Abstract
We present the oldest genetically identified dog in the Americas, directly dated to 9,260±170 Cal. B.P. The DNA was extracted from an occipital condyle imbedded in a human paleofecal sample from Hinds Cave in southwest Texas. A 368 base pair fragment of the mitochondrial genome control region was sequenced. These data were analyzed with comparable data, which included other ancient dogs and extant dogs, wolves and coyotes from around the world. Compiled with published data, our results characterize ancient American dogs within clades rooted by Eurasian wolves. In the Americas, these data provide no evidence of local interbreeding with wolves. This is a departure from the genetic pattern in other areas of the world where interbreeding with local wolf populations is apparent. Our discovery of domestic dog bone in a human paleofecal sample provides the earliest direct evidence for human consumption of dogs in the New World. These data support the hypothesis that dogs were a food source for early Paleoamericans.
Keywords: canine domestication, peopling of America, Paleoamerican, Paleoindian, aDNA
It is now established that extant domestic dogs (Canis lupus familiaris) were domesticated from Middle Eastern gray wolves (Canis lupus), with interbreeding with local wolf populations for specific lineages (Vonholdt et al., 2010). The old idea that domestic dogs in the Americas descend from a Eurasian wolf (Olsen and Olsen, 1977) is now widely accepted (Anderson et al., 2009; Boyko et al., 2009; Leonard et al., 2002; Malmstrom et al., 2005; Savolainen et al., 2002; Vilà et al., 1997; Vonholdt et al., 2010). We further evaluate the relationship between Paleoamerican dogs and humans from an ancient DNA analysis of an extraordinary sample. A portion of a domestic dog skull was recovered from the interior of an intact human paleofecal sample (BE-20) excavated from Hinds Cave in southwest Texas (Archaeological Survey Number: 41VV456) (Shafer et al., 1975; Shafer and Bryant Jr., 1977). Accelerator Mass Spectrometry (AMS) directly dated the BE-20 sample to 9260±170 B.P. (2σ calibration).
We sequenced a fragment of the mitochondrial DNA (mtDNA) control region for the BE-20 bone sample. We compared the genealogical relationship of the mtDNA fragment to previously published data for ancient dogs and extant dogs, wolves and coyotes from around the world. The results fill an important gap in the geographic distribution of ancient dog genetics. In the Americas, the previously published mtDNA data originate from archaeological sites in Alaska, southern Mexico, and South America (Leonard et al., 2002), while BE-20 originates from south central North America.
MATERIALS AND METHODS
The BE-20 paleofecal sample was identified as human based on its archaeological context and by established criteria (Bryant Jr., 1974; Bryant Jr. and Williams-Dean, 1975). The paleofecal sample was AMS dated by Beta Analytic Incorporated (reference number Beta-273755). Dissection and sorting of the paleofecal content was conducted within the Sobolik Lab at University of Maine.
The BE20 bone was observed within the paleofecal sample during dissection and sorting. The BE20 bone sample was handled with gloves at all times. The sample never came into direct contact with any canid material during this study. The BE20 bone was identified as a canid right occipital condyle by a comparison with reference specimens from the University of Maine’s zooarchaeological collection. The BE-20 bone was then taken to the Harvard Museum of Comparative Zoology and morphologically compared to ancient domestic dogs, wolves and coyotes.
Ancient DNA extraction and analysis was performed at the University of Oklahoma’s Molecular Anthropology Ancient DNA Laboratory. The laboratory is a positive pressure clean room isolated from the modern genetics lab. Incoming air passes through ISO 7 (class 10,000) HEPA-filtration. The room is equipped with UVC lighting. Sterile disposable gowns, gloves, hairnets and masks are worn while working in the laboratory.
Two extractions were performed on the BE20 bone averaging 0.25 grams of material each. The bone fragments were immersed in 6% sodium hypochlorite for 10 minutes to remove any surface contamination, followed by three rinses with molecular grade water. The bone fragments were decalcified by a 72 hour soak in EDTA 0.5M. DNA extraction followed a salting out procedure described previously (Tito et al., 2008), with the modification of using ammonium acetate for ionic strength and QIAmp Mini kits (Qiagen) for purification and concentration.
PCR setups were performed in the Ancient DNA Laboratory in order to avoid external contamination. A 368 base pair fragment of the mitochondrial DNA control region was amplified by PCR using four different combinations of primers (Table 1). In addition to the DNA extract, an extraction blank, two PCR blanks, and a modern control were included in each PCR setup. Each 30ul PCR reaction contained the following: 1 × PCR Buffer (Invitrogen), 1.5 mM MgC12 (Invitrogen), 85nM of each primer (IDT), 0.1 × SYBR green (Molecular Probes), 1 unit of Platinum® Taq (Invitrogen) and 3 µl of either ancient DNA extract, extraction blank solution, modern dog DNA extract and two PCR blanks by using molecular grade water. The modern dog extract was added outside of the ancient DNA laboratory after all other PCR tubes were sealed.
Table 1.
Primer sets used in the study of BE-20 bone sample.
| Primer | Position* | Primer sequence | length (bp)** | Reference |
|---|---|---|---|---|
| mt-U1 | 15412-15432 | CCACCATCAGCACCCAAAGCT | 103 | (Nakamura et al., 2009) |
| dogDL-5 | 15555-15533 | CATTAATGCACGACGTACATAGG | (Leonard et al., 2002) | |
| dL15590MGB | 15568-15590 | CATATAAGCATGTACATAATATT | 99 | (Malmstrom et al., 2005) |
| dH15669R | 15692-15669 | CATGGTGATTAAGCCCTTATTGGA | (Malmstrom et al., 2005) | |
| H15422 | 15404-15422 | CTCTTGCTCCACCATCAGC | 112 | (Boyko et al., 2009) |
| dogDL-5 | 15555-15533 | CATTAATGCACGACGTACATAGG | (Leonard et al., 2002) | |
| dogDL-4 | 15656-15679 | GCATATCACTTAGTCCAATAAGGG | 109 | (Leonard et al., 2002) |
| mt-U2 | 15809-15789 | TGGCCCTGAAGTAAGAACCAG | (Nakamura et al., 2009) |
Position based on the reference dog sequence deposited in the UCSC Genome Browser (Kent et al., 2002).
The expected length of the amplicon after the removal of the primer sequence. The actual sequence data may differ because of insertion/deletion polymorphisms and sequence quality.
The temperature profile for the PCR reaction included an initial activation of the enzyme at 94°C for 2 minutes, followed by 60 cycles of the following: 94°C for 15 seconds, 55°C - the annealing temperature for all pairs of primers - for 15 seconds and 72°C for 15 seconds. The melting curve was obtained measuring the fluorescence intensity of the PCR product in a linear denaturation ramp from 55 to 95°C, increasing 0.5°C every 6 seconds.
Agarose gels and ethidium bromide staining were used for initial visualization of the PCR products. For the ancient sample, an estimation of the number of copies of canine mtDNA molecules was performed by quantitative PCR as described in Tito et al. (2008); the total extract was estimated to have 1,000 to 2,000 copies.
PCR products were purified with exonuclease I - shrimp alkaline phosphatase and sequenced in an ABI 3730XL capillary sequencer. Sequencing of at least two independent PCR products was attempted for each fragment per extraction. These raw data were compiled using Sequencher 4.10.1. The final sequences were deposited in GenBank: HQ585886 and HQ585887, for the BE-20 bone and modern control, respectively.
A Neighbor-Joining tree (Saitou and Nei, 1987) was generated using the program Mega 4 (Tamura et al., 2007), which included data for the BE20 bone sample and the modern lab control aligned to previously published data (Deguilloux et al., 2009; Leonard et al., 2002; Malmstrom et al., 2005; Verginelli et al., 2005; Vilà et al., 1997; Vilà et al., 1999). We assume a Tamura-Nei evolutionary model (1993). The phylogeny was rooted using the coyote haplotypes as the outgroup. Relevant aspects of the topology were robust to the other tree building algorithms, such as Minimum Evolution and Maximum Parsimony and other substitution models, including an equal rate model.
RESULTS AND DISCUSSION
The BE-20 bone morphology most closely matched MCZ Sample #41173, a short-nosed Indian Dog from New Mexico, when it was compared to the materials housed at the Harvard Museum of Comparative Zoology. However, morphologically differentiating wolves and ancient dogs is challenging (Wang and Tedford, 2008). This is particularly true for putative early Holocene remains.
Fiedel (2005) argued that putative early dog samples have disputable dating, context, identification and/or they are fragmentary. Despite several studies providing important contributions to the study of Holocene dogs (Aikens, 1970; Frison and Stanford, 1982; Grayson, 1988; Jennings, 1957; McMillan, 1970; Morey and Wiant, 1992; Sablin and Khlopachev, 2002), we were unable to reject Fiedel's claim. Consequently, an accurate identification of early Holocene domestic dogs may require both direct absolute dating and molecular evidence, as is provided by our study.
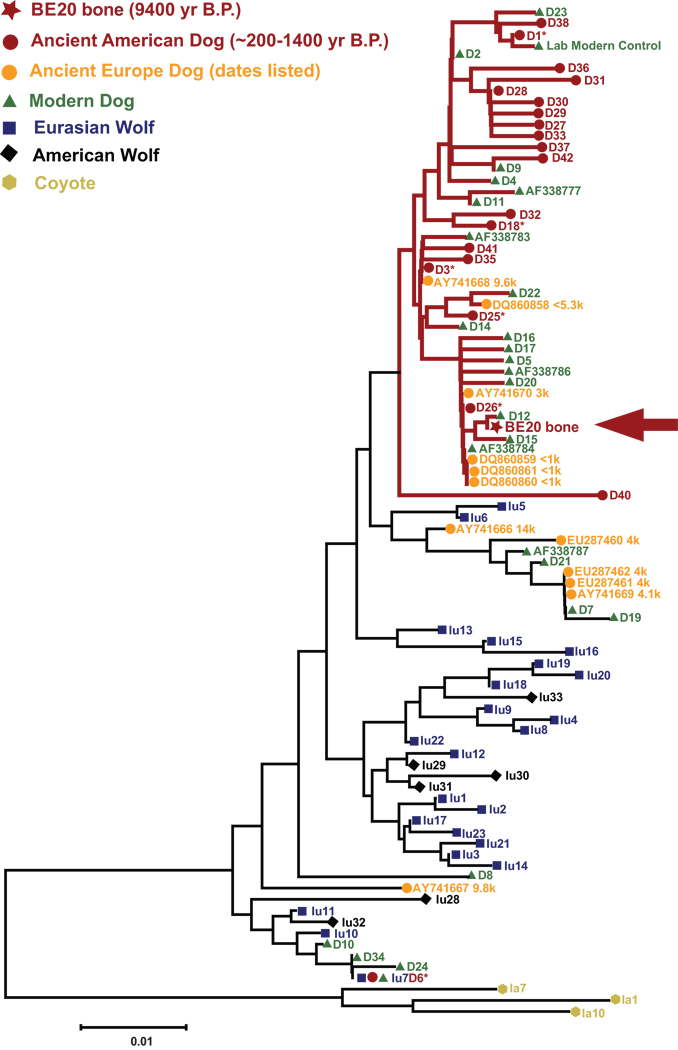
Figure 1 provides the phylogeny of the mtDNA sequence for BE-20 bone and the available comparative data. The BE-20 bone belongs to a different haplotype than our modern dog positive control sample, excluding the possibility of cross contamination. Sequence data from independent extractions and PCRs were consistent with a domestic dog haplotype (see Supporting Information online). The haplotype was observed previously in the modern dog D12 (Vilà et al., 1997). This haplotype belongs to Clade I (Vilà et al., 1997), which includes pre-Columbian dogs in the Americas and modern and ancient dogs throughout the world. Clade I excludes the studied wolves and coyotes, providing strong support that the BE-20 bone sample is indeed a domestic dog.
FIG. 1.
Neighbor-Joining tree of mtDNA control region sequences including the BE-20 bone sample. Samples within Clade I are highlighted in red. The asterisk identifies haplotypes found in both modern and ancient samples.
Coupled with the morphological data, this sample represents the oldest incontrovertible evidence for domestic dog in the New World. The genetic data predates that previously available in the Americas by 7800 years (Leonard et al., 2002). When considering the available comparative data presented in Figure 1, only three other possible dog samples in the world have similar ages (Verginelli et al., 2005).
The results provide clear evidence that pre-Columbian dogs, from widely dispersed areas in the Americas, were domesticated in Eurasia. However, it is common knowledge that domestic dogs are still capable of interbreeding with wolves. For example, hybridization with dogs has impacted the American gray wolf gene diversity (Anderson et al., 2009; Muñoz-Fuentes et al., 2010). While local interbreeding between dogs and wolves is expected, our compiled data provide no example of introgression of American wolf DNA into the domestic dog population. This may be attributed to an ascertainment bias where hybrids are misidentified as wolves in archaeological context (Wang and Tedford, 2008). Further research on the diversity of ancient North American wolves and dogs may provide more clarity on this issue.
The presence of a domestic dog bone in the BE-20 human paleofecal sample provides the earliest evidence of human-dog relationships in the Americas. From this, we can infer that domestic dogs were most likely associated with humans during the peopling of the Americas. We raise the question: To what degree were the first Americans dependent on domestic dogs?
It is intuitive that dogs could assist Paleoindian hunters in tracking, immobilizing and killing prey. Dogs could provide protection for Paleoindians, particularly by alerting the group against formidable Holocene predators (Turner, 2002). Additionally, the contemporary use of dogs as pack animals may reflect a Paleoindian homology (Turner, 2002).
The Paleoindian use of dogs as a food source is apparent from this study. The consumption of dogs is a relatively common practice throughout Eurasia and the Americas (Morey, 2006; Park, 1989). An ethnographic account of Polar Eskimos provides an anecdote:
“Dogs are also esteemed excellent food, and are bred as live stock, as well as for drawing the sledge; but they are only eaten in winter, when no other food can be obtained” (Ross, 1819:180)
To what extent the first Americans consumed dogs remains speculative. Early Paleoindians may have eaten dogs only in emergencies, but this idea may undervalue the pervasiveness of dogs in the Paleoindian diet. Currently, a new set of genomic tools are available to explore this issue further. For example, the human paleofecal composition can be explored using metagenomics (Tito et al., 2008), which may lead to an exciting new level of prehistoric dietary reconstruction in the near future. In the mean time, our discovery of the BE-20 bone provides the earliest direct evidence for human consumption of dogs in the Americas.
Supplementary Material
Acknowledgements
The University of Oklahoma’s Molecular Anthropology Laboratories provided support for this research. These labs are currently supported by the National Science Foundation (NSF#0845314) and the National Institutes of Health (NHGRI/NIH R01 HG005172-01). The College of Liberal Arts and Sciences, the Department of Anthropology, and the Climate Change Institute at the University of Maine provided additional support for this research. The Getty Archaeological Studies Fund administered through the Department of Anthropology at the University of Maine provided the funding for AMS analysis. We thank those providing published datasets, and Harvard’s Museum of Comparative Zoology for access to their comparative collection.
Grant sponsorship: The University of Oklahoma’s Molecular Anthropology Laboratories provided support for this research.
Literature Cited
- Aikens MC. Hogup Cave. Salt Lake City: University Utah Press; 1970. [Google Scholar]
- Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, Stahler DR, Smith DW, Padhukasahasram B, Randi E, Leonard JA, Bustamante CD, Ostrander EA, Tang H, Wayne RK. Molecular and evolutionary history of melanism in North American gray wolves. Science. 2009;323:1339–1343. doi: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Boyko RH, Boyko CM, Parker HG, Castelhano M, Corey L, Degenhardt JD, Auton A, Hedimbi M, Kityo R, Ostrander EA, Schoenebeck J, Todhunter RJ, Jones P, Bustamante CD. Complex population structure in African village dogs and its implications for inferring dog domestication history. Proc Natl Acad Sci USA. 2009;106:13903–13908. doi: 10.1073/pnas.0902129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant VM., Jr Prehistoric diet in southwest Texas: the coprolite evidence. Am Antiq. 1974;39:407–420. [Google Scholar]
- Bryant VM, Jr, Williams-Dean G. The coprolites of man. Sci Am. 1975;232:100–109. [Google Scholar]
- Deguilloux MF, Moquel J, Pemonge MH, Colombeau G. Ancient DNA supports lineage replacement in European dog gene pool: insight into Neolithic southeast France. J Archaeol Sci. 2009;36:513–519. [Google Scholar]
- Fiedel SJ. Man’s best friend—Mammoth’s worst enemy? A speculative essay on the role of dogs in Paleoindian colonization and megafaunal extinction. World Archaeol. 2005;37:670–698. [Google Scholar]
- Frison GC, Stanford DJ. The Agate Basin site: a record of Paleoindian occupation of the Northwestern High Plains (Studies in Archaeology) New York: Academic Press; 1982. [Google Scholar]
- Grayson DK. Danger Cave, Last Supper Cave, and Hanging Rock Shelter: The Faunas. New York: American Museum of Natural History; 1988. [Google Scholar]
- Jennings JD. Danger Cave. Salt Lake City: University Utah Press; 1957. [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JA, Wayne RK, Wheeler J, Valadez R, Guillen S, Vilà C. Ancient DNA evidence for Old World origin of New World dogs. Science. 2002;298:1613–1616. doi: 10.1126/science.1076980. [DOI] [PubMed] [Google Scholar]
- Malmstrom H, Stora J, Dalen L, Holmlund G, Gotherstrom A. Extensive human DNA contamination in extracts from ancient dog bones and teeth. Mol Biol Evol. 2005;22:2040–2047. doi: 10.1093/molbev/msi195. [DOI] [PubMed] [Google Scholar]
- McMillan RB. Early canid burial from the western Ozark highland. Science. 1970;167:1246–1247. doi: 10.1126/science.167.3922.1246. [DOI] [PubMed] [Google Scholar]
- Morey DF. Burying key evidence: the social bond between dogs and people. J Archaeol Sci. 2006;33:158–175. [Google Scholar]
- Morey DF, Wiant MD. Early Holocene domestic dog burials from the North American Midwest. Curr Anthropol. 1992;33:224–229. [Google Scholar]
- Muñoz-Fuentes V, Darimont CT, Paquet PC, Leonard JA. The genetic legacy of extirpation and re-colonization in Vancouver Island wolves. Conserv Genet. 2010;11:547–556. [Google Scholar]
- Nakamura H, Muro T, Imamura S, Yuasa I. Forensic species identification based on size variation of mitochondrial DNA hypervariable regions. Int J Legal Med. 2009;123:177–184. doi: 10.1007/s00414-008-0306-7. [DOI] [PubMed] [Google Scholar]
- Olsen SJ, Olsen JW. The Chinese wolf, ancestor of New World dogs. Science. 1977;197:533–535. doi: 10.1126/science.197.4303.533. [DOI] [PubMed] [Google Scholar]
- Park R. Dog remains from Devon Island, N.W.T.: archaeological and osteological evidence for domestic dog use in the Thule culture. Arctic. 1989;40:184–190. [Google Scholar]
- Ross J. A voyage of discovery, made under the orders of the Admiralty, in His Majesty's ships Isabella and Alexander, for the purpose of exploring Baffin's Bay, and inquiring into the probability of a north-west passage. London: John Murray; 1819. [Google Scholar]
- Sablin MV, Khlopachev GA. The earliest Ice Age dogs: evidence from Eliseevichi 1. Curr Anthropol. 2002;43:795–819. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Shafer HJ, Dering P, Williams-Dean G, Bryant VM., Jr . Texas Anthropological Laboratory Report. College Station: Texas A&M University; 1975. A preliminary report of Hinds Cave: Val Verde County Texas. [Google Scholar]
- Shafer HJ, Bryant VM., Jr . Texas A&M University Anthropological Laboratory Special Series 1. College Station: Texas A&M University; 1977. Archaeological and botanical studies at Hinds Cave, Val Verde County Texas. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tito RY, Macmil S, Wiley G, Najar F, Cleeland L, Qu C, Wang P, Romagne F, Leonard S, Ruiz AJ, Reinhard K, Roe BA, Lewis CM., Jr Phylotyping and functional analysis of two ancient human microbiomes. PLoS One. 2008;3:e3703. doi: 10.1371/journal.pone.0003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CGI. Teeth, needles, dogs, and Siberia: bioarchaeological evidence for the colonization of the New World. In: Jablonski NG, editor. The first Americans: the Pleistocene colonization of the New World. San Francisco: Memoirs of the California Academy of Sciences; 2002. pp. 123–158. [Google Scholar]
- Verginelli F, Capelli C, Coia V, Musiani M, Falchetti M, Ottini L, Palmirotta R, Tagliacozzo A, De Grossi Mazzorin I, Mariani-Costantini R. Mitochondrial DNA from prehistoric canids highlights relationships between dogs and South-East European wolves. Mol Biol Evol. 2005;22:2541–2551. doi: 10.1093/molbev/msi248. [DOI] [PubMed] [Google Scholar]
- Vilà C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, Honeycutt RL, Crandall KA, Lundeberg J, Wayne RK. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- Vilà C, Amorim IR, Leonard JA, Posada D, Castroviejo J, Petrucci-Fonseca F, Crandall KA, Ellegren H, Wayne RK. Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus. Mol Ecol. 1999;8:2089–2103. doi: 10.1046/j.1365-294x.1999.00825.x. [DOI] [PubMed] [Google Scholar]
- Vonholdt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, Reynolds A, Bryc K, Brisbin A, Knowles JC, Mosher DS, Spady TC, Elkahloun A, Geffen E, Pilot M, Jedrzejewski W, Greco C, Randi E, Bannasch D, Wilton A, Shearman J, Musiani M, Cargill M, Jones PG, Qian Z, Huang W, Ding ZL, Zhang YP, Bustamante CD, Ostrander EA, Novembre J, Wayne RK. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tedford RH. Dogs: their fossil relatives and evolutionary history. Cambridge: Cambridge University Press; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.