Abstract
DIVEIN is a web interface that performs automated phylogenetic and other analyses of nucleotide and amino acid sequences. Starting with a set of aligned sequences, DIVEIN estimates evolutionary parameters and phylogenetic trees while allowing the user to choose from a variety of evolutionary models, it then reconstructs the consensus, Most Recent Common Ancestor (MRCA) and Center of Tree (COT) sequences. DIVEIN also provides tools for further analyses, including condensing sequence alignments to show only informative sites or private mutations, computing phylogenetic or pairwise divergence from any user-specified sequence (MRCA, Consensus, COT, or existing sequence from the alignment), computing and outputting all genetic distances in column format, calculating summary statistics of diversity and divergence from pairwise distances, and graphically representing the inferred tree and plots of divergence, diversity, and distance distribution histograms. DIVEIN is available at http://indra.mullins.microbiol.washington.edu/DIVEIN.
Keywords: phylogeny, divergence, diversity, informative sites, center of tree, maximum likelihood
Fast and accurate estimation of phylogenies and determination of genetic and phylogenetic divergence and diversity of molecular sequences are essential components of biological research. For a set of sequences, a typical phylogenetic analysis involves several steps, including multiple sequence alignment, phylogenetic reconstruction, visualization of the inferred tree, and calculation of evolutionary measures. A large number of phylogenetic analysis resources have been developed, as catalogued by Dr. Joseph Felsenstein (http://evolution.genetics.washington.edu/phylip/software.html). Web servers for phylogenetic analysis provide an easy route to address specific evolutionary questions. For example, PhyML Online (1) performs maximum likelihood (ML) phylogenetic estimation under a wide range of evolutionary models. Phylemon (2) provides experts with a suite of online programs and a Java interface to build a phylogeny pipeline. Dereeper et al. Recently made available Phylogeny.fr (3), a combination of an easy-to-use interface with a phylogeny pipeline designed for the non-specialist with up-to-date programs that are frequently reserved for experts. These tools provide excellent interfaces to phylogenetic reconstruction; however, there is an increasing demand by researchers for a tool that performs not only typical phylogenetic reconstructions, as most existing web servers do capably, but also enables downstream results processing and interpretation. For example, calculating divergence and diversity measurements and genetic distance distributions from the phylogenetic output are usually very time-consuming processes that require caution if conducted manually to ensure that calculations are carried out correctly and that data has not been altered in the transfer between the several necessary software packages.
Furthermore, reducing an alignment to only its phylogenetically informative sites, a position at which there are at least two different character states and each of those states occurs in at least two of the sequences, has proven to be a useful approach in recombination analysis (4–6) and visualizing extended alignments. Calculation of central sequences and comparison of a set of sequences to a consensus (CON), most recent common ancestor (MRCA), or Center of Tree (COT; an ancestral state that minimizes the phylogenetic distance from the specified sequences) (7–9), have been used in a variety of studies of sequence evolution, structure, function, and rational vaccine design.
We therefore identified a need for a unified web interface to integrate useful tools and perform automated phylogenetic and other genetic analyses including summaries and visualization of the resulting data, leading us to develop DIVEIN.
DIVEIN has four major components:
A pipeline to automatically guide a set of aligned sequences through phylogenetic tree estimation under a variety of evolutionary models, and visualization of the inferred tree
An interface to reconstruct MRCA/COT/CON sequences and reconstruct and visualize trees re-rooted by MRCA and COT sequences.
Calculation of genetic distance distributions, pairwise diversity and divergence from the MRCA/COT/CON.
An interface to detect, visualize and numerically summarize phylogenetically informative sites as well as private mutations (found only in a single sequence) in an alignment.
DIVEIN runs on an Apache web server. The web interfaces are implemented via Perl CGI and JavaScript. Data manipulation and presentation employ standard Perl and BioPerl (10) modules. Maximum likelihood phylogenetic reconstructions use PhyML v3.0 (11), which applies a hill-climbing algorithm that adjusts tree topology and branch lengths simultaneously. The inferred tree can be viewed and edited through the included Archaeopteryx v0.954 beta Java applet (http://www.phylosoft.org/archaeopteryx/). The MRCA and COT sequences are reconstructed using a joint maximum likelihood procedure (12) via HyPhy v2.0 (13), a scriptable software package for performing a wealth of evolutionary sequence analyses. Distance distribution histograms and divergence and diversity plots are generated using the open source Gnuplot graphing package (http://www.gnuplot.info/). DIVEIN is hosted on a Linux computer with two quad-core Intel Xeon 2.5 GHz processors (8 cores) and 8 GB of RAM. It has a queuing system for user-submitted projects, configured to run up to eight projects simultaneously. Any additional projects are queued for later execution. Bootstrap replicates are limited to 100 because of computational resource limitations.
Given a collection of sequences, the divergence is derived by calculating the mean distance of all sequences from a reference or founder sequence and the diversity is given as the mean distance between all sequences (14). Using d(i,j) to denote either the path length between nodes i and j in the reconstructed phylogenetic tree or a genetic distance between sequences i and j, we measure divergence and diversity for a collection of N sequences as follows:
| (1) |
| (2) |
DIVEIN accepts aligned nucleotide or amino acid sequences in NEXUS, PHYLIP or FASTA format. For phylogenetic analyses, users can perform ML estimation alone or include divergence/diversity analyses. They can select the option to calculate divergence from any or all of: MRCA, COT, CON, or any sequence in the alignment. When calculating MRCA, a file listing sequence name(s) that belong to the outgroup must be provided. Users can optionally provide a file assigning input sequences to multiple groups and calculate divergence and diversity for each of those groups. If a group file is not provided, DIVEIN will assign all sequences to a single group, excluding the defined outgroup sequences. For COT analysis, users may upload a tree to reconstruct its COT. If the tree is not provided, DIVEIN will estimate one using the general time reversible (GTR) (15) substitution model (nucleotides) or an improved general amino acid replacement matrix (called LG) (16) (amino acids).
We have also included an informative sites module in DIVEIN that is useful for condensing sequence data to allow users to quickly identify sites that are changing within an alignment and more easily obtain an overview of complex and large data sets. To detect phylogenetically informative sites (those found in more than one sequence, and thus contributing to branch ordering), users can include a reference sequence at the top of the alignment, or DIVEIN will calculate the consensus of the alignment as the reference. Example data sets are provided to familiarize users with the correct input formats and expected output results. DIVEIN also provides the functionality to retrieve finished results via a previously assigned project id.
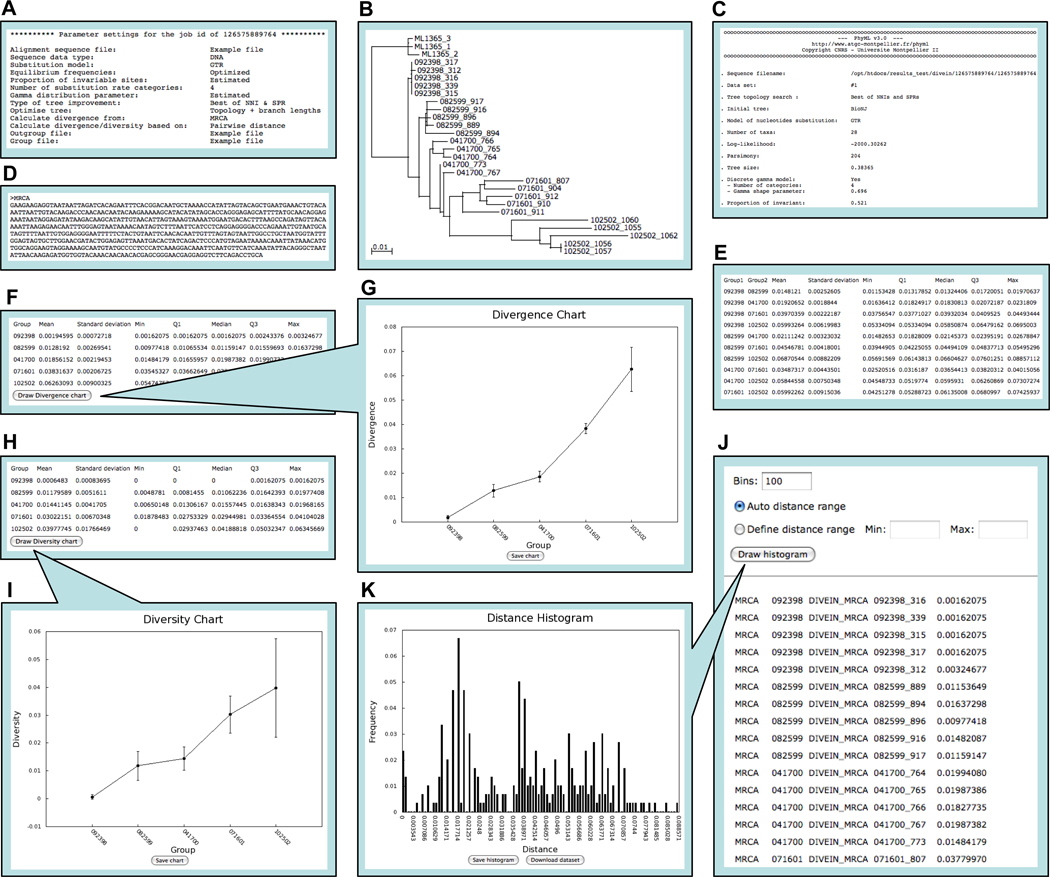
When an analysis is finished, a link is sent to the user by email to view and download results. The results are accessible on the server for 2 days (after which they are deleted) via a randomly generated URL known only to the user initiating the analysis. Users can locally visualize and edit phylogenetic trees and dynamically generate and download graphs of distance distribution histograms and divergence and diversity (if applicable). Sample screen shots of DIVEIN output (phylogeney/divergence/diversity) are shown in Figure 1. Using an example alignment of 28 DNA sequences with 624 sites (available on the DIVEIN website), it takes less than 30 seconds to finish the entire analysis process of phylogeny/divergence/diversity. For the analysis of phylogenetically informative sites, the states at each informative site are displayed as an alignment and in a table.
Figure 1.
Screen shots of phylogeny/divergence/diversity output in DIVEIN. (A) Parameter settings for running the program. (B) Phylogenetic tree viewed through the Archaeopteryx Java applet. (C) Estimated evolutionary parameters. (D) Reconstructed MRCA sequence. (E) Summarized distances between groups. (F) Summarized divergence from MRCA for each group. (G) Divergence plot generated by clicking the Draw divergence chart button. (H) Summarized diversity for each group. (I) Diversity plot generated by clicking the Draw diversity chart button. (J) Distance matrix. (K) Distance distribution histogram generated by clicking the Draw histogram button.
In conclusion, DIVEIN performs fast, accurate, and automated phylogenetic analyses, including (i) informative sites detection, (ii) ML tree estimation under a variety of evolutionary models, (iii) MRCA, COT and consensus reconstruction, (iv) distance distribution calculation, and (v) distance- and phylogenetic-based divergence and diversity measurements, along with resulting data summarization and visualization. Future versions will add the option to select the best-fit evolutionary model via ModelTest (17) and ProtTest (18) to reconstruct the phylogeny. Furthermore, we will incorporate other widely used phylogenetic analysis programs, e.g., MrBayes (19), into DIVEIN to allow users to have easy access to other state-of-the-art molecular evolution analysis programs.
ACKNOWLEDGEMENTS
We thank Dr. John E. Mittler for discussions. This work was supported by grants from the US Public Health Services [AI047734, AI057005], including to the Computational Biology Core of the University of Washington Center for AIDS Research [AI27757].
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
REFERENCES
- 1.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic acids research. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tarraga J, Medina I, Arbiza L, Huerta-Cepas J, Gabaldon T, Dopazo J, Dopazo H. Phylemon: a suite of web tools for molecular evolution, phylogenetics and phylogenomics. Nucleic acids research. 2007;35:W38–W42. doi: 10.1093/nar/gkm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eshleman SH, Gonzales MJ, Becker-Pergola G, Cunningham SC, Guay LA, Jackson JB, Shafer RW. Identification of Ugandan HIV type 1 variants with unique patterns of recombination in pol involving subtypes A and D. AIDS Res Hum Retroviruses. 2002;18:507–511. doi: 10.1089/088922202317406655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottlieb GS, Heath L, Nickle DC, Wong KG, Leach SE, Jacobs B, Gezahegne S, van ’t Wout AB, et al. HIV-1 variation before seroconversion in men who have sex with men: analysis of acute/early HIV infection in the multicenter AIDS cohort study. J Infect Dis. 2008;197:1011–1015. doi: 10.1086/529206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell MS, Gottlieb GS, Hawes SE, Nickle DC, Wong KG, Deng W, Lampinen TM, Kiviat NB, Mullins JI. HIV-1 superinfection in the antiretroviral therapy era: are seroconcordant sexual partners at risk? PloS one. 2009;4:e5690. doi: 10.1371/journal.pone.0005690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nickle DC, Jensen MA, Gottlieb GS, Shriner D, Learn GH, Rodrigo AG, Mullins JI. Consensus and ancestral state HIV vaccines. Science. 2003;299:1515–1518. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- 8.Nickle DC, Rolland M, Jensen MA, Pond SL, Deng W, Seligman M, Heckerman D, Mullins JI, Jojic N. Coping with viral diversity in HIV vaccine design. PLoS Comput Biol. 2007;3:e75. doi: 10.1371/journal.pcbi.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rolland M, Jensen MA, Nickle DC, Yan J, Learn GH, Heath L, Weiner D, Mullins JI. Reconstruction and function of ancestral center-of-tree human immunodeficiency virus type 1 proteins. J. Virol. 2007;81:8507–8514. doi: 10.1128/JVI.02683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 12.Pupko T, Pe'er I, Shamir R, Graur D. A fast algorithm for joint reconstruction of ancestral amino acid sequences. Molecular biology and evolution. 2000;17:890–896. doi: 10.1093/oxfordjournals.molbev.a026369. [DOI] [PubMed] [Google Scholar]
- 13.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 14.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tavare S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. American mathematical Society: Lectures on Mathematics in the Life Sciences. 1986;17:57–86. [Google Scholar]
- 16.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Molecular biology and evolution. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 17.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 18.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 19.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]