Abstract
Congenital muscular dystrophies have a broad spectrum of genotypes and phenotypes and there is a need for a better biochemical understanding of this group of diseases in order to aid diagnosis and treatment. Several mutations resulting in these diseases cause reduced O-mannosyl glycosylation of glycoproteins, including α-dystroglycan. The enzyme POMGnT1 (protein-O-mannose N-acetylglucosaminyltransferase 1; EC 2.4.1.-) catalyses the transfer of N-acetylglucosamine to O-linked mannose of α-dystroglycan. In the present paper we describe the biochemical characterization of 14 clinical mutants of the glycosyltransferase POMGnT1, which have been linked to muscle-eye-brain disease or similar conditions. Truncated mutant variants of the human enzyme (recombinant POMGnT1) were expressed in Escherichia coli and screened for catalytic activity. We find that three mutants show some activity towards mannosylated peptide substrates mimicking α-dystroglycan; the residues affected by these mutants are predicted by homology modelling to be on the periphery of the POMGnT1 surface. Only in part does the location of a previously described mutated residue on the periphery of the protein structure correlate with a less severe disease mutant.
Keywords: congenital muscular dystrophy, glycopeptide, muscleeye-brain disease, protein-O-mannose N-acetylglucosaminyltransferase 1 (POMGnT1)
Abbreviations: CK, creatine kinase; CMD, congenital muscular dystrophy; α-DG, α-dystroglycan; β-DG, β-dystroglycan; FKRP, fukutin-related protein gene; FKTN, fukutin gene; Fmoc, fluoren-9-ylmethoxycarbonyl; GnTI, N-acetylglucosaminyltransferase I; IPTG, isopropyl β-D-thiogalactopyranoside; LARGE, like-acetylglucosaminyltransferase gene; LB, Luria–Bertani; LGMD, limb-girdle muscular dystrophy; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; MEB, muscle-eye-brain disease; POMGnT1, protein-O-mannose N-acetylglucosaminyltransferase 1; POMT1, protein-O-mannosyltransferase 1; POMT2, protein-O-mannosyltransferase 2; rPOMGnT1, recombinant protein-O-mannose N-acetylglucosaminyltransferase 1; SPSS, solid-phase peptide synthesis; WWS, Walker–Warburg syndrome
INTRODUCTION
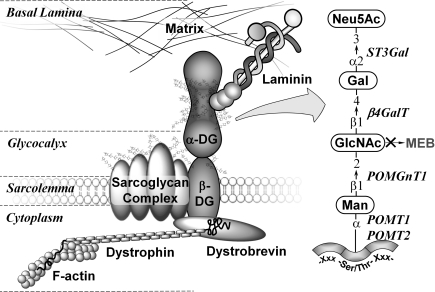
The cell-surface receptor α-DG (α-dystroglycan) is, alongside β-DG (β-dystroglycan), a central component of the dystrophin–glycoprotein complex, which connects the extracellular matrix to the cytoskeleton. α-DG is a 650 amino acid protein containing three distinct domains, two globular domains on the N- and the C-termini, and a large central mucin-type domain essential for the binding to proteins in the basal lamina, such as laminin, agrin, biglycan, neurexin and perlecan [1–5]. The proper glycosylation of the mucin-type domain of α-DG with O-linked specific carbohydrates is key for maintenance of membrane stability. The mature human α-DG glycoprotein has three N-glycosylation sites, up to five predicted glycosaminoglycan-attachment sites [6], and more than 50 other serine and threonine residues which can potentially carry O-glycans. Although the predicted protein mass is only 71 kDa, native glycosylated α-DG migrates as a diffuse band between 120 and 250 kDa on reducing SDS/PAGE [7,8]. Other than the mucin-like O-glycan structures, another less common O-glycan structure, Neu5Ac(α2,3)Gal(β1,4)GlcNAc(β1,2)Man-O, has been detected on mammalian α-DG and may play a role in the binding of α-DG to extracellular protein ligands [9–11]. Originally regarded to be a yeast-specific modification, O-mannosylation in mammals is only present on a limited number of proteins in neuronal and muscular tissue [12]. Although various O-mannose glycopeptides could be identified from total brain tissue [13,14], α-DG still remains the only mammalian glycoprotein clearly identified to contain this rare modification (Figure 1).
Figure 1. α-DG plays a key role in the formation of the dystrophin glycoprotein complex.
Laminin in the basal lamina binds to O-mannose-linked carbohydrates of α-DG. In muscle cells, α-DG binds non-covalently to β-DG in the sarcolemma, which is linked to the F-actin (filamentous actin) cytoskeleton and signalling proteins (dystrobrevin) via dystrophin. Loss of POMGnT1 function results in the truncation of the saccharide structures down to a single O-linked mannose; clinically this loss correlates with MEB and similar conditions. The sugar phosphate modification of the mannose residue, potentially affected by mutation of LARGE, is not shown.
The essential nature of glycosylation for the function of α-DG has been confirmed by studying muscle tissue of patients with α-dystroglycanopathies. This heterogenous group of autosomal recessive disorders encompasses a broad spectrum of conditions, with clinical phenotypes ranging from mild LGMD (limb-girdle muscular dystrophy) without cognitive impairment, to MEB (muscle-eye-brain disease) and WWS (Walker–Warburg syndrome) with severe brain and eye abnormalities [15,16]. Immunostaining of muscle samples using the polyclonal GT20ADG antibody raised against hypoglycosylated α-DG showed that α-DG expression levels of α-dystroglycanopathy patients are comparable with these of control samples [10]. In contrast, the same patient samples have reduced or absent immunostaining with IIH6 and VIA4-1 monoclonal antibodies, whose binding requires native glycosylation of α-DG. The loss of glycosylation is significant, reducing the molecular mass of the mature protein by 50 kDa or more [17].
Molecular testing of patients with clinical signs of CMD (congenital muscular dystrophy) revealed that mutations in six genes encoding known or putative glycosyltransferases are responsible for hypoglycosylation of α-DG, supposedly due to the reduced or lost enzymatic function. The mutated forms of the POMT1 (protein-O-mannosyltransferase 1), POMT2 (protein-O-mannosyltransferase 2), POMGnT1 (protein O-mannose N-acetylglucosaminyltransferase 1), FKTN (fukutin), FKRP (fukutin-related protein) and LARGE (like-acetylglucosaminyltransferase) genes were originally reported in patients with distinct phenotypes [18–22], and therefore defects of those glycosyltransferases were initially associated with different clinical conditions. In recent years, a wider spectrum of phenotypes with different severities have been reported for mutations in the POMT1 [23,24], POMT2 [25,26] and the FKTN gene [27,28], whereas mutations affecting FKRP [29,30], POMGnT1 [31–33] and LARGE [34,35] have been described to have less phenotypical variation. However, a study of 81 CMD patients from Italy could only detect 43 homozygous or heterocompound mutations (53%) in those six genes [34], which allows the presumption that even more, as yet unidentified, genes are involved in the post-translational modification of α-DG.
Biochemically, POMT1, POMT2 and POMGnT1 are known to transfer the first two residues (mannose and GlcNAc) of the O-mannosyl modification [22,36]. Recently, LARGE was shown to be involved in the phosphorylation of O-linked mannose on α-DG [37], whereas the biochemical functions of fukutin and FKRP are still unknown. The aim of the present study was to characterize the enzymatic properties of 14 clinical mutations of POMGnT1 which have been linked to MEB, LGMD and similar conditions. Therefore a simple expression and purification protocol for POMGnT1 in the prokaryotic host Escherichia coli had to be developed; a series of mannopeptides derived from α-DG were synthesized and tested as acceptor substrate for the recombinant, partially purified wild-type enzyme. The optimal substrate was then used for (i) testing recombinantly expressed POMGnT1 mutant enzymes for activity, (ii) determining the kinetic parameters of the active POMGnT1 variants, and (iii) evaluating the possibility of correlating genotype with the obtained biochemical data.
EXPERIMENTAL
Subcloning of POMGnT1 from a bacterial cDNA clone
The cDNA clone containing the complete POMGnT1 gene was obtained from Geneservice (GenBank® accession number BC001471). The plasmid was isolated (Qiagen Miniprep kit) and used as a DNA template for a PCR amplification. The primer pair Fw (forward)/EcoRI (5′-ACGAATTCAGTGAAGCCAATGAAGACCC-3′) and Rv (reverse)/XhoI (5′-GACTCGAGCTATGTCTGTTCTGGGGCTC-3′) was used in combination with KOD DNA Polymerase (Roche). The primers were designed to amplify the sequence encoding amino acids 66–660, i.e. a form lacking the N-terminal putative transmembrane domain. The PCR fragment was cut with the relevant enzymes and ligated into pET30a which contains a sequence encoding an N-terminal His tag; clones which displayed no mutation after full sequencing of the insert were used for further experiments.
Expression and purification of POMGnT1 mutants
Complementary oligonucleotide primers containing the desired mutation were used to generate POMGnT1 mutant genes (see Supplementary Table S1 at http://www.BiochemJ.org/bj/436/bj4360447add.htm) according to the standard protocol for QuikChange® site-directed mutagenesis (Stratagene); plasmid DNA of transformants was sequenced to verify that selected clones contained the desired mutation. Mutant plasmids were transformed into E. coli BL21(DE3) competent cells (Invitrogen) and plated on LB (Luria–Bertani) agar plates containing 50 μg/ml kanamycin. Alternatively, plasmids were transformed into E. coli ArcticExpress™ (DE3) competent cells (Stratagene) and plated on LB agar plates containing 50 μg/ml kanamycin and 20 μg/ml gentamicin. For each transformed mutant, 400 ml of LB medium (Sigma) was inoculated with 2 ml of starter culture and cells were grown to a D600 value of 0.6. Induction was carried out with IPTG (isopropyl β-D-thiogalactopyranoside) at a final concentration of 500 μM, and cells were typically incubated at 13 °C for 16 h. The cells were harvested at 4000 g for 15 min and the supernatant discarded. The resulting cell pellet (approximately 500 mg) was resuspended in 20 ml of lysis buffer [50 mM Tris/HCl and 500 mM NaCl (pH 8.0)] and sonicated for 20 min (20 on/off cycles with 15 μm amplitude for 30 s at 4 °C). The lysates were centrifuged at 24000 g for 30 min at 4 °C, the supernatant was syringe-filtered (0.45 μM pore size, Sartorius) and the resulting protein solution was loaded on to a 1 ml Ni2+-chelation column (HiTrap, GE Healthcare). The column was washed with 20 ml of binding buffer [50 mM Tris/HCl, 10 mM imidazole and 500 mM NaCl (pH 8.0)] to remove unbound proteins from the column. The partially purified recombinant protein (rPOMGnT1; see also Figure 2a) was eluted with 2 ml of elution buffer [50 mM Tris/HCl, 500 mM imidazole and 500 mM NaCl (pH 8.0)], and concentrated to 100 μl using centrifugal concentrators [Sartorius, 30 kDa MWCO (molecular mass cut-off)].
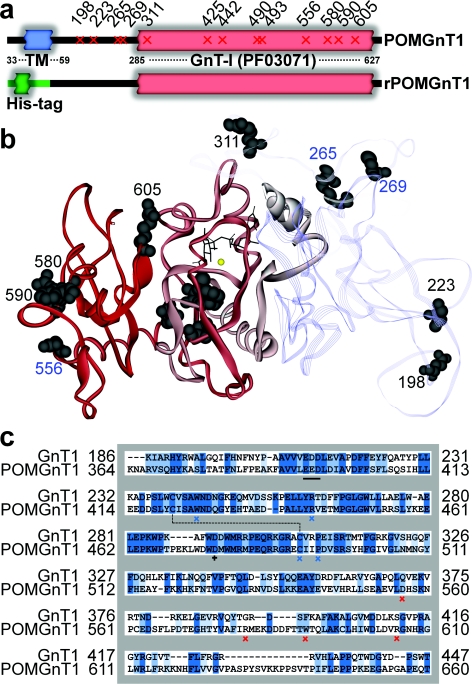
Figure 2. Native POMGnT1 in comparison with the recombinant form and to GnT1.
(a) Schematic diagram of native POMGnT1 shows an N-terminal transmembrane domain (TM) and a C-terminal GnT-I domain (Pfam PF03071). The position of mutations introduced are marked with a slanted cross. For expressing the recombinant protein rPOMGnT1, the transmembrane domain was substituted with a His tag. (b) Homology model of POMGnT1 based on the known crystal structure of GnT1 (PDB code 1FOA). The modelled C-terminal GnT-I domain is modelled as a red solid ribbon, whereas the non-homologous N-terminal part is displayed as a transparent ribbon loop. Inactive mutants are numbered in black, and mutants with catalytic activity are numbered in blue. UDP-GlcNAc is shown in stick form. (c) Sequence alignment of POMGnT1 with PDB code 1FOA using BLOSOM (default parameters=multiple alignment gap penalty=10.0, gap extension penalty=0.05, open delay divergent=40.0); comparison of putative catalytic subunits POMGnT1 (Arg367–Thr660) and GnT1 (Lys82–Thr343) gives a sequence identity (dark blue shading) of 33.2% and a sequence similarity (light blue shading) of 50.3%. Single amino acid changes found in clinical mutants are marked with blue (conserved in GnT1) and red (not conserved in GnT1) crosses; the catalytic amino acid Asp476 is marked with a black cross; the modified DXD-motif (Glu393–Asp395) is underlined with a black bar.
SDS/PAGE and Western blot analysis
The partially purified mutant rPOMGnT1 proteins were incubated with SDS/PAGE buffer at 95 °C for 10 min, separated by SDS/PAGE and visualized by Coomassie Blue staining. Protein concentrations of the expressed mutant rPOMGnT1 ranged from 50 to 250 μg/ml, calculated by densitometric comparison with a 70 kDa protein standard (300 μg/ml) using ImageJ analysis software [38]. For Western blot analysis, protein gels were transferred on to nitrocellulose using a semi-dry blotting apparatus. After blocking with 0.5% BSA, the membranes were incubated with horseradish peroxidase-conjugated mouse anti-polyHistidine antibody (anti-His, 1:1000 dilution, Sigma); after washing, recombinant protein was visualized chromogenically using the SigmaFAST™ DAB (3,3-diaminobenzidine) reagent.
Assay of recombinant rPOMGnT1 variants
The mannopeptide acceptor substrates were derived from the sequence of human α-DG (UniProt accession number Q14118), a glycoprotein that plays a role in the formation of a transmembrane link between laminin and dystrophin [39]. Sequences corresponding to serine/threonine-rich regions of the protein were selected for SPSS (solid-phase peptide synthesis) (see Figures 3a–3c; chemical synthesis is described in the Supplementary Experimental section at http://www.BiochemJ.org/bj/436/bj4360447add.htm).
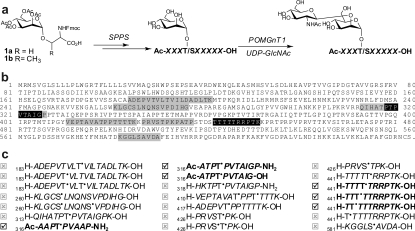
Figure 3. Chemical synthesis of mannopeptides derived from α-DG.
(a) SPPS using Fmoc-protected mannosyl serine (1a) and mannosyl threonine (1b) building blocks for the evaluation of enzyme substrates of POMGnT1. (b) Synthesized peptides corresponding to portions of the amino acid sequence of α-DG are highlighted (grey, no enzymatic activity observed; black, enzymatic activity observed). (c) The glycoforms of the mannopeptide synthesized are shown with the glycosylamino acids indicated with a superscript dot; those which acted as POMGnT1 acceptor substrates are shown with a tick, whereas those which did not function are shown with a cross.
For testing the enzymatic activity of the wild-type rPOMGnT1 variant towards the chemically synthesized acceptor substrates, 5 μl of partially purified enzyme solution was added to 5 μl of 1 mM acceptor, 2 μl of 0.4 M Mes (pH 7), 2 μl of 0.2 M MnCl2 and 6 μl of 1 mM UDP-GlcNAc, and incubated for 16 h at 37 °C; a volume of 0.2 μl from the reaction mix was analysed using a Voyager-DE STR MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) mass spectrometer (Applied Biosystems) with 2,5-dihydroxybenzoic acid as matrix.
The HPLC-based enzymatic assay for testing the activity of the different rPOMGnT1 variants was performed with the synthetic mannopeptide Ac-ATPT•PVTAIG-OH (T• indicates the α-mannosylated threonine); in contrast with the other mannopeptides tested, this acceptor substrate allowed, after optimizing of the solvent gradient and the type of reverse-phase column, baseline separation between substrate and product peak. The different retention times of the mannopeptide substrate and GlcNAc-mannopeptide product (17.7 and 18.5 min respectively) could be achieved using a C8 reversed-phase column (Phenomenex Sphereclone, 250 mm × 2 mm, 3 μm pore size) with a flow rate of 0.2 ml/min. Solvent A was 0.1% trifluoroacetic acid in H2O, solvent B was 0.1% trifluoroacetic acid in acetonitrile. The programme used was as follows: 0–10 min, 100% solvent A; 10–40 min, linear gradient to 50% solvent and then returning to solvent A (reconditioning for 20 min). The elution of the glycopeptides was monitored via UV detection at 214 nm and electrospray MS detection (Agilent System 1100 LC/MSD SL).
For the determinations of the Km values of both UDP-GlcNAc and the mannopeptide Ac-ATPT•PVTAIG-OH, an enzymatic assay was performed with a series of different substrate concentrations. For determination of the Km value of the acceptor substrate for an active rPOMGnT1 mutant, assays were performed for 4 h at 37 °C using variable concentrations of the mannopeptide (2–10 mM), and a constant UDP-GlcNAc concentration of 10 mM. After heat-inactivation at 95 °C, the reaction mixture was analysed using the HPLC method described above. All experiments were performed in triplicate, and the kinetic data were calculated by using the relative peak areas corresponding to acceptor substrate and product.
Homology modelling of POMGnT1
As the crystal structure of POMGnT1 has not yet been solved, a homology model of POMGnT1 (UniProt accession number Q8WZA1) was built based on the known crystal structure of rabbit GnTI (N-acetylglucosaminyltransferase I, UniProt accession number P27115) encoded by the MGAT1 gene [40]. The catalytic domains of human POMGnT1 and GnTI show 45% homology over 159 amino acid residues [41]. All modelling was performed using Accelrys Discovery Studio 2.0 with default parameters (cut overhangs, no disulfides, no ligands and no loop refining). In order to identify an homologous structure, a BLAST search of the POMGnT1 amino acid sequence was performed against all sequences in the PDB database [42]. The top hit was found to be PDB code 1FOA, which corresponds to the catalytic fragment of GnTI. The sequence of POMGnT1 was then aligned with the sequence of GnTI using the ‘Align Multiple Sequences’ protocol. The overall sequence identity was found to be 18.8% and the sequence similarity was 33.5%. The homology model was then generated based on the sequence alignment and structure of GnTI using the ‘Build Homology Models’ protocol. A final alignment was created in which the POMGnT1 sequence was aligned with the sequence of the homology model.
RESULTS
Human rPOMGnT1 can be expressed catalytically active in E. coli
Although the recombinant expression of POMGnT1 has been typically performed either as a full-length protein in mammalian cell culture [22,41], or in soluble form in insect cells, lacking the first 226 residues [43], our strategy was to assess the expression capabilities of the prokaryotic host E. coli using a form lacking the first 65 residues. Initial experiments were performed using transformed BL21(DE3) competent cells, the most common host strain for protein expression. Culture samples were taken after 3, 8 and 16 h of induction with 500 μM IPTG at 16, 25 and 30 °C. The highest expression levels were observed after 8 h of induction at 16 °C, judged by anti-His Western blot analysis. The conditions described above were used for a purification experiment using Ni2+-chelation chromatography, and the elution fraction analysed by Western blot showed a band migrating between 70 and 100 kDa, indicating the soluble expression of the recombinant rPOMGnT1 protein. Although a whole series of activity tests were performed, which systematically tested different mannopeptides, and substrate and metal ion concentrations, no catalytic activity could be determined for rPOMGnT1 expressed in BL21(DE3) cells. Therefore a new attempt was made using ArcticExpress™ (DE3) cells, which are in comparison with other strains re-engineered for protein expression at lower temperatures, thereby potentially facilitating increased yield and activity of the recombinant protein [44].
Expression experiments were performed at 13 °C using 500 μM IPTG and cells were harvested 16 h after induction. Anti-His Western blot analysis of the cell lysate and from the eluate after Ni2+-chelation chromatography both showed a band migrating at approximately 80 kDa, indicating soluble expression of rPOMGnT1 (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/436/bj4360447add.htm). In contrast with the expression in BL21(DE3) cells, partially purified rPOMGnT1 from ArcticExpress™ (DE3) cells showed catalytic activity. Activity tests with a final concentration of 250 μM of the mannopeptide Ac-ATPT•PVTAIG-OH and a 1.2-fold excess of the donor substrate UDP-GlcNAc showed 20–25% conversion into product within 4 h. More than 95% conversion could be achieved after 48 h of incubation.
Wild-type rPOMGnT1 was active towards some, but not all, synthesized mannopeptides
In order to establish that peptide sequences in α-DG are indeed substrates for POMGnT1, a library of mannopeptides was generated which corresponded to sequences of the mucin-like stem region of α-DG (Figure 3b). Thus wild-type POMGnT1 was screened against a library of 21 mannopeptides produced by SPPS, which was optimized for the introduction of Fmoc (fluoren-9-ylmethoxycarbonyl)-protected mannosyl serine and mannosyl threonine building blocks (Figure 3a). Of these protein sequences, 20 where chosen according to the serine/threonine-rich sequence of human α-DG, including three mannopeptides based on the region surrounding Thr319. In total, three variants corresponding to this latter region were used: (i) Ac-ATPT•PVTAIG-OH, (ii) Ac-ATPT•PVTAIGP-NH2, and (iii) the modified Ac-AAPT•PVAAP-NH2, featuring an exchange of Thr317 for alanine, adopted by Takahashi et al. [45] in order to avoid modification by a background O-GlcNAc transferase activity and by proteases present in brain extracts (Figure 3c). All three peptides based on the region around Thr319 acted as substrates in a comparable manner; in particular, only one GlcNAc residue was transferred and no proteolysis was observed. On the basis of this finding, and also as E. coli is known to have no endogenous O-GlcNAc transferase activity, the kinetic studies were performed with the first of these peptides, i.e. the peptide without the threonine to alanine and C-terminal modifications.
Surprisingly, rPOMGnT1 only displayed catalytic transfer towards six of the screened mannopeptide substrates. None of the mannopeptides containing mannosyl serine residue were glycosylated, and all of the positive substrates contained a proline residue which was C-terminal of the acceptor mannosyl threonine residue, although such sequences were also observed in the mannopeptides which were not substrates for POMGnT1. Moreover, small variations in the mannopeptide sequences could affect the substrate specificity of POMGnT1. For example, for mannosylated isomers of the H-TTTTTRRPTK-OH peptide, only modifications in positions 2, 3 and 4 of the penta-threonine motif, but unexpectedly not positions 1 and 5, were accepted as substrates (Figure 3c). Nevertheless, these studies allowed us to select a natural mannopeptide fragment of α-DG which could be used for further kinetic studies of POMGnT1 mutant activities.
Some rPOMGnT1 variants containing clinically relevant mutations show in vitro activity
A number of clinically relevant POMGnT1 mutations have been described in the literature, ranging from nonsense/frameshift mutations to approximately 14 different point mutations which encode full-length proteins, as well as truncated proteins caused by nonsense mutations within the C-terminal region [22,31–33,46–53]. Table 1 gives an overview of homozygous and compound heterozygous mutations described to date in the literature.
Table 1. Description of clinically relevant POMGnT1 mutations.
CK, creatine kinase measured and given in units/l (normal values in the range of 24–240 for children from 1 to 18 years of age [54]); FCMD, Fukuyama CMD; f, female; m, male; nd, information is not available for these patients.
| Mutation | Age (sex) | 1st allele | 2nd allele | CK | Clinical diagnosis | Biopsy | Reference(s) |
|---|---|---|---|---|---|---|---|
| 1+9 | 7 | S198R | C490Y | 1000 | MEB | Yes | [47] |
| 2 | 12 (m) | E223K | Phe149 frameshift 167 stop | 2365 | WWS or MEB | nd | [33] |
| 2 | 8 (f) | E223K | Mutation in intron 17 | 1844 | MEB or FCMD | nd | [33] |
| 3 | 7 (f) | C269Y | Val328 frameshift 338 stop | 852 | MEB or severe FCMD | nd | [33] |
| 4/5+7 | 11 month (m) | R265H | R442C | 495 | Postnatal diagnosis of hydrocephalus | nd | [53] |
| R311Q | |||||||
| 4/5+7 | 9 (f) | R265H | R442C | 2327 | MEB | nd | [53] |
| R311Q | |||||||
| 4/5+13 | 20 month (f) | R265H | R605H | >3000 | MEB | Yes | [51] |
| R311Q | |||||||
| 6 | 17 (m) | W425S | Mutation in intron 21 | nd | MEB | Yes | [48,50] |
| 7 | 15 months (m) | R442C | R442C | 2697 | MEB | nd | [32] |
| 7 | 3 months (m) | R442C | R442C | 1798 | MEB | nd | [32] |
| 7 | 14 (f) | R442C | R442C | 724 | MEB | nd | [32] |
| 7+8 | 2 (m) | R442C | R442H | 1000 | MEB | nd | [32] |
| 9 | nd | C490Y | Mutation in intron 7 | 1000 | MEB or FCMD | Yes | [49] |
| 9 | nd | C490Y | C490Y | nd | MEB | nd | [48] |
| 10 | nd (m) | P493R | Val262 frameshift 633 stop | nd | MEB | Yes | [22] |
| 9+11 | nd | Trp590 stop | C490Y | nd | MEB | nd | [48] |
| 12+13 | 5 (f) | Arg580 stop | R605H | 1953 | MEB | Yes | [32] |
| 13 | 16 (f) | R605H | R605H | 9311 | MEB | Yes | [46,52] |
| 14 | 21 (f) | D556N | D556N | 12000 | MEB | Yes | [31,49] |
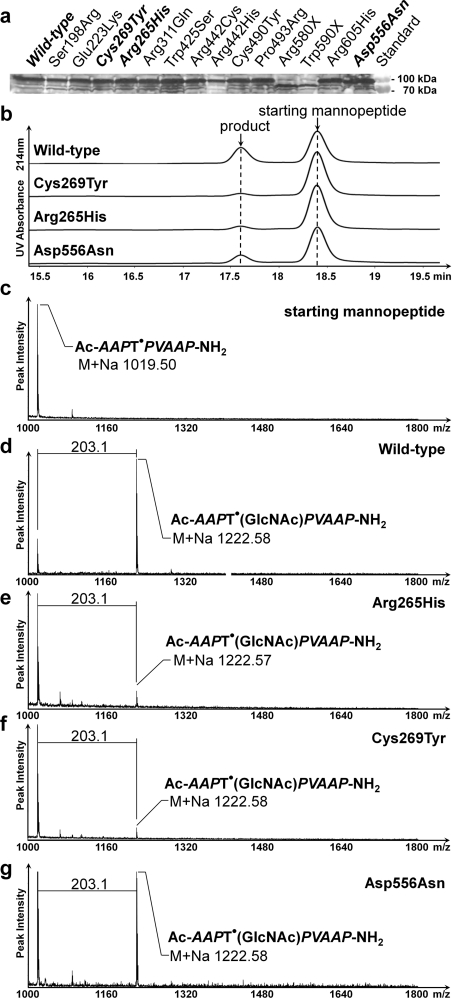
When expressed in E. coli, the 14 point-mutant variants and the wild-type rPOMGnT1 showed similar expression levels ranging from approx. 20 to 100 μg of rPOMGnT1 per g of wet cell weight (Figure 4a, see also Supplementary Table S2 at http://www.BiochemJ.org/bj/436/bj4360447add.htm). Activity tests showed that three mutants (R265H, C269Y and D556N) still had catalytic activity, whereas all other mutations resulted in no activity at all. In comparison with the specific enzymatic activity for wild-type rPOMGnT1 (25.5±5.8 m-units/mg), R265H and C269Y showed a 5-fold decrease (5.3±1.4 and 4.4±0.3 m-units/mg respectively), and D556N showed a 2.5-fold decrease (10.8±1.6 m-units/mg) in activity (Figures 4b–4g). Further kinetic analysis of the wild-type and those three active POMGnT1 mutants showed similar Km values for UDP-GlcNAc of between 2 and 3 mM. Thus these mutations do not seem to affect the binding to the donor substrate. However, the Km values for the mannopeptide Ac-ATPT•PVTAIG-OH varied from ≈6 mM for the wild-type enzyme and the D556N mutant, to >25 mM for the mutants R265H and C269Y.
Figure 4. Clinically relevant POMGnT1 mutations of MEB patients and activity studies of the recombinant enzyme.
(a) Western blot analysis of His-tagged rPOMGnT1 mutants expressed in E. coli. Molecular mass standards are shown on the right-hand side. (b) Assay of active rPOMGnT1 mutants by liquid chromatography. The mannopeptide substrate Ac-ATPT•PVTAIG-OH was incubated for 4 h with partially purified POMGnT1 wild-type and mutant enzymes in the presence of UDP-GlcNAc. The peaks eluting at 17.7 and 18.5 min were verified by MS to contain the reaction product and substrate respectively. (c–g) Characterization of the acceptor substrate Ac-ATPT•PVTAIG-NH2 (c) and the product after enzymatic reaction of rPOMGnT1 wild-type (d) and mutants (e–g) by MALDI–TOF MS (see more spectra in Supplementary Figure S2 at http://www.BiochemJ.org/bj/436/bj4360447add.htm).
Homology modelling suggests locations of mutations in protein structure
POMGnT1 was first identified as a homologue of the N-glycan-modifying enzyme GnTI and was indeed initially named in one study as GnTI.2 [22,41]. Sequence alignment showed that the homology of POMGnT1 to GnTI was much higher in the putative catalytic domain than in the N- and C- terminal regions, a finding compatible with a previous study which delineated a minimal catalytic region between residues 299 and 642 [43]. Using this sequence alignment, a three-dimensional model of the putative catalytic subunit of POMGnT1 was constructed based on the crystal structure of GnT1. The distribution of the clinically relevant mutations in the enzyme structure was examined (Figures 2b and 2c), and no disease-specific ‘hot-spot’ on the protein structure was identified; some mutations were close to the putative active site, others some distance away. Nevertheless, Arg265, Cys269 and Asp556 are predicted to be on the periphery of the structure of the protein.
DISCUSSION
Since the correlation between mutations of the glycosyltransferase POMGnT1 in patients with MEB was discovered almost a decade ago [22], a continuous stream of case studies have described more than 25 independent homozygous mutations distributed throughout the 21 exonic regions of the POMGnT1 locus [32]. Some of the phenotypes studied widened the spectrum of the disease caused by these mutations to the milder form of LGMD [31], whereas other POMGnT1 mutations result in symptoms overlapping with the more severe form of WWS [32,33]. The 14 POMGnT1 mutations analysed for catalytic activity in the present study are derived from gene analyses described in 11 different previously published studies (see Table 1). All of these studies showed a diverse subset of diagnostic methods for the determination of the phenotypical findings, such as the evaluation of psychomotoric and mental development, muscle biopsies with immunostaining or immunoblotting, MRI (magnetic resonance imaging), CK (creatine kinase) analysis or biochemical assays for each patient examined. To the best of our knowledge, the present study gives the only comprehensive biochemical evaluation of all clinically relevant POMGnT1 point mutations known to date.
One of the open questions was whether the full loss of enzyme activity is required for the phenotype, or if reduced activities also have the potential for pathogenic alterations. In good correlation with the genotypes observed, we show that three out of the 14 mutants tested display between 17 and 42% residual activities compared with the wild-type enzyme, whereas the remaining 11 showed no activity. As noted by others, the D556N mutation (42% activity as compared with the wild-type) introduces a putative N-glycosylation site into POMGnT1, which cannot be glycosylated in our E. coli expression system; the introduction of such a modification into the human enzyme might account for the comparably less severe phenotype observed for this mutation in vivo [31]. Previously this D556N mutant was assayed from crude extracts of fibroblasts from patients.
The mutation C269Y (17% activity) was observed in one allele of a patient who is a compound heterozygote; the other allele displays a frameshift at base 1077. Thus it appears that POMGnT1 containing this mutation is not able to function properly and, despite a reasonable degree of activity in vitro, still results in symptoms; it has been proposed by others that Cys269 lies within the so-called stem region, and expression of a full-length form of this mutant resulted in more-or-less no activity in transfected cell lysates [33], whereas a soluble form had 10% activity [43]. In the case of the other patients, the combination of mutant alleles does not allow us to draw a conclusion as to the in vivo effect of the specific mutation which is still compatible with in vitro activity. The R265H mutation (21% activity) was observed in a patient together with mutation R311Q (not active in our assay) on the same allele in addition to mutation R442C (not active) on the other allele. Although the R265H mutation resulted in residual activity, one would expect to observe a severe phenotype in this patient, due to the other mutation observed [53].
The distribution of the analysed POMGnT1 mutations within the enzyme was visualized using a model of POMGnT1. Therefore the crystal structure of GnTI, a homologous glycosyltransferase also from GT family 13, was used as a template. Interestingly, the three mutants are shown in the present study to be active affect side chains (residues 265, 269 and 556) on the periphery of our homology model. Furthermore, the six mutations in the region between Arg311 and Pro493, which result in an apparent total loss of activity, correspond to residues also conserved in human GnTI; indeed, on the basis of homology, Arg311 itself is postulated to form a key contact with the O-2a atom of the α-phosphate of the UDP-GlcNAc donor substrate [41]. On the other hand, essential residues affected by three mutations of GnT1 described in the literature (Chinese hamster ovary lec1 and Arabidopsis thaliana cgl) are not represented among the POMGnT1 variants [55,56]. However, other than the peripheral location of the three ‘active’ mutations, no absolute disease-specific correlation between biochemical data and the position of a mutation on the protein structure could be determined. Thus it was concluded that neither genetic data nor structural modelling would allow us to correlate point mutations with severity of disease, and that a biochemical analysis is required in each case.
Possibly significant, though, is the location of a predicted cystine bridge, based on that revealed in the crystal structure of GnTI, between Cys421 and Cys490 of POMGnT1 [41]. The reduced activity of the rPOMGnT1 C490Y mutant tested in the present study suggests that Cys–Cys bonding of these residues is essential for the catalytic function of POMGnT1. As disulfide-bond formation in the cytoplasm of E. coli has been described to be a rare event [57], the presence of this disulfide could explain the initial difficulties of expressing catalytically active wild-type rPOMGnT1 in BL21(DE3) cells. POMGnT1 had previously been expressed in insect and mammalian cell culture by recombinant methods [22,41], but full-length expression in mammalian cells has the disadvantage of endogenous GlcNAc transferase activity, which leads to high background and reduced sensitivity. In eukaryotic expression systems, the role of cofactor proteins in vivo that may play a regulatory role cannot be excluded and, indeed, fukutin may interact intracellularly with POMGnT1 [58]. However, as wild-type POMGnT1 has no putative N-glycosylation sites, it was a good candidate for expression in E. coli. When induced at low temperature, the enzyme was well expressed in a soluble form in bacteria and was shown to be fully active, with Km values similar to those reported for the enzyme expressed in other systems [22,31]. The use of a higher-throughput soluble expression system also allows us to focus on the effect of the mutations on catalytic function without considering the role of the cytosolic, transmembrane or stem regions in intracellular trafficking.
Previous biochemical data indicate that POMGnT1 transfers GlcNAc to mannosylated sites on α-DG [41,45]. These in vitro studies with either Man(α1-)O-benzyl or a synthetic glycopeptide such as NH2-CYAT•AV-OH (not found in α-DG) and Ac-AAPT•PVAAP-NH2 (similar to α-DG region 316–321) were performed; in the present study, we greatly extended the range of substrates tested. From the 20 mannopeptide sequences chosen, only six were shown to be POMGnT1 substrates. It seems that there is some selectivity of glycosylation by POMGnT1 and perhaps not all potential glycosylation sites on α-DG are modified by this enzyme, in agreement with the reports on other O-glycans found in α-DG [37]. Two recent reports describe either three [59] or five [60] O-mannosylated glycan sites in tryptically digested α-DG peptides isolated from skeletal muscle. These two papers do not, however, describe the same glycosylation sites; this raises the question as to exactly how many sites are mannosylated in vivo, as well as whether there are species differences. Certainly further synthesis of mannopeptides to find potential POMGnT1 glycosylation sites is an essential part of further studies to uncover the substrate specificities of enzymes involved in the O-glycosylation of α-DG.
Kinetic analysis of the wild-type and three active POMGnT1 mutants showed Km values for UDP-GlcNAc between 2 and 3 mM, similar to those reported in the literature using other methods [31]. Thus these mutations do not seem to affect the binding to the donor substrate. However, the Km values for the mannopeptide Ac-ATPT•PVTAIG-OH varied from ≈6 mM for the wild-type enzyme and D556N mutant, to >25 mM for the mutants R265H and C269Y. Km values of the two other acceptor substrates Man(α1-)O-benzyl and the mannopeptide H-CYA T•AV-OH reported in the literature were determined at >15 mM and ≈12 mM respectively [41]. Perhaps because of using natural substrate sequences, the present kinetic data compare favourably with those previously reported, and show that strategies based on glycopeptides sequences derived from protein substrates can lead to significantly improved chemical probes.
In conclusion, the biochemical data obtained from studying clinical POMGnT1 mutants with synthetic glycopeptides derived from glycoprotein substrate sequences should be an important tool to understand the mechanism and severity of disease. This methodology should also be applicable to other congenital disorders of glycosylation (such as those caused by mutations of LARGE [37] which may use similar mannopeptide substrates) and might help us in future to generate structure–activity models of the enzyme, by extending our understanding of the molecular basis of these diseases.
AUTHOR CONTRIBUTION
Josef Voglmeir initiated the study, designed, performed and optimized the biological experiments, analysed data and wrote the manuscript; Sara Kaloo performed and optimized biological experiments, and built the homology model; Nicolas Laurent designed and performed the mannopeptide synthesis and analysed data; Marco Meloni performed the mannopeptide synthesis; Lisa Bohlmann performed biological experiments; Iain Wilson conceived parts of the study and edited the manuscript prior to submission; Sabine Flitsch, the scientific supervisor, co-ordinated the study, and also edited and approved the final manuscript.
ACKNOWLEDGEMENTS
We thank Professor Harry Schachter (Toronto) and Dr Katharina Paschinger for helpful comments on an earlier version of the manuscript.
FUNDING
This work was supported by the Engineering and Physical Sciences Research Council (EPSRC) [grant numbers EP/G037604/1, EP/G037604/1 and EP/I016716/1 (to S.L.F. and J.V.)]; the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/G024685/1 (to S.L.F.)]; and the Royal Society (Wolfson Merit Award to S.L.F.).
References
- 1.Bowe M. A., Mendis D. B., Fallon J. R. The small leucine-rich repeat proteoglycan biglycan binds to α-dystroglycan and is upregulated in dystrophic muscle. J. Cell Biol. 2000;148:801–810. doi: 10.1083/jcb.148.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gee S. H., Montanaro F., Lindenbaum M. H., Carbonetto S. Dystroglycan-α, a dystrophin-associated glycoprotein, is a functional agrin receptor. Cell. 1994;77:675–686. doi: 10.1016/0092-8674(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 3.Peng H. B., Ali A. A., Daggett D. F., Rauvala H., Hassell J. R., Smalheiser N. R. The relationship between perlecan and dystroglycan and its implication in the formation of the neuromuscular junction. Cell Adhes. Commun. 1998;5:475–489. doi: 10.3109/15419069809005605. [DOI] [PubMed] [Google Scholar]
- 4.Smalheiser N. R., Schwartz N. B. Cranin: a laminin-binding protein of cell membranes. Proc. NAtl. Acad. Sci. U.S.A. 1987;84:6457–6461. doi: 10.1073/pnas.84.18.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugita S., Saito F., Tang J., Satz J., Campbell K., Sudhof T. C. A stoichiometric complex of neurexins and dystroglycan in brain. J. Cell Biol. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patnaik S. K., Stanley P. Mouse large can modify complex N- and mucin O-glycans on α-dystroglycan to induce laminin binding. J. Biol. Chem. 2005;280:20851–20859. doi: 10.1074/jbc.M500069200. [DOI] [PubMed] [Google Scholar]
- 7.Ervasti J. M., Campbell K. P. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- 8.Ibraghimov-Beskrovnaya O., Milatovich A., Ozcelik T., Yang B., Koepnick K., Francke U., Campbell K. P. Human dystroglycan: skeletal muscle cDNA, genomic structure, origin of tissue specific isoforms and chromosomal localization. Hum. Mol. Genet. 1993;2:1651–1657. doi: 10.1093/hmg/2.10.1651. [DOI] [PubMed] [Google Scholar]
- 9.Chiba A., Matsumura K., Yamada H., Inazu T., Shimizu T., Kusunoki S., Kanazawa I., Kobata A., Endo T. Structures of sialylated O-linked oligosaccharides of bovine peripheral nerve α-dystroglycan. The role of a novel O-mannosyl-type oligosaccharide in the binding of α-dystroglycan with laminin. J. Biol. Chem. 1997;272:2156–2162. doi: 10.1074/jbc.272.4.2156. [DOI] [PubMed] [Google Scholar]
- 10.Michele D. E., Barresi R., Kanagawa M., Saito F., Cohn R. D., Satz J. S., Dollar J., Nishino I., Kelley R. I., Somer H., et al. Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature. 2002;418:417–422. doi: 10.1038/nature00837. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki T., Yamada H., Matsumura K., Shimizu T., Kobata A., Endo T. Detection of O-mannosyl glycans in rabbit skeletal muscle α-dystroglycan. Biochim. Biophys. Acta. 1998;1425:599–606. doi: 10.1016/s0304-4165(98)00114-7. [DOI] [PubMed] [Google Scholar]
- 12.Endo T. O-mannosyl glycans in mammals. Biochim. Biophys. Acta. 1999;1473:237–246. doi: 10.1016/s0304-4165(99)00182-8. [DOI] [PubMed] [Google Scholar]
- 13.Krusius T., Reinhold V. N., Margolis R. K., Margolis R. U. Structural studies on sialylated and sulphated O-glycosidic mannose-linked oligosaccharides in the chondroitin sulphate proteoglycan of brain. Biochem. J. 1987;245:229–234. doi: 10.1042/bj2450229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smalheiser N. R., Haslam S. M., Sutton-Smith M., Morris H. R., Dell A. Structural analysis of sequences O-linked to mannose reveals a novel Lewis X structure in cranin (dystroglycan) purified from sheep brain. J. Biol. Chem. 1998;273:23698–23703. doi: 10.1074/jbc.273.37.23698. [DOI] [PubMed] [Google Scholar]
- 15.Endo T. Dystroglycan glycosylation and its role in α-dystroglycanopathies. Acta Myol. 2007;26:165–170. [PMC free article] [PubMed] [Google Scholar]
- 16.Hewitt J. E. Abnormal glycosylation of dystroglycan in human genetic disease. Biochim. Biophys. Acta. 2009;1792:853–861. doi: 10.1016/j.bbadis.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Saito F., Blank M., Schroder J., Manya H., Shimizu T., Campbell K. P., Endo T., Mizutani M., Kroger S., Matsumura K. Aberrant glycosylation of α-dystroglycan causes defective binding of laminin in the muscle of chicken muscular dystrophy. FEBS Lett. 2005;579:2359–2363. doi: 10.1016/j.febslet.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 18.Beltran-Valero de Bernabe D., Currier S., Steinbrecher A., Celli J., van Beusekom E., van der Zwaag B., Kayserili H., Merlini L., Chitayat D., Dobyns W. B., et al. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am. J. Hum. Genet. 2002;71:1033–1043. doi: 10.1086/342975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi K., Nakahori Y., Miyake M., Matsumura K., Kondo-Iida E., Nomura Y., Segawa M., Yoshioka M., Saito K., Osawa M., et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- 20.Longman C., Brockington M., Torelli S., Jimenez-Mallebrera C., Kennedy C., Khalil N., Feng L., Saran R. K., Voit T., Merlini L., et al. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of α-dystroglycan. Hum. Mol. Genet. 2003;12:2853–2861. doi: 10.1093/hmg/ddg307. [DOI] [PubMed] [Google Scholar]
- 21.van Reeuwijk J., Janssen M., van den Elzen C., Beltran-Valero de Bernabe D., Sabatelli P., Merlini L., Boon M., Scheffer H., Brockington M., Muntoni F., et al. POMT2 mutations cause α-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J. Med. Genet. 2005;42:907–912. doi: 10.1136/jmg.2005.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida A., Kobayashi K., Manya H., Taniguchi K., Kano H., Mizuno M., Inazu T., Mitsuhashi H., Takahashi S., Takeuchi M., et al. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev. Cell. 2001;1:717–724. doi: 10.1016/s1534-5807(01)00070-3. [DOI] [PubMed] [Google Scholar]
- 23.Akasaka-Manya K., Manya H., Endo T. Mutations of the POMT1 gene found in patients with Walker-Warburg syndrome lead to a defect of protein O-mannosylation. Biochem. Biophys. Res. Commun. 2004;325:75–79. doi: 10.1016/j.bbrc.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 24.D'Amico A., Tessa A., Bruno C., Petrini S., Biancheri R., Pane M., Pedemonte M., Ricci E., Falace A., Rossi A., et al. Expanding the clinical spectrum of POMT1 phenotype. Neurology. 2006;66:1564–1567. doi: 10.1212/01.wnl.0000216145.66476.36. [DOI] [PubMed] [Google Scholar]
- 25.Murakami T., Hayashi Y. K., Ogawa M., Noguchi S., Campbell K. P., Togawa M., Inoue T., Oka A., Ohno K., Nonaka I., Nishino I. A novel POMT2 mutation causes mild congenital muscular dystrophy with normal brain MRI. Brain Dev. 2009;31:465–468. doi: 10.1016/j.braindev.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagisawa A., Bouchet C., Van den Bergh P. Y., Cuisset J. M., Viollet L., Leturcq F., Romero N. B., Quijano-Roy S., Fardeau M., Seta N., Guicheney P. New POMT2 mutations causing congenital muscular dystrophy: identification of a founder mutation. Neurology. 2007;69:1254–1260. doi: 10.1212/01.wnl.0000268489.60809.c4. [DOI] [PubMed] [Google Scholar]
- 27.Godfrey C., Escolar D., Brockington M., Clement E. M., Mein R., Jimenez-Mallebrera C., Torelli S., Feng L., Brown S. C., Sewry C. A., et al. Fukutin gene mutations in steroid-responsive limb girdle muscular dystrophy. Ann. Neurol. 2006;60:603–610. doi: 10.1002/ana.21006. [DOI] [PubMed] [Google Scholar]
- 28.Murakami T., Hayashi Y. K., Noguchi S., Ogawa M., Nonaka I., Tanabe Y., Ogino M., Takada F., Eriguchi M., Kotooka N., et al. Fukutin gene mutations cause dilated cardiomyopathy with minimal muscle weakness. Ann. Neurol. 2006;60:597–602. doi: 10.1002/ana.20973. [DOI] [PubMed] [Google Scholar]
- 29.Brockington M., Yuva Y., Prandini P., Brown S. C., Torelli S., Benson M. A., Herrmann R., Anderson L. V., Bashir R., Burgunder J. M., et al. Mutations in the fukutin-related protein gene (FKRP) identify limb girdle muscular dystrophy 2I as a milder allelic variant of congenital muscular dystrophy MDC1C. Hum. Mol. Genet. 2001;10:2851–2859. doi: 10.1093/hmg/10.25.2851. [DOI] [PubMed] [Google Scholar]
- 30.Louhichi N., Triki C., Quijano-Roy S., Richard P., Makri S., Meziou M., Estournet B., Mrad S., Romero N. B., Ayadi H., et al. New FKRP mutations causing congenital muscular dystrophy associated with mental retardation and central nervous system abnormalitiesIdentification of a founder mutation in Tunisian families. Neurogenetics. 2004;5:27–34. doi: 10.1007/s10048-003-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clement E. M., Godfrey C., Tan J., Brockington M., Torelli S., Feng L., Brown S. C., Jimenez-Mallebrera C., Sewry C. A., Longman C., et al. Mild POMGnT1 mutations underlie a novel limb-girdle muscular dystrophy variant. Arch. Neurol. 2008;65:137–141. doi: 10.1001/archneurol.2007.2. [DOI] [PubMed] [Google Scholar]
- 32.Hehr U., Uyanik G., Gross C., Walter M. C., Bohring A., Cohen M., Oehl-Jaschkowitz B., Bird L. M., Shamdeen G. M., Bogdahn U., et al. Novel POMGnT1 mutations define broader phenotypic spectrum of muscle-eye-brain disease. Neurogenetics. 2007;8:279–288. doi: 10.1007/s10048-007-0096-y. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi K., Kobayashi K., Saito K., Yamanouchi H., Ohnuma A., Hayashi Y. K., Manya H., Jin D. K., Lee M., Parano E., et al. Worldwide distribution and broader clinical spectrum of muscle-eye-brain disease. Hum. Mol. Genet. 2003;12:527–534. doi: 10.1093/hmg/ddg043. [DOI] [PubMed] [Google Scholar]
- 34.Mercuri E., Messina S., Bruno C., Mora M., Pegoraro E., Comi G. P., D'Amico A., Aiello C., Biancheri R., Berardinelli A., et al. Congenital muscular dystrophies with defective glycosylation of dystroglycan: a population study. Neurology. 2009;72:1802–1809. doi: 10.1212/01.wnl.0000346518.68110.60. [DOI] [PubMed] [Google Scholar]
- 35.van Reeuwijk J., Grewal P. K., Salih M. A., Beltran-Valero de Bernabe D., McLaughlan J. M., Michielse C. B., Herrmann R., Hewitt J. E., Steinbrecher A., Seidahmed M. Z., et al. Intragenic deletion in the LARGE gene causes Walker-Warburg syndrome. Hum. Genet. 2007;121:685–690. doi: 10.1007/s00439-007-0362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manya H., Chiba A., Yoshida A., Wang X., Chiba Y., Jigami Y., Margolis R. U., Endo T. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:500–505. doi: 10.1073/pnas.0307228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida-Moriguchi T., Yu L. P., Stalnaker S. H., Davis S., Kunz S., Madson M., Oldstone M. B. A., Schachter H., Wells L., Campbell K. P. O-Mannosyl phosphorylation of α-dystroglycan is required for laminin binding. Science. 2010;327:88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abramoff M. D., Magalhaes P. J., Ram S. J. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 39.Henry M. D., Campbell K. P. Dystroglycan inside and out. Curr. Opin. Cell Biol. 1999;11:602–607. doi: 10.1016/s0955-0674(99)00024-1. [DOI] [PubMed] [Google Scholar]
- 40.Gordon R. D., Sivarajah P., Satkunarajah M., Ma D., Tarling C. A., Vizitiu D., Withers S. G., Rini J. M. X-ray crystal structures of rabbit N-acetylglucosaminyltransferase I (GnT I) in complex with donor substrate analogues. J. Mol. Biol. 2006;360:67–79. doi: 10.1016/j.jmb.2006.04.058. [DOI] [PubMed] [Google Scholar]
- 41.Zhang W., Betel D., Schachter H. Cloning and expression of a novel UDP-GlcNAc:α-D-mannoside β1,2-N-acetylglucosaminyltransferase homologous to UDP-GlcNAc:α-3-D-mannoside β1,2-N-acetylglucosaminyltransferase I. Biochem. J. 2002;361:153–162. doi: 10.1042/0264-6021:3610153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akasaka-Manya K., Manya H., Kobayashi K., Toda T., Endo T. Structure-function analysis of human protein O-linked mannose β1,2-N-acetylglucosaminyltransferase 1, POMGnT1. Biochem. Biophys. Res. Commun. 2004;320:39–44. doi: 10.1016/j.bbrc.2004.05.129. [DOI] [PubMed] [Google Scholar]
- 44.Ferrer M., Chernikova T. N., Yakimov M. M., Golyshin P. N., Timmis K. N. Chaperonins govern growth of Escherichia coli at low temperatures. Nat. Biotechnol. 2003;21:1266–1267. doi: 10.1038/nbt1103-1266. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi S., Sasaki T., Manya H., Chiba Y., Yoshida A., Mizuno M., Ishida H., Ito F., Inazu T., Kotani N., et al. A new β-1,2-N-acetylglucosaminyltransferase that may play a role in the biosynthesis of mammalian O-mannosyl glycans. Glycobiology. 2001;11:37–45. doi: 10.1093/glycob/11.1.37. [DOI] [PubMed] [Google Scholar]
- 46.Balci B., Morris-Rosendahl D. J., Celebi A., Talim B., Topaloglu H., Dincer P. Prenatal diagnosis of muscle-eye-brain disease. Prenatal Diagn. 2007;27:51–54. doi: 10.1002/pd.1622. [DOI] [PubMed] [Google Scholar]
- 47.Biancheri R., Bertini E., Falace A., Pedemonte M., Rossi A., D'Amico A., Scapolan S., Bergamino L., Petrini S., Cassandrini D., et al. POMGnT1 mutations in congenital muscular dystrophy: genotype-phenotype correlation and expanded clinical spectrum. Arch. Neurol. 2006;63:1491–1495. doi: 10.1001/archneur.63.10.1491. [DOI] [PubMed] [Google Scholar]
- 48.Diesen C., Saarinen A., Pihko H., Rosenlew C., Cormand B., Dobyns W. B., Dieguez J., Valanne L., Joensuu T., Lehesjoki A. E. POMGnT1 mutation and phenotypic spectrum in muscle-eye-brain disease. J. Med. Genet. 2004;41:e115. doi: 10.1136/jmg.2004.020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Godfrey C., Clement E., Mein R., Brockington M., Smith J., Talim B., Straub V., Robb S., Quinlivan R., Feng L., et al. Refining genotype phenotype correlations in muscular dystrophies with defective glycosylation of dystroglycan. Brain. 2007;130:2725–2735. doi: 10.1093/brain/awm212. [DOI] [PubMed] [Google Scholar]
- 50.Pascual-Castroviejo I., Pascual-Pascual S. I., Gutierrez-Molina M., Saarinen A., Joensuu T. H., Bayes M., Cormand B. [Muscle-eye-brain diseasePresentation of one case with genetic study.] Neurologia (Barcelona, Spain) 2005;20:261–266. [PubMed] [Google Scholar]
- 51.Shenoy A. M., Markowitz J. A., Bonnemann C. G., Krishnamoorthy K., Bossler A. D., Tseng B. S. Muscle-Eye-Brain disease. J. Clin. Neuromuscular Dis. 2010;11:124–126. doi: 10.1097/CND.0b013e3181c5054d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Topaloglu H., Cila A., Tasdemir A. H., Saatci I. Congenital muscular dystrophy with eye and brain involvementThe Turkish experience in two cases. Brain Dev. 1995;17:271–275. doi: 10.1016/0387-7604(95)00047-f. [DOI] [PubMed] [Google Scholar]
- 53.Vervoort V. S., Holden K. R., Ukadike K. C., Collins J. S., Saul R. A., Srivastava A. K. POMGnT1 gene alterations in a family with neurological abnormalities. Ann. Neurol. 2004;56:143–148. doi: 10.1002/ana.20172. [DOI] [PubMed] [Google Scholar]
- 54.Soldin S. J., Murthy J. N., Agarwalla P. K., Ojeifo O., Chea J. Pediatric reference ranges for creatine kinase, CKMB, troponin I, iron, and cortisol. Clin. Biochem. 1999;32:77–80. doi: 10.1016/s0009-9120(98)00084-8. [DOI] [PubMed] [Google Scholar]
- 55.Strasser R., Stadlmann J., Svoboda B., Altmann F., Glossl J., Mach L. Molecular basis of N-acetylglucosaminyltransferase I deficiency in Arabidopsis thaliana plants lacking complex N-glycans. Biochem. J. 2005;387:385–391. doi: 10.1042/BJ20041686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W., Unligil U. M., Rini J. M., Stanley P. Independent Lec1A CHO glycosylation mutants arise from point mutations in N-acetylglucosaminyl-transferase I that reduce affinity for both substrates. Molecular consequences based on the crystal structure of GlcNAc-T1. Biochemistry. 2001;40:8765–8772. doi: 10.1021/bi015538b. [DOI] [PubMed] [Google Scholar]
- 57.Derman A. I., Prinz W. A., Belin D., Beckwith J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 58.Xiong H., Kobayashi K., Tachikawa M., Manya H., Takeda S., Chiyonobu T., Fujikake N., Wang F., Nishimoto A., Morris G. E., et al. Molecular interaction between fukutin and POMGnT1 in the glycosylation pathway of α-dystroglycan. Biochem. Biophys. Res. Commun. 2006;350:935–941. doi: 10.1016/j.bbrc.2006.09.129. [DOI] [PubMed] [Google Scholar]
- 59.Nilsson J., Nilsson J., Larson G., Grahn A. Characterization of site-specific O-glycan structures within the mucin-like domain of α-dystroglycan from human skeletal muscle. Glycobiology. 2010;20:1160–1169. doi: 10.1093/glycob/cwq082. [DOI] [PubMed] [Google Scholar]
- 60.Stalnaker S. H., Hashmi S., Lim J. M., Aoki K., Porterfield M., Gutierrez-Sanchez G., Wheeler J., Ervasti J. M., Bergmann C., Tiemeyer M., Wells L. Site mapping and characterization of O-glycan structures on α-dystroglycan isolated from rabbit skeletal muscle. J. Biol. Chem. 2010;285:24882–24891. doi: 10.1074/jbc.M110.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]