Abstract
The complexity of microbiota inhabiting the intestine is increasingly apparent. Delicate balance of numerous bacterial species can affect development of the immune system, how susceptible a host is to pathogenic organisms, and the auto-inflammatory state of the host. In the last decade, with the increased use of germ-free mice, gnotobiotic mice, and animal models in which a germ-free animal has been colonized with a foreign microbiota such as humanized mice, it has been possible to delineate relationships that specific bacteria have with the host immune system and to show what role they may play in overall host health. These models have not only allowed us to tease out the roles of individual species, but have also allowed the discovery and characterization of functionally unknown organisms. For example, segmented filamentous bacteria (SFB) have been shown to play a vital role in expansion of IL-17 producing cells. Prior to linking their key role in immune system development, little was known about these organisms. Bacteroides fragilis can rescue some of the immune defects of gnotobiotic mice after mono-colonization and have anti-inflammatory properties that can alleviate colitis and experimental allergic encephalitis in murine models. Additionally, Clostridium species have most recently been shown to expand regulatory T-cell populations leading to anti-inflammatory conditions. This review will highlight and summarize some of the major findings within the last decade concerning the role of select groups of bacteria including SFB, Clostridium, Bacteroides, Bifidobacterium, and Lactobacillus, and their impact on host mucosal immune systems.
Keywords: mono-colonization, homeostasis, microbiome, germ-free, segmented filamentous bacteria, Bacteroides fragilis, Clostridium, probiotics
Introduction
Humans play host to 500–1000 different species of bacteria in the intestine and 100 times more bacterial cells than eukaryotic cells (Whitman et al., 1998). Increased appreciation for the crucial role that the intestinal microbiota plays in host health and immunity is continually surfacing. While mammalian hosts provide a nutrient rich niche for these bacteria, the bacteria provide the host with much more including: aid in digestion, protection against pathogenic enteric pathogens, and development of the immune system (Hooper and Gordon, 2001; Macpherson and Harris, 2004; Sekirov et al., 2008; Sekirov and Finlay, 2009). Indeed, a skewed microbiota balance can affect health in numerous ways and can lead to conditions that promote diseases such as obesity, diabetes, inflammatory bowel disorders, and multiple sclerosis (Round and Mazmanian, 2009; Ochoa-Reparaz et al., 2010b). Additionally, a skewed microbiota can leave a host susceptible to infection (Manichanh et al., 2006; Turnbaugh et al., 2006, 2009; Frank et al., 2007; Garrett et al., 2007; Peterson et al., 2008; Wen et al., 2008). The complexity of the mammalian intestinal microbiota has long been appreciated; however, our knowledge of this area has greatly expanded in recent years as more advanced sequencing methods have become available. Due to the harsh conditions of the intestine, many of the organisms which dwell there are fastidious and cannot be cultured in vitro. Modern sequencing techniques have enabled us to begin cataloging in detail the microbial life that exists within the intestine whether or not it can be cultured ex vivo (Turnbaugh et al., 2007). The importance of commensal bacteria in host development and health is most clearly demonstrated by germ-free mice, raised in the absence of any bacteria. These mice exhibit numerous developmental defects, which can be compensated for by microbial colonization (Macpherson and Harris, 2004).
Developmental problems and defects faced by germ-free mice are, in part, centered around immune system development and function (Smith et al., 2007). While not an exhaustive list, some of the immunological defects seen in germ-free mice include immature lymphoid follicles, an enlarged cecum, reduced plasma cells and reduced production of mucosal immunolobulin A (IgA), anti-microbial peptides, and adenosine tri-phosphate (ATP). The number of CD8+ intestinal epithelial cells (IELs) and αβ T-cell receptor (TCR) IELs is reduced as well as Thy1 expression and cytolytic activity. IEL expression of major histocompatibility complex (MHC) II, Toll-like receptor (TLR) 9, and interleukin (IL) 25 is also reduced. CD4+ T-cells in the lamina propria (LP), Foxp3+ regulatory cells in the colonic LP, and CD4+CD25+ T-cells in the mesenteric lymph nodes (MLNs) are reduced. Immune structures in germ-free animals are also compromised. The Peyer’s patches are small compared to conventional animals, and the spleens and MLNs have depletion of lymphocyte zones (Lefrancois and Goodman, 1989; Rothkotter and Pabst, 1989; Shroff et al., 1995; Umesaki et al., 1995; Smith et al., 2007; Round and Mazmanian, 2009). Finally, germ-free mice have increased susceptibility to infection from enteric pathogens. It has been shown that they have less resistance than conventional mice to infection with: Shigella flexneri, Listeria monocytogenes, Clostridium difficile, and Salmonella enterica (Sprinz et al., 1961; Zachar and Savage, 1979; Nardi et al., 1989). When germ-free mice are colonized with as little as one commensal bacterial species, the susceptibility to infection is reduced. This feat is not accomplished by every intestinal commensal organism, however, indicating that colonization resistance (CR) ability is specific (Maier and Hentges, 1972; Round and Mazmanian, 2009; Ivanov and Littman, 2010).
Given the number of bacterial species residing in the intestine, it is hard to imagine teasing out specific contributions by individual species. However, mono-colonization of germ-free mice has allowed exactly this and we have gained important knowledge by examining which components of the faulty germ-free mouse immune system can be re-constituted with the addition of one bacterial species. This review highlights several groups of bacterial species that have been shown to play an important role in immune development and homeostasis and were characterized in this manner. While not an exhaustive description of enteric bacteria that affect host immune systems, the groups we have picked form a representative sample of species that show varied impacts on their hosts. Each of these groups has been historically well studied; however, recent advances make them particularly relevant.
Bacteroides Fragilis
Although members of the Bacteroidales order are the most prominent gram negative bacteria in the intestine, the colonic bacteria Bacteroides fragilis make up only one percent of intestinal microbiota. While not numerically dominant amongst the Bacteroidetes in the intestine, this species of Bacteroides has been shown to have important effects on host health, both beneficial and detrimental (Polk and Kasper, 1977; Troy and Kasper, 2010). Normally symbiotic when contained within the intestine, in the event of bowel perforation, B. fragilis becomes pathogenic, inducing abscess formation throughout the peritoneal cavity (Polk and Kasper, 1977). B. fragilis is the most common clinically isolated anaerobic bacterial species, but in recent years, has become more well-known for the extent of positive effects it exerts on the immune system (Polk and Kasper, 1977; Mazmanian et al., 2005, 2008; Lassmann et al., 2007). B. fragilis and other Bacteroides have the genetic capability to produce multiple capsular polysaccharides, with B. fragilis producing eight. In B. fragilis these polysaccharides are important for commensal colonization of the intestine (Krinos et al., 2001; Coyne et al., 2008; Liu et al., 2008). At least two of these polysaccharides, polysaccharide A (PSA) and polysaccharide B (PSB) contain both positive and negative charges, making them zwitterionic (Tzianabos et al., 1993). Contrary to traditional characterization of carbohydrates as T-cell independent (Gonzalez-Fernandez et al., 2008), PSA and other zwitterionic carbohydrates evoke both CD4+ T-cell dependent and T-cell independent immune responses (Tzianabos et al., 2000; Cobb et al., 2004). Many characterized zwitterionic carbohydrates from bacteria have been shown to have immunomodulatory properties (Tzianabos et al., 1993, 2000, 2001; Cobb et al., 2004); however, PSA might be the most well-characterized microbial factor involved in commensalism. When germ-free mice are colonized with B. fragilis, many of the defects seen in these mice are corrected almost to the level of conventionally colonized mice (Mazmanian et al., 2005). Colonization of germ-free mice with B. fragilis promotes expansion of CD4+ T-cells, corrects depletion of splenic lymphocytic zones, and corrects Th1/Th2 imbalances by reduced IL-4 production and increased interferon (IFN)-γ production. Importantly, these rescue effects have been narrowed down to the activity of zwitterionic PSA. When B. fragilis defective in production of PSA is used to colonize germ-free mice, no correction of Th1/Th2 imbalance is seen. This was one of the first instances that showed correction of germ-free immunological defects by not just a commensal organism, but a specific surface molecule or symbiosis factor (Mazmanian et al., 2005). B. fragilis’s immunomodulatory capabilities have been shown to directly play a role in health and disease. Numerous studies have shown that an imbalance of microbiota can lead to intestinal inflammation due to lack of mucosal immune tolerance (Frank et al., 2007; Round and Mazmanian, 2009; Ivanov and Littman, 2011). B. fragilis has been shown to be protective against both immune (CD4+CD45Rb transfer with Helicobacter hepaticus inoculation) and chemically (trinitrobenzene sulfonic acid, TNBS) induced colitis (Mazmanian et al., 2008). In addition, purified PSA itself is protective against chemically induced colitis. This protection is dependent on CD4+ production of IL-10, induced by PSA. In animals defective for IL-10 production, no protective effect is seen by B. fragilis (Mazmanian et al., 2008). B. fragilis has subsequently been shown to alleviate chemically induced colitis in mice post colitis induction, showing that it can have both a preventative and therapeutic role. IL-10 production induced by B. fragilis results from increased numbers of Foxp3+ T-regulatory cells and mono-colonization of mice with B. fragilis has shown that these bacteria alone are capable of mediating the development of Foxp3+ T-regulatory cells from CD4+ T-cells (Round and Mazmanian, 2010). The anti-inflammatory effects of B. fragilis are not limited to a role in mouse models of models of colitis, but also additional models of inflammatory disease. For example, it was recently shown that B. fragilis is protective against an experimental autoimmune encephomyelitis (EAE), a mouse model mimicking human multiple sclerosis (Ochoa-Reparaz et al., 2010b). In an initial study, Ochoa-Reparaz et al., showed that purified B. fragilis PSA given to mice orally could protect against EAE. The administration of PSA to these mice enhanced a population of dendritic cells which express CD103 and these cells were seen accumulating in the cervical lymph nodes. Similar to the proposed mechanism for PSA’s protection against colitis, in IL-10 deficient mice, PSA offered no protection (Ochoa-Reparaz et al., 2010a). In an exciting follow-up study, the same group showed that oral colonization of mice with the entire organism, B. fragilis, could also protect against EAE and when a PSA deficient strain of B. fragilis was used, no protection was seen. Similar to purified PSA, the addition of B. fragilis to these mice stimulated the numbers of Foxp3+ T-regulatory cells accumulating in the cervical lymph nodes (Ochoa-Reparaz et al., 2010a,b). These studies are particularly ground-breaking since they show one of the first examples of systemic effects of B. fragilis’s immunomodulatory capabilities. B. fragilis and PSA’s anti-inflammatory properties are currently being tested in other models of inflammation to determine how universal their effects are.
Segmented Filamentous Bacteria
Segmented filamentous bacteria (SFB) or Candidatus Arthromitus (Snel et al., 1995) were originally observed and morphologically characterized over three decades ago in the small intestine of rats, mice, and chickens. These segmented organisms uniquely attach to the apical epithelium of primarily the small intestine through an attachment point in which the host cell epithelium and apical cytoplasm is modified (Hampton and Rosario, 1965; Savage, 1969; Fuller and Turvey, 1971; Davis and Savage, 1974). The biology of these organisms is intriguing as they appear in numerous vertebrates after birth and disappear shortly after adulthood (Davis and Savage, 1974). Early reports indicate many influences over the colonization of SFB within a host organism including age, sex, and immune status (Davis and Savage, 1974; Jiang et al., 2001). The timing of the appearance of these organisms in their vegetative state has led to numerous studies as well as speculation about the environmental signal that induces sporulation or overall disappearance of SFB from their host. It is possible that this could be related to the establishment of microbial colonization in young vertebrates or possibly it is related to nutritional changes once mammals are weaned from their mothers. Microscopically, SFB have been observed in numerous laboratory and non-laboratory vertebrates, including chickens, dogs, cats, pigs, fowl, macaque, and humans (Klaasen et al., 1993); however, they have also been shown to be extremely species specific. Tannock et al. (1984) showed that when ileal homogenates containing SFB from mice were given to germ-free rats, the SFB failed to attach to the epithelium. Likewise, when ileal homogenates containing SFB from rats were given to germ-free mice, the SFB also failed to adhere to the epithelium, whereas when source SFB were given to the same species, they were able to adhere to the epithelium (Tannock et al., 1984). This indicates a co-evolutionary relationship between SFB and their hosts. Although SFB were visualized by light microscopy to be part of the human microbiota, definitive genetic identification of these microbes in humans has not been reported (Klaasen et al., 1993). Many of the aforementioned characteristics of SFB led researchers to believe they had an important role in the immunology of their hosts; however, the un-culturability of SFB has made many studies of this organism’s unique features impossible. A break-through came with the ability to mono-colonize mice with SFB. Several groups were able to take fecal homogenates and limit them to only spore-forming bacteria using chloroform and ethanol washes (Klaasen et al., 1990). These mixes of SFB and some Clostridium species could be diluted to only contain SFB and were illeo-inoculated into mice establishing a mono-colonized species (Klaasen et al., 1991). The study of mice mono-colonized with SFB in combination with high powered sequencing methods has allowed many groups to observe the profound effects that SFB have on their host’s immunity (Klaasen et al., 1991; Umesaki et al., 1995; Ivanov et al., 2008, 2009; Gaboriau-Routhiau et al., 2009).
Mono-colonization of mice with SFB leads to many immunological changes to germ-free mice particularly within the small intestine. In the small intestine, there is an induction of MHCII molecules on IECs as well as fucosylation of GM1 glyco-lipids (Umesaki et al., 1995). Significant changes are also seen on IELs such as an expansion of IELs bearing both αβ and γδ TCRs, increased CD8+ T-cells, increased cytolytic activity, and increased Thy1 expression. Crypt cell proliferation is induced as well as production of columnar cells. Upon mono-colonization, germinal center reactions in the Peyer’s patches are stimulated and CD4+ and CD45RBlow T-cells increase until they reach levels of conventional mice (Umesaki et al., 1995; Talham et al., 1999). IgA is also produced in significant amounts after SFB colonization of germ-free mice (Umesaki et al., 1995; Suzuki et al., 2004). While these changes are substantial, the levels of αβ TCRS do not fully come up to conventional levels nor are many of the morphological characteristics of germ-free mice such as an enlarged cecum normalized (Umesaki et al., 1995; Talham et al., 1999). Interestingly, in mice deficient in IgA, a prominent and persistent expansion of SFB is seen that returns to normal when the mice are compensated with IgA (Suzuki et al., 2004). IgA is seen as a major mechanism for maintaining intestinal homeostasis among commensal organisms (Duerkop et al., 2009), and its absence in these mice is compensated for by large amounts of IgM and normal expression of defensins and angiogenins (Suzuki et al., 2004). This reaffirms the paradigm in which an IgA feedback loop is a method of keeping commensal organisms near the epithelial surface in check (Hooper, 2009).
Most recently, several studies have pinpointed dramatic effects of SFB colonization based on their presence in some host species and not in others. A major indication of SFB’s role in the induction of pro-inflammatory factors came with the report that mice from differing sources (Taconic Farms versus Jackson Laboratory) had differing levels of IL-17 producing cells and animals that had decreased Th17 cells had increased Foxp3+ regulatory cells (Ivanov et al., 2008). In a subsequent report, the authors were able to show, using 16S rRNA phylochip analysis, that SFB which were present in mice from some sources and not others accounted for the difference seen in Th17 cells. When the authors mono-colonized germ-free mice with SFB, they saw induction of Th17 cells that produce IL-17 and IL-22 in the lamina propria as well as up-regulation of genes associated with inflammation and anti-microbial defenses (Ivanov et al., 2009). In a similar deductive study, Gaboriau-Routhiau et al. noticed that they were able to conventionalize the transcriptome of germ-free mice with a murine microbiota, but not with a human fecal microbiota or cultured murine microbiota. This led them to believe an un-culturable organism must be the missing link to reconstituting conventional level gene responses in their germ-free mice. They deduced that SFB may be the missing link and upon mono-colonizing mice with these organisms, they saw expression of many mucosal genes equal to those of conventional mice including: RegIIIγ, IFNγ, IL-1β, IL-10, IL-17, inducible nitric oxide synthase (iNOS), and IL-12p40. They also saw local induction of IFNγ, IL-10, and IL-17. SFB increased production of IFNγ by CD4+ T-cells, IL-17 production by CD4+ T-cells and increased the total number of CD4+CD25+Foxp3+ T-regulatory cells in the lamina propria of the small intestine and colon (Gaboriau-Routhiau et al., 2009). This induction of both pro-inflammatory and anti-inflammatory factors is extremely interesting; however, in general, SFB have been more frequently associated with pro-inflammatory Th17 cells (Ivanov et al., 2008, 2009; Wu et al., 2010). Importantly, while SFB were initially suspected to be a sole factor needed to conventionalize mice, none of these factors were increased to conventional levels, indicating that SFB works in concert with other commensal organisms (Gaboriau-Routhiau et al., 2009). Due to their stimulation of the immune system and immediate proximity to the host epithelium, many groups have hypothesized that SFB are involved in CR to host enteric pathogens. Several studies have already shown that in the presence of SFB, hosts are immune to organisms they might otherwise by susceptible to. For example, Heczko et al. (2000) showed that rabbits colonized with SFB were less likely to be infected by rabbit enteropathogenic Escherichia coli (REPEC) and conversely, all rabbits who did not have SFB were colonized by REPEC. SFB has also been shown to have protective effects against Salmonella enteritidis in rats and Citrobacter rodentium in mice (Garland et al., 1982; Ivanov et al., 2009). The culmination of these studies and the observation that SFB are present at a young age strongly supports a role for SFB in CR, possibly selective, during the establishment of a host’s intestinal microbiota.
It is well-known that the composition of the microbiota can have strong impacts on health and any imbalance can cause either susceptibility to infection or on the other side can lead to auto-inflammatory conditions. Although SFB have been designated as commensal organisms due to their generally non-pathogenic characteristics, it is possible that this classification may be pre-emptive as we learn more about the effects they exert on their hosts. Given SFB’s strong pro-inflammatory capabilities, when not reined in by other regulatory factors, their effects could lead to diseased states. For example, recently it was shown that in a mouse model of autoimmune arthritis, for germ-free mice, arthritis is attenuated. Arthritis is quickly re-induced; however, with the addition of solely SFB (Wu et al., 2010). Similar phenomenon have been observed with SFB in EAE (Lee et al., 2011). Alternatively, SFB may exhibit pathogenic qualities in the presence of other bacteria. Work by Stepankova et al. indicates that SFB may require additional microbiota to exert its pro-inflammatory effects. This group showed that SFB could trigger chronic inflammation in SCID mice, which received CD45RBhigh CD4+ T-cells. However, colitis was only triggered in germ-free SCID mice which received a cocktail of specific pathogen free (SPF) microbiota as well as SFB and not in the mice which received either SPF microbiota or SFB alone (Stepankova et al., 2007).
No studies thus far have looked at the long-term effects of mono-colonization of mice with SFB. We are just uncovering the surface in characterization of these organisms and their mechanisms. While they have been identified in humans through light microscopy (Klaasen et al., 1993), they have not been identified in humans using 16S rRNA sequencing. Understanding the true niche of this organism will help elucidate their role in host biology. Current work is underway to sequence the genome of these species from fecal DNA isolation. In addition, numerous efforts to culture this organism in vitro are underway in multiple labs. Armed with the genome sequence and a way to grow these organisms to high numbers in culture, there is potential to understand the molecular mechanisms behind the fascinating SFB interaction with mammalian epithelial cells and the mechanisms responsible for their effects.
Clostridium
While SFB, located primarily in the small intestine of their hosts, induce effector T-cell function and pro-inflammatory conditions, a recent report shows that members of the genus Clostridium, most commonly located in the large intestine, do the exact opposite (Atarashi et al., 2011). This is may indicate that the commensal microbiota of the small and large intestine have compartmentalized effects on the resident T-cells. The effect of mixed Clostridium species on germ-free mice has been examined previously. In earlier, less controlled studies, the effect of colonizing germ-free mice with chloroform treated or, Clostridium rich feces from conventional mice was examined. Under these conditions, normalization of the enlarged germ-free cecum was seen and the mix of 46 Clostridium species was defined (Itoh and Mitsuoka, 1980, 1985). These studies were some of the original work to show that Clostridium species alone can have a big impact on the intestinal status of a mouse and were part of the initial efforts to tease apart the roles of individual genuses in the intestine. More recent studies build upon those initial efforts by examining specific parameters induced by this defined mix of Clostridium species. In the recent study by Atarashi et al., a combination of 46 spore-forming Clostridium species mainly composed of clusters IV (leptum) and XIVa (coccoides) induced a strong anti-inflammatory response in the intestine through expansion of Foxp3+ regulatory T-cells. This effect was partially mediated by the release of TGF-β from IELs. The group showed that pattern recognition receptors such as Myd88, Rip2, and Card9 were not involved. A similar regulatory T-cell accumulation was seen when the Clostridium mix was enhanced in normal mice with a healthy immune system and these mice were also more resistant to animal models of inflammation (Atarashi et al., 2011). This response invites the use of Clostridium as an anti-inflammatory probiotic. In future studies, it will be interesting to see how individual species of Clostridium have an effect on mucosal immunology especially considering the opposite effect exerted by very closely related SFB. Compartmentalized effects of Clostridium species and SFB species have been illustrated by the mono-colonization of germ-free mice with SFB or the aforementioned 46 species of Clostridium or dual colonized with both. In the SFB mice, αβ IELs and MHC II were increased only in the small intestine while in the Clostridium mice, CD8+ T-cells and αβ IELs were increased only in the large intestine. In the co-colonized mice, the mice more closely resembled conventional mice indicating a distinct balance and localization of the effects of each group of species (Umesaki et al., 1999).
Lactobacillus and Bifidobacterium
The beneficial health effects of the endogenous intestinal bacterial genera Lactobacillus and Bifidobacterium are reflected through their frequent use as probiotics. Species within these bacterial genera have anti-inflammatory properties as well as many other health benefits for hosts such as a contribution toward CR against pathogens, and aid in improved digestion, nutrient adsorption, and increased availability of nutrients in the intestine (Sanchez et al., 2010; Turpin et al., 2010). The genomes of Bifidobacterium species reflect a large propensity for carbohydrate uptake and metabolism as well as the presence of many enzymes for the break-down of complex carbohydrates. These traits are thought to give Bifidobacterium species a competitive advantage within the intestine (Schell et al., 2002; Ryan et al., 2005; Kim et al., 2009). Meanwhile, Lactobacillus species encode numerous transporters and have a large capacity for sugar internalization and break-down as well as numerous mucus binding cell surface proteins (Kleerebezem et al., 2003; Boekhorst et al., 2006a,b; Siezen et al., 2006). Bifidobacterium were originally isolated from human baby feces and were identified as a substantial portion of the normal microbiota of humans. Their positive effects were seen through bottle fed babies that lacked Bifidobacterium and subsequently suffered from more diarrhea (Kleerebezem and Vaughan, 2009). Both Bifidobacterium and Lactobacillus species are among the first subsets of bacteria to colonize the human colon after birth and decrease in number into adulthood (Favier et al., 2003; Vaughan et al., 2005). Much of the characterization of Bifidobacterium and Lactobacillus and their effects on the mammalian host has come through the mono and co-colonization of germ-free mice and observation of immune and physiological responses from the host as well as bacterial transcriptome changes as they adapted to different niches within a host (Sonnenburg et al., 2006; Denou et al., 2007, 2008; Menard et al., 2008; Kleerebezem and Vaughan, 2009) and also through their probiotic effects on humans (Ouwehand, 2007; Kleerebezem and Vaughan, 2009). Among the many positive effects these groups of bacteria have on hosts is the ability to reduce inflammation. Skewed levels of microbiota are one important factor in inflammatory bowel disease as well as other inflammatory conditions like rheumatoid arthritis (Frank et al., 2007; Gueimonde et al., 2007; Round and Mazmanian, 2009). In mouse models of colitis, under germ-free conditions or after treatment of mice with antibiotics, intestinal inflammation cannot be readily induced (Bamias et al., 2002; Strober et al., 2002). Bifidobacterium and Lactobacillus are both important in the natural balance of the intestinal community and in cases of inflammatory bowel disease (IBD), both groups of bacteria are seen at decreased levels in fecal samples as opposed to Enterococcus and Bacteroides, which are seen elevated in the mucosa of patients (Frank et al., 2007). Both Bifidobacterium lactis and Bifidobacterium infantis have been shown to be protective against inflammation caused by chemically and Salmonella induced colitis respectively (Round and Mazmanian, 2009). Both species of bacteria can suppress the transcription of the inflammatory factors: IL-1β, tumor necrosis factor (TNF)-α, NFκB and translation of IL-1β and IL-6 (Turpin et al., 2010). Treatment of colitic mice with Bifidobacterium infantis induces the production of CD4+CD25+ regulatory T-cells and these cells can be adoptively transferred to another mouse and prevent activation of inflammatory factors (O’Mahony et al., 2008). Additionally, numerous species of Lactobacillus have exerted protective effects against chemically and IL-10−/− induced models of colitis (Round and Mazmanian, 2009). It has been suggested that Lactobacillus rhamnosus can also induce regulatory T-cell activity. Bone marrow dendritic cells (BMDCs) incubated with Lactobacillus rhamnosus offer protection from induction of intestinal inflammation in a CD4+CD25+ regulatory T-cells dependent fashion (Foligne et al., 2007).
In addition to their anti-inflammatory properties, Bifidobacterium and Lactobacillus species have been shown to play a role in exclusion of enteric pathogens. For example, the inflammatory effects of disease seen after infection of mice with Salmonella serotype Typhimurium can be countered by treating the mice with Bifidobacterium infantis through the induction of Foxp3+ T-regulatory cells (O’Mahony et al., 2008).
While the precise mechanisms behind the beneficial effects of Lactobacillus and Bifidobacterium are largely unknown, a significant amount of their activity can be attributed to cell surface associated structures and extracellular protein interaction with mucosal immune cells (Kleerebezem et al., 2010). Such cell surface structures include but are not limited to: exopolysaccharides, bacteriocins, lipoteichoic acid, and extracellular proteins (Sanchez et al., 2010). Many of these proteins from both Bifidobacterium and Lactobacillus are primarily identified using bioinformatics and most have yet to be fully characterized. For example, Lactobacillus plantarum are capable of adhering to mannosyl moieties on human mucosa and in doing so prevent ETEC infection (Adlerberth et al., 1996); however, the responsible mannose specific adhesion (Msa), a sortase dependent cell surface protein was only recently discovered thanks to bioinformatics (Pretzer et al., 2005). Informatics searches for potential adhesions, mucin binding domains, and secretory sequences have been very successful (Buck et al., 2005; Boekhorst et al., 2006a; Sanchez et al., 2008; Barinov et al., 2009). Secreted surface molecules have been shown to play a role in Bifidobacterium and Lactobacillus CR through the enhancement of the mucosal barrier and tight junctions, induction of anti-microbial peptides, and some secreted proteins are thought to interact directly with host epithelial cells possibly blocking niches for pathogenic bacteria (Sanchez et al., 2010). Schlee et al. (2008) showed that numerous species of Lactobacillus are able to induce anti-microbial peptide production, which in turn contributes to CR of pathogens. Pre-conditioned media from Lactobacillus rhamnosus GG contains peptides with anti-microbial activity against: E. coli EAEC, Salmonella typhi and Staphylococcus aereus (Lu et al., 2009). Anti-microbial peptide production by Lactobacillus salivarius can protect mice from Listeria monocytogenes while non-bacteriocin producing Lactobacillus salivarius do not confer protection (Corr et al., 2007). The Lactobacillus crispatus S-layer protein (SlpA) interacts directly with collagen on host epithelial cells (Antikainen et al., 2002) and has been shown to block the binding of pathogens such as Escherichia coli O157:H7 and Salmonella typhimurium (Chen et al., 2007), indicating that the CR ability of some probiotics may be directly mediated by adhesion molecules. As well as playing a role in CR, SlpA was shown by Konstantinov et al. to play a role in the induction of host immune responses. They showed that SlpA interacts directly with the non-integrin DC-SIGN, inducing IL-10 production and low IL-12p70 and that in an slpA mutant strain, which overexpresses slpB, the immune reaction is skewed toward a more pro-inflammatory response (Konstantinov et al., 2008). Another extracellular protein immune modulator is the serine protease inhibitor (Serpin) present in many species of Bifidobacterium. Ivanov et al. (2006) showed through a series of in vitro studies that serpin could inhibit pancreatic neutrophil elastases, thereby modulating host inflammatory responses. Bifidobacterium can play a role in mucosal health partially through strengthening of tight junctions. Ewaschuck et al. have shown that Bifidobacterium infantis pre-conditioned media (BiCM) can increase production of epithelial cell tight junction proteins and increase transepithelial resistance (TER). This BiCm also had drastic effects in vivo, attenuating inflammation and colonic permeability in IL-10 deficient mice in part mediated through the Mapk pathway (Ewaschuk et al., 2008). Extracellular proteins secreted by Lactobacillus species also play a role in mucosal barrier maintenance through MapKs. Two of the better characterized Lactobacillus extracellular proteins are p40 (a hypothetical cell surface antigen) and p75 (a hypothetical cell wall-associated hydrolase). These proteins, when purified, promoted growth in human and murine colonic epithelial cells through activating protein kinase (akt) and were able to reduce TNF-α induced colonic injury in tissue explants (Yan et al., 2007; Seth et al., 2008).
While many Lactobacillus and Bifidobacterium species are considered culturable, there are still many uncultured species of each within the intestine indicating that we still have much to discover about Bifidobacterium and Lactobacillus mechanisms of action and surface molecules (Heilig et al., 2002; Ben-Amor et al., 2005). Also, while many extracellular and secreted proteins from Bifidobacterium and Lactobacillus have been characterized in vitro, their roles have yet to be confirmed in vivo (Kleerebezem et al., 2010). Full characterization of secreted and surface proteins from these groups of bacteria could further advance therapeutics in intestinal diseases and our knowledge of immune regulation by commensal bacteria.
Discussion
In this review, we have highlighted several bacterial groups and specific species that have an immunomodulatory impact on their hosts (summarized in Figure 1). There are an incredible 1014 bacteria in the intestine and the mammalian immune system must be able to sustain these constant visitors without eliciting a strong reaction, yet at the same time, be primed to react to incoming and invading pathogens. We have described several different instances in which intestinal bacteria prime responses that mirror and enhance this vital balance by either promoting inflammatory (SFB and Th17 cells) or anti-inflammatory conditions (Clostridium, Bacteroides fragilis, Bifidobacterium, and Lactobacillus).
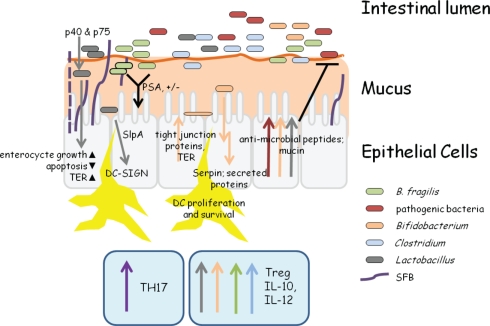
Figure 1.
Snapshot of bacterial–host activity in the intestine. This extremely simplistic view of intestinal activity highlights some of the major roles of different bacteria within the intestine. SFB attach to epithelial cells, induce pro-inflammatory responses, and expand TH17 cells (Davis and Savage, 1974; Gaboriau-Routhiau et al., 2009; Ivanov and Littman, 2010). Bacteroides fragilis and many Clostridium species induce IL-10 production and the expansion of T-regulatory cells (Mazmanian et al., 2005,2008; Atarashi et al., 2011). In B. fragilis, this is mediated through the surface polysaccharide, PSA (Mazmanian et al., 2008). Both Lactobacillus and Bifidobacterium can induce anti-inflammatory cytokine production, anti-microbial peptide, and mucin production, and may adhere to epithelial cells (Adlerberth et al., 1996; Pretzer et al., 2005; Kleerebezem et al., 2010; Sanchez et al., 2010; Turpin et al., 2010). The secreted proteins, p40 and p75 from many Lactobacillus species promote cell growth through a PI-3K and AKT pathway, inhibit apoptosis by causing decreased TNFα levels, and increase transepithelial resistance (TER) through increased tight junction protein production (Yan et al., 2007; Seth et al., 2008). Lactobacillus S-layer protein A (SlpA) binds to DC-SIGN which leads to increased IL-10 production. SlpA can bind directly to epithelial cells, which may play a role in colonization resistance (CR) against pathogenic bacteria (Antikainen et al., 2002; Chen et al., 2007). The Bifidobacterium serine protease inhibitor Serpin inhibits neutrophil elastase, thereby modulating acute inflammation in the intestine (Ivanov et al., 2006). Finally, undefined secreted proteins from Bifidobacterium species cause an increase in tight junction protein production and thereby TER, contributing to CR (Sanchez et al., 2010).
It is interesting that closely related groups of bacteria such as Clostridium and SFB can exert such different effects. This is also seen in the case of the pathogen Clostridium difficile. These differing effects could possibly be due to uncharacterized effector molecules present on the surface of SFB species versus Clostridium. The effects of B. fragilis on host immunology are clearly shown to be mediated through the symbiosis factor PSA, which has been well-characterized over the last two decades. Not as much is known about the active molecules from many other symbiotic bacteria. Discovery and characterization of these molecules will be extremely important for fully understanding immune system–bacterial cell interactions. In the case of SFB, the potential molecules that allow its association with epithelial cells and possible effector proteins or carbohydrates that elicit host immune responses from SFB are tantalizing. One of the proposed mechanisms of homeostasis within the intestine that allows residence of bacteria without a hyper-activated immune response is the sequestration of commensal bacteria in the mucus layer of epithelial cells and in the intestinal lumen (Hooper, 2009). SFB clearly break this rule. Actin accumulation by eukaryotic epithelial cells has been seen underneath the attachment points (Jepson et al., 1993) of SFB to eukaryotic cells. This is highly reminiscent of the manner in which many enteric pathogens such as enterohemorrhagic E. coli attach to host cells. These pathogens contain a secretion system that enable them to “inject” effector proteins into host cells (Coburn et al., 2007). Is it possible that SFB may have a similar system of their own? This also begs the question as to whether SFB truly are commensal, non-pathogenic organisms. The ability to grow these organisms in vitro as well as obtaining the genome sequence of these organisms will allow for studies that will begin to answer these questions.
While we have merely scratched the surface and new roles of different species are continually being discovered, the studies highlighted here give us an idea of how powerful the effects of single bacterial species can be within the intestinal framework. Given an environment of over 500 species, it is remarkable that individual species could have such strong effects. Although mono-colonization of germ-free mice is an excellent way to observe species specific immune modulation, it is also an extremely simplified view of an extremely complex ecosystem and as additional roles of individual species are discovered, investigating how bacteria interact with each other within the intestine will be the next direction. Taken from the simple environment of mono-colonization, it is hard to believe that the roles of bacteria would remain the same once placed in a melting pot of hundreds of species all competing for nutrients and space. However in many cases, such as with Clostridium, Bifidobacterium, Lactobacillus, SFB, and Bacteroides, the effects of these bacteria can be seen when their populations are increased within a conventional animal, not solely within a mono-colonized animal (Dasgupta et al., unpublished data) (Sonnenburg et al., 2005, 2006; Mazmanian et al., 2005, 2008; Stepankova et al., 2007; Kleerebezem and Vaughan, 2009; Round and Mazmanian, 2010). In some cases, such as SFB, immunomodulatory effects are seen more readily in the context of a complete microbiota rather than a mono-colonized animal (Stepankova et al., 2007). With such great success at teasing apart the individual contributions of many resident intestinal bacteria through mono-colonization experiments, another body of information will come from determining dynamic interactions between multiple groups of a host’s natural inhabitants and trying to determine how they use symbiosis factors to communicate with not only their host tissues, but also each other. Already, many groups have looked at the effects of colonizing mice with a select microbiota that represents many of the abundant species normally present in the mammalian intestine (Hooper et al., 2001; Macpherson and Harris, 2004; Sonnenburg et al., 2006; Round and Mazmanian, 2009; Stecher and Hardt, 2011).
As described earlier, SFB from a rat cannot attach to the epithelium of mouse and vice versa (Tannock et al., 1984; Hooper et al., 2001; Macpherson and Harris, 2004; Sonnenburg et al., 2006). This is one example illustrating that colonization of germ-free animals by any bacteria is not always sufficient to conventionalize the animal. Rather, specific bacterial colonization is necessary. This implies a co-evolutionary relationship between a host and its microbiota. The study of humanized mice (germ-free mice colonized with human fecal samples) is one tool that has been utilized to answer whether hosts require a host-specific microbiota. While humanized mice have a “complete” microbiota, it is foreign. At least two studies have shown that humanized mice may more closely resemble germ-free mice in many immunological traits than conventionalized mice and that colonizing mice with a foreign microbiota cannot completely restore immune defects seen in germ-free mice, nor can it restore many other germ-free defects such as metabolism (Chung et al. submitted; Gaboriau-Routhiau et al., 2009). When comparing the composition of microbiota from mice colonized with human fecal matter and mice colonized with mouse cecal matter, the two groups of microbiota had a high degree of resemblance through the genus level. The majority of differences were seen on the species level (Chung et al. submitted) again emphasizing the dramatic effects on the host carried out by individual species.
The interest in the dynamics of microbes in the intestine has existed for decades, but has gained and lost momentum as new technology comes and goes. As we are starting to see definite trends in the composition of the intestine, gain knowledge through sequencing, and identify the diversity of intestinal bacteria through the Microbiome project, we are in a position to understand the gut like we never have before. This knowledge will expand our understanding of bacterial symbiosis factors, host regulation of the commensal microbiota, and likewise bacteria–host communication and bacterial–bacterial communication within the intestine. This will lead to a better understanding of and characterization of intestinal bacterial imbalances that lead to diseased states, giving us a better grasp of this previously mysterious world and the tools to greatly impact intestinal health in the future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Nicola C. Reading was supported by a training grant from NIAID (T32AI007061) and Dennis L. Kasper by a Senior Investigator Award from the Crohns Colitis Foundation of America. Unfortunately, due to space limitations, it is not possible to site the work of numerous authors who have contributed in these fields.
References
- Adlerberth I., Ahrne S., Joansson M. L., Molin G., Hanson L. A., Wold A. E. (1996). A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl. Environ. Microbiol. 62, 2244–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antikainen J., Anton L., Sillanpää J., Korhonen T. K. (2002). Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol. Microbiol. 46, 381–394 10.1046/j.1365-2958.2002.03180.x [DOI] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., Taniguchi T., Takeda K., Hori S., Ivanov I. I., Umesaki Y., Itoh K., Honda K. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamias G., Marini M., Moskaluk C. A., Odashima M., Ross W. G., Rivera-Nieves J., Cominelli F. (2002). Down-regulation of intestinal lymphocyte activation and Th1 cytokine production by antibiotic therapy in a murine model of Crohn’s disease. J. Immunol. 169, 5308–5314 [DOI] [PubMed] [Google Scholar]
- Barinov A., Loux V., Hammani A., Nicolas P., Langella P., Ehrlich D., Maguin E., van de Guchte M. (2009). Prediction of surface exposed proteins in Streptococcus pyogenes, with a potential application to other Gram-positive bacteria. Proteomics 9, 61–73 10.1002/pmic.200800195 [DOI] [PubMed] [Google Scholar]
- Ben-Amor K., Heilig H., Smidt H., Vaughan E. E., Abee T., de Vos W. M. (2005). Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl. Environ. Microbiol. 71, 4679–4689 10.1128/AEM.71.8.4679-4689.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhorst J., Helmer Q., Kleerebezem M., Siezen R. J. (2006a). Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 152(Pt 1): 273–280 10.1099/mic.0.29217-0 [DOI] [PubMed] [Google Scholar]
- Boekhorst J., Wels M., Kleerebezem M., Siezen R. J. (2006b). The predicted secretome of Lactobacillus plantarum WCFS1 sheds light on interactions with its environment. Microbiology 152(Pt 11): 3175–3183 10.1099/mic.0.29217-0 [DOI] [PubMed] [Google Scholar]
- Buck B. L., Altermann E., Svingerud T., Klaenhammer T. R. (2005). Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71, 8344–8351 10.1128/AEM.71.12.8344-8351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu J., Shuai J., Chen J., Zhang Z., Fang W. (2007). The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int. J. Food Microbiol. 115, 307–312 10.1016/j.ijfoodmicro.2006.11.007 [DOI] [PubMed] [Google Scholar]
- Cobb B. A., Wang Q., Tzianabos A. O., Kasper D. L. (2004). Polysaccharide processing and presentation by the MHCII pathway. Cell 117, 677–687 10.1242/jcs.00898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B., Sekirov I., Finlay B. B. (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20, 535–549 10.1128/CMR.00013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corr S. C., Li Y., Riedel C. U., O’Toole P. W., Hill C., Gahan C. G. (2007). Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U.S.A. 104, 7617–7621 10.1073/pnas.0700440104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne M. J., Chatzidaki-Livanis M., Paoletti L. C., Comstock L. E. (2008). Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. U.S.A. 105, 13099–13104 10.1073/pnas.0804220105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. P., Savage D. C. (1974). Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect. Immun. 10, 948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denou E., Berger B., Barretto C., Panoff J. M., Arigoni F., Brüssow H. (2007). Gene expression of commensal Lactobacillus johnsonii strain NCC533 during in vitro growth and in the murine gut. J. Bacteriol. 189, 8109–8119 10.1128/JB.00991-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denou E., Pridmore R. D., Berger B., Panoff J. M., Arigoni F., Brüssow H. (2008). Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190, 3161–3168 10.1128/JB.01637-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop B. A., Vaishnava S., Hooper L. V. (2009). Immune responses to the microbiota at the intestinal mucosal surface. Immunity 31, 368–376 10.1016/j.immuni.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Ewaschuk J. B., Diaz H., Meddings L., Diederichs B., Dmytrash A., Backer J., Looijer-van Langen M., Madsen K. L. (2008). Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G1025–G1034 10.1152/ajpgi.90227.2008 [DOI] [PubMed] [Google Scholar]
- Favier C. F., de Vos W. M., Akkermans A. D. (2003). Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe 9, 219–229 10.1016/j.anaerobe.2003.07.001 [DOI] [PubMed] [Google Scholar]
- Foligne B., Zoumpopoulou G., Dewulf J., Ben Younes A., Chareyre F., Sirard J. C., Pot B., Grangette C. (2007). A key role of dendritic cells in probiotic functionality. PLoS ONE 2, e313. 10.1371/journal.pone.0000313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. N., Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R., Turvey A. (1971). Bacteria associated with the intestinal wall of the fowl (Gallus domesticus). J. Appl. Bacteriol. 34, 617–622 [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V., Rakotobe S., Lécuyer E., Mulder I., Lan A., Bridonneau C., Rochet V., Pisi A., De Paepe M., Brandi G., Eberl G., Snel J., Kelly D., Cerf-Bensussan N. (2009). The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 10.1016/j.immuni.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Garland C. L., Lee A., Dickson M. R. (1982). Segmented filamentous bacteria in the rodent small intestine: their colonization of growing animals and possible role in host resistance to Salmonella. Microb. Ecol. 8, 181–190 10.1007/BF02010451 [DOI] [PubMed] [Google Scholar]
- Garrett W. S., Lord G. M., Punit S., Lugo-Villarino G., Mazmanian S. K., Ito S., Glickman J. N., Glimcher L. H. (2007). Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 10.1016/j.cell.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Fernandez A., Faro J., Fernández C. (2008). Immune responses to polysaccharides: lessons from humans and mice. Vaccine 26, 292–300 10.1016/j.vaccine.2007.11.042 [DOI] [PubMed] [Google Scholar]
- Gueimonde M., Ouwehand A., Huhtinen H., Salminen E., Salminen S. (2007). Qualitative and quantitative analyses of the bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J. Gastroenterol. 13, 3985–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton J. C., Rosario B. (1965). The attachment of microorganisms to epithelial cells in the distal ileum of the mouse. Lab. Invest. 14, 1464–1481 [PubMed] [Google Scholar]
- Heczko U., Abe A., Finlay B. B. (2000). Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. J. Infect. Dis. 181, 1027–1033 10.1086/315348 [DOI] [PubMed] [Google Scholar]
- Heilig H. G., Zoetendal E. G., Vaughan E. E., Marteau P., Akkermans A. D., de Vos W. M. (2002). Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68, 114–123 10.1128/AEM.68.1.114-123.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L. V. (2009). Do symbiotic bacteria subvert host immunity? Nat. Rev. Microbiol. 7, 367–374 10.1038/nrmicro2114 [DOI] [PubMed] [Google Scholar]
- Hooper L. V., Gordon J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115–1118 10.1126/science.1058709 [DOI] [PubMed] [Google Scholar]
- Hooper L. V., Wong M. H., Thelin A., Hansson L., Falk P. G., Gordon J. I. (2001). Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884 10.1126/science.291.5505.881 [DOI] [PubMed] [Google Scholar]
- Itoh K., Mitsuoka T. (1980). Production of gnotobiotic mice with normal physiological functions. I. Selection of useful bacteria from feces of conventional mice. Z. Versuchstierkd. 22, 173–178 [PubMed] [Google Scholar]
- Itoh K., Mitsuoka T. (1985). Characterization of clostridia isolated from faeces of limited flora mice and their effect on caecal size when associated with germ-free mice. Lab. Anim. 19, 111–118 10.1258/002367785780887464 [DOI] [PubMed] [Google Scholar]
- Ivanov D., Emonet C., Foata F., Affolter M., Delley M., Fisseha M., Blum-Sperisen S., Kochhar S., Arigoni F. (2006). A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J. Biol. Chem. 281, 17246–17252 10.1074/jbc.M601678200 [DOI] [PubMed] [Google Scholar]
- Ivanov I. I., Atarashi K., Manel N., Brodie E. L., Shima T., Karaoz U., Wei D., Goldfarb K. C., Santee C. A., Lynch S. V., Tanoue T., Imaoka A., Itoh K., Takeda K., Umesaki Y., Honda K., Littman D. R. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Frutos Rde L., Manel N., Yoshinaga K., Rifkin D. B., Sartor R. B., Finlay B. B., Littman D. R. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Littman D. R. (2010). Segmented filamentous bacteria take the stage. Mucosal Immunol. 3, 209–212 10.1038/mi.2010.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Littman D. R. (2011). Modulation of immune homeostasis by commensal bacteria. Curr. Opin. Microbiol. 14, 106–114 10.1016/j.mib.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson M. A., Clark M. A., Simmons N. L., Hirst B. H. (1993). Actin accumulation at sites of attachment of indigenous apathogenic segmented filamentous bacteria to mouse ileal epithelial cells. Infect. Immun. 61, 4001–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. Q., Bos N. A., Cebra J. J. (2001). Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect. Immun. 69, 3611–3617 10.1128/IAI.69.3.1244-1255.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. F., Jeong H., Yu D. S., Choi S. H., Hur C. G., Park M. S., Yoon S. H., Kim D. W., Ji G. E., Park H. S., Oh T. K. (2009). Genome sequence of the probiotic bacterium Bifidobacterium animalis subsp. lactis AD011. J Bacteriol 191, 678–679 10.1128/JB.00044-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaasen H. L., Koopman J. P., Van den Brink M. E., Bakker M. H., Poelma F. G., Beynen A. C. (1993). Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab. Anim. 27, 141–150 10.1258/002367793780810441 [DOI] [PubMed] [Google Scholar]
- Klaasen H. L., Koopman J. P., Van den Brink M. E., Van Wezel H. P., Beynen A. C. (1991). Mono-association of mice with non-cultivable, intestinal, segmented, filamentous bacteria. Arch. Microbiol. 156, 148–151 10.1007/BF00290989 [DOI] [PubMed] [Google Scholar]
- Klaasen H. L. B. M., Koopman J. P., Van Den Brink M. E., Van Wezel H. P. N., Scholten P. M., Beynen A. C. (1990). Colonisation of germ-free mice by segmented filamentous bacteria after oral administration of various murine intestinal wall preparations. Microb. Ecol. Health Dis. 3, 281–284 10.3109/08910609009140123 [DOI] [Google Scholar]
- Kleerebezem M., Boekhorst J., van Kranenburg R., Molenaar D., Kuipers O. P., Leer R., Tarchini R., Peters S. A., Sandbrink H. M., Fiers M. W., Stiekema W., Lankhorst R. M., Bron P. A., Hoffer S. M., Groot M. N., Kerkhoven R., de Vries M., Ursing B., de Vos W. M., Siezen R. J. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U.S.A. 100, 1990–1995 10.1073/pnas.0337704100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M., Hols P., Bernard E., Rolain T., Zhou M., Siezen R. J., Bron P. A. (2010). The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 34, 199–230 10.1111/j.1574-6976.2009.00208.x [DOI] [PubMed] [Google Scholar]
- Kleerebezem M., Vaughan E. E. (2009). Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu. Rev. Microbiol. 63, 269–290 10.1146/annurev.micro.091208.073341 [DOI] [PubMed] [Google Scholar]
- Konstantinov S. R., Smidt H., de Vos W. M., Bruijns S. C., Singh S. K., Valence F., Molle D., Lortal S., Altermann E., Klaenhammer T. R., van Kooyk Y. (2008). S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U.S.A. 105, 19474–19479 10.1073/pnas.0810305105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinos C. M., Coyne M. J., Weinacht K. G., Tzianabos A. O., Kasper D. L., Comstock L. E. (2001). Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414, 555–558 10.1038/35107092 [DOI] [PubMed] [Google Scholar]
- Lassmann B., Gustafson D. R., Wood C. M., Rosenblatt J. E. (2007). Reemergence of anaerobic bacteremia. Clin. Infect. Dis. 44, 895–900 10.1086/512197 [DOI] [PubMed] [Google Scholar]
- Lee Y. K., Menezes J. S., Umesaki Y., Mazmanian S. K. (2011). Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4615–4622 10.1073/pnas.1000082107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois L., Goodman T. (1989). In vivo modulation of cytolytic activity and Thy-1 expression in TCR-gamma delta + intraepithelial lymphocytes. Science 243, 1716–1718 10.1126/science.2564701 [DOI] [PubMed] [Google Scholar]
- Liu C. H., Lee S. M., Vanlare J. M., Kasper D. L., Mazmanian S. K. (2008). Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc. Natl. Acad. Sci. U.S.A. 105, 3951–3956 10.1073/pnas.0712306105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Fasano S., Madayiputhiya N., Morin N. P., Nataro J., Fasano A. (2009). Isolation, identification, and characterization of small bioactive peptides from Lactobacillus GG conditional media that exert both anti-Gram-negative and Gram-positive bactericidal activity. J. Pediatr. Gastroenterol. Nutr. 49, 23–30 10.1097/MPG.0b013e31817e6f95 [DOI] [PubMed] [Google Scholar]
- Macpherson A. J., Harris N. L. (2004). Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4, 478–485 10.1038/nri1373 [DOI] [PubMed] [Google Scholar]
- Maier B. R., Hentges D. J. (1972). Experimental Shigella infections in laboratory animals. I. Antagonism by human normal flora components in gnotobiotic mice. Infect. Immun. 6, 168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., Roca J., Dore J. (2006). Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55, 205–211 10.1136/gut.2005.073817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S. K., Liu C. H., Tzianabos A. O., Kasper D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Round J. L., Kasper D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- Menard O., Butel M. J., Gaboriau-Routhiau V., Waligora-Dupriet A. J. (2008). Gnotobiotic mouse immune response induced by Bifidobacterium sp. strains isolated from infants. Appl. Environ. Microbiol. 74, 660–666 10.1128/AEM.01276-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi R. M., Silva M. E., Vieira E. C., Bambirra E. A., Nicoli J. R. (1989). Intragastric infection of germfree and conventional mice with Salmonella typhimurium. Braz. J. Med. Biol. Res. 22, 1389–1392 [PubMed] [Google Scholar]
- Ochoa-Reparaz J., Mielcarz D. W., Ditrio L. E., Burroughs A. R., Begum-Haque S., Dasgupta S., Kasper D. L., Kasper L. H. (2010a). Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 185, 4101–4108 10.4049/jimmunol.1001443 [DOI] [PubMed] [Google Scholar]
- Ochoa-Reparaz J., Mielcarz D. W., Wang Y., Begum-Haque S., Dasgupta S., Kasper D. L., Kasper L. H. (2010b). A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 3, 487–495 10.1038/mi.2010.29 [DOI] [PubMed] [Google Scholar]
- O’Mahony C., Scully P., O’Mahony D., Murphy S., O’Brien F., Lyons A., Sherlock G., MacSharry J., Kiely B., Shanahan F., O’Mahony L. (2008). Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 4, e1000112. 10.1371/journal.ppat.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwehand A. C. (2007). Antiallergic effects of probiotics. J. Nutr. 137(3 Suppl. 2), 794S–797S [DOI] [PubMed] [Google Scholar]
- Peterson D. A., Frank D. N., Pace N. R., Gordon J. I. (2008). Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3, 417–427 10.1016/j.chom.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polk B. F., Kasper D. L. (1977). Bacteroides fragilis subspecies in clinical isolates. Ann. Intern. Med. 86, 569–571 [DOI] [PubMed] [Google Scholar]
- Pretzer G., Snel J., Molenaar D., Wiersma A., Bron P. A., Lambert J., de Vos W. M., van der Meer R., Smits M. A., Kleerebezem M. (2005). Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187, 6128–6136 10.1128/JB.187.17.6128-6136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkotter H. J., Pabst R. (1989). Lymphocyte subsets in jejunal and ileal Peyer’s patches of normal and gnotobiotic minipigs. Immunology 67, 103–108 [PMC free article] [PubMed] [Google Scholar]
- Round J. L., Mazmanian S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 10.1038/nri2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L., Mazmanian S. K. (2010). Inducible Foxp3 + regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 107, 12204–12209 10.1073/pnas.0909122107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S. M., Fitzgerald G. F., van Sinderen D. (2005). Transcriptional regulation and characterization of a novel beta-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 71, 3475–3482 10.1128/AEM.71.9.4992-4997.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez B., Champomier-Verges M. C., Anglade P., Baraige F., de Los Reyes-Gavilán C. G., Margolles A., Zagorec M. (2008). A preliminary analysis of Bifidobacterium longum exported proteins by two-dimensional electrophoresis. J. Mol. Microbiol. Biotechnol. 14, 74–79 10.1159/000106085 [DOI] [PubMed] [Google Scholar]
- Sanchez B., Urdaci M. C., Margolles A. (2010). Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa-bacteria interactions. Microbiology 156(Pt 11), 3232–3242 10.1099/mic.0.044057-0 [DOI] [PubMed] [Google Scholar]
- Savage D. C. (1969). Localization of certain indigenous microorganisms on the ileal villi of rats. J. Bacteriol. 97, 1505–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Karmirantzou M., Snel B., Vilanova D., Berger B., Pessi G., Zwahlen M. C., Desiere F., Bork P., Delley M., Pridmore R. D., Arigoni F. (2002). The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U.S.A. 99, 14422–14427 10.1073/pnas.212527599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M., Harder J., Köten B., Stange E. F., Wehkamp J., Fellermann K. (2008). Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin. Exp. Immunol. 151, 528–535 10.1111/j.1365-2249.2007.03587.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Finlay B. B. (2009). The role of the intestinal microbiota in enteric infection. J. Physiol. (Lond.) 587(Pt 17), 4159–4167 10.1113/jphysiol.2009.172742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Tam N. M., Jogova M., Robertson M. L., Li Y., Lupp C., Finlay B. B. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736 10.1128/IAI.00319-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A., Yan F., Polk D. B., Rao R. K. (2008). Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 294(4): G1060-9. 10.1152/ajpgi.00202.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff K. E., Meslin K., Cebra J. J. (1995). Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 63, 3904–3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen R., Boekhorst J., Muscariello L., Molenaar D., Renckens B., Kleerebezem M. (2006). Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics 7, 126. 10.1186/1471-2164-7-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., McCoy K. D., Macpherson A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69 10.1016/j.smim.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Snel J., Heinen P. P., Blok H. J., Carman R. J., Duncan A. J., Allen P. C., Collins M. D. (1995). Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus.” Int. J. Syst. Bacteriol. 45, 780–782 10.1099/00207713-45-4-780 [DOI] [PubMed] [Google Scholar]
- Sonnenburg J. L., Chen C. T., Gordon J. I. (2006). Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4, e413. 10.1371/journal.pbio.0040413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg J. L., Xu J., Leip D. D., Chen C. H., Westover B. P., Weatherford J., Buhler J. D., Gordon J. I. (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 10.1126/science.1109051 [DOI] [PubMed] [Google Scholar]
- Sprinz H., Kundel D. W., Dammin G. J., Horowitz R. E., Schneider H., Formal S. B. (1961). The response of the germfree guinea pig to oral bacterial challenge with Escherichia coli and Shigella flexneri. Am. J. Pathol. 39, 681–695 [PMC free article] [PubMed] [Google Scholar]
- Stecher B., Hardt W. D. (2011). Mechanisms controlling (pathogen) colonization of the gut. Curr. Opin. Microbiol. 14, 82–91 10.1016/j.mib.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Stepankova R., Powrie F., Kofronova O., Kozakova H., Hudcovic T., Hrncir T., Uhlig H., Read S., Rehakova Z., Benada O., Heczko P., Strus M., Bland P., Tlaskalova-Hogenova H. (2007). Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4 + T cells. Inflamm. Bowel Dis. 13, 1202–1211 10.1002/ibd.20221 [DOI] [PubMed] [Google Scholar]
- Strober W., Fuss I. J., Blumberg R.S. (2002). The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 20, 495–549 10.1146/annurev.immunol.20.100301.064816 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Meek B., Doi Y., Muramatsu M., Chiba T., Honjo T., Fagarasan S. (2004). Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. U.S.A. 101, 1981–1986 10.1073/pnas.0402822101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talham G. L., Jiang H. Q., Bos N. A., Cebra J. J. (1999). Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect. Immun. 67, 1992–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock G. W., Miller J. R., Savage D. C. (1984). Host specificity of filamentous, segmented microorganisms adherent to the small bowel epithelium in mice and rats. Appl. Environ. Microbiol. 47, 441–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy E. B., Kasper D. L. (2010). Beneficial effects. (of) Bacteroides fragilis polysaccharides on the immune system. Front. Biosci. 15, 25–34 10.2741/3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., Sogin M. L., Jones W. J., Roe B. A., Affourtit J. P., Egholm M., Henrissat B., Heath A. C., Knight R., Gordon J. I. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. L. (2007). The human microbiome project. Nature 449, 804–810 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardisand E. R. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Turpin W., Humblot C., Thomas M., Guyot J. P. (2010). Lactobacilli as multifaceted probiotics with poorly disclosed molecular mechanisms. Int. J. Food Microbiol. 143, 87–102 10.1016/j.ijfoodmicro.2010.07.032 [DOI] [PubMed] [Google Scholar]
- Tzianabos A. O., Finberg R. W., Wang Y., Chan M., Onderdonk A. B., Jennings H. J., Kasper D. L. (2000). T cells activated by zwitterionic molecules prevent abscesses induced by pathogenic bacteria. J. Biol. Chem. 275, 6733–6740 10.1074/jbc.275.10.6733 [DOI] [PubMed] [Google Scholar]
- Tzianabos A. O., Onderdonk A. B., Rosner B., Cisneros R. L., Kasper D. L. (1993). Structural features of polysaccharides that induce intra-abdominal abscesses. Science 262, 416–419 10.1126/science.8211161 [DOI] [PubMed] [Google Scholar]
- Tzianabos A. O., Wang J. Y., Lee J. C. (2001). Structural rationale for the modulation of abscess formation by Staphylococcus aureus capsular polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 98, 9365–9370 10.1073/pnas.161175598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesaki Y., Okada Y., Matsumoto S., Imaoka A., Setoyama H. (1995). Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol. Immunol. 39, 555–562 [DOI] [PubMed] [Google Scholar]
- Umesaki Y., Setoyama H., Matsumoto S., Imaoka A., Itoh K. (1999). Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 67, 3504–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan E. E., Heilig H. G., Ben-Amor K., de Vos W. M. (2005). Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiol. Rev. 29, 477–490 10.1016/j.fmrre.2005.04.009 [DOI] [PubMed] [Google Scholar]
- Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., Stonebraker A. C., Hu C., Wong F. S., Szot G. L., Bluestone J. A., Gordon J. I., Chervonsky A. V. (2008). Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455, 1109–1113 10.1038/nature07336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Coleman D. C., Wiebe W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. J., Ivanov I. I., Darce J., Hattori K., Shima T., Umesaki Y., Littman D. R., Benoist C., Mathis D. (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 10.1016/j.immuni.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Cao H., Cover T. L., Whitehead R., Washington M. K., Polk D. B. (2007). Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132, 562–575 10.1053/j.gastro.2006.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z., Savage D. C. (1979). Microbial interference and colonization of the murine gastrointestinal tract by Listeria monocytogenes. Infect. Immun. 23, 168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]