Abstract
Biochemical and biophysical characterization of CFTR (the cystic fibrosis transmembrane conductance regulator) is thwarted by difficulties to obtain sufficient quantities of correctly folded and functional protein. Here we have produced human CFTR in the prokaryotic expression host Lactococcus lactis. The full-length protein was detected in the membrane of the bacterium, but the yields were too low (< 0.1% of membrane proteins) for in vitro functional and structural characterization, and induction of the expression of CFTR resulted in growth arrest. We used isobaric tagging for relative and absolute quantitation based quantitative proteomics to find out why production of CFTR in L. lactis was problematic. Protein abundances in membrane and soluble fractions were monitored as a function of induction time, both in CFTR expression cells and in control cells that did not express CFTR. Eight hundred and forty six proteins were identified and quantified (35% of the predicted proteome), including 163 integral membrane proteins. Expression of CFTR resulted in an increase in abundance of stress-related proteins (e.g. heat-shock and cell envelope stress), indicating the presence of misfolded proteins in the membrane. In contrast to the reported consequences of membrane protein overexpression in Escherichia coli, there were no indications that the membrane protein insertion machinery (Sec) became overloaded upon CFTR production in L. lactis. Nutrients and ATP became limiting in the control cells as the culture entered the late exponential and stationary growth phases but this did not happen in the CFTR expressing cells, which had stopped growing upon induction. The different stress responses elicited in E. coli and L. lactis upon membrane protein production indicate that different strategies are needed to overcome low expression yields and toxicity.
The human cystic fibrosis transmembrane conductance regulator CFTR1 is an atypical member of the superfamily of ATP binding cassette (ABC) transporters, because it is a channel (for chloride ions) rather than a transporter. Mutations in CFTR cause cystic fibrosis (1–3), the most common genetic disease among Caucasians. Mechanistic studies on CFTR and attempts to rationally design drugs to treat cystic fibrosis are hampered by difficulties to produce the protein in amounts needed for biochemical and biophysical studies, such as x-ray crystallography. A major bottleneck is a lack of suitable overexpression systems to produce recombinant CFTR, a problem which is often encountered for human membrane proteins (4–11). In an attempt to find suitable hosts for recombinant production of CFTR the cystic fibrosis foundation has funded a project to express CFTR in the bacterium Lactococcus lactis.
L. lactis is a Gram-positive bacterium for which expression plasmids and inducible promoters are available (12). Several cases have been reported in which functional overexpression of membrane proteins could be achieved in L. lactis, but not in E. coli (e.g. the human KDEL receptor, Na+/tyrosine transporter (Tyt1) of Fusobacterium nucleatum and several membrane proteins from Arabidopsis) (5, 13–17). The use of L. lactis as host for (eukaryotic) membrane protein expression has been reviewed by Kunji et al. (14). Among the potential advantages of L. lactis are its growth rate of ∼1 doubling per hour, which is much slower than E. coli and could be beneficial for expression of proteins that do not fold easily. Also the presence of a different repertoire of chaperones, e.g. two copies of the integral membrane chaperone YidC (18–20), could facilitate insertion and assembly of heterologous membrane proteins. Other factors such as different membrane lipids and cytosolic environment could play a role as well.
Here we have used L. lactis for the expression of the human cystic fibrosis transmembrane conductance regulator CFTR. We were able to express full length (1480 amino acids long) CFTR in the bacterial host, but the expression levels were too low for to pursue structural studies, and expression was toxic to the cells. To understand this toxicity and to identify potential remedies to improve expression levels, we investigated the physiological responses that were elicited in L. lactis upon CFTR expression by performing a global quantitative proteomics study.
EXPERIMENTAL PROCEDURES
DNA Manipulations and Cloning of CFTR
Human cftr cDNA (gift from Christine Bear, Toronto) (accession nr M28668) was cloned into the E. coli vectors pREnLIC, and pREcLIC (supplemental Table 1), which introduce the sequences coding for N- and C-terminal His10-tags, respectively, by Ligation Independent Cloning (LIC) as described by Geertsma et al. (21), yielding plasmids pREnCFTR and pREcCFTR. Plasmid pREncCFTR, which contains the cftr coding region fused to sequences coding for both an N- and C-terminal His10-tag, was constructed by exchanging the NcoI-XhoI fragment of pREnCFTR with the NcoI-XhoI fragment of pREcCFTR. pRE_MBP-CFTR was constructed by amplifying cftr by PCR and subsequent cloning in the NcoI and SpeI sites of pRE_MBP (supplemental Table 1). The pRE vectors were converted into pNZ8048-related vectors for L. lactis by Vector Backbone Exchange as described by Geertsma et al. (21).
Expression of CFTR in L. lactis and Sample Preparation
L. lactis NZ9000 transformed with pNZ8048-derived plasmids was cultivated in M17 medium (Oxoid, Basingstoke, UK) containing 1% glucose, and 5 μg/ml chloramphenicol. To test for expression of CFTR L. lactis was grown in 10-ml cultures (inoculated with O/N cultures that were diluted 1:50) to an OD600 of 0.5 at 30 °C, after which Nisin A (1:5000 dilution of the culture supernatant of the nisin producing strain L. lactis NZ9700 (5)) was added and the cells were incubated for another 2 h. A volume of culture containing the equivalent amount of cells as 1 ml of OD600 of 5 was spun down (20,000 × g, 2 min, room temperature) and the pellet was resuspended in 400 μl of 50 mm potassium phosphate buffer (KPi) pH 7.5, 10% glycerol. Phenyl methanesulphonyl fluoride (1 mm) was added and the cells were disrupted in a Fastprep machine (Bio101, Vista, CA) by vigorous shaking in the presence of glass beads (two times at force 6.0 for 30 s, with 10 min incubation on ice in between the two runs). The crude cell extracts were supplemented with EDTA (15 mm final concentration) and centrifuged for 15 min at 20,000 × g at 4 °C. The supernatant was subsequently centrifuged at 300,000 × g (30 min, 4 °C) to obtain the membranes.
SDS-sample buffer was added and samples were incubated at 37 °C for 5 min before loading on SDS-PAGE. His-tag specific antibodies (GE healthcare) and CFTR C terminus specific antibodies (clone 24–1, R&D systems) were used for western hybridizations.
Purification of His-MBP-CFTR
L. lactis NZ9000 pNZ_MBP-CFTR was grown in a fermenter (Applikon) in 2 L M17 supplemented with glucose (1%) and chloramphenicol (5 μg/ml) as described later. The cells were harvested 2 h after induction by centrifugation (6800 × g, 15 min, 4 °C) and membranes were prepared as described later. The membranes were stored at −80 °C in 50 mm KPi pH7.5, 10% glycerol at a concentration of 10 mg/ml protein.
Membranes containing 10 mg protein were resuspended in 10 ml of 50 mm Tris-HCl pH 8.0, 300 mm NaCl, 20% glycerol, 10 mm Imidazole. n-Dodecyl-β-d-maltoside (DDM) was added (1% final concentration) and the proteins were solubilized on ice for 1 h. Solubilized membranes were centrifuged at 100,000 × g for 30 min at 4 °C. The supernatant was incubated with Ni-Sepharose resin (GE Healthcare) for 1 h (400 μl slurry, which had been pre-equilibrated with solubilization buffer), with gentle rotation. The resin was washed with 10 ml of the same buffer containing 0.05% DDM and 50 mm imidazole and finally proteins were eluted with buffer containing 500 mm imidazole and 0.05% DDM (100, 200, 200 μl fractions).
In-gel Trypsin Digest
Bands were excised from a Coomassie Blue-stained SDS-PAGE and cut into ∼1 mm3 pieces. Gel slices were incubated 3–4 times for 15 min in 150 μl of destaining solution (50% acetonitrile, 50 mm ammonium bicarbonate). The gel slices were dehydrated in 150 μl of 100% acitonitrile for 10 min, the supernatant was discarded and the gel slices were dried by evaporation. The reduction of cysteine residues was performed by incubating the gel slices in 30 μl of 10 mm dithiotreitol in 50 mm ammonium bicarbonate for 45 min at 55 °C. The supernatant was removed, 30 μl of 55 mm iodoacetamide in 50 mm ammonium bicarbonate was added to each gel slice and incubated for 30 min at RT. The gel slices were dehydrated as above. To each dried gel slice, 7–10 μl of 10 ng/μl trypsin gold (Cat.: V5280, Promega, Madison, WI) in 40 mm ammonium bicarbonate/10% acetonitrile were added and allowed to re-swell for ∼20 min at 37 °C. The gel slices were overlaid with 20 μl of 40 mm ammonium bicarbonate, 10% acetonitrile and incubated overnight at 37 °C. The peptides were extracted by adding 50 μl of 2% trifluoroacetic acid (TFA) to each gel slice without removing the overlay. The extraction was repeated twice with 33% acetonitrile/1.3% TFA and 63% acetonitrile/0.7% TFA. The extracted peptides were combined and the peptide mixture was dried. The peptide mixture was resuspended in 10 μl of 0.1% TFA and subjected to tandem MS (MS/MS) analysis directly by the mixing 1:2 with 20 mg/ml α-cyano-4-hydroxycinnamic acid matrix solution (LaserBio Labs, Sophia-Antipolis, France) onto a matrix assisted laser desorption ionization (MALDI) target.
Proteomics: Growth and Preparation of Samples
Growth in Fermenters
L. lactis NZ9000 pNZ8048 and L. lactis NZ9000 pNZncCFTR were grown in 3 L fermenters (Applikon) in M17 medium supplemented with glucose (1%) and chloramphenicol (5 μg/ml). The temperature was set at 30 °C and the pH was maintained at 6.5 during growth by addition of KOH. At an OD600 of 0.5 900 ml of the culture was removed and to the remaining culture Nisin A was added (1:5000 dilution of the supernatant of a culture of L. lactis NZ9700). After 1 h and 4 h of induction 900 ml of the culture was collected. Cell were spun down (6800 × g for 15 min, 4 °C), and pellets were washed once with 10 mm KPi pH 7.5. The washed cell pellets were frozen in liquid nitrogen and stored at −80 °C.
Isolation of Membrane and Soluble Protein Fractions
The cell pellets were resuspended in 10 mm KPi pH 7.5 at an OD600 of 50. To 6 ml of the suspension MgCl2 was added (1 mm final concentration) and the cells were disrupted at 39 kPsi with a Constant Systems cell disrupter. The cells were passed through the disrupter cell twice. EDTA was added (15 mm) to the suspensions and they were incubated on ice for 15 min.
To remove nonbroken cells the crude cell lysates were centrifuged for 15 min at 12,000 × g at 4 °C. The supernatant was carefully recovered and subsequently centrifuged at 267,000 × g for 15 min at 4 °C. The supernatant, containing the soluble protein fraction was carefully pipetted off and stored at −80 °C. Residual supernatant was completely removed from the membranes pellet. The membranes were washed once with 1 ml 10 mm KPi containing 10% glycerol. The pellets were finally resuspended in 500 μl 10 mm KPi, 10% glycerol and stored at −80 °C. Protein concentrations were determined with the BCA kit (Pierce/Thermo Fisher, Waltham, MA).
Sample Preparation for Strong Cation Exchange/ Reverse Phase-Liquid Chromatography (SCX/RP-LC) and iTRAQ Labeling
For trypsinization, 100 μg of protein (when used for 4-plex iTRAQ labeling, experiment A) or 50 μg (when used for 8-plex iTRAQ labeling, experiment B) was resuspended in 20 μl of 500 mm TEAB, 2% acetonitrile plus 0.08% SDS. Reduction of disulfide bonds with Tris(2-carboxyethyl) phosphine hydrochloride, cysteine-modification with methyl-methanethiosulfonate (MMTS) were performed according to the manufacturer's protocol for iTRAQ (Applied Biosystems, Foster City, CA). For enzymatic digestion, trypsin gold (Cat.: V5280, Promega) was reconstituted in 500 mm TEAB and 5 mm calcium chloride, and used in 1:6 (μg/μg) trypsin-to-protein ratio. Digestion was performed over night at 37 °C. Undigested material was spun down for 10 min at 14,000 × g. The pellets were suspended in TEAB/acetonitrile/SDS solution as before and digested for 5 h at 37 °C with 0.8 μg trypsin per sample. The corresponding samples from two digests were combined, freeze-dried and suspended in 15 μl 500 mm TEAB. The 8-plex iTRAQ labeling was performed according to the manufacturer's protocol with a few modifications. Each label was reconstituted in 210 μl 100% isopropanol and to each sample of 15 μl, 100 μl reconstituted label was added, so that each label was used for two samples. The four-plex iTRAQ labeling was performed according to the manufacturer's protocol except that each label was resuspended in 200 μl ethanol and combined with 20 μl tryptic digest. The samples were incubated for at least 2.5 h at room temperature and stored at –20 until required. Organic solvent (isopropanol or ethanol) was removed by evaporation. Each sample was suspended in 100 μl water. From each sample, 50 μl were combined (200 μg) and concentrated to a volume of 250 μl. The same volume of twofold concentrated SCX buffer A (see below) was added, the pH was adjusted to 2.7 with phosphoric acid. The peptide mixture was subjected to chromatography and mass spectrometry analysis.
Prefractionation of Peptides on SCX
For off-line peptide pre-fractionation, a silica-based Polysulfoethyl Aspartamide SCX column was used (Cat.: 202SE0502 PolyLC Inc., Columbia USA). The column was run at a flow rate of 200 μl/min on an AKTA purifier (GE Healthcare). Gradient solutions A: 10 mm triethylammonium phosphate, pH 2.7, 25% acetonitrile; B: 10 mm triethylammonium phosphate, pH 2.7, 25% acetonitrile, 500 mm KCl. Gradient conditions: column equilibration with five column volumes (CV) (1 CV = 0.7 ml) of 100% A. Peptides were loaded in 100% A and the column was washed with 10 CV at 100% A. Peptides were eluted: 1) 0 to 5% B in 5 CV; 2) followed by 12 to 30% B in 10 CV; and 3) 24–60% B in 5 CV. Fractions of elution steps 1 and 2 were collected every 45 s, and fractions of the elution step 3 were collected every 1 min in a 96-well plate. Eluted peptides were dried in a vacuum centrifuge and resuspended in 50 μl of 0.1% TFA. Depending on the complexity, either separate fractions or pools of two fractions were analyzed by RP-LC MALDI-time-of-flight (TOF)/TOF.
RP-LC and MALDI-TOF/TOF analysis
Peptides were trapped on a pre-column (300 μm x 5 mm, C18 PepMap300, LC Packing) and then separated on a C18 capillary column (C18 PepMap 300, 75 μm × 150 mm, 3 μm particle size, LC-Packing) mounted on the Dionex Ultraflex 3000 LC system (LC Packings, Amsterdam, The Netherlands). Mobile phase solutions contained A: 0.05% TFA; B: 0.05% TFA, 80% acetonitrile. Gradient conditions: equilibration of column, binding and washing of peptides was performed with 3% B, elution with 3 to 50% B in 60 min at a flow rate of 300 nL/min. The eluting peptides were mixed 1:4 with 2.2 mg/ml α-cyano-4-hydroxycinnamic acid matrix (LaserBio Labs, Sophia-Antipolis, France) and spotted directly onto a MALDI target (12 s x 260 spots), using a Probot system (LC Packings, Amsterdam, The Netherlands). Peptides were analyzed with a 4800 Proteomics analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems).
The MALDI-TOF/TOF was operated in reflectron positive ionization mode in the m/z range 900–4000. The 15 most intense peaks above the signal-to-noise threshold of 120 from each MS spectrum of odd-numbered RP-LC runs were selected for MS/MS fragmentation in the m/z range from 900 to 2000. The 10 most intense peaks above the signal-to-noise of 50 were selected from each MS spectrum of even-numbered RP-LC runs in the m/z range from 2000 to 4000. The MS/MS spectra were acquired using 2 kV acceleration voltage and air as collision gas at 5 × 10−7 Torr. The precursor mass transmission window was set to 300 (full width at half maximum, FWHM) for peptides in the m/z range of 900–2000, and to 200 (FWHM) in the range of 2000–4000 m/z. The peak-lists of the acquired MS/MS spectra were generated, using default settings and the S/N threshold of 10. The MS spectra were calibrated in the plate model mode, using 4700 calibration mixture (Applied Biosystems). MS/MS calibration of the instrument was performed when required, using ACTH 18–39 (m/z = 2465.199) fragment ions.
Database Search and Criteria for Protein Identification
MS/MS peak-lists were extracted by the ProteinPilot software, version 2.0, using default parameters and were automatically submitted to a database search. All MS/MS spectra were analyzed using Mascot (Matrix Science, London, UK; version 2.0) and X!Tandem (www.thegpm.org; version 2007.01.01.1). Mascot and X!Tandem were set up to search a combined L. lactis sp. cremoris MG1363 database, allowing one missed cleavage of the digestion by trypsin. The database was created by combining forward and reversed entries of the L. lactis proteome (release version 31.08.07) and included sequences of porcine trypsin (NCBI accession: P00761), human keratins (P35908, P35527, P13645, NP_006112), chloramphenicol acetyltransferase (P00485), replication protein A (Q04138), and the human CFTR (NCBI accession: NP_000483) containing in total 4902 protein entries. Mascot and X!Tandem searches were performed with a fragment ion mass tolerance of 0.30 Da and a parent ion tolerance of 200 ppm. MMTS modification of cysteine and Applied Biosystems 4-plex or 8-plexed iTRAQ quantitation chemistry of lysine and the N terminus were specified in Mascot and X!Tandem as fixed modifications. Deamidation of asparagine and glutamine, oxidation of methionine and Applied Biosystems 4-plex or 8-plexed iTRAQ quantitation chemistry of tyrosine were specified in Mascot and X!Tandem as variable modifications.
Scaffold (version Scaffold-2_02_03, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (22). Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 uniquely identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (23). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principle of parsimony. Those peptides were removed from the dataset when quantification was performed. The false positive rate was calculated by dividing 2 times the number of proteins identified in the reversed database by 4902, the sum of all proteins identified in forward and reversed versions of the database. In all measured samples, no hits from the reversed database were detected, using the criteria described above.
Relative Quantification of Protein Expression
The relative quantification was based on peptides that were chemically labeled with isobaric reagents, using the 4-plex or 8-plex iTRAQ technique. The quantification information was obtained from the peak areas of the reporter ions (m/z 112.2, 113.2, 114.2, 115.2, 116.2, 117.2, 118.2, 119.2, and 121.2). The peak areas were extracted from the MS/MS spectra by the ProteinPilot software using default settings as specified by the ProteinPilot for the 4800 MALDI instruments (Applied Biosystems). The peak areas were corrected for isotopic impurities by the ProteinPilot using the information provided by the manufacturer in the Certificate of Analysis for each iTRAQ batch. To select quantification data, those ratios were removed where the peak area of one reporter ion was below the signal-to-noise threshold of 10.
The global bias correction was performed for all identified peptides. The bias correction factor for a given iTRAQ ratio (e.g. 113/114) was calculated as the sum of all reporter peak areas in all measured spectra from one iTRAQ reagent (e.g. 114) divided by the sum of reporter peak areas of another reagent (e.g. 113). To obtain the bias-corrected peptide iTRAQ ratios, all measured ratios (in this example all 113/114 ratios) were multiplied by the correction factor. The bias-corrected peptide ratios of the same protein were weight-averaged and protein iTRAQ ratios were obtained according to the method used by the ProteinPilot software (Applied Biosystems). Peptides that matched to multiple proteins were excluded from quantification.
The relevant protein and peptide data and given in supplemental Tables S5 and S6.
Statistical Analysis
Identifying Significantly Changed Protein Abundances
To identify proteins with significantly changed abundances two different methods were used depending on the number of available replicate values. Rank Sum analysis was used for the comparison of the CFTR expressing strain versus the control strain at the 4 h time point, where four independent replicates were available. Rank Sum is a nonparametric statistical method based on the Rank Product analysis (24, 25), which allows the data from biological replicates to be analyzed in a robust way. For the Rank Sum analysis the weighted protein ratios for each of the four replicate samples were calculated as described above and sorted in descending order. Ranks were assigned to each protein, so that the protein with the highest ratio had rank 1, and the protein with the lowest ratio had a rank corresponding to the total number of identified proteins. To combine the protein ranks of all four measured replicates, the sum of ranks across replicates was calculated, sorted in descending order and ranked again. The p value for each protein was calculated by comparing its rank sum with the result of 1000 permutations of the list using the RankProd package for R (26). The resulting p values were then corrected for multiple testing using the adaptive false discovery rate (FDR) control method (27), giving the so-called q-values. This was done using the fdrtool R package (28). An FDR rate of 10% was used as the threshold for selecting proteins with significantly changed expression. The lists of proteins sorted by the RankSum were used as input for iterative Group Analysis (29) as described before (30) to analyze the ribosomal proteins.
For all the other comparisons (0 h–1 h; 0 h–4 h; 1 h–4 h for both the CFTR expressing or control strain; 0 h–0 h and 1 h–1 h CFTR expressing versus control strain) only two biological replicates were available, and therefore a different analysis was done. For each of these comparisons the iTRAQ log ratios from the two biological replicates were averaged and the distribution of the values was compared with the distribution of the iTRAQ ratios obtained from the comparison between the two biological controls that should be identical (Control strain 1 h after induction from two different replicate fermentations, supplemental Fig. S2). As described in the results section the threshold for selecting proteins with significantly changed expression was chosen based on this comparison.
RESULTS
CFTR Expression by L. lactis
The cftr gene was fused to the coding sequence for an N-terminal (nHis-CFTR) or C-terminal (CFTR-cHis) His-tag, or both (nHis-CFTR-cHis) and was cloned in plasmid pNZ8048 for expression in L. lactis. Expression of genes from this plasmid is under control of the Nisin A inducible promoter. The cftr containing plasmids were transformed to L. lactis expression strain NZ9000. No mutations or gene rearrangements were observed in the expression plasmid, even after many generations of growth of the transformed strains, indicating that the gene was stable and well tolerated by L. lactis, in contrast to what has been reported for E. coli (31, 32).
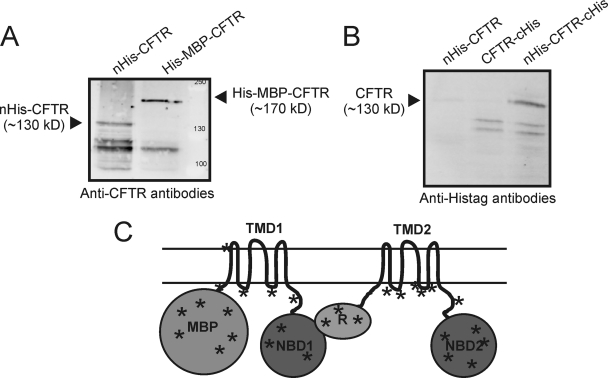
L. lactis strains with the plasmids for nHis-CFTR, CFTR-cHis or nHis-CFTR-cHis were cultivated and cftr expression was induced with Nisin A in the exponential growth phase (OD600 of 0.5). Induction of the expression with Nisin A severely affected growth, and at the time of harvest (2 hours after induction) the cultures of the cftr expression strains had reached much lower cell densities than control cultures (supplemental Fig. S1). Cells were lysed, membranes were isolated, and the proteins in the membrane fraction were separated by SDS-PAGE. CFTR expression was examined by Western blot analysis, using two different monoclonal antibodies, which recognized the engineered His-tags or the native C terminus of the CFTR protein. The nHis-CFTR protein could be detected by both antibodies as a band that migrated at an apparent molecular mass of 130 kDa, showing that both the N terminus (His-tag) and the C terminus (epitope of the CFTR specific antibodies) were present, and thus full-length protein had been produced (Fig. 1A and 1B). For detection of CFTR constructs with C-terminal His-tags (CFTR-cHis and nHis-CFTR-cHis), only the anti-His-tag antibodies could be used, because the His-tag on the C terminus prevented detection of the protein by the CFTR specific antibodies. Full-length CFTR-cHis was not detected, but nHis-CFTR-cHis was detected and migrated at approximately the same apparent molecular weight as nHis-CFTR on SDS-PAGE, which again confirmed that the full-length CFTR had been produced by L. lactis. In addition to the full-length proteins, a number of smaller fragments were detected which are likely to be degradation products (Fig. 1).
Fig. 1.
Expression of human CFTR in L. lactis. A, Membranes of L. lactis expressing His-MBP-CFTR and nHis-CFTR were analyzed by SDS-PAGE/Western blotting. CFTR was detected using anti-CFTR antibodies (clone 24–1, R&D systems). Ten micrograms protein was loaded per lane. B, Membranes of L. lactis expressing nHis-CFTR, CFTRcHis and nHis-CFTR-cHis were analyzed as described above, but now using anti-His-tag antibodies. C, Topology model of His-MBP-CFTR indicating the different domains, and showing positions of the tryptic peptides identified with LC-MS/MS. Peptides derived from all soluble domains of His-MBP-CFTR were found. For a complete list see supplemental Table S2 .
Apparently, the presence of an N-terminal His-tag was necessary for production/detection of full-length CFTR. To investigate further how N-terminal modification affected the production of full-length CFTR, a construct was made with an MBP domain plus a His-tag fused at the N terminus (His-MBP-CFTR). His-MBP-CFTR was detected in L. lactis membranes with the anti-CFTR antibodies and had an apparent molecular weight of 170 kDa (Fig. 1A), as expected for the full-length protein. To confirm that the full-length protein was produced, and to obtain an estimate of the expression levels, His-MBP-CFTR was partially purified. Membranes containing His-MBP-CFTR were solubilized with the detergent DDM and subjected to Ni-Sepharose chromatography. On a Coomassie-stained gel a very faint band corresponding to a protein with an apparent mass of 170 kDa was visible in the elution fraction (not shown). Based on the intensity of the Coomassie stained band we estimated that His-MBP-CFTR represented <0.1% of the proteins in the membrane fraction. The stained 170 kDa protein band was excised from the gel, peptides were generated by trypsin hydrolysis, and the peptides were analyzed by MALDI tandem mass spectrometry. Forty-four peptides were identified covering 28% of the protein sequence and including peptides from both the N-terminal domain (MBP) and the most C-terminally located domain (NBD2), again confirming that L. lactis had produced the full His-MBP-CFTR fusion (Fig. 1C, and supplemental Table S2).
Consequences of CFTR Overexpression
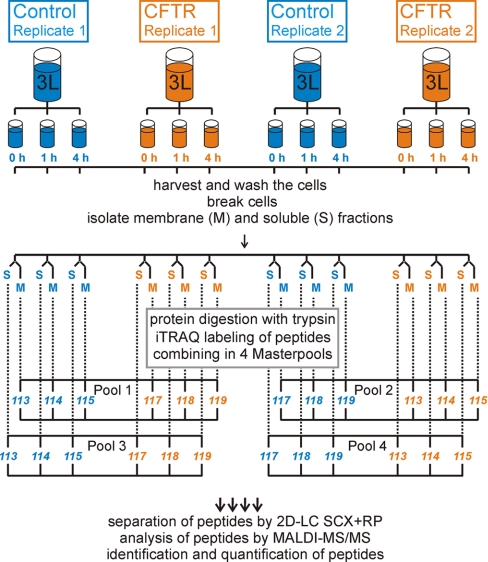
The above experiments show that full-length human CFTR was produced in L. lactis membranes. Although this result is extremely encouraging, the expression levels were too low for functional or structural characterization; in addition expression of CFTR severely affected the growth of L. lactis (supplemental Fig. S1). To investigate the effect of CFTR expression on the physiology of L. lactis, and to identify why L. lactis produced only small amounts of CFTR, a proteomic approach was followed using quantitative mass spectrometry. The experimental setup is outlined in Fig. 2. Two replicate fermentations (Replicate 1 and 2 in Fig. 2) were carried out both of L. lactis containing the expression plasmid for nHis-CFTR-cHis, and of L. lactis containing the empty plasmid pNZ8048. All cultures were grown in fermenters of 3 liter volume, with temperature (30 °C) and pH control (6.5). The inducer Nisin A was added to both the control and the expression strains in the mid-exponential growth phase (OD600 ∼0.5, supplemental Fig. S1). 900 ml of cells was harvested at each of three timepoints: just before the addition of Nisin A (time point 0 h), and at 1 h and 4 h after induction, yielding a total of 12 cell-samples (3 timepoints per fermentation) (Fig. 2 and supplemental Fig. S1).
Fig. 2.
Workflow of the proteomics study. To compare relative protein abundance in the control and CFTR-expression strains, the cells were grown in fermenters under controlled conditions. Each strain was grown twice (biological replicates). To follow the relative changes in expression in the CFTR and the control strain in time, three cell aliquots (900 ml each) were harvested immediately before induction of CFTR-expression, and 1 h and 4 h after induction, resulting in 12 cell samples. The cells were lysed and the cell lysate was fractionated into the membrane (M) und soluble (S) fractions by differential centrifugation. Thus 24 protein samples were obtained from four cultures. Each protein sample was digested with trypsin to create peptide samples and the peptides were labeled with the isobaric iTRAQ reagents. Eight different iTRAQ reagents were used, termed 113, 114, 115, 116, 117, 118, 119, 121 (for the mass of the reporter fragment). Six peptide samples labeled with different reagents (three from the control strain, and three from the CFTR-expression strain) were combined in one “Master pool” (for details see supplemental Fig. S2). The 24 labeled peptide samples were combined in four Master pools. Each Master pool of labeled peptides was subjected to two-dimensional chromatography separation, using off-line Strong Cation Exchanger in combination with the Reversed-phase Chromatography. Separated peptides were collected on a MALDI-target and analyzed by tandem MS/MS. The obtained MS/MS spectra were analyzed by Mascot and ProteinPilot software which provided identification and quantification information for each spectrum. The identified and quantified peptides derived from the same protein were integrated, resulting in identification and quantification information of proteins.
Each of the 12 cell-samples was lysed, and membrane and soluble fractions were isolated (abbreviated as M and S, respectively in Fig. 2), resulting in 24 protein samples. The separation of membrane and soluble fractions was done to facilitate the identification of low-abundance membrane proteins, and to follow the possible redistribution of proteins between the membrane soluble fractions upon overexpression (see Discussion). The membrane and soluble fractions were kept separate during the subsequent analysis.
The 24 protein samples were digested with trypsin, yielding 24 peptide-samples, which were divided into four sets, each containing six different peptide-samples (Fig. 2). Three of these peptide-samples were derived from control cells (corresponding to the three timepoints 0 h, 1 h and 4 h of the same fermentation), and the other three from the CFTR expression cells. This was done separately for the membrane and the soluble fractions, and separately for the replicate fermentations.
Each peptide-sample in the set of six was labeled with a different isotope label, for which isobaric iTRAQ reagents from the 8-plex iTRAQ kit were used, and the six differentially labeled samples were combined into a “Master pool.” The Master-pool was supplemented with two more peptide-samples (labeled with yet two different iTRAQ labels from the 8-plex iTRAQ kit): (1) a “technical control,” which was a peptide sample identical to one of the six peptide-samples already present, but labeled with a different isotope label; (2) a “biological control,” which was related to one of the six peptide-samples already present but obtained from the replicate fermentation. The iTRAQ labeling scheme is shown in supplemental Fig. S2. Different label combinations (label swaps) were used in the different Master pools.
The labeled peptide mixtures in the four Master pools were fractionated using cation exchange and reversed phase chromatography, and the eluting peptides were analyzed by MALDI-MS/MS, which provided both identification and quantification data. The fragmentation spectrum was used to identify each peptide, and the areas of the eight different reporter peaks from the iTRAQ labels were measured for later comparative quantification. The identification and quantification data of different peptides originating from the same protein were integrated, and the resulting protein data from the different replicates were combined, yielding two lists of proteins (744 from the membrane fractions and 688 from the soluble fractions) that were fully quantified using iTRAQ in both replicates (supplemental Tables S3 and S4).
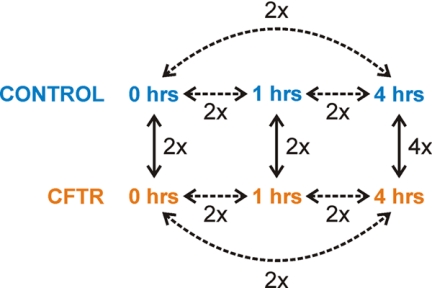
The experimental design allowed for the comparison of the relative protein abundances (ratios of the quantified iTRAQ reporter peaks) for each pair of samples present in the Master-pool. Because there were eight peptide samples in the Master pool, a total of 28 different iTRAQ pairs (ratios) could be calculated (8!/2!(8–2)!) for each protein. Only nine of these ratios were biologically relevant: (a) the changes in protein levels during the time course of the expression (0 h versus 1 h; 1 h versus 4 h; 0 h versus 4 h) for both the control strain and the CFTR expression strain (six iTRAQ ratios in total; Fig. 3, dashed lines); and (b) the differences in protein levels between the control strain and the CFTR expression strain at each of the three time-points (0 h, 1 h, 4 h; three iTRAQ ratios; Fig. 3, solid lines). The technical and biological controls which had been taken along in each 8-plex iTRAQ experiment resulted in two more relevant ratios for control purposes (supplemental Fig. S2).
Fig. 3.
Schematic representation of a Master pool and an outline of the meaningful comparisons of the relative protein abundances. Each Master pool contained peptide samples representing three time points of the control strain and three time points of the CFTR-expression strain (0 h, 1 h, 4 h after induction expression). The expression levels of proteins could be followed as function of the time within the control and CFTR-expression strains (dashed lines). Furthermore, the relative changes in protein abundance could also be calculated between the control and the CFTR-expression strains at three of the time points (solid lines). All the comparisons were based on two independent biological replicates (Fig. 2), indicated by 2×. However, the comparison between the CFTR expressing strain and the control strain at the 4 h time point was repeated two more times in order to obtain four replicate values (4x).
In supplemental Tables S3 and S4, the averaged iTRAQ ratios from the two biological replicate (expressed as logarithms, log10-ratios) for each of the nine biologically relevant comparisons are given for all of the identified proteins. Also the averaged iTRAQ ratios for the two biological controls samples and the technical controls are listed.
Control Experiments: Technical and Biological Replicates
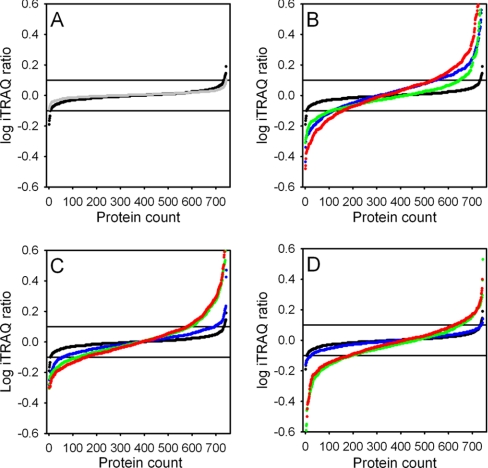
To determine which proteins had changed in abundance, the distributions of iTRAQ log ratios in the nine biologically relevant comparisons (Fig. 3 and supplemental Tables S3 and S4) were evaluated against the distribution in the biological control (Fig. 4). The biological control consisted of two samples from the same condition (1 h of induction; control cells) but from two different fermentations (supplemental Fig. 2, indicated with asterisk) and thus should be identical. Ninety-eight percent of the iTRAQ log-ratios of the biological control fell in the window between –0.1 and 0.1 (Fig. 4A). The distribution of the iTRAQ log-ratios of the biological control was similar to the distribution of the technical control (gray line in Fig. 4A) showing that the biological noise was low. In contrast, the distribution of the iTRAQ log ratios was different when the protein abundances in the samples taken after 1 h and 4 h of induction were compared with the time point 0 h, both in the control and the CFTR-expressing cells, and both in the membrane and soluble fractions (Fig. 4B and 4C for the membrane fraction). The distribution of iTRAQ ratios was also different from the biological control when the protein abundances were compared between CFTR expressing cells and control cells at the timepoints 1 h and 4 h (Fig. 4D). For example, at the 4 h time point (column M in supplemental Tables S3 and S4) ∼40% of the iTRAQ log ratios were higher than 0.1 or lower than –0.1 (red dots in Fig. 4D). In Fig. 5 and supplemental Tables S3 and S4, numbers in green and red indicate iTRAQ log ratios above 0.1 and below –0.1 respectively. These thresholds were used to analyze trends in the data as described below.
Fig. 4.
Distribution of iTRAQ log ratios of the 744 proteins identified in the membrane fractions in the various comparisons (cf Fig. 3). The averaged values of two biological replicates are shown. Horizontal reference lines are drawn at the cutoff values of –0.1 and 0.1. A, Technical (Gray) and biological controls (Black). B, Biological controls (Black), comparisons 1 h–0 h (Blue), 4 h–1 h (Green) and 4 h–0 h (red) for the control cells. C, Biological controls (Black), comparisons 1 h–0 h (Blue), 4 h–1 h (Green) and 4 h–0 h (red) for the CFTR expressing cells. D, Biological controls (Black), comparisons CFTR expressing cells versus control cells at timepoints 0 h (Blue), 1 h (Green), and 4 h (red).
Fig. 5.
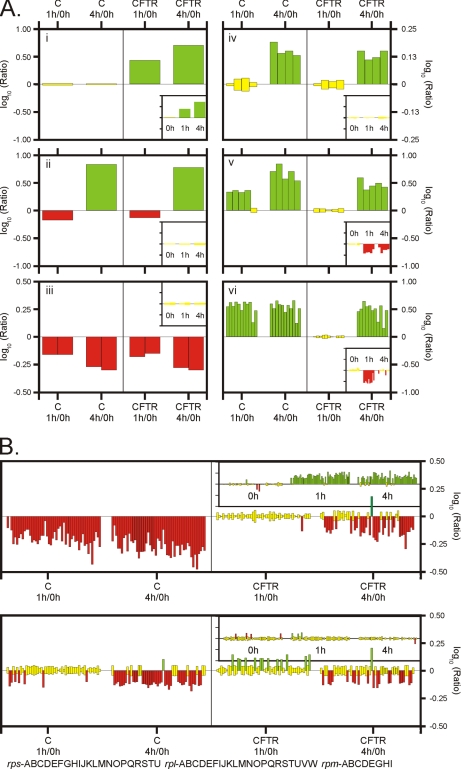
Examples of proteins changing abundance upon induction. Each bar in the panels and insets represents one protein. The height of the bar indicates relative difference in abundance of protein (logarithm of the iTRAQ ratio). On the left-hand side of each panel, the relative changes in protein expression within the control strain upon induction are presented (comparing 0 h and 1 h, and 0 h and 4 h). On the right-hand side of each panel, the relative changes in protein expression in the CFTR-expression strain are presented. The apparent changes in protein abundance between the CFTR- and the control strain (at time point 0 h, 1 h, and 4h) are shown in the insets. Color: red, green and yellow colors indicate significantly decreased, increased and unchanged protein abundance, respectively. A, Panel i: CFTR; Panel ii: RibA (riboflavin biosynthesis protein); Panel iii: Proteins encoded by the fructose operon: FruC (fructose 1-phosphate kinase) and FruR (transcriptional regulator of the fructose operon). Panel iv: Proteins encoded by the pyruvate dehydrogenase operon: PdhA, PdhB and PdhC are the subunits of the pyruvate dehydrogenase complex; PdhD is dihdrolipoamide dehydrogenase. Panel v: Proteins encoded by of the oligopeptide transport system operon: OppA, OppB, OppD and OppF are subunits of the oligopeptide ABC-transporter (OppC was not identified); PepO is the endopeptidase O. Panel vi: Proteins encode by the purin operon. PurC, D, E,F, H, K,L, M, Q, R, S. B, Apparent changes in abundance of ribosomal proteins in the membrane and soluble fractions. Top panel: membrane fraction, bottom panel: soluble fraction. The following ribosomal subunits are shown: rpsABCDEFGHIJKLMNOPQRSTU, rplABCDEFIJKLMNOPQRSTUVW, rpmABCDEGHI.
Time Series: Patterns
We identified several patterns of change as a function of induction time when the samples taken after 1 h and 4 h of induction were compared with the time point 0 h, both in the control and the CFTR-expressing cells, and both in the membrane and soluble fractions (supplemental Tables S3 and S4). When the expression level of a protein increased or decreased in time, the increment or decline could follow several patterns: (1) “gradual,” i.e. it increased or decreased after 1 h of expression when compared with the zero time point, and increased or decreased even more after 4 h of expression (e.g. FruC and FruD, Fig. 5A); (2) “leveling off,” i.e. the change within 1 h was followed by constant levels upon further expression (e.g. the Pur proteins in the control cells, Fig. 5A); (3) “delayed,” i.e. no changes after 1 h followed by a change after 4 h (e.g. the Pur proteins in the CFTR expressing cells or the subunits of the pyruvate dehydrogenase complex in both the control and expression strains, Fig. 5A); (4) “opposite,” i.e. a change in expression after 1 h is followed by an opposite change after 4 h (e.g. RibA in Fig. 5A).
Importantly, the patterns of change in time for different subunits of known complexes, or for different proteins coded by the same operon, were very similar, indicating a high level of consistency in the results. For instance, the four subunits of the pyruvate dehydrogenase complex mentioned above (Pdh proteins), all showed the “delayed” pattern of change (Fig. 5A). Similarly, the subunits of the Opp system, an ABC transporter for peptides, showed the same pattern of expression (“gradual” or “delayed” in the control and expression strains, respectively, Fig. 5A). In addition, PepO, which is coded by the same operon as the opp genes, also displayed the same pattern of changes. Another example of proteins coded by an operon are the Pur proteins, all of which showed the same pattern of change (“leveling off” and “delayed” in the control cells and expression cells, respectively, Fig. 5A).
Control versus CFTR-expressing Cells
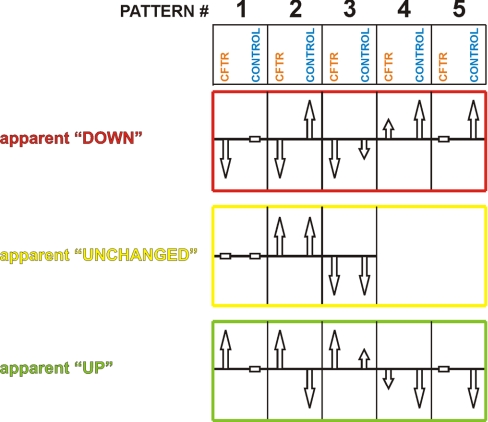
The relative protein abundances between control cells and CFTR expressing cells at the timepoints 1 h and 4 h after induction were also compared. Differences resulted from unequal patterns of time-dependent changes in protein levels between the control and the CFTR-expression cells. At each time point thirteen different combinations of changes in proteins levels in the control and expression cells are possible (Fig. 6). If no differences are observed for the abundance of a protein at a time point, this could be the result of the absence of time-dependent changes in both the control and CFTR-expressing cultures (Fig. 6, middle row, pattern #1), but it also could result from similar extents of up- or down-regulation of in both strains (Fig. 6, middle row, patterns #2 and 3, respectively). An example of pattern #2 is the pyruvate dehydrogenase complex where all subunits were up-regulated to a similar extent in both the control- and the expression-strain, resulting in an apparently unchanged expression when comparing the two strains with each other.
Fig. 6.
Patterns of relative changes. At any given time point, the abundance of a protein in the CFTR expression strain can be the same (yellow), increased (green) or decreased (red) compared with the control strain. These apparent effects depend on the changes in abundance of the proteins as function of the time within the control and CFTR-expression strains. Arrows pointing upward and downward indicate that protein abundance respectively increased or decreased in time. A horizontal bar indicated unchanged levels in time. An apparently unchanged expression level in the CFTR/control comparison is the result of equal changes in the control and CFTR-expression strains, respectively (middle panel). An apparent down-regulation in the CFTR/control comparison at a time point (upper panel) can be caused by decreased protein levels in the CFTR-expression strain and either unchanged levels in the control strain (upper panel, pattern #1), increased levels in the control strain (upper panel, pattern #2), or decreased levels in the control strain but of a lower magnitude than in the CFTR-strain (upper panel, pattern #3). Furthermore, an apparent down-regulation can be the consequence of increased protein expression levels in the control-strain, combined with either unchanged levels in the CFTR-strain (upper panel, pattern #5), or increased levels in the CFTR strain of a lower magnitude than in the control strain (upper panel, pattern #4). An apparent increase in protein abundance between the CFTR- and control strains (bottom panel) is caused by the opposite effects as an apparent decrease.
An apparent elevation of a protein level as a consequence of CFTR-overexpression could be the result of up-regulation in the CFTR-strain and concomitantly either no changes (Fig. 6, bottom row, pattern #1), down-regulation (Fig. 6, #2), or up-regulation to a lesser extent (Fig. 6, #3) in the control strain. An apparent up-regulation also could be a result of down-regulation in the control strain and either no changes in the CFTR expression strain (Fig. 6, #5) or a weaker down-regulation (Fig. 6, #4). An example of pattern #1 is, of course, the CFTR protein itself: its abundance “gradually” increased in time in the CFTR-expressing strain, whereas the protein was absent in the control stain (Fig. 5A). In theory, the iTRAQ ratio, when comparing CFTR abundance in the expression strain and in the control strain, should be infinite (division by zero). However, this was not the case because iTRAQ quantification tends to dampen to ratio of proteins that are of very low abundance in one of the two strains (33). Nonetheless, the CFTR protein had one of the highest iTRAQ ratios found.
Apparent down-regulation at the 1 h and 4 h time-points also could be the results of several scenarios (Fig. 6, upper row). For instance, the proteins of the oligopeptide transport system Opp followed pattern #4: in the control strain the abundances of all subunits were increased after 1 h of expression and further increased after 4 h (“gradual”, Fig. 5A). In contrast, the same proteins had not changed in abundance in the CFTR strain after 1 h, but did increase in abundance after 4 h (“delayed”). So, even though the levels of the Opp proteins increased in time in the CFTR expression strain, they increased at a lower rate than in the control strain, resulting in an apparent decrease of protein abundances when comparing the two strains at any given time point. Another example for the apparent lowering of protein levels as the consequence of CFTR-overexpression was seen for the proteins of the purine metabolism. PurB, PurC, PurD, PurH, PurK, and Purl, were up-regulated in the control cells when comparing time point 1 h with time point 0 h, but remained constant when comparing timepoints 4 h with time point 1 h (“leveling off,” Fig. 5A). In contrast, in the CFTR-expressing cells the protein levels remained constant or slightly increased during the first hour of induction, and further increased during the next 3 h to levels comparable to the control cells (“gradual” or “delayed,” Fig. 5A). Therefore, when comparing the CFTR-expression strain to the control, the relative abundances of these proteins were apparently decreased at 1 h (Fig. 6, upper row, pattern #4 or #5), but were unchanged after 4 h (Fig. 6. middle row, pattern #2).
The data described above, which showed consistent time-dependent changes in protein abundances for the various comparisons, were based on two biological replicates only. Therefore, to improve the confidence of the analyses and to be able to apply more rigorous statistical criteria to find significantly up-or down-regulated proteins we repeated the comparison between CFTR expressing cells versus control cells at the time point 4 h two more times, so as to get four replicates (Fig. 4 and supplemental Fig. S2). Time point 4 h was chosen for the extra replicates, because we had noticed that the relative abundances of almost all proteins (CFTR expression strain versus the control strain) were qualitatively similar at timepoints 1 h or 4 h after induction (down- or up-regulation or no change), but the extent of change (absolute values of the iTRAQ ratios) were generally larger, and thus more reliable, at the 4 h time point. The extra replicates values were obtained essentially in the same way as described in Fig. 2, but in this case the 4plex iTRAQ reagents were used. For the iTRAQ labeling scheme see supplemental Fig. S2 and for the values see supplemental Tables S3, S4 and S5. Seven hundred and nine and 644 proteins were identified and quantified in the membrane and soluble fractions in all four replicates of the 4 h time point. To find proteins with significantly different abundances the RankSum and FDR algorithms were used as described in the methods section. At the 4 h time point, 147 proteins had significantly changed in abundance in the membrane fraction (70 up and 77 down) and 202 proteins in the soluble fraction (104 up and 98 down) (FDR-corrected p values <0.1; supplemental Tables S3 and S4).
Only the proteins that were found to be significantly changed in abundance at the 4 h time point using the RankSum/FDR criterion have been included in the discussion on the physiology below. The observed time-dependent changes in the protein abundances based on two biological replicates (Figs. 3, 4, and 5 and supplemental Tables S3 and S4) have been used in the discussion only for those proteins that had significantly changed in abundance according to the stringent analysis based on four replicates.
DISCUSSION
Production of sufficient amounts of well-folded membrane protein is a major bottleneck in membrane protein research. CFTR is no exception, and biochemical/biophysical studies on the protein are hampered by low yields of correctly folded and stable protein. Here we have used the prokaryotic expression host L. lactis to express full-length human CFTR. To the best of our knowledge this is the first report of bacterial expression of full-length human CFTR. Although the results are encouraging, the yields of CFTR were too low (<0.1% of membrane protein) for functional/structural characterization. In addition, growth of the cells was severely compromised when expression of CFTR was induced, resulting in low biomass yield and indicating toxicity to the cell. Low yields and growth arrest have been observed upon expression of numerous human membrane proteins in L. lactis, also for proteins that could be assayed for function. For instance the human KDEL receptor was shown to be functional in the membrane of L. lactis by a ligand binding assay, despite low levels of expression and growth arrest (5). The ligand binding assay for the low-abundant KDEL receptor was possible because a high-affinity radiolabeled ligand was available. Such ligands are not available for CFTR.
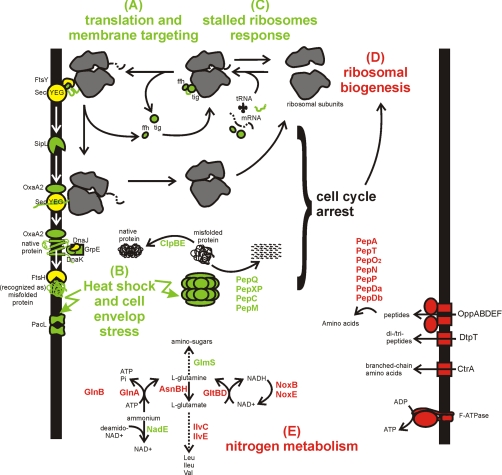
In an attempt to understand why the CFTR yields were low, and possibly to remedy the expression bottlenecks, we used quantitative proteomics to characterize the response of L. lactis to expressing CFTR in its plasma membrane. In the combined membrane and soluble fractions we identified and quantified a total of 846 proteins, representing 35% of the predicted L. lactis proteome. Among the identified proteins were 163 integral membrane proteins, which were strongly enriched in the isolated membrane fractions. The large number of identified proteins allows reliable analysis of the physiological responses of L. lactis to the expression of CFTR. The major responses are summarized in Fig. 7, and will be discussed below. To our knowledge this is the first study in which the stress response of L. lactis upon membrane protein production is systematically analyzed.
Fig. 7.
The physiological responses of the bacterium Lactococcus lactis to the production of the human CFTR. Red and green colors indicate proteins that are lower and higher in abundance respectively in the CFTR expressing strain compared with the control strain at time point 4 h after induction. Yellow proteins have the same abundance. A, Nascent chains of membrane proteins emerging from the ribosome are recognized by trigger factor (tig) and the signal recognition particle (containing the ffh protein) and the complex is targeted to the membrane via the receptor FtsY. The nascent chain is co-translationally inserted into the membrane upon docking on the SecYEG translocon. B, The proper folding of the protein is assisted by membrane-integral (OxaA) and/or cytosolic (DnaJK) chaperones. Misfolded proteins trigger heat-shock and cell envelope stress responses. C, Upon production of CFTR, the stalled ribosomes response is triggered. Also proteins involved in general and oxidative stress responses are up-regulated in response to CFTR production (not shown). D, CFTR production leads to growth arrest which correlates with the reduced ribosomal biogenesis. E, Proteins that become de-repressed in response to limited nitrogen were collectively lower in abundance in the CFTR expressing strain than in the control strian (Opp, DtpT, CtrA, Pep proteases, IlvC, IlvE, GlnA, GlnB, and GltBD). The reduced abundance was caused by a strong up-regulation of these proteins in the control strain.
Stress from Protein Misfolding
For the majority of the proteins that had higher abundance in the CFTR-expression strain than in the control strain at the 4 h time point (supplemental Table S3 and S4), the abundance had increased as a function of time in the expression strain, but remained unchanged in the control strain (Fig. 6, bottom row, pattern #1). Almost all of the proteins following this pattern were found to be stress related. A striking example is PacL, a putative cation transporting P-type ATPase (34). When comparing the expression and control strains at the 4 h time point, PacL displayed the highest level of up-regulation (highest iTRAQ ratio) of all identified proteins (supplemental Table S3 and S4). The biological role of PacL is not known, but expression of the protein is under control of the CesSR two-component system, which orchestrates the response of L. lactis to cell envelope stress caused by, for instance, anti-bacterial peptides (35, 36). CesSR regulates the expression of numerous proteins, many of which were also affected in the CFTR expression strain: the membrane protein chaperone/insertase OxaA2 (YidC homologue), the peptidyl-prolyl isomerase PpiB, CesR itself, and a number of proteins of unknown function were all up-regulated, indicating cell envelope stress.
A second group of stress related proteins that was collectively up-regulated comprised proteins of the heat shock response (37). The chaperones DnaK, GrpE, GroEL, and GroES were all up-regulated in the CFTR expression strain, indicating a response to misfolded proteins. Similarly, ClpB and ClpE, involved in the degradation of misfolded proteins (38), were up-regulated but, surprisingly, DnaJ was not, and its expression pattern was very different from the other heat-shock related proteins. In contrast to the heat shock proteins, the cold shock protein CspE decreased in abundance in the CFTR expression cells.
Other proteins that have been associated with stress responses also increased in abundance upon CFTR expression, including the major stress regulator SpxA, NAD synthase NadE, and endo-1,4-β-xylanase D XynD (involved in cell wall stress). In addition, a number of universal stress proteins, hydrolases and peptidases (PepQ, PepXP, PepC, and PepM) were up-regulated, indicating that these proteins may also be stress-related.
Two proteins that have been reported to be involved in the oxidative stress response (thioredoxin TrxH and thioredoxin reductase TrxB1 (39, 40)) were also up-regulated in the CFTR-expressing strain, whereas their expression levels were nearly unchanged in the control sample. In contrast, many other proteins involved in the oxidative stress response were very differently regulated. In control cells numerous proteins involved in the response to oxidative stress were up-regulated at the 1 h and 4 h time-points, including SodA (superoxide dismutase), NoxB (NADH dehydrogenase), NoxE (NADH oxidase), Rex (redox sensing transcriptional regulator), and the manganese transporters MntT and MtsAB (37, 41). In the CFTR expression cells these proteins were not changed, up-regulated to a lesser extent, or downregulated, resulting in an apparent strong down-regulation when comparing the CFTR expression cells with the control cells (Fig. 6, upper row, patterns #2, 4 or 5). This finding suggests that in the late-exponential and stationary growth phases L. lactis normally up-regulates the proteins involved in oxidative stress, even in the absence of excess oxygen (the cultures were grown semi-anaerobically), but that this response is largely absent in the CFTR expression cells.
To sum up, multiple stress responses were observed in L. lactis upon CFTR expression, the most obvious of which are the responses to heat shock (misfolded protein) and cell envelope stress. Similar responses to the expression of (nonnative) proteins have also been observed in other organisms (e.g. E. coli or Bacillus subtilis (42, 43)). The stress experienced by L. lactis could be related directly to the presence of CFTR, i.e. CFTR misfolds, and the misfolded protein affects the integrity of the membrane leading to cell envelope stress. Alternatively, the stress responses may be an indirect consequence of CFTR production, similar to the cellular responses that have been observed in E. coli as a consequence of membrane protein overexpression. Overexpression of membrane proteins in E. coli caused overloading of the membrane proteins insertion machinery (the Sec machinery), resulting in misfolding/aggregation of endogenous proteins targeted for secretion (42). To distinguish between the two possibilities, we examined the effects of CFTR expression on the translation and targeting and membrane insertion machinery.
Translation and Targeting
The majority of integral membrane proteins are targeted to the Sec translocase as ribosome-bound nascent chains, which are cotranslationally inserted into the membrane upon docking on the Sec translocon (44–46). Higher rates of membrane protein translation may result in a higher fraction of ribosomes associated with the membrane (47), e.g. induced synthesis of bacteriorohodopsin increases the amounts of ribosomes isolated with the membrane fraction (48). In E. coli an increased fraction of membrane bound ribosomes caused by membrane protein overexpression resulted in overloading of the Sec insertion machinery, and consequently toxicity to the cell (42). In L. lactis the situation is very different: The abundance of ribosomal proteins in the membrane fraction decreased as a function of time in both the control cells and in the CFTR-expressing cells, albeit at different pace (Fig. 5B). Also in the soluble fraction the amounts of ribosomal proteins went down in time, both in the control and CFTR-expressing cells (Fig. 5B). Thus, the absolute amounts of ribosomes decreased, both in the control and in the CFTR expression cells, and both in the membrane and in the soluble fractions. In contrast, the abundance of translocon channel SecY, as well as SecA, the motor protein for protein secretion, remained unchanged in the membrane fraction. As the majority of integral membrane proteins are targeted to the Sec translocase as ribosome-bound nascent chain/Ffh/FtsY complex (49), this result indicates that it is very unlikely that the Sec translocon became overloaded in L. lactis upon CFTR expression. The unchanged level of SecY in itself also points at the absence of jammed translocons, because jammed translocons are degraded rapidly (50). Therefore, we tentatively conclude that the misfolded protein response is not triggered by secondary effects (Sec overloading), but rather directly by CFTR expression.
When the abundances of ribosomal protein were compared between the control and expression cells at the 1 and 4 h time point, we found that many ribosomal proteins had apparently increased in abundance in the membrane fraction of the CFTR expressing cells compared with the control cells whereas their levels in the soluble fraction remained unchanged (Fig. 5B). The apparent up-regulation of ribosomal proteins in the membrane fraction was caused by a stronger down-regulation in the control cells in comparison to the CFTR-expressing cells (Fig. 6, top row, pattern #4), and the unchanged levels of ribosomal proteins in the soluble fraction resulted from their down-regulation in the control and CFTR-expressing cells to the same extent (Fig. 6, middle row, pattern #2). Statistical analysis using iterative Group Analysis (iGA, (30)) confirmed that almost all ribosomal subunits cluster among the proteins with the highest iTRAQ ratios in the membrane fraction but not in the soluble fraction. These results show that the distribution of ribosomes over the membrane and soluble fractions becomes different in the control and CFTR expressing cells as a function of induction time. The redistribution takes place predominantly in the control cells, where the fraction of membrane bound ribosomes decreases to a much larger extent than in soluble fraction. In the CFTR-expressing cells the distribution remains approximately the same. This finding is surprising, and shows that normal (control) L. lactis cells entering the late exponential or stationary growth phase specifically decrease the amounts of membrane bound ribosomes, perhaps indicating a lower need for integral membrane and secreted proteins.
The signal recognition particle protein Ffh increased in abundance at the membrane on CFTR expression. Because the receptor FtsY remained unchanged, and membrane associated ribosomal proteins decreased upon expression, a possible explanation for the increased Ffh abundance is that the ribosome-nascent chain complexes stayed attached for longer with the signal recognition particle after targeting to the membrane. Increased association times at the membrane could be indicative of hindered translation, in which case the ribosomes, mRNA and other components of the translation machinery have to be recycled from the stalled ribosomes. In particular peptidyl-tRNAs must be degraded because they are toxic to the cell (51, 52). Several proteins involved in ribosome recycling, including Pth (peptidyl tRNA hydrolase), Frr (ribosomal recycling factor), InfC (initiation factor), and RelA (GTP pyrophosphokinase), increased in abundance upon CFTR expression (53–58). We therefore tentatively conclude that CFTR expression leads to a higher extent of stalled ribosomes, which necessitates their rescue.
Ribosomal biogenesis was reduced when comparing the CFTR expressing cells with the control cells (Fig. 6, top row, pattern #5), as indicated by apparent down-regulation of almost all polypeptides of the ribosomal RNA methyltransferase, the ribosomal biogenesis GTPases Era and llmg_1175, the ribosome maturation factor RimM and the ribonuclease Rnc, which is involved into rRNA processing together with the ribosome maturation factor RimM (59–61). The apparent reduction of ribosome biogenesis correlates with the observed growth stasis (Fig. 7).
Metabolism
Nitrogen Metabolism
CFTR expression strongly affected nitrogen metabolism. CodY is a global repressor of genes that become expressed only when nitrogen sources are limiting (62, 63). Proteins of which the expression is regulated by CodY were collectively lower in abundance in the CFTR expression strain than in the control strain, indicative of higher intracellular levels of branched-chain amino acids (corepressors of CodY) and presumably higher levels of amino acids in general. Among the proteins with the strongest down-regulation were the subunits of the oligopeptide transport system Opp (Fig. 5A), the peptidase PepO, enzymes of the brached-chain amino acid synthetic pathway (IlvC, IlvE), glutamate synthase GltBD, the branched chain amino acid transporter CtrA, and asparagine synthase AsnB. Without exception, the apparent down-regulation of these proteins was caused by strong up-regulation in the control cells, which did not occur in the CFTR expressing cells (Fig. 6, upper row, patterns #4 and #5, see also Fig. 5A for the Opp proteins). Also other proteins regulated by nitrogen limitation were apparently down-regulated, such as the nitrogen regulatory protein GlnB (P-II), the glutamine synthetase regulator GlnR, glutamine synthetase GlnA, the di- and tripeptide transporter DtpT, and the numerous peptidases involved in the breakdown of imported peptides (PepA, PepT, PepO2, PepN, PepP, PepDA, and PepDB). The data indicates that the control cells become starved for nitrogen at timepoints 1 h and 4 h, but that the CFTR expressing cells experience no shortage of nitrogen supply. This is consistent with the fast growth of the control cells (supplemental Fig. 1), which deplete the available nitrogen compounds for biomass production, and the growth arrest of the CFTR expressing cells, which lowers the need for nitrogen compounds. It is remarkable that the strong up-regulation in the control cells is noticeable already after 1 h of induction when the cells still appear to be growing exponentially (albeit in the late exponential phase). Clearly L. lactis begins to experience nitrogen shortage in this phase already.
Sugar Metabolism
The effects of CFTR expression on the sugar metabolism are not as clear-cut as in the case of the nitrogen metabolism, possibly because many of the enzymes involved in glycolysis, and pathways downstream of pyruvate are not primarily regulated at the level of expression, but rather by allosteric and feedback regulation using molecules that sample the energetic status (such as the NADH/NAD+ and ADT/ADP ratio). Nonetheless there are indications that the control cells become starved for sugar, as opposed to the CFTR expression cells. For example, the PTS transport systems for alternative sugars (cellobiose and mannose) become more abundant as function of time in the control cells, but not in the expression cells. Similarly, AdhE (alcohol acetaldehyde dehydrogenase) increases in abundance in the control cells only, indicating that these cells are switching to mixed acid fermentation to produce more ATP. Shortage of ATP is also indicated by up-regulation of the F-type ATPase in the control cells only.
Taken together the data indicates that the direct effects of CFTR expression on metabolism are minor. The main difference between the control cells and the expression cells is that the control cells continue to grow rapidly after induction, thereby depleting their sugar and nitrogen supplies, whereas the expressing cells stop growing, and, initially, do not experience energy and nitrogen shortage. In E. coli the situation is very different: Upon overexpression of membrane proteins the Sec translocon becomes overloaded, which negatively affects the levels of respiratory enzymes in the membrane, and leads to activation of the Arc two-component system, which mediates adaptive responses to changing respiratory states. The acetate-phosphotransacetylase pathway for ATP production was induced and the tricarboxylic acid cycle was down-regulated. E. coli thus switches to a less efficient energy metabolism, which strongly affects biomass production (42).
Outlook
The different responses of E. coli and L. lactis to stress caused by membrane protein expression imply that different strategies must be used to remedy expression bottlenecks. In E. coli, careful tuning of the expression levels (to prevent overloading of the Sec machinery) has been used successfully to optimize expression levels (64). What can be done to improve the expression of CFTR in L. lactis? The answer hinges on two possibilities. 1) It is possible that CFTR was folded properly upon expression in L. lactis, but that the protein was recognized as a misfolded protein (because it is non-endogenous). In that case the answer could be to trick L. lactis and force it not to use the stress responses and thus to prevent growth arrest, e.g. by deleting heat shock proteins (65). 2) If CFTR was not properly folded, then the protein could be helped to fold properly, e.g. by including (human) chaperones, or by mutagenesis, such as changing all the phophorylation sites in the R-domain into negatively charged residues. Chaperone co-expression has been used with mixed success to improve heterologous expression in E. coli (66–68). Also, the production of recombinant proteins under thermal stress could be improved by co-expression of GroESL in E. coli (69). The fact that the expression of CFTR improved at higher temperatures (supplemental Fig. S3) indeed suggests that the up-regulation of heat shock proteins helps L. lactis to deal better with the expression stress. Again, this is very different from what is normally observed in E. coli, where lower temperatures, and thus lower expression rates, usually improve production, possibly because overloading of the Sec machinery is prevented.
Acknowledgments
We thank Peter Maloney (Johns Hopkins Medical School), Mohabir Ramjeesingh (University of Toronto) and Bert Poolman (University of Groningen) for their helpful comments.
Footnotes
* This work was supported by the Netherlands Organisation for Scientific Research NWO (VIDI fellowships to DJS and RB), the Netherlands Proteomics Centre (NPC), the Cystic Fibrosis Foundation Therapeutics, Inc. (CFFT), the European Union (EDICT program), the EFRO (Europees Fonds voor Regionale Ontwikkeling) and the Province of Groningen (IAG-2).
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S6.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S6.
1 The abbreviations used are:
- CFTR
- cystic fibrosis transmembrane conductance regulator
- DDM
- n-dodecyl-β-D-maltoside
- iTRAQ
- isobaric tag for relative and absolute quantification
- MALDI
- matrix assisted laser desorption/ionisation
- MBP
- maltose binding protein
- MMTS
- methyl methanethiosulfonate
- PMSF
- phenyl methanesulphonyl fluoride
- TEAB
- triethylammonium bicarbonate buffer
- TFA
- trifluoroacetic acid
- TOF
- time-of-flight.
REFERENCES
- 1. Rommens J. M., Iannuzzi M. C., Kerem B., Drumm M. L., Melmer G., Dean M., Rozmahel R., Cole J. L., Kennedy D., Hidaka N., Zsiga M., Buchwald M., Riordan J. R., Lap C. T., Collins F. S. (1989) Identification of the cystic fibrosis gene: chromosome walking and jumping. Science 245, 1059–1065 [DOI] [PubMed] [Google Scholar]
- 2. Kerem B., Rommens J. M., Buchanan J. A., Markiewicz D., Cox T. K., Chakravarti A., Buchwald M., Tsui L. C. (1989) Identification of the cystic fibrosis gene: genetic analysis. Science 245, 1073–1080 [DOI] [PubMed] [Google Scholar]
- 3. Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L., et al. (1989) Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 4. Surade S., Klein M., Stolt-Bergner P. C., Muenke C., Roy A., Michel H. (2006) Comparative analysis and “expression space” coverage of the production of prokaryotic membrane proteins for structural genomics. Protein Sci. 15, 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kunji E. R., Slotboom D. J., Poolman B. (2003) Lactococcus lactis as host for overproduction of functional membrane proteins. Biochim. Biophys. Acta. 1610, 97–108 [DOI] [PubMed] [Google Scholar]
- 6. Tate C. G., Haase J., Baker C., Boorsma M., Magnani F., Vallis Y., Williams D. C. (2003) Comparison of seven different heterologous protein expression systems for the production of the serotonin transporter. Biochim. Biophys. Acta 1610, 141–153 [DOI] [PubMed] [Google Scholar]
- 7. Tate C. G., Grisshammer R. (1996) Heterologous expression of G-protein-coupled receptors. Trends Biotechnol. 14, 426–430 [DOI] [PubMed] [Google Scholar]
- 8. Grisshammer R., Tate C. G. (1995) Overexpression of integral membrane proteins for structural studies. Q. Rev. Biophys. 28, 315–422 [DOI] [PubMed] [Google Scholar]
- 9. Cereghino J. L., Cregg J. M. (2000) Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 24, 45–66 [DOI] [PubMed] [Google Scholar]
- 10. Bonander N., Bill R. M. (2009) Relieving the first bottleneck in the drug discovery pipeline: using array technologies to rationalize membrane protein production. Expert Rev. Proteomics 6, 501–505 [DOI] [PubMed] [Google Scholar]
- 11. Zweers J. C., Wiegert T., van Dijl J. M. (2009) Stress-responsive systems set specific limits to the overproduction of membrane proteins in Bacillus subtilis. Appl. Environ. Microbiol. 75, 7356–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Ruyter P. G., Kuipers O. P., de Vos W. M. (1996) Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62, 3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frelet-Barrand A., Boutigny S., Moyet L., Deniaud A., Seigneurin-Berny D., Salvi D., Bernaudat F., Richaud P., Pebay-Peyroula E., Joyard J., Rolland N. Lactococcus lactis, an alternative system for functional expression of peripheral and intrinsic Arabidopsis membrane proteins. PLoS One 5, e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kunji E. R., Chan K. W., Slotboom D. J., Floyd S., O'Connor R., Monné M. (2005) Eukaryotic membrane protein overproduction in Lactococcus lactis. Curr. Opin. Biotechnol. 16, 546–551 [DOI] [PubMed] [Google Scholar]
- 15. Monné M., Chan K. W., Slotboom D. J., Kunji E. R. (2005) Functional expression of eukaryotic membrane proteins in Lactococcus lactis. Protein Sci. 14, 3048–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monné M., Robinson A. J., Boes C., Harbour M. E., Fearnley I. M., Kunji E. R. (2007) The mimivirus genome encodes a mitochondrial carrier that transports dATP and dTTP. J. Virol. 81, 3181–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quick M., Javitch J. A. (2007) Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc. Natl. Acad. Sci. U.S.A. 104, 3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luirink J., Samuelsson T., de Gier J. W. (2001) YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 501, 1–5 [DOI] [PubMed] [Google Scholar]
- 19. Zweers J. C., Barák I., Becher D., Driessen A. J., Hecker M., Kontinen V. P., Saller M. J., Vavrová L., van Dijl J. M. (2008) Towards the development of Bacillus subtilis as a cell factory for membrane proteins and protein complexes. Microb. Cell Fact. 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Funes S., Hasona A., Bauerschmitt H., Grubbauer C., Kauff F., Collins R., Crowley P. J., Palmer S. R., Brady L. J., Herrmann J. M. (2009) Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc. Natl. Acad. Sci. U.S.A. 106, 6656–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Geertsma E. R., Poolman B. (2007) High-throughput cloning and expression in recalcitrant bacteria. Nat. Methods 4, 705–707 [DOI] [PubMed] [Google Scholar]
- 22. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 23. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 24. Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Letts. 573, 83–92 [DOI] [PubMed] [Google Scholar]
- 25. Breitling R., Herzyk P. (2005) Rank-based methods as a non-parametric alternative of the T-statistic for the analysis of biological microarray data. J. Bioinform. Comput. Biol. 3, 1171–1189 [DOI] [PubMed] [Google Scholar]
- 26. Hong F., Breitling R., McEntee C. W., Wittner B. S., Nemhauser J. L., Chory J. (2006) RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22, 2825–2827 [DOI] [PubMed] [Google Scholar]
- 27. Benjamini Y., Hochberg Y. (2000) On the adaptive control of the false discovery fate in multiple testing with independent statistics. J. Educ. Behav. Stat. 25, 60–83 [Google Scholar]
- 28. Strimmer K. (2008) fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24, 1461–1462 [DOI] [PubMed] [Google Scholar]
- 29. Breitling R., Amtmann A., Herzyk P. (2004) Iterative Group Analysis (iGA): A simple tool to enhance sensitivity and facilitate interpretation of microarray experiments. BMC Bioinformatics 5, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wiederhold E., Gandhi T., Permentier H. P., Breitling R., Poolman B., Slotboom D. J. (2009) The yeast vacuolar membrane proteome. Mol. Cell. Proteomics 8, 380–392 [DOI] [PubMed] [Google Scholar]
- 31. Drumm M. L., Pope H. A., Cliff W. H., Rommens J. M., Marvin S. A., Tsui L. C., Collins F. S., Frizzell R. A., Wilson J. M. (1990) Correction of the cystic fibrosis defect in vitro by retrovirus-mediated gene transfer. Cell 62, 1227–1233 [DOI] [PubMed] [Google Scholar]
- 32. Gregory R. J., Cheng S. H., Rich D. P., Marshall J., Paul S., Hehir K., Ostedgaard L., Klinger K. W., Welsh M. J., Smith A. E. (1990) Expression and characterization of the cystic fibrosis transmembrane conductance regulator. Nature 347, 382–386 [DOI] [PubMed] [Google Scholar]
- 33. Ow S. Y., Salim M., Noirel J., Evans C., Rehman I., Wright P. C. (2009) iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly”. J. Proteome Res. 8, 5347–5355 [DOI] [PubMed] [Google Scholar]
- 34. Berkelman T., Garret-Engele P., Hoffman N. E. (1994) The pacL gene of Synechococcus sp. strain PCC 7942 encodes a Ca(2+)-transporting ATPase. J. Bacteriol. 176, 4430–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martínez B., Zomer A. L., Rodríguez A., Kok J., Kuipers O. P. (2007) Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the Lactococcal two-component system CesSR. Mol. Microbiol. 64, 473–486 [DOI] [PubMed] [Google Scholar]
- 36. Roces C., Campelo A. B., Veiga P., Pinto J. P., Rodriguez A., Martinez B. (2009) Contribution of the CesR-regulated genes llmg0169 and llmg2164–2163 to Lactococcus lactis fitness. Int. J. Food Microbiol. 133, 279–285 [DOI] [PubMed] [Google Scholar]
- 37. van de Guchte M., Serror P., Chervaux C., Smokvina T., Ehrlich S. D., Maguin E. (2002) Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82, 187–216 [PubMed] [Google Scholar]
- 38. Ingmer H., Vogensen F. K., Hammer K., Kilstrup M. (1999) Disruption and Analysis of the clpB, clpC, and clpE Genes in Lactococcus lactis: ClpE, a New Clp Family in Gram-Positive Bacteria. J. Bacteriol. 181, 2075–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prieto-Alamo M. J., Jurado J., Gallardo-Madueno R., Monje-Casas F., Holmgren A., Pueyo C. (2000) Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 275, 13398–13405 [DOI] [PubMed] [Google Scholar]
- 40. Jobin M. P., Garmyn D., Diviès C., Guzzo J. (1999) Expression of the Oenococcus oeni trxA gene is induced by hydrogen peroxide and heat shock. Microbiology 145, 1245–1251 [DOI] [PubMed] [Google Scholar]
- 41. Miyoshi A., Rochat T., Gratadoux J. J., Le Loir Y., Oliveira S. C., Langella P., Azevedo V. (2003) Oxidative stress in Lactococcus lactis. Gen. Mol. Res. 2, 348–359 [PubMed] [Google Scholar]
- 42. Wagner S., Baars L., Ytterberg A. J., Klussmeier A., Wagner C. S., Nord O., Nygren P. A., van Wijk K. J., de Gier J. W. (2007) Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell. Proteomics 6, 1527–1550 [DOI] [PubMed] [Google Scholar]
- 43. Mogk A., Völker A., Engelmann S., Hecker M., Schumann W., Völker U. (1998) Nonnative Proteins Induce Expression of the Bacillus subtilis CIRCE Regulon. J. Bacteriol. 180, 2895–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Valent Q. A., Scotti P. A., High S., de Gier J. W., von Heijne G., Lentzen G., Wintermeyer W., Oudega B., Luirink J. (1998) The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17, 2504–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Gier J. W., Luirink J. (2001) Biogenesis of inner membrane proteins in Escherichia coli. Mol. Microbiol. 40, 314–322 [DOI] [PubMed] [Google Scholar]
- 46. Urbanus M. L., Scotti P. A., Froderberg L., Saaf A., de Gier J. W., Brunner J., Samuelson J. C., Dalbey R. E., Oudega B., Luirink J. (2001) Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bisle B., Schmidt A., Scheibe B., Klein C., Tebbe A., Kellermann J., Siedler F., Pfeiffer F., Lottspeich F., Oesterhelt D. (2006) Quantitative Profiling of the Membrane Proteome in a Halophilic Archaeon. Mol. Cell. Proteomics 5, 1543–1558 [DOI] [PubMed] [Google Scholar]
- 48. Gropp R., Gropp F., Betlach M. C. (1992) Association of the halobacterial 7S RNA to the polysome correlates with expression of the membrane protein bacterioopsin. Proc. Natl. Acad. Sci. U.S.A. 89, 1204–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Luirink J., von Heijne G., Houben E., de Gier J. W. (2005) Biogenesis of Inner Membrane Proteins in Escherichia coli. Ann. Rev. Microbiol. 59, 329–355 [DOI] [PubMed] [Google Scholar]
- 50. van Stelten J., Silva F., Belin D., Silhavy T. J. (2009) Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science 325, 753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Menninger J. R. (1979) Accumulation of peptidyl tRNA is lethal to Escherichia coli. J. Bacteriol. 137, 694–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Atherly A. G., Menninger J. R. (1972) Mutant E. coli strain with temperature sensitive peptidyl-transfer RNA hydrolase. Nat. New Biol. 240, 245–246 [DOI] [PubMed] [Google Scholar]
- 53. Heurgué-Hamard V., Karimi R., Mora L., MacDougall J., Leboeuf C., Grentzmann G., Ehrenberg M., Buckingham R. H. (1998) Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J. 17, 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Karimi R., Pavlov M. Y., Heurgué-Hamard V., Buckingham R. H., Ehrenberg M. (1998) Initiation factors IF1 and IF2 synergistically remove peptidyl-tRNAs with short polypeptides from the P-site of translating Escherichia coli ribosomes. J. Mol. Biol. 281, 241–252 [DOI] [PubMed] [Google Scholar]
- 55. Menninger J. R., Caplan A. B., Gingrich P. K., Atherly A. G. (1983) Tests of the ribosome editor hypothesis. II. Relaxed (relA) and stringent (relA+) E. coli differ in rates of dissociation of peptidyl-tRNA from ribosomes. Mol. Gen. Genet. 190, 215–221 [DOI] [PubMed] [Google Scholar]
- 56. Rao A. R., Varshney U. (2001) Specific interaction between the ribosome recycling factor and the elongation factor G from Mycobacterium tuberculosis mediates peptidyl-tRNA release and ribosome recycling in Escherichia coli. EMBO J. 20, 2977–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kossel H., RajBhandary U. L. (1968) Studies on polynucleotides. LXXXVI. Enzymic hydrolysis of N-acylaminoacyl-transfer RNA. J. Mol. Biol. 35, 539–560 [DOI] [PubMed] [Google Scholar]
- 58. Singh N. S., Varshney U. (2004) A physiological connection between tmRNA and peptidyl-tRNA hydrolase functions in Escherichia coli. Nucleic Acids Res. 32, 6028–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Comartin D. J., Brown E. D. (2006) Non-ribosomal factors in ribosome subunit assembly are emerging targets for new antibacterial drugs. Curr. Opin. Pharmacol. 6, 453–458 [DOI] [PubMed] [Google Scholar]
- 60. Dunn J. J., Studier F. W. (1973) T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc. Natl. Acad. Sci. U.S.A. 70, 3296–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Drider D., Bolotine A., Renault P., Prevost H. (2002) Functional study of Lactococcus lactis RNase III in Escherichia coli. Plasmid 47, 246–250 [DOI] [PubMed] [Google Scholar]
- 62. den Hengst C. D., van Hijum S. A., Geurts J. M., Nauta A., Kok J., Kuipers O. P. (2005) The Lactococcus lactis CodY Regulon: Identification of A Conserved Cis-Regulatory Element. J. Biol. Chem. 280, 34332–34342 [DOI] [PubMed] [Google Scholar]
- 63. Guédon E., Sperandio B., Pons N., Ehrlich S. D., Renault P. (2005) Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151, 3895–3909 [DOI] [PubMed] [Google Scholar]
- 64. Wagner S., Klepsch M. M., Schlegel S., Appel A., Draheim R., Tarry M., Högbom M., van Wijk K. J., Slotboom D. J., Persson J. O., de Gier J. W. (2008) Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. U.S.A. 105, 14371–14376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Skretas G., Georgiou G. (2009) Genetic analysis of G protein-coupled receptor expression in Escherichia coli: inhibitory role of DnaJ on the membrane integration of the human central cannabinoid receptor. Biotechnol. Bioeng. 102, 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen Y., Song J., Sui S. F., Wang D. N. (2003) DnaK and DnaJ facilitated the folding process and reduced inclusion body formation of magnesium transporter CorA overexpressed in Escherichia coli. Protein Exp. Purif. 32, 221–231 [DOI] [PubMed] [Google Scholar]
- 67. Link A. J., Skretas G., Strauch E. M., Chari N. S., Georgiou G. (2008) Efficient production of membrane-integrated and detergent-soluble G protein-coupled receptors in Escherichia coli. Protein Sci. 17, 1857–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kolaj O., Spada S., Robin S., Wall J. G. (2009) Use of folding modulators to improve heterologous protein production in Escherichia coli. Microbial Cell Factories 8, 9–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim S. Y., Ayyadurai N., Heo M. A., Park S., Jeong Y. J., Lee S.G. (2009) Improving the Productivity of Recombinant Protein in Escherichia coli Under Thermal Stress by Coexpressing GroELS Chaperone System. J. Microbiol. Biotechnol. 19, 72–77 [PubMed] [Google Scholar]