionized H2O) and solvent B (0.1% TFA in ACN) at flow rate of 0.7 mL/min. After a sample injection (5 mL) the column was washed with 3% solvent B for 5 min. The column was then eluted with a 35-min gradient from 5–30% solvent B, followed by a 35-min gradient from 30–55% solvent B and a 10-min gradient from 55–100% solvent B. The column was reequilibrated with an isocratic flow (3% solvent B) for 10 min. Concentration of the eluted proteins was monitored at A214 and A280. Fractions were collected in 1-min intervals during the entire chromatographic run and dried under the vacuum in a speedvac.
Proteolytic digestion
Trypsin digestion in can-dried samples were resuspended in 50 μL organic digestion buffer (80% ACN; 50 mM Tris buffer, pH7.8; 10 mM CaCl2). Trypsin (1 μg/fraction) was added and samples were incubated at 37°C for 1 h.
Trypsin digestion in urea/SDS
Similarly dried samples were redissolved in 50 μL of ammonium bicarbonate buffer (50 mM; pH8.0) containing 0.01% SDS and 1 M Urea to assist digestion. Following solubilization, trypsin (1 μg/fraction) was added for overnight incubation at 37°C.
Glu-C digestion in urea/SDS
Protein samples from each dried fraction were resuspended in 50 μL of ammonium bicarbonate buffer (50 mM; pH8.0) containing 0.01% SDS and 1 M Urea, Glu-C (1 μg/fraction) was added and samples were incubated overnight at 25°C.
Sample desalting and spotting
Digestion was stopped with 0.5 μL of 10% Trifluoroacetic acid (TFA), samples were desalted using ZipTip-C18 (Millipore Co., Billerica, MA) and directly spotted on a stainless steel 384-well MALDI plate by mixing with an equal volume of matrix solution (5 mg/mL of α-cyano-4-hydroxy-cinnamic acid in 60% ACN and 0.1% TFA).
MALDI-MS/MS analysis
MALDI-TOF/TOF data were acquired in MS reflector positive ion mode using an ABI 4800 Proteomics Analyzer mass spectrometer (Applied BioSystems). The laser intensity was set for 3800 Hz for MS and 4300 Hz for MS/MS acquisition. Laser shots, 500, were accumulated in MS mode and up to 2,000 laser shots were accumulated for MS/MS. Spectra were acquired for a mass range of 800–4,000 Da. The signal-to-noise threshold was set at 20 for MS and 10 for MS/MS. Mass tolerance was set at ± 0.03 (m/z). The 25 strongest precursor ions per spot were chosen automatically for MS/MS analysis.
The files acquired on LC-MALDI-TOF/TOF were analyzed using ProteinPilot 2.0 software which uses the Pro Group algorithm (Applied Biosystems) for protein identification. The parameters of the search were as follows: no more than three tryptic missed cleavages allowed, cysteine searched as carbamidomethyl cysteine, 50 ppm peptide tolerance, and 0.3 Da MS/MS mass tolerance. Proteins were identified with minimum value 0.7 or 1.3 of unused cutoff for a model protein, and 2 for carbonylated protein identification. The minimum acceptance criterion was applied to 95 or 99% confidence level for model protein or carbonylated proteins, respectively. Searches were also performed using Mascot protein identification software and the NCBI database. The peak lists were exported using the TS2Mascot application, enabling accurate iTRAQ quantification, and submitted manually through the Mascot Web interface. The following parameters were used in the MASCOT analysis: fixed modification, iTRAQ4plex(K); variable modification, carbamidomethyl(C), oxidation(M), iTRAQ4plex(Y), Argbiotinhydrazide(R), Lysbiotinhydrazide(K), Probiotinhydrazide(P), Thrbiotinhydrazide(T); peptide tolerance, 1.2 Da; and MS/MS tolerance, 0.6 Da. In addition, the Mascot search for protein modifications was applied to the subdatabase of the proteins identified in ProteinPilot with a significance threshold of p < 0.05. Identified proteins were further investigated according to their functions and localizations using MIPS Saccharomyces cerevisiae genome database (http://mips.gsf.de/genre/proj/yeast/).
Distribution of carbonylated proteins
Differential centrifugation in combination with 2,4-dinitrophenylhydrazine derivatization (Maisonneuve et al., 2008a) was used to determine protein carbonyls content and distribution of carbonylated proteins. Cell collection and disruption was performed as previously described (Kim et al., 2010). Following disintegration, the homogenate was centrifuged at 500 × g for 3 min to remove cell debris and unbroken cells. The supernatant was transferred into a new tube and the 250-μL aliquot removed and stored in −80°C until further analysis [total carbonyl content (TCC)]. The rest of the supernatant was centrifuged at 14,000 × g for an additional 10 min. The supernatant was moved to the new centrifugation tube and the pellet [large protein aggregates (LPA)] was redissolved in 250 μL carbonyl lysis buffer (3.8 mM NaH2PO4·H2O, 49.4 mM Na2HPO4·H2O, 48.4 mM NaCl, 5 mM KCl, 20% glycerol, 1% 2-mercaptoethanol, 0.3% IGEPAL CA-630, “protease inhibitor cocktail” from Sigma) and stored in −80°C until further analysis. Then the supernatant was centrifuged at 20,000 × g for 30 min, resulting in soluble proteins (SP) and small protein aggregates (SPA) fractions in supernatant and pellet, respectively. Proteins obtained from pellet fractions were dissolved in carbonyl lysis buffer as described above. Protein concentration was measured by the Bradford protein assay (Bio-Rad, Hercules, CA). Finally, 250 μL 12% SDS was added to TCC, SP, and SPA and LPA samples, mixed with equal volumes of 2,4-dinitrophenylhydrazine (2,4-DNPH; 10 mM in 2 N HCl) and concentrations of protein carbonyls were determined as described by Stadtman and colleagues (Levine et al., 1990).
Pathway and structural analysis
Metabolic and physiological pathway analysis and comparisons were performed using the Web-accessible programs DAVID version 6.0 (The Database for Annotation, Visualization and Integrated Discovery) from NIAID/NIH Bioinformatics Resources (http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003) and Kyoto Encyclopedia of Genes and Genomes (http://www.genome.ad.jp/kegg/) (Kanehisa and Goto, 2000). KEGG pathway maps were identified with a high relevance of the entity to the dataset (p < 0.05).
Structural data of the proteins were downloaded from RCSB PDB (Protein Data Bank) (http://www.rcsb.org/pdb/home/home.do) and extracted using 3D-JIGSAW software (http://bmm.cancerresearchuk.org/∼3djigsaw/). Visualization of 3D structures, amino acid positions, and distances were estimated using RASMOL version 2.7.4.2.
Results and Discussion
Evaluation of iTRAQ labeling and LC method
Similar to other posttranslation modifications, the accurate identification and quantification of oxidized proteins remains challenging due to their low concentration and the interpretation of complicated fragmentation pattern from the carbonylated peptides (Kim et al., 2010; Regnier and Julka, 2006). This is particularly true for “bottom-up” approaches in which proteins are first converted into a mixture of proteolytic peptides that are then analyzed by LC-MS/MS techniques. The improvement in protein identification can be achieved through the separation of intact proteins followed by proteolytic digestion of individual fractions and their analysis using LC-MS/MS. This technique, however, suffers large variation and isotope coding has to be implemented for accurate quantification. Recently, a protocol for labeling intact proteins using commercially available iTRAQ multiplex reagents has been published (Wiese et al., 2007). The method is gel based and the maximum amount of proteins per labeling reaction is 100 μg only for model proteins. Although the method suggests promise, in-solution digestion is generally less time-consuming and has better protein coverage than in-gel digestion (Roe and Griffin, 2006).
For these reasons, we have developed a new iTRAQ derivatization technique coupled with LC fractionation of intact proteins that allows for labeling up to 100 μg proteins per reaction and analysis of four samples in a single chromatographic run. One of the major concerns during the development process was compatibility of solvents and buffers for labeling, separation and proteolytic digestion. For example, protein coding with iTRAQ reagents requires 60% organic solvent. High organic solvent content, on the other hand, may interfere with separation of proteins in reversed-phase chromatography. Therefore, to reduce concentration of the organic solvent, the samples must be diluted 10 × with the dilution buffer following the labeling reaction. Another problem was solubilization of fractionated and dried proteins for proteolytic digestion. Traditionally, ammonium bicarbonate buffer containing 1 M urea was insufficient to redissolve the proteins and 0.01% SDS had to be added. An alternative, and very attractive solution to the problem, was the tryptic digestion in buffer containing 80% ACN, 20% of 50 mM Tris buffer (pH 7.8), and 10 mM CaCl2 as described by Hurst et al. (Strader et al., 2006). This would allow for proteolysis directly in the collected fractions and eliminate the drying step. Both methods were evaluated using iTRAQ labeled BSA as a model protein. Two aliquots of BSA standard solution were labeled with iTRAQ reagent 116 and 117, mixed in 1:1 ratio, and split in half. Samples were then dried, resuspended in either urea or can-based buffer, and digested with trypsin. Both samples were directly placed on a MALDI plate and analyzed by MALDI-MS/MS. A representative MS and MS/MS spectra are shown in Supplementary Figure S1. The ratios of all isotopic pairs were calculated from the peak area in the MALDI-MS/MS spectrum and averaged. The average ratios (n = 6) and SD observed in ACN and urea-based digestion buffers were 1.01 ± 0.05 SD and 1.03 ± 0.04 SD, respectively, indicating a high experimental accuracy. The average sequence coverages of BSA protein (% amino acids sequence covered by identified peptides) obtained by peptide mass mapping were 16.4% and 15.5% using can- and urea-based digestion, respectively. Overall there were no significant differences between organic and aqueous buffers.
Labeling efficiency was evaluated by visual inspection of MS spectra. The ratio between the unlabeled and the labeled peptides was determined as ≤0.03 and potential derivatization of Tyr residues was estimated to be ≤10% out of total identified peptides. Based on these results, it was concluded that the labeling efficacy was satisfactory and did not compromise the overall quantification (Asara et al., 2006). In addition, no unlabeled lysine containing peptides were observed.
BSA standard labeled with iTRAQ reagent 116 and 117 and mixed in 1:1 ratio was also used to examine the reproducibility of LC fractionation. The average ratio among labeled peptides was 0.97 ± 0.042 (n = 3). Interestingly, two more peptides were identified with addition of the reversed-phase chromatography step (Table 1) that increased the coverage to 21.95%. This may be attributed to the elimination of compounds that potentially interfere with trypsin digestion or to a decrease in sample complexity and better ionization. Most peptides were detected within three fractions and the total number of unique peptides (average 11) and lysine containing peptides (average 8) identified from all fractions were similar to those obtained without LC run (10 and 7, respectively).
Table 1.
Comparison of Predicted and Observed BSA Peptides
| Position of cleavage site | Peptide sequences | Without LC | With LC | Observed modifications |
|---|---|---|---|---|
| 19 | MKWVTFISLLLLFSSAYSR | – | ✓ | – |
| 34 | DTHKSEIAHR | – | – | – |
| 122 | ETYGDMADCCEKQEPER | ✓ | ✓ | Carbamidomethyl [9,10],Lysine(K)_iTRAQ[12] |
| ✓ | ✓ | Carbamidomethyl (C)[9,10],Lysine(K)_iTRAQ[12], Tyrosine_iTRAQ(var)[3] | ||
| 218 | EKVLASSAR | ✓ | ✓ | Lysine(K)_iTRAQ[2] |
| 232 | CASIQKFGER | ✓ | ✓ | Carbamidomethyl (C)[1], Lysine(K)_iTRAQ[6] |
| 241 | ALKAWSVAR | ✓ | ✓ | Lysine(K)_iTRAQ[3] |
| 371 | (R)HPEYAVSVLLR | ✓ | ✓ | – |
| ✓ | ✓ | Tyrosine_iTRAQ(var)[5] | ||
| 451 | KVPQVSTPTLVEVSR | ✓ | ✓ | Lysine(K)_iTRAQ[1] |
| 459 | SLGKVGTR | ✓ | ✓ | Lysine(K)_iTRAQ[4] |
| 468 | CCTKPESER | ✓ | ✓ | Carbamidomethyl (C)[1,2],Lysine(K)_iTRAQ[4] |
| 482 | MPCTEDYLSLILNR | ✓ | ✓ | Carbamidomethyl (C)[3] |
| ✓ | ✓ | Carbamidomethyl (C)[3], Oxidation (M) [1] | ||
| ✓ | ✓ | Carbamidomethyl (C)[3], Tyrosine_iTRAQ [7] | ||
| 507 | LCVLHEKTPVSEKVTKCCTESLVNR | – | ✓ | Carbamidomethyl (C)[2,17,18],Lysine(K)_iTRAQ[7,13,16] |
Protein identification and quantification in Δyfh1 strain exposed to H2O2
In our previous study (Kim et al., 2010), we investigated the differences in carbonyl content of Saccharomyces cerevisiae wild-type and Δyfh1 mutant strains exposed to oxidative stress. Mutant initially had a significant increase in growth relative to the wild type followed by a decline in growth 4 h following H2O2 addition. Both the carbonyls and ROS levels reached their maximum at 3 h, whereas the viability of the mutant cells significantly decreased after 4 h following H2O2 treatment. Compared to the wild type, mutant cells exhibited a significant increase in growth. This can be explained by the fact that the Δyfh1 strain is already adapted to OS as the cells are experiencing higher levels of OS prior to H2O2 addition. In addition, the cells are diluting the damaged biomolecules by accelerated division. However, 4 h following exposure, the Δyfh1 strain reach a “no return” point at which the cells cannot defend themselves and die.
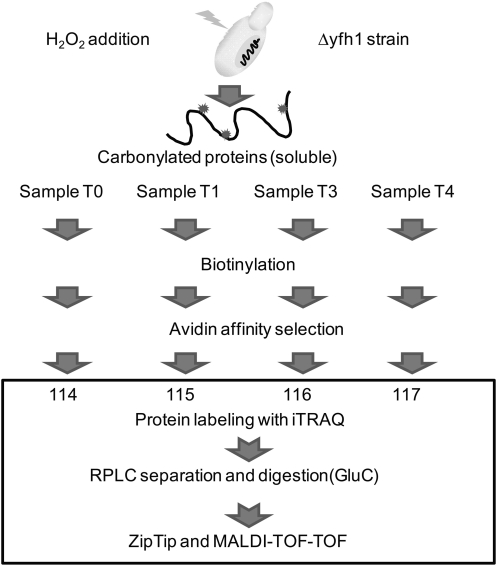
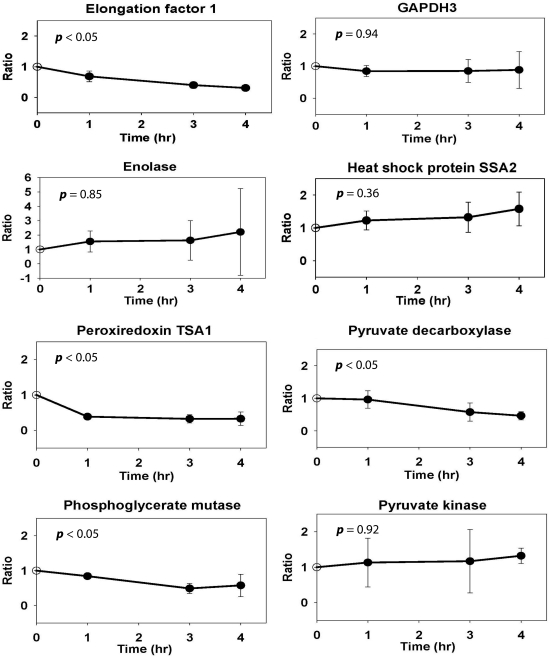
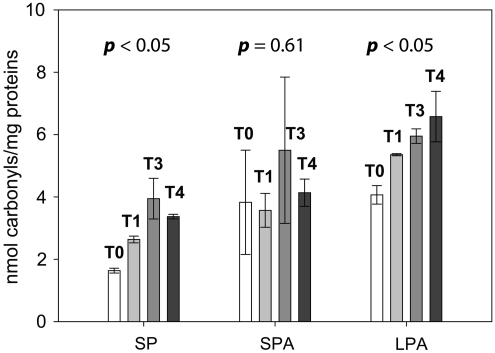
To elucidate the dynamics of this process, mutant cells were grown in YEPD media at 30°C to OD of ∼160 KU, H2O2 was added and TCC was determined before (T0,) and 1 (T1), 3 (T3), and 4 (T4) h following exposure (Kim et al., 2010). Both the standard curve and TCC levels were consistent with previously published data (Kim et al., 2010) (Fig. 1). The same time points were also used to identify and quantify the specific oxidatively damaged proteins. The unique properties of iTRAQ reagents allow simultaneous analysis of four different samples. Prior to mixing, samples are first labeled individually with corresponding iTRAQ tags that have an exclusive reporter group exhibiting a specific peak upon MS/MS, and then analyzed a in single chromatographic run. This approach minimizes technical variation as all samples are treated exactly the same during the separation and digestion steps. Figure 2 shows a strategy implemented for determination of oxidized proteins in the Δyfh1 strain exposed to H2O2. The carbonylated proteins from individual samples were first biotinylated with biotinhydrazide and then isolated using avidin affinity selection. To increase the protein amount for analysis, this step was repeated twice for each time point and corresponding fractions were combined prior to iTRAQ labeling. Interestingly, the peak areas representing biotinylated proteins obtained from T0, T1, T3, and T4 were similar and do not follow TCC patterns. This suggests that part of the carbonylated proteins is soluble while the rest may be aggregated into the partially soluble or insoluble aggregates. Another explanation could be that multiple carbonylation appears on the same proteins during exposure. This would lead to the increase of TCC levels (Fig. 1) while maintaining a similar concentration of affected proteins. Following the affinity selection, samples from T0, T1, T3, and T4 were derivatized with iTRAQ reagents 114, 115, 116, and 117, respectively, combined, and proteins separated on RPLC. Forty fractions were collected and subjected to proteolytic digestion. Although the original protocol was developed for trypsin, we have experienced difficulty in MS/MS identification of oxidized proteins using this enzyme. This is because lysine residues are either labeled with iTRAQ reagent or potentially modified by oxidation and biotinylated. Digestion of such proteins generates relatively large peptides with complicated fragmentation patterns. Therefore, another enzyme, Glu-C recognizing either glutamic acid (E) or aspartic acid (D) residues, was tested and found to be compatible with the urea/SDS buffer (no change in Glu-C activity was found using urea/SDS digestion protocol). Individual fractions were digested with Glu-C, spotted on the plate, and analyzed by a MALDI-TOF-TOF mass spectrometer. A total of 31 proteins were identified with 99% peptide confidence and unused cutoff value of 2 using ProteinPilot software (Supplementary Table S1). Of those, 27 proteins were quantified (Supplementary Table S1) and the abundance expressed as a fold change relative to T0 (iTRAQ 114). The changes in carbonylation of representative proteins are shown in Figure 3. Overall, three patterns were found including proteins with either gradually increasing, constant, or decreasing levels of damage. An increase in protein damage was expected, and it is consistent with the increasing carbonylation levels observed during exposure (Fig. 1). Proteins with a constant level probably reflect a dynamic balance between the translation of unaffected proteins, oxidation, and degradation of the damaged proteins. The most interesting group is represented by a decrease in the concentration of oxidized proteins following exposure. One explanation could be that the damaged proteins are degraded and gradually replaced with unaffected ones. Alternatively, the results may also suggest a possibility of aggregation of carbonylated protein. In this case, the proteins are oxidized on multiple sites, which leads to changes in quaternary structures and assembly into the large aggregates. This theory is also supported by the experiment in which the distribution of carbonylation between soluble proteins, and proteins associated in small and large aggregates was measured. As indicated in Figure 4, carbonylation of large protein aggregates gradually increases with time, while the levels of oxidation in SP and SPA reach the maximum at the third hour after exposure and then decrease. In recent studies of oxidized proteins using Escherichia coli and 2D-ELFO (2D Electrophoresis) analysis, most of the soluble carbonylated proteins were also found in an aggregate state. In addition the total amount of these proteins remains at similar levels during growth arrest (Maisonneuve et al., 2008a, 2008b).
FIG. 1.
Growth and total carbonyl content (TCC) of the (yfh1 strain exposed to oxidative stress. Solid and dotted graphs represent growth curves of the (yfh1 strain without and with H2O2 addition, respectively. After cell disruption followed by cell debris removal, TCC levels (bar graph) in the cell lysate were measured before (T0) and 1 (T1), 3 (T3), and 4 (T4) h following H2O2 addition. TCC levels were defined as total carbonyl amount (nmoles) divided by total protein amount (mg) in the cell lysate. Arrows indicate the sampling time.
FIG. 2.
Strategy for determination of carbonylated proteins in the Δyfh1 strain exposed to H2O2. After cell disruption followed by centrifugation at 20,000 × g for 30 min, SP (soluble proteins) at each time point T0, T1, T3, and T4 were biotinylated using 5 mM biotinhydrazide, respectively. Then, carbonylated proteins were selected by using avidin affinity column. Proteins from individual time points were labeled with different iTRAQ labeling reagents, combined, and separated using reversed-phase chromatography. Collected fractions were digested and analyzed by MALDI-TOF-TOF.
FIG. 3.
Time profiles of representative carbonylated proteins. The individual ratios were calculated from three protein labeling experiments. The p-values ≤0.05 represent a statistically significant differences.
FIG. 4.
Distribution of carbonyl content. The carbonyls were assessed in SP (soluble proteins), SPA (small protein aggregates), and LPA (large protein aggregates) fraction before (T0) and 1 (T1), 3 (T3), and 4 (T4) h following H2O2 addition. Fractions SP, SPA, and LPA were generated by centrifugation as described in the text. Carbonyl content and protein concentration were determined by 2,4-dinitrophenylhydrazine reaction and Bradford protein assay, respectively.
Protein classification and pathway analysis
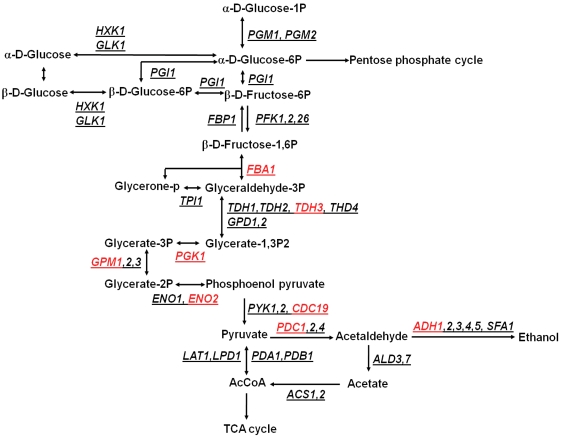
In order to elucidate potential pathways affected by iron-mediated oxidative stress in the mutant strain, identified proteins were analyzed by their function and location through DAVID and KEGG pathway analysis software. These proteins were associated with various cellular functions including metabolism (32.3%), transcription (9.7%), cell defense/transport/folding (38.7%), and protein synthesis (16.1%) (Supplementary Table S2). The majority of the proteins were involved in energy metabolism and the cellular defense. As suggested by Irazusta et al. (2008), the main targets of oxidative damage represent metal binding proteins that are related to glycolysis, such as enolase (ENO2), pyruvate kinase (CDC19), and pyruvate decarboxylase (PDC1). We observed eight carbonylated proteins associated with glycolysis, and it was also the most affected pathway with p-value = 3.7 × 10−6 (Fig. 5). Interestingly, we found many proteins such as heat shock protein (SSC1), ketol-acid reductoisomerase (ILV5), spermidine synthase (SPE3), or superoxide dismutase (SOD1) (Supplementary Table S2). These proteins are well known to play a key role in increased tolerance to oxidative stress by maintaining cellular redox balance, and by protecting or repairing other proteins. One of the explanation could be that the significant decrease in viability of the mutant cells observed 4 h following H2O2 treatment may be caused by a drop in energy production and reducing power of the cells due to the damage of glycolytic enzymes and by insufficient enzymatic support to protein protection and regeneration. This may also explain accelerated growth at the early stages of exposure. Because the Δyfh1 strain experiences higher levels of stress prior to H2O2 addition (Kim et al., 2010), we can expect a decrease in activities of antioxidant defense enzymes due to their carbonylation. This, in turn, leads to protein aggregation. In order to restore the free radical/antioxidant balance, cells then have to dilute the damage by accelerating division which, however, is limited by energy production and results in cellular death.
FIG. 5.
Oxidized proteins involved in glycolysis. Affected proteins are indicated in red color. The detailed information for gene names and functions are summarized in Supplementary Table S2.
Determination of oxidation sites
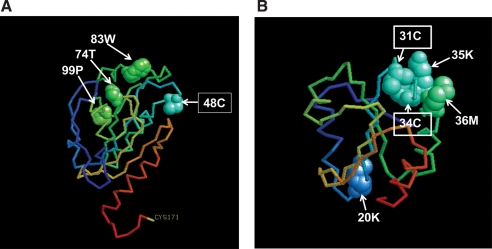
Although a few cases of successfully identified oxidation sites have been reported (Chavez et al., 2006; Mirzaei and Regnier, 2005), an accurate determination of oxidized amino acid residues in affected proteins remains challenging (Jensen, 2004; Mann and Jensen, 2003) due to the complicated interpretation of MS/MS spectra. In this study we were able to identify the oxidation sites in seven proteins (Table 2). Although most of the carbonylation occurs on the random sites, it has been also reported that carbonylation can be site specific causing functional changes of enzymes (Cabiscol et al., 2000) and aggregation of structural proteins (Irazusta et al., 2008). To estimate the position of oxidation sites and a potential impact on the protein activity, predicted 3D structures of peroxiredoxin (TSA1) and thioredoxin II (TRX2) were visualized and analyzed through the Web-based PDB viewer RASMOL and comparative modeling software, 3D-JIGSAW (Fig. 6). Peroxiredoxin is an important enzyme involved in detoxification of H2O2. In yeast, a reduced form of cysteine in position 48 is oxidized by H2O2 and rapidly reacts with reduced cysteine in position 171 of another subunit to form an intermolecular disulfide (Jara et al., 2007). All three oxidized amino acid residues (74T, 83W, 99P) of peroxiredoxin identified in our study revealed close proximity to the 48C suggesting that they may interfere with peroxiredoxin activity. Similarly, two oxidation sites (35K and 36M) found close to the active sites (31C and 34C) of thioredoxin may have an effect on activity (Fig. 6B).
Table 2.
Oxidation Sites of Representative Proteins
| |
|
|
Protein pilot confidence |
|
|
|---|---|---|---|---|---|
| Accession | Names | Length (a.a.) | [Mascot Score] | Sequences | Oxidation sites |
| P02994|EF1A_YEAST | Elongation factor 1-alpha | 458 | 99 | RGITIDIALWKFE | 80F |
| 99 | TVPFVPISGWNGDNMIE | 199M | |||
| 98 | AFSEYPPLGRF | 413F | |||
| [9] | KRTIE | 37R | |||
| [20] | AIDAIEQPSRPTDKPLRLPLQD | 242K | |||
| P34760|TSA1_YEAST | Peroxiredoxin TSA1 | 196 | 99 | YSLLAWTNIPRKE | 83W |
| [11] | GGLGPINIPLLADTNHSLSRDYGVLIE | 99P | |||
| [10] | QGAQVLFASTD | 74T | |||
| P06169|PDC1_YEAST | Pyruvate decarboxylase isozyme 1 | 563 | 99 | IMLPVFDAPQNLVE | 539M |
| [101] | IMLPVFDAPQNLVE | 539M | |||
| P07284|SYSC_YEAST | Seryl-tRNA synthetase | 462 | [6] | WVKTRFELDELNKKFNKLQKD | 44K, 45T, 58K,61K |
| [8] | KKYVHCLNSTLAATQRALCCILE | 410R | |||
| P09436 |SYIC_YEAST | Isoleucyl-tRNA synthetase | 1072 | [11] | IVPRYATMTGHHVE | 68P, 73M, 74T |
| P10592|HSP72_YEAST | Heat shock protein SSA2 | 639 | [7] | RAKRTLSSSAQTSVEIDSLFEGID | 269R |
| P22803|TRX2_YEAST | Thioredoxin II(Thioredoxin1) | 104 | [7] | KLVVVDFFATWCGPCKMIAPMIE | 20K, 35K, 36M |
FIG. 6.
Predicted 3D structure of peroxiredoxin TSA1 (A) and thioredoxin II TRX2 (B). Boxed amino acids represent active sites of enzymes. Nonboxed amino acids indicate identified oxidation sites. The 3D structures were obtained using 3D-JIGSAW software and visualized with RASMOL version 2.7.4.2.
Conclusions
Protein oxidation was investigated in an iron accumulating Δyfh1 yeast strain using biotinhydrazide–avidin affinity selection of carbonylated proteins and isotope coding quantification method. Results indicated that the affected proteins were primarily involved in glycolysis and cellular defense system. The position of oxidized amino acid residues close to active sites implicated a direct impact on protein function. One possible conclusion is that the decrease in viability of the mutant cells could be caused by drop in energy production/reducing power of the cells by unsatisfactory enzymatic support to protein protection and regeneration. It is important to note, however, that the method described in this article quantifies soluble proteins only. To fully understand the dynamics of the process, other approaches covering protein degradation and aggregation must be developed and implemented.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Aging (Grant Number 5R01AG025362-02) and by the National Cancer Institute (Clinical Proteomics Technology Assessment for Cancer Program—U24 CA 126480).
Author Disclosure Statement
No conflict of interests exist.
References
- Adamec J. Rusnak F. Owen W.G. Naylor S. Benson L.M. Gacy A.M., et al. Iron-dependent self assembly of recombinant yeast frataxin: implications for Friedreich ataxia. Am J Hum Genet. 2000;67:549–562. doi: 10.1086/303056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal K. Choe L.H. Lee K.H. 6th Siena Meeting on From Genome to Proteome. Siena; Italy: 2004. Quantitative analysis of protein expression using amine-specific isobaric tags in Escherichia coli cells expressing rhsA elements; pp. 2297–2308. [DOI] [PubMed] [Google Scholar]
- Akman S. Critical Reviews of Oxidative Stress and Aging; Advances in Basic Science, Diagnosis, and Intervention. World Scientific Publishing; Hackensack, NJ: 2003. Overview of oxidative stress and cancer; pp. 925–954. [Google Scholar]
- Aliev G. Smith M.A. Seyidova D. Neal M.L. Lamb B.T. Nunomura A., et al. The role of oxidative stress in the pathophysiology of cerebrovascular lesions in Alzheimer's disease. Brain Pathol. 2002;12:21–35. doi: 10.1111/j.1750-3639.2002.tb00419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asara J.M. Zhang X. Zheng B. Maroney L.A. Christofk H.R. Wu N., et al. In-gel stable isotope labeling for relative quantification using mass spectrometry. Nat Protoc. 2006;1:46–51. doi: 10.1038/nprot.2006.7. [DOI] [PubMed] [Google Scholar]
- Babcock M. deSilva D. Oaks R. DavisKaplan S. Jiralerspong S. Montermini L., et al. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- Berg D. Gerlach M. Youdim M.B.H. Double K.L. Zecca L. Riederer P., et al. Brain iron pathways and their relevance to Parkinson's disease. J Neurochem. 2001;79:225–236. doi: 10.1046/j.1471-4159.2001.00608.x. [DOI] [PubMed] [Google Scholar]
- Boehm A.M. Putz S. Altenhofer D. Sickmann A. Falk M. Precise protein quantification based on peptide quantification using iTRAQ (TM) BMC Bioinformatics. 2007;8:18. doi: 10.1186/1471-2105-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G.H.H. Harbour M. Hester S. Lilley K.S. Robinson M.S. Comparative proteomics of clathrin-coated vesicles. J Cell Biol. 2006;175:571–578. doi: 10.1083/jcb.200607164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabiscol E. Piulats E. Echave P. Herrero E. Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- Campuzano V. Montermini L. Molto M.D. Pianese L. Cossee M. Cavalcanti F., et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Chavez J. Wu J.Y. Han B.N. Chung W.G. Maier C.S. New role for an old probe: affinity labeling of oxylipid protein conjugates by N′-aminooxymethylcarbonylhydrazino D-biotin. Anal Chem. 2006;78:6847–6854. doi: 10.1021/ac0607257. [DOI] [PubMed] [Google Scholar]
- Chong P.K. Gan C.S. Pham T.K. Wright P.C. Isobaric tags for relative and absolute quantitation (iTRAQ) reproducibility: Implication of multiple injections. J Proteome Res. 2006;5:1232–1240. doi: 10.1021/pr060018u. [DOI] [PubMed] [Google Scholar]
- Dennis G. Sherman B.T. Hosack D.A. Yang J. Gao W. Lane H.C., et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- England K. Cotter T. Identification of carbonylated proteins by MALDI-TOF mass spectroscopy reveals susceptibility of ER. Biochem Biophys Res Commun. 2004;320:123–130. doi: 10.1016/j.bbrc.2004.05.144. [DOI] [PubMed] [Google Scholar]
- Foury F. Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- Gan C.S. Chong P.K. Pham T.K. Wright P.C. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ) J Proteome Res. 2007;6:821–827. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- Hardt M. Witkowska H.E. Webb S. Thomas L.R. Dixon S.E. Hall S.C., et al. Assessing the effects of diurnal variation on the composition of human parotid saliva: quantitative analysis of native peptides using iTRAQ reagents. Anal Chem. 2005;77:4947–4954. doi: 10.1021/ac050161r. [DOI] [PubMed] [Google Scholar]
- Huynen M.A. Snel B. Bork P. Gibson T.J. The phylogenetic distribution of frataxin indicates a role in iron–sulfur cluster protein assembly. Hum Mol Genet. 2001;10:2463–2468. doi: 10.1093/hmg/10.21.2463. [DOI] [PubMed] [Google Scholar]
- Irazusta V. Moreno-Cermeno A. Cabiscol E. Ros J. Tamarit J. Major targets of iron-induced protein oxidative damage in frataxin-deficient yeasts are magnesium-binding proteins. Free Radic Biol Med. 2008;44:1712–1723. doi: 10.1016/j.freeradbiomed.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Jamieson D.J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Jara M. Vivancos A.P. Calvo I.A. Moldón A. Sansó M. Hidalgo E. The peroxiredoxin Tpx1 is essential as a H2O2 scavenger during aerobic growth in fission yeast. Mol Biol Cell. 2007;18:2288–2295. doi: 10.1091/mbc.E06-11-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O.N. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2004;8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Jeong S.Y. David S. Age-related changes in iron homeostasis and cell death in the cerebellum of ceruloplasmin-deficient mice. J Neurosci. 2006;26:9810–9819. doi: 10.1523/JNEUROSCI.2922-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H. Sedlak M. Gao Q. Riley C.P. Regnier F.E. Adamec J. Oxidative stress studies in yeast with a frataxin mutant: a proteomics perspective. J Proteome Res. 2010;9:730–736. doi: 10.1021/pr900538e. [DOI] [PubMed] [Google Scholar]
- Korolainen M.A. Goldsteins G. Alafuzoff I. Koistinaho J. Pirttila T. Proteomic analysis of protein oxidation in Alzheimer's disease brain. Electrophoresis. 2002;23:3428–3433. doi: 10.1002/1522-2683(200210)23:19<3428::AID-ELPS3428>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kristensen B.K. Askerlund P. Bykova N.V. Egsgaard H. Moller I.M. Identification of oxidised proteins in the matrix of rice leaf mitochondria by immunoprecipitation and two-dimensional liquid chromatography-tandem mass spectrometry. Phytochemistry. 2004;65:1839–1851. doi: 10.1016/j.phytochem.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Levine R.L. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic Biol Med. 2002;32:790–796. doi: 10.1016/s0891-5849(02)00765-7. [DOI] [PubMed] [Google Scholar]
- Levine R.L. Garland D. Oliver C.N. Amici A. Climent I. Lenz A.G., et al. Oxygen Radicals in Biological Systems, Part B. Oxygen Radicals and Antioxidants. Determination of carbonyl content in oxidatively modified proteins. In: Packer L., editor; Glazer A.N., editor. Methods in Enzymology. Vol. 186. Academic Press, Inc.; San Diego, CA: 1990. pp. 464–478. [DOI] [PubMed] [Google Scholar]
- Levine R.L. Stadtman E.R. Oxidative modification of proteins during aging. Exp Gerontol. 2001;36:1495–1502. doi: 10.1016/s0531-5565(01)00135-8. [DOI] [PubMed] [Google Scholar]
- Lund T.C. Anderson L.B. McCullar V. Higgins L. Yun G.H. Grzywacz B., et al. iTRAQ is a useful method to screen for membrane-bound proteins differentially expressed in human natural killer cell types. J Proteome Res. 2007;6:644–653. doi: 10.1021/pr0603912. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E. Fraysse L. Lignon S. Capron L. Dukan S. Carbonylated proteins are detectable only in a degradation-resistant aggregate state in Escherichia coli. J Bacteriol. 2008a;190:6609–6614. doi: 10.1128/JB.00588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E. Fraysse L. Moinier D. Dukan S. Existence of abnormal protein aggregates in healthy Escherichia coli cells. J Bacteriol. 2008b;190:887–893. doi: 10.1128/JB.01603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. Jensen O.N. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- Maritim A.C. Sanders R.A. Watkins J.B. Diabetes, oxidative stress, and antioxidants: a review. J Biochem Mol Toxicol. 2003;17:24–38. doi: 10.1002/jbt.10058. [DOI] [PubMed] [Google Scholar]
- Mirzaei H. Regnier F. Affinity chromatographic selection of carbonylated proteins followed by identification of oxidation sites using tandem mass spectrometry. Anal Chem. 2005;77:2386–2392. doi: 10.1021/ac0484373. [DOI] [PubMed] [Google Scholar]
- Nystrom T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24:1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky D.C. Babcock M.C. Kaplan J. The yeast frataxin homologue mediates mitochondrial iron efflux—evidence for a mitochondrial, iron cycle. J Biol Chem. 1999;274:4497–4499. doi: 10.1074/jbc.274.8.4497. [DOI] [PubMed] [Google Scholar]
- Regnier F.E. Julka S. Primary amine coding as a path to comparative proteomics. Proteomics. 2006;6:3968–3979. doi: 10.1002/pmic.200500553. [DOI] [PubMed] [Google Scholar]
- Reinders Y. Schulz I. Graf R. Sickmann A. Identification of novel centrosomal proteins in Dictyostelium discoideum by comparative proteomic approaches. J Proteome Res. 2006;5:589–598. doi: 10.1021/pr050350q. [DOI] [PubMed] [Google Scholar]
- Roe M.R. Griffin T.J. Gel-free mass spectrometry-based high throughput proteomics: tools for studying biological response of proteins and proteomes. Proteomics. 2006;6:4678–4687. doi: 10.1002/pmic.200500876. [DOI] [PubMed] [Google Scholar]
- Ross P.L. Huang Y.L.N. Marchese J.N. Williamson B. Parker K. Hattan S., et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Sayre L.M. Perry G. Smith M.A. Oxidative stress and neurotoxicity. Chem Res Toxicol. 2008;21:172–188. doi: 10.1021/tx700210j. [DOI] [PubMed] [Google Scholar]
- Schmidt A. Kellermann J. Lottspeich F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics. 2005;5:4–15. doi: 10.1002/pmic.200400873. [DOI] [PubMed] [Google Scholar]
- Sies H. Oxidative stress: oxidants and antioxidants. Exp Physiol. 1997;82:291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- Soreghan B.A. Yang F. Thomas S.N. Hsu J. Yang A.J. High-throughput proteomic-based identification of oxidatively induced protein carbonylation in mouse brain. Pharm Res. 2003;20:1713–1720. doi: 10.1023/b:pham.0000003366.25263.78. [DOI] [PubMed] [Google Scholar]
- Strader M.B. Tabb D.L. Hervey W.J. Pan C.L. Hurst G.B. Efficient and specific trypsin digestion of microgram to nanogram quantities of proteins in organic-aqueous solvent systems. Anal Chem. 2006;78:125–134. doi: 10.1021/ac051348l. [DOI] [PubMed] [Google Scholar]
- Talent J.M. Kong Y.L. Gracy R.W. A double stain for total and oxidized proteins from two-dimensional fingerprints. Anal Biochem. 1998;263:31–38. doi: 10.1006/abio.1998.2752. [DOI] [PubMed] [Google Scholar]
- Tamarit J. Cabiscol E. Ros J. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem. 1998;273:3027–3032. doi: 10.1074/jbc.273.5.3027. [DOI] [PubMed] [Google Scholar]
- Thomas S.N. Soreghan B.A. Nistor M. Sarsoza F. Head E. Yang A.J. Reduced neuronal expression of synaptic transmission modulator HNK-1/neural cell adhesion molecule as a potential consequence of amyloid beta-mediated oxidative stress: a proteomic approach. J Neurochem. 2005;92:705–717. doi: 10.1111/j.1471-4159.2004.02892.x. [DOI] [PubMed] [Google Scholar]
- Unwin R.D. Smith D.L. Blinco D. Wilson C.L. Miller C.J. Evans C.A., et al. Quantitative proteomics reveals posttranslational control as a regulatory factor in primary hematopoietic stem cells. Blood. 2006;107:4687–4694. doi: 10.1182/blood-2005-12-4995. [DOI] [PubMed] [Google Scholar]
- Wiese S. Reidegeld K.A. Meyer H.E. Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- Yoo B.-S. Regnier F.E. Proteomic analysis of carbonylated proteins in two-dimensional gel electrophoresis using avidin–fluorescein affinity staining. Electrophoresis. 2004;25:1334–1341. doi: 10.1002/elps.200405890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.