Abstract
Hepatocellular carcinoma (HCC) is a common malignancy in the world with high morbidity and mortality rate. Identification of novel biomarkers in HCC remains impeded primarily because of the heterogeneity of the disease in clinical presentations as well as the pathophysiological variations derived from underlying conditions such as cirrhosis and steatohepatitis. The aim of this study is to search for potential metabolite biomarkers of human HCC using serum and urine metabolomics approach. Sera and urine samples were collected from patients with HCC (n = 82), benign liver tumor patients (n = 24), and healthy controls (n = 71). Metabolite profiling was performed by gas chromatography time-of-flight mass spectrometry and ultra performance liquid chromatography-quadrupole time of flight mass spectrometry in conjunction with univariate and multivariate statistical analyses. Forty three serum metabolites and 31 urinary metabolites were identified in HCC patients involving several key metabolic pathways such as bile acids, free fatty acids, glycolysis, urea cycle, and methionine metabolism. Differentially expressed metabolites in HCC subjects, such as bile acids, histidine, and inosine are of great statistical significance and high fold changes, which warrant further validation as potential biomarkers for HCC. However, alterations of several bile acids seem to be affected by the condition of liver cirrhosis and hepatitis. Quantitative measurement and comparison of seven bile acids among benign liver tumor patients with liver cirrhosis and hepatitis, HCC patients with liver cirrhosis and hepatitis, HCC patients without liver cirrhosis and hepatitis, and healthy controls revealed that the abnormal levels of glycochenodeoxycholic acid, glycocholic acid, taurocholic acid, and chenodeoxycholic acid are associated with liver cirrhosis and hepatitis. HCC patients with alpha fetoprotein values lower than 20 ng/ml was successfully differentiated from healthy controls with an accuracy of 100% using a panel of metabolite markers. Our work shows that metabolomic profiling approach is a promising screening tool for the diagnosis and stratification of HCC patients.
Hepatocelluar carcinoma (HCC)1 is the fifth most common cancer (1) and the third leading cause of cancer-related death (2) with a five-year survival rate of less than 7% (3). The morbidity of HCC in Southeast Asia and sub-Saharan Africa is greater than 20 cases per 100,000 population, whereas in North America and Western Europe is much lower, less than 5 per 100,000 population (4). However, a dramatically increasing incidence of HCC in the world, especially in the United States has been reported in recent years, primarily because of chronic alcohol use and chronic hepatitis C infection (5). Diabetic and metabolic diseases of the liver have been known to contribute to an increased incidence of HCC in recent years (6, 7). Despite significant progress in cancer diagnosis and treatment, the morbidity and mortality rate of liver cancer remains high because early diagnosis is still a challenge. Early and accurate diagnosis of HCC is of central importance for timely treatment and five-year survival rate (38.1% at stage I, 3.9% at stage IV) (8). Therefore, considerable efforts have been devoted to search for biomarkers for early diagnosis of HCC and patient stratification. Glypican-3, a cell surface-linked heparan sulfate proteoglycan, is one of the potential biomarkers in serum currently under investigation for HCC (9). At present, the most clinically used serum biomarker for HCC is alpha fetoprotein (AFP); however, clinicians are unsatisfied with it because of its high false positive and false negative rates (10).
Genomics and proteomics have merged as biochemical profiling tools to provide important insight into the biology of various cancers (11). Although these profiling approaches focus on upstream genetic and protein variations, metabolomics captures the global metabolic changes that occur in response to pathological, environmental or lifestyle factors (12). Consequently, metabolomics complements the information obtained by genomics and proteomics (13) and has already shown promise in identifying metabolite-based biomarkers in prostate (14), breast (15), ovarian (16), brain (17), and oral (18) cancers. Recently metabolomic study of HCC has been performed by high resolution magic-angle spinning 1H nuclear magnetic resonance spectroscopy (19) and a panel of 13 differential tissue metabolites, including alanine, leucine and glucose were identified. Several serum and urine metabolites as potential markers in a small number of HCC patients (n = 20) were identified by gas chromatography mass spectrometry (GC-MS, LC-MS) (20, 21), including nucleosides, butanoic acid, ethanimidic acid, glycerol, isoleucine, valine, aminomalonic acid, glycine, tyrosine, threonine, etc.
It is generally accepted that a single analytical technique could only identify a limited number of the metabolites, and therefore, multiple complementary analytical platforms are needed for an enhanced metabolic visualization. We reported an enhanced metabolomic profiling study using a combined GC-MS and LC-MS analytical platform in 2007 on the metabolic disruption associated with nephrotoxicity by aristolochic acid intervention in a rat model (22). We have recently demonstrated that a combination of gas chromatography time-of-flight mass spectrometry (GC-TOFMS) and ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC-QTOFMS) significantly increased the number of serum metabolite markers identified in a clinical metabolomic study of colorectal cancer (23).
In this study, we conducted a comprehensive analysis of the serum and urine metabolites in 177 participants (71 healthy individuals, 24 benign liver tumor patients, and 82 HCC patients diagnosed as stage I, II, III, and IV, detailed information is listed in Table I) using GC-TOFMS and UPLC-QTOFMS. The metabolic variations in HCC patients with different cancer stages were comprehensively investigated. The differential metabolites identified in HCC patients were cross checked by the two analytical methods as well as by the results from two biological specimens, serum, and urine.
Table I. Clinical information of study cohorts.
| HCC patients (n = 82) | Benign liver tumor patientsa (n = 24) | Healthy control (n = 71) | |
|---|---|---|---|
| Age (Mean, range) | 55, 29–76 | 44, 18–65 | 55, 42–65 |
| Male/Female | 55/27 | 13/11 | 39/32 |
| Stage Ib | 33 (M21/F12) | / | / |
| Stage IIb | 20 (M16/F4) | / | / |
| Stage IIIb | 22 (M13/F9) | / | / |
| Stage IVb | 7 (M5/F2) | / | / |
| AFP value (mean, range)c | 5010.84, 1.30–60500.00 | 60.74, 1.20–288.20 | / |
| ALT | 47.13 | 33.87 | / |
| AST | 52.73 | 39.04 | / |
| Liver cirrhosis (%) | 80.77 | 25.00 | / |
| HBsAg (positive %) | 66.67 | 45.80 | / |
a 24 benign liver tumor patients include 8 with hemangioma, 6 with focal nodular hyperplasia of liver, 4 with liver cirrhosis, 2 with liver cyst, 1 with intrahepatic bile duct stone and 1 with recurrent hemangioma after surgery.
b TNM Classification.
c AFP values were provided for 52 (M34/F18) among a total of 82 HCC patients, and 9(M5/F4) among the 24 benign liver tumor patients, others were labeled as “Negative” or “Positive” but without a specific AFP value; M, male; F, female.
EXPERIMENTAL PROCEDURES
Clinical Samples
A total of 82 HCC patients, 52 males and 30 females, aged 29 to 76 years old, and 24 benign, 13 males and 11 females, aged 18 to 65 years old, were enrolled in this study. The proportion of females in this cohort is higher than the national average number (the ratio of males/females is about 3:1) in favor of males. No significance is attached to the high proportion of females in the study population because the patients were taken from sequentially presenting patients in a single unit. Patient characteristics, staging of disease and other parameters are shown in Table I. The clinical diagnosis and pathological reports of all the patients were obtained from Zhongshan Hospital, Fudan University, Shanghai, China. Control samples were collected from a total of 71 healthy volunteers (39 males and 32 females, aged 42 to 65 years old) using the same sample collection protocol, and any subjects with inflammatory conditions, steatohepatitis, or gastrointestinal tract disorders were excluded. The average level of serum AFP in the HCC group is 5010.84 ng/ml ranging from 1.3 to 60,500 ng/ml, any AFP values higher than 60,500 ng/ml were recorded as 60,500 ng/ml. Ten serum enzyme levels correlating to liver function for HCC patients and benign liver tumor patients were measured (detailed information is provided in supplemental Table S1 and S2). Tumor invasion of neighboring organs, lesion nature and dimension, and presence of angiolymphatic or perineural invasion were also recorded. Serum and urine samples were collected in the morning before breakfast from all the participants. Serum samples were placed into clean tubes and kept at −80 °C until analysis. The collected urine samples were centrifuged at 3000 rpm for 10 min at 4 °C to remove suspended debris, and the resulting supernatants were immediately stored at −80 °C without any preservatives. The protocol was approved by the Zhongshan Hospital Institutional Review Board and written consents were signed by all participants before the study.
Serum Sample Preparation and Analysis by GC-TOFMS
Serum samples were derivatized and subsequently analyzed by GC-TOFMS following our previously published protocols (23). A 100 μl aliquot of serum sample was spiked with two internal standards (10 μl l-2-chlorophenylalanine in water, 0.3 mg/ml; 10 μl heptadecanoic acid in methanol, 1 mg/ml) and vortexed for 10 s. The mixed solution was extracted with 300 μl of methanol/chloroform (3:1) and vortexed for 30 s. After storing for 10 min at −20 °C, the samples were centrifuged at 10,000 rpm for 10 min. An aliquot of the 300 μl supernatant was transferred to a glass sampling vial to vacuum dry at room temperature. The residue was derivatized using a two-step procedure. First, 80 μl methoxyamine (15 mg/ml in pyridine,) was added to the vial and kept at 30 °C for 90 min followed by 80 μl BSTFA (1%TMCS) at 70 °C for 60 min.
Each 1 μl aliquot of the derivatized solution was injected in spitless mode into an Agilent 6890N gas chromatography coupled with a Pegasus HT time-of-flight mass spectrometer (Leco Corporation, St Joseph, MI). Separation was achieved on a DB-5MS capillary column (30 m × 250 μm I.D., 0.25-μm film thickness; (5%-phenyl)-methylpolysiloxane bonded and crosslinked; Agilent J&W Scientific, Folsom, CA) with helium as the carrier gas at a constant flow rate of 1.0 ml/min. The temperature of injection, transfer interface, and ion source was set to 270 °C, 260 °C, and 200 °C, respectively. The GC temperature programming was set to 2 min isothermal heating at 80 °C, followed by 10 °C/min oven temperature ramps to 180 °C, 5 °C/min to 240 °C, and 25 °C/min to 290 °C, and a final 9 min maintenance at 290 °C. Electron impact ionization (70 eV) at full scan mode (m/z 30–600) was used, with an acquisition rate of 20 spectrum/second in the TOFMS setting.
Urine Sample Preparation and Analysis by GC-TOFMS
Urine samples for GC-TOFMS analysis was processed according to our previously published protocol (24). Each 600 μl aliquot of standard mixture or diluted urine sample (urine/water = 1:1, v/v) was added to a screw-top glass tube. After adding 100 μl of l-2-chlorophenylalanine (0.1 mg/ml), 400 μl of anhydrous ethanol, and 100 μl of pyridine to the urine sample, 50 μl of ethyl chloroformate was added for first derivatization at 20.0 ± 0.1 °C. The pooled mixtures were sonicated at 40 kHz for 60 s. Subsequently, extraction was performed using 300 μl of chloroform, with the aqueous layer pH carefully adjusted to 9–10 using 100 μl of NaOH (7 m). The derivatization procedure was repeated with the addition of 50 μl ethyl chloroformate into the aforementioned products. After the two successive derivatization steps, the overall mixtures were vortexed for 30 s and centrifuged for 3 min at 3,000 rpm. The aqueous layer was aspirated off, whereas the remaining chloroform layer containing derivatives were isolated and dried with anhydrous sodium sulfate and subsequently subjected to GC-TOFMS analysis.
The derivatized extracts were analyzed with an Agilent 6890N gas chromatography coupled with a Pegasus HT time-of-flight mass spectrometer (Leco Corporation). A 1-μl extract aliquot of the extracts was injected into a DB-5MS capillary column coated with 5% diphenyl cross-linked 95% dimethylpolysiloxane (30m × 250 μm i.d., 0.25-μm film thickness; Agilent J&W Scientific, Folsom, CA) in the split mode (3:1). Either the injection temperature or the interface temperature was set to 260 °C; and the ion source temperature was adjusted to 200 °C. Initial GC oven temperature was 80 °C; 2 min after injection, the GC oven temperature was raised to 140 °C with 10 °C/min, to 240 °C at a rate of 10 °C/min, to 290 °C with 15 °C/min again, and finally held at 290 °C for 3 min. Helium was the carrier gas with a flow rate set at 1 ml/min. The measurements were made with electron impact ionization (70 eV) in the full scan mode (m/z 30–550).
Serum Sample Preparation and Analysis by UPLC-QTOFMS
Serum sample preparation and analysis with UPLC-QTOFMS was performed according to our published report (23). Each 100 μl of serum was used for metabolite extraction before UPLC-QTOFMS analysis. The metabolite extraction procedure was carried out after adding 100 μl of water (containing 0.1 mg/ml l-2-chlorophenylalanine as the internal standard) and 400 μl of a mixture of methanol and acetonitrile (5:3) to 100 μl of serum. After vortexing for 2 min, the mixture was stored at room temperature for 10 min, centrifuged at 12,000 rpm for 20 min. The supernatant was filtered through a syringe filter (0.22 μm) and transferred into the sampling vial pending UPLC-QTOFMS analysis. A 5 μl aliquot of the filtrate was subjected at a random order into a 100 mm × 2.1 mm, 1.7 μm BEH C18 column (Waters, Milford, MA) held at 40 °C using an ultra performance liquid chromatography system (Waters). The column was eluted with a linear gradient of 1–20% B over 0–1 min, 20–70% B over 1–3 min, 70–85% B over 3–8 min, 85–100% B over 8–9 min, the composition was held at 100% B for 0.5 min. For positive ion mode (ES+) where A = water with 0.1% formic acid and B = acetonitrile with 0.1% formic acid, whereas A = water and B = acetonitrile for negative ion mode (ES-). The flow rate was 0.4 ml/min. All the samples were kept at 4 °C during the analysis.
The mass spectrometric data were collected using a Waters Q-TOF premier (Manchester, UK) equipped with an electrospray source operating in either positive or negative ion mode. The source temperature was set at 120 °C with a cone gas flow of 50 L/h, a desolvation gas temperature of 300 °C with a desolvation gas flow of 600 L/h. In the case of positive and negative ion mode the capillary voltage was set to 3.2 kV and 3 kV, and the cone voltage of 35 V and 50 V, respectively. Centroid data were collected from 50 to 1000 m/z with a scan time of 0.3 s and interscan delay of 0.02 s over a 9.5 min analysis time. MassLynx software (Waters) was used for system controlling and data acquisition. Leucine enkephalin was used as the lock mass (m/z 556.2771 in ES+ and 554.2615 in ES-) at a concentration of 100 ng/ml and flow rate of 0.2 ml/min for all analyses.
Urine Sample Preparation and Analysis by UPLC-QTOFMS
Urine sample preparation was processed according to our previous work (25). The collected urine samples were centrifuged at 13,000 rpm for 10 min at 4 °C, and the resulting supernatants were immediately stored at −80 °C pending UPLC-QTOFMS analysis. Ultrapure water (500 μl) was added to urine (500 μl) and vortexed for 1 min, and then filtered through a syringe filter (0.22 μm) for UPLC-QTOFMS analysis.
A 5 μl aliquot of the filtrate was injected into a 100 mm × 2.1 mm, 1.7 μm BEH C18 column (Waters) held at 40 °C using an ultra performance liquid chromatography system (Waters). The column was eluted with a linear gradient of 1–20% B over 0–1 min, 20–70% B over 1–3 min, 70–85% B over 3–8 min, 85–100% B over 8–9 min, the composition was held at 100% B for 0.5 min. For positive ion mode (ES+) where A = water with 0.1% formic acid and B = acetonitrile with 0.1% formic acid, whereas A = water and B = acetonitrile for negative ion mode (ES-). The flow rate was 0.4 ml/min. All the samples were kept at 4 °C during the analysis.
The mass spectrometric data was collected using a Waters Q-TOF premier (Manchester, UK) equipped with an electrospray ion source operating in either positive or negative ion mode. The source temperature was set at 120 °C with a cone gas flow of 50 L/h, a desolvation gas temperature of 300 °C with a desolvation gas flow of 600 L/h. In the case of positive and negative ion modes the capillary voltage was set to 3.2 kV and 3 kV, and the cone voltage of 35 V and 50 V, respectively. Centroid data was collected from 50 to 1000 m/z with a scan time of 0.3 s and interscan delay of 0.02 s over a 9.5 min analysis time. Leucine enkephalin was used as the lock mass (m/z 556.2771 in ES+ mode and 554.2615 in ES- mode) at a concentration of 100 ng/ml and flow rate of 0.2 ml/min for all analyses.
Quantitative Analysis of Bile Acids in Serum and Urine Samples by UPLC-QTOFMS
To verify the linearity, the spiked standard solution including chenodeoxycholic acid, deoxycholic acid, taurocholic acid, cholic acid, glycochenodeoxycholic acid, lithocholic acid, and glycocholic acid was prepared and diluted to appropriate concentration ranges for the establishment of calibration curves. The limit of detection (signal to noise ratio (S/N) = 3) and limit of quantitation (S/N = 9) were determined, respectively. Serum and urine samples were prepared as the method for UPLC-QTOFMS metabolomics analysis described in above section. The concentration of each metabolite was subsequently determined from the corresponding calibration curve.
Data Analysis
The acquired MS data from GC-TOFMS and UPLC-QTOFMS were analyzed according to our previously published work (23, 26). The acquired MS data from GC-TOFMS analysis were exported to NetCDF format by ChromaTOF software (version 3.30; Leco Co.). CDF files were extracted using custom scripts (revised MATLAB toolbox hierarchical multivariate curve resolution (H-MCR), developed by Par Jonsson et al. (27, 28)) in the MATLAB 7.0 (The MathWorks, Natick, MA) for data pretreatment procedures such as baseline correction, denoising, smoothing, peak alignment, time-window splitting, and multivariate curve resolution (based on the multivariate curve resolution algorithm). The resulting three dimension data sets include sample information, peak retention time and peak intensities. Internal standards and any known pseudo positive peaks, such as peaks caused by noise, column bleed and BSTFA derivatization procedure, were removed from the data set.
The UPLC-QTOFMS ES+ and ES- raw data were analyzed by the MarkerLynx Applications Manager version 4.1 (Waters, Manchester, U.K.) using the following parameters. The parameters used were retention-time range 0–9.5 min, mass range 50–1000 Da, mass tolerance 0.02 Da, internal standard detection parameters were deselected for peak retention time alignment, isotopic peaks were excluded for analysis, noise elimination level was set at 10.00, minimum intensity was set to 15% of base peak intensity, maximum masses per RT was set at 6 and, finally, RT tolerance was set at 0.01 min. A list of the ion intensities of each peak detected was generated, using retention time and the m/z data pairs as the identifier for each ion. The resulting three-dimensional matrix contains arbitrarily assigned peak index (retention time-m/z pairs), sample names (observations), and ion intensity information (variables). To obtain consistent differential variables, the resulting matrix was further reduced by removing any peaks with missing value (ion intensity = 0) in more than 80% samples. The internal standard was used for data quality control (reproducibility) and data normalization. The ion peaks generated by the internal standard were also removed.
The three data sets resulting from GC-TOFMS, UPLC-QTOFMS ES+, and ES- (expressed as G, P, and N, respectively) were analyzed and validated by uni- and multivariate statistical methods, separately (the raw data sets were supplied as a supplemental Table (Raw data sets-urine-serum.xls)).
Each data set was imported into SIMCA-P 12.0 software package (Umetrics, Umeå, Sweden). Principle component analysis (PCA) and orthogonal partial least squares-discriminant analysis (OPLS-DA) were carried out to visualize the metabolic alterations between HCC patients and healthy controls after mean centering and unit variance scaling. In this study, the default 7-round cross-validation was applied with 1/7th of the samples being excluded from the mathematical model in each round, to guard against over-fitting. The variable importance in the projection (VIP) values of all the peaks from the 7-fold cross-validated OPLS-DA model were taken as a coefficient for peak selection. VIP ranks the overall contribution of each variable to the OPLS-DA model, and those variables with VIP > 1.0 are considered relevant for group discrimination (29). Herein, VIP statistics and S-plot were applied to obtain the significant variables for subsequent metabolic pathway analysis (30, 31). Besides the multivariate approaches, one univariate method, the Student's t test, was selected to measure the significance of each metabolite in separating HCC patients from healthy controls. Several peaks responsible for the differentiation of the metabolic profiles of diseased individuals and healthy controls could be obtained by comprehensive consideration of these two coefficients. The corresponding up- and down-regulated trend shows how these selected differential metabolites varied between the HCC and the healthy controls.
Metabolites identification from these selected peaks was performed separately. GC-TOFMS metabolites were identified by comparing the mass fragments with NIST 05 Standard mass spectral databases in NIST MS search 2.0 (NIST, Gaithersburg, MD) software with a similarity of more than 70% and finally verified by available reference compounds. Metabolites obtained from POS and NEG mode of UPLC-QTOFMS analysis were identified with the aid of available reference standards in our lab and the web-based resources such as the Human Metabolome Database (http://www.hmdb.ca/).
We used 55 HCC patients, 16 benign tumor patients, and 47 healthy controls (sample information are provided in supplemental Table S3) to establish the OPLS-DA model for selecting the differential metabolites in HCC patients, HCC patients (stage I+II), and HCC patients (stage III+IV), relative to healthy controls. Then, the performance of the OPLS-DA model and the selected differential metabolites are tested in a different sample set comprising 27 HCC patients, 8 benign tumor patients, and 24 healthy controls (see supplemental Table S4). Potential differential metabolites selected and identified from the three data sets were normalized, and combined for comprehensive analysis. Aiming to exploring the natural interrelation between HCC patients and the healthy controls, unsupervised PCA model was build. The original set of metabolites was reduced to a new set of principal components that retain the variance-covariance structure of the data, but use less (one or two only) dimensions of data space. Its stability and performance was validated by both permutation and new samples test.
Statistical analysis of ANOVA was performed on SPSS PASW Statistics 18.
RESULTS
Serum Metabolite Profiles and Markers of HCC
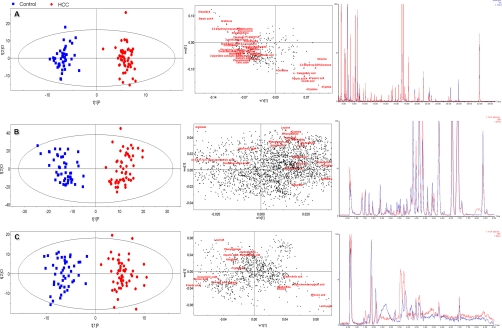
Clinical characteristics of HCC patients and other study subjects are detailed in Table I. After data normalization, PCA was performed on the dataset, which showed a trend of inter-group separation on the scores plot (Figs. not provided). Fig. 1A–1C illustrate scores plots and loadings plots of the OPLS-DA model of 55 HCC patients (red dots) and 47 healthy controls (blue squares) based on spectral data of (Fig. 1A) GC-TOFMS; (Fig. 1B) UPLC-QTOFMS positive ion mode; and (Fig. 1C) UPLC-QTOFMS negative ion mode (sample information is provided in supplemental Tables S3–S4). Representative total ion current chromatograms of GC-TOFMS and base peak intensity chromatograms in positive ion mode (ES+) and negative ion mode (ES-) of UPLC-QTOFMS obtained from a HCC patient, a benign liver tumor patient (hemangioma) and a healthy control are shown in supplemental Fig. S1, where marked variations can be visually observed among the three serum chromatograms. A total of 324 peaks were obtained from GC-TOFMS spectra (expressed as G data set), whereas 2626 peaks were obtained from UPLC-QTOFMS ES+ mode (expressed as P data set) and 925 peaks obtained from ES- mode (expressed as N data set). The OPLS-DA scores plots (supplemental Fig. S1 A–S1C) showed three clusters of HCC patients, benign liver tumor patients and healthy controls. HCC and benign liver tumor patients were clearly separated from healthy controls. An OPLS-DA model based on the total spectral data of GC-TOFMS, UPLC-QTOFMS positive ion mode, and negative ion mode between 55 HCC patients and 16 benign liver tumor patients was established in supplemental Fig. S2. HCC patients and benign liver tumor patients can be successfully differentiated by PC1 (the first principal component of the model) with statistical significance (supplemental Fig. S2). To further test the performance of this model, another group of 27 HCC patients and 8 benign liver tumor patients were used as testing samples. supplemental Fig. S2 shows the prediction results of the 32 testing samples (green squares and blue stars) using the model established with the 71 training samples. All the test samples are correctly classified as HCC or benign liver tumor patients and clear separation was achieved between benign and HCC. The permutation test (1000 times) of the OPLS-DA model corresponding to PCA model including correlation coefficient between the original Y and the permuted Y versus the cumulative R2 and Q2, with the regression line was shown in supplemental Fig. S2B. The intercept (R2 and Q2 when correlation coefficient is zero) which is correlated with the extent of overfitting is rather small (R2 = 0.51 and Q2 = −0.19) and the model is satisfactory. The OPLS-DA model of data from G, P, and N demonstrated distinctly different metabolite profiles of HCC patients, HCC patients (stage I+ II), HCC patients (stage III+ IV), and benign liver tumor patients, from healthy controls, with satisfactory modeling and predictive abilities using one predictive component and three orthogonal components (supplemental Fig. S3).
Fig. 1.
OPLS-DA scores plots and loadings plots of 55 HCC patients (red dots) and 47 healthy controls (blue squares) based on serum spectral data of (A) GC-TOFMS; (B) UPLC-QTOFMS positive ion mode; and (C) UPLC-QTOFMS negative ion mode. On the right side of the three scores plots, three representative chromatograms of a HCC (red) and a healthy control sample (blue) derived from GC-TOFMS, UPLC-QTOFMS positive ion mode, and negative ion mode, respectively.
Fifty-one most significantly altered serum metabolites (supplemental Table S5) in HCC patients relative to healthy controls were identified from a two-component OPLS-DA model of the G, P, and N spectral datasets, annotated by the mass of molecular and fragment ions, among which 38 were further validated by reference standards available in our laboratory. A summary of metabolite markers identified and compared among HCC patients at stage I and II, HCC at stage III and IV, and HCC group (all stages) is provided in supplemental Table S5.
Urinary Metabolite Profiles and Markers of HCC
Urine samples obtained from healthy controls, and HCC patients were analyzed following the procedures described in Experimental Procedures. Thirty-three urinary differential metabolites were identified in HCC patients relative to healthy controls from the G and P datasets using the same statistical criterion for serum metabolites selection (supplemental Table S6 and supplemental Fig. S4). A summary of differential metabolites in the urine samples of HCC patients (stage I and II), HCC (stage III and IV), and HCC (all stages), relative to healthy controls, is provided in supplemental Table S6.
As listed in supplemental Tables S5 and S6, five differential metabolites are found both in serum and urine samples (supplemental Table S7) in HCC patients. Among them, phenylalanine, altered in different directions, presumably because of the different metabolic process involving gut microflora in urine. Because a great portion of HCC subjects are accompanied with liver cirrhosis and hepatitis, some of the metabolite markers are associated with liver cirrhosis and hepatitis, rather than HCC (see the following paragraph). Table II showed a corrected list of the serum and urinary metabolite markers for HCC.
Table II. Summary of the differentially expressed serum and urine metabolites in patients of HCC (all stages) relative to healthy controls. An empty cell in the table means the alteration of metabolite level is statistically insignificant. G: data obtained from GC-TOFMS, P: data obtained from positive ion mode of UPLC-QTOFMS, N: data obtained from negative ion mode of UPLC-QTOFMS.
| Metabolites | Metabolic pathway | HCC (Serum) |
HCC (Urine) |
||||
|---|---|---|---|---|---|---|---|
| VIPc | Pd | FC ± S.D.e | VIPc | Pd | FC ± S.D.e | ||
| Deoxycholic acid, N | Bile acid metabolism | 1.49 | 2.43E-03 | 0.36 ± 0.1 | |||
| Glycochenodeoxycholic acida N | Bile acid metabolism | 1.53 | 1.80E-03 | 6.67 ± 1.59 | |||
| Glycocholic acida,‖P | Bile acid metabolism | 1.18 | 3.52E-03 | 6.4 ± 1.66 | 0.63 | 9.33E-04 | 45 ± 14.96 |
| Taurocholic acidaN, € | Bile acid metabolism | 1.17 | 1.87E-02 | 25.36 ± 6.41 | |||
| Lithocholic acidaP | Bile acid metabolism | 0.12 | 1.45E-02 | 0.93 ± 0.21 | |||
| Arachidonic acid,G | Fatty acid metabolism | 1.44 | 5.38E-08 | 0.84 ± 0.01 | |||
| cis-5,8,11,14, 17-Eicosapentaenoic acidaP | Fatty acid metabolism | 1.68 | 5.70E-04 | 0.43 ± 0.08 | |||
| Docosahexaenoic acida,bG | Fatty acid metabolism | 1.27 | 2.43E-06 | 0.82 ± 0.03 | |||
| Glycerol,G | Fatty acid metabolism | 1.16 | 2.00E-05 | 0.85 ± 0.02 | |||
| Myristic acidaN | Fatty acid metabolism | 1.84 | 1.45E-04 | 0.66 ± 0.06 | |||
| Nervonic acida,bN | Fatty acid metabolism | 1.39 | 4.75E-03 | 0.56 ± 0.09 | |||
| Alaninea,bG | Methionine Metabolism | 2.12 | 2.68E-02 | 0.71 ± 0.09 | |||
| Cysteinea,b‖G | Methionine Metabolism | 1.29 | 1.81E-06 | 1.25 ± 0.03 | 1.57 | 3.34E-03 | 0.35 ± 0.12 |
| Cystinea,b‖G | Methionine Metabolism | 1.91 | 1.54E-14 | 1.44 ± 0.04 | 1.09 | 4.34E-02 | 1.32 ± 0.11 |
| GlycineaG | Methionine Metabolism | 1.01 | 2.37E-04 | 0.86 ± 0.02 | |||
| Serinea,bG | Methionine Metabolism | 1.16 | 2.06E-05 | 0.84 ± 0.02 | |||
| TaurineaP | Methionine Metabolism | 0.91 | 2.48E-02 | 1.45 ± 0.15 | 1.35 | 1.22E-02 | 1.43 ± 0.13 |
| Aspartic acida,bG | Urea cycle | 1.26 | 3.06E-06 | 0.89 ± 0.01 | |||
| Citrulline.bG | Urea cycle | 1.51 | 9.47E-09 | 0.87 ± 0.01 | |||
| Glutamic acida,b G | Urea cycle | ||||||
| Ornithinea,b G | Urea cycle | 1.55 | 3.55E-09 | 0.96 ± 0.03 | |||
| Fumaric acidaG | TCA cycle | 1.39 | 2.17E-07 | 1.28 ± 0.04 | |||
| Succinic acid,G | TCA cycle | 1.97 | 4.02E-02 | 0.63 ± 0.12 | |||
| α−Ketoglutaric acid,G | TCA cycle | 1.18 | 1.58E-05 | 1.11 ± 0.01 | |||
| Lactic acida,bG | Glycolysis | 1.08 | 7.97E-05 | 1.16 ± 0.03 | |||
| Pyruvic acidbG | Glycolysis | 1.26 | 3.46E-06 | 1.18 ± 0.03 | |||
| 4-Hydroxyphenylacetate,G | Gut flora metabolism | 1.39 | 4.51E-02 | 0.69 ± 0.13 | |||
| Trimethylamine N-oxideaP | Gut flora metabolism | 1.74 | 1.09E-03 | 0.54 ± 0.07 | |||
| Kynureninea,bG | Tryptophan Metabolism | 1.24 | 4.76E-06 | 0.77 ± 0.03 | |||
| Tryptophana,bG | Tryptophan metabolism | 1.12 | 4.00E-05 | 0.76 ± 0.03 | |||
| DopamineaP | Tyrosine Metabolism | 2.2 | 2.53E-05 | 1.64 ± 0.11 | |||
| Homovanillate,G | Tyrosine Metabolism | 2.73 | 4.10E-03 | 0.65 ± 0.06 | |||
| NormetanephrineaP | Tyrosine Metabolism | 1.48 | 5.69E-03 | 0.44 ± 0.12 | |||
| AdenineaP | Purine Metabolism | 1.45 | 7.09E-03 | 0.44 ± 0.1 | |||
| AdenosineaP | Purine Metabolism | 1.92 | 2.97E-04 | 1.6 ± 0.11 | |||
| Hypoxanthinea,bP,€ | Purine Metabolism | 1.42 | 8.18E-03 | 1.42 ± 0.13 | |||
| Inosinea,bN | Purine Metabolism | 0.93 | 1.27E-10 | 40.62 ± 19.37 | |||
| Uric acidaP | Purine Metabolism | 1.5 | 5.15E-03 | 1.61 ± 0.16 | |||
| XanthineaP | Purine Metabolism | 1.64 | 2.18E-03 | 1.61 ± 0.16 | |||
| Carnosine,P | Histidine Metabolism | 1.28 | 1.81E-02 | 0.83 ± 0.04 | |||
| Lysinea,bG | Lysine Degradation | 1.32 | 8.93E-07 | 0.79 ± 0.03 | |||
| Nicotinic acidaP | Nicotinate and Nicotinamide Metabolism | 1.33 | 1.37E-02 | 0.24 ± 0.1 | |||
| Glucosamine,G | Amino Sugar Metabolism | 1.53 | 6.16E-09 | 0.62 ± 0.03 | |||
| N-Acetyl-l-Aspartic acidaP | Aspartate metabolism | 1.09 | 4.38E-02 | 1.37 ± 0.14 | |||
| d-(+)-galactose,G | Galactose Metabolism | ||||||
| Pyroglutamic acidaN | Glutathione Metabolism | 0.88 | 1.53E-03 | 0.9 ± 0.02 | |||
| Phenylalaninea,b‖G | Phenylalanine and Tyrosine Metabolism | 1.06 | 1.22E-04 | 0.85 ± 0.02 | 1.41 | 8.64E-03 | 1.26 ± 0.07 |
| CytidineaN | Pyrimidine Metabolism | 1.02 | 4.05E-02 | 0.55 ± 0.13 | |||
| DihydrouracilaP | Pyrimidine Metabolism | 2 | 1.50E-04 | 1.41 ± 0.07 | |||
| Cysteic acida,bP | Taurine Metabolism | 1.84 | 5.33E-04 | 0.4 ± 0.05 | |||
| HypotaurineP | Taurine Metabolism | 1.55 | 3.89E-03 | 1.53 ± 0.16 | |||
| Threoninea,bP | Threonine metabolism | 2.34 | 7.18E-06 | 1.77 ± 0.13 | |||
| Leucinea,bN | Valine, Leucine and Isoleucine Degradation | 1.21 | 1.45E-02 | 0.83 ± 0.05 | |||
| β-alaninea,bG € | β-Alanine Metabolism | 0.85 | 2.45E-03 | 0.8 ± 0.05 | |||
| PyridoxalaP | Vitamine | 1.33 | 1.32E-02 | 1.99 ± 0.32 | |||
| α-tocopherolaG | Vitamine | 1.37 | 3.19E-07 | 0.96 ± 0 | |||
| 2,3-dihydroxy-2(3H)-furanone,G | Others | 1.63 | 3.37E-10 | 1.43 ± 0.04 | |||
| 2,3-dihydroxyl-propanoic acid,G | Others | 1.93 | 6.53E-15 | 0.79 ± 0.02 | |||
| 2-pyrrolidone-5-carboxylic acidaP | Others | 1.2 | 2.65E-02 | 0.52 ± 0.11 | |||
| 3-amino-2-piperidoneaG | Others | 1.56 | 2.71E-09 | 0.85 ± 0.01 | |||
| 4-ketoglucose,G | Others | 1.74 | 8.19E-12 | 0.6 ± 0.03 | |||
| 6-Aminocaproic acidaP | Others | 2.36 | 5.49E-06 | 0.09 ± 0.02 | |||
| AgmatineaP | Others | 1.38 | 1.06E-02 | 1.5 ± 0.17 | |||
| Arabinose,G | Others | 1.98 | 6.70E-16 | 0.45 ± 0.03 | |||
| CarnitineaP | Others | 1.39 | 5.16E-04 | 1.36 ± 0.07 | |||
| Creatininea,bG | Others | 1.6 | 8.81E-10 | 0.77 ± 0.02 | |||
| CreatineaP, € | Others | 2 | 1.49E-04 | 0.46 ± 0.07 | |||
| N-Acetyl-neuraminic acidaP | Others | 2.29 | 1.08E-05 | 2.43 ± 0.26 | |||
| OleamideΔ,G | Others | 2.67 | 7.62E-48 | 0.7 ± 0.01 | |||
| O-Phospho-l-serineaP | Others | 1.22 | 2.33E-02 | 1.36 ± 0.11 | |||
| Phosphoric acid,G | Others | 1.48 | 2.36E-08 | 0.88 ± 0.01 | |||
a Metabolites validated by reference standards.
b Metabolites that can be identified by GC-TOFMS and UPLC-TOFMS.
c Variable importance in the projection (VIP) was obtained from OPLS with a threshold of 1.0.
d P means p value obtained from Student's t-test.
e The Fold change (FC) with a value larger than 1.0 indicates a significantly higher level of the serum or urine metabolite in patients while a FC value lower than 1.0 indicates a lower level, relative to healthy controls.
Metabolite Markers Associated with Liver Cirrhosis and Hepatitis
As listed in Table III, X differentially expressed metabolites in liver cirrhosis and hepatitis condition including inositol, 2,2′-bipyridine, methionine, arginine, stearic acid, palmitic acid, citric acid, 2-piperidine carboxylic acid, 5-hydroxy-tryptophan, and tyrosine were obtained by comparison among healthy controls, benign liver tumor patients with liver cirrhosis and hepatitis, HCC with liver cirrhosis and hepatitis, and HCC without liver cirrhosis and hepatitis. All 10 metabolites were of statistical significance (p < 0.05) and all of them have the same direction of perturbation (up- or down-regulation) both in liver cirrhosis and hepatitis patients and HCC patients with cirrhosis and hepatitis but not in the HCC patients without cirrhosis and hepatitis. They can be considered potential markers specific for liver cirrhosis and hepatitis, and therefore were removed from the list of HCC markers.
Table III. Summary of the metabolite markers associated with liver cirrhosis and hepatitis (p < 0.05).
| Metabolite | Liver cirrhosis+hepatitis |
HCC (cirrhosis+hepatitis) |
HCC |
|||
|---|---|---|---|---|---|---|
| FC ± S.D.a | Pb | FC ± S.D.a | Pb | FC ± S.D.a | Pb | |
| Inositol | 0.73 ± 0.07 | 4.41E-03 | 0.87 ± 0.06 | 4.44E-02 | 0.86 ± 0.14 | 4.21E-01 |
| 2,2′-Bipyridine | 0.85 ± 0.03 | 9.34E-04 | 0.90 ± 0.02 | 2.50E-04 | 0.95 ± 0.04 | 3.11E-01 |
| Methionine | 0.70 ± 0.08 | 6.31E-03 | 0.78 ± 0.03 | 1.56E-04 | 0.81 ± 0.02 | 5.48E-02 |
| Arginine | 0.41 ± 0.12 | 3.18E-02 | 0.48 ± 0.08 | 7.72E-04 | 0.60 ± 0.17 | 2.87E-01 |
| Stearic acid | 0.60 ± 0.01 | 1.96E-02 | 0.71 ± 0.01 | 4.94E-03 | 0.82 ± 0.005 | 3.12E-01 |
| Palmitic acid | 0.80 ± 0.03 | 8.95E-03 | 0.82 ± 0.04 | 5.95E-04 | 0.88 ± 0.06 | 1.02E-01 |
| Citric acid | 0.61 ± 0.15 | 2.81E-03 | 0.79 ± 0.05 | 2.84E-04 | 0.81 ± 0.08 | 5.16E-02 |
| 2-piperidine carboxylic acid | 0.86 ± 0.06 | 5.29E-03 | 0.92 ± 0.03 | 1.66E-03 | 0.93 ± 0.06 | 2.86E-01 |
| 5-Hydroxy-tryptophan | 2.23 ± 0.31 | 3.64E-02 | 2.05 ± 0.26 | 9.40E-03 | 2.95 ± 0.28 | 3.06E-01 |
| Tyrosine | 0.24 ± 0.03 | 4.04E-02 | 0.31 ± 0.10 | 8.60E-04 | 0.72 ± 0.15 | 3.12E-01 |
a FC with a value larger than 1.0 indicates a relatively higher concentration present in HCC patients (or HCC patients accompanied with cirrhosis and hepatitis, or benign liver tumor patients with cirrhosis and hepatitis) while a FC value lower than 1.0 means a relatively lower concentration as compared to the healthy controls.
b P means p value obtained from Student's t-test.
Bile Acid Markers in Serum and Urine Samples
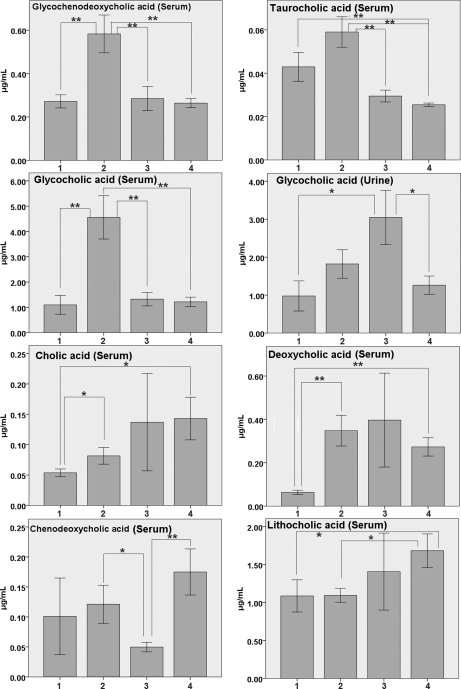
As shown in Table II, higher levels of conjugated bile acids, glycocholic acid was found in serum and urine and glycochenodeoxycholic acid and taurocholic acid were found in the serum of HCC subjects compared with healthy controls. Unconjugated bile acid, lithocholic acid, and deoxycholic acid on the other hand, was at lower level in HCC patients compared with healthy controls. Other unconjugated bile acids such as cholic acid and chenodeoxycholic acid were also shown at lower levels in HCC patients (Fig. 2). However, the levels of glycocholic acid in serum and urine, and the level of glycochenodeoxycholic acid in serum in Table II were inconsistent with the results in Fig. 2, primarily because of the fact that we used a subset of samples in Fig. 2.
Fig. 2.
Bar charts of quantitative analysis results of bile acids in serum and urine samples (Mean ± S. E., μg/ml) (* p < 0.05; **, p < 0.01). (1. HCC patients; 2. HCC patients with cirrhosis and hepatitis; 3. liver cirrhosis and hepatitis patients; 4. Healthy controls).
The alteration of bile acid levels seem to be affected by the condition of liver cirrhosis. Using our optimized UPLC-QTOFMS method, a high regression coefficient (r > 0.99) value of each calibration curve from the spiked seven standards was obtained, indicating an good linearity in this study (supplemental Fig. S5 and supplemental Table S8). Bile acid levels of chenodeoxycholic acid, deoxycholic acid, taurocholic acid, cholic acid, glycochenodeoxycholic acid, lithocholic acid, and glycocholic acid were quantitatively determined and compared among benign liver tumor patients with liver cirrhosis and hepatitis, HCC with cirrhosis and hepatitis, HCC without cirrhosis and hepatitis, and healthy controls, as shown in Fig. 2. Deoxycholic acid was elevated in subjects with liver cirrhosis and hepatitis by a factor of 1.45 whereas decreased in HCC patients without cirrhosis and hepatitis by a factor of 0.23, as compared with the healthy controls. Cholic acid level decreased in liver cirrhosis patients by 0.95-fold, but decreased in HCC patients by 0.37-fold (Fig. 2).
AFP Prediction
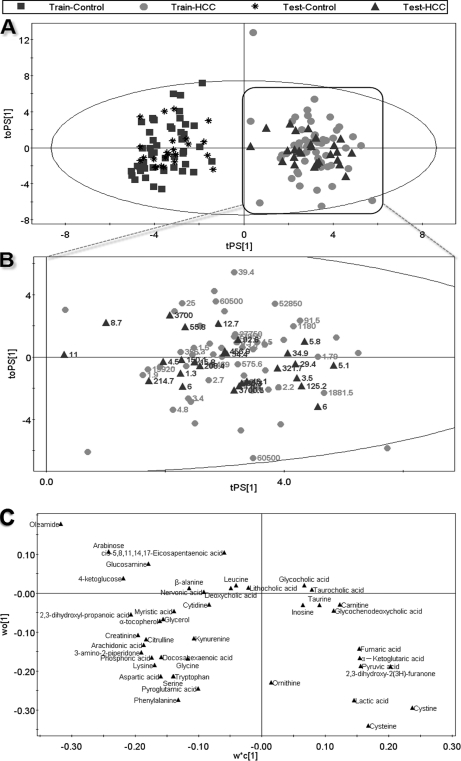
OPLS-DA model score plots of 55 serum samples both from G (Fig. 1A), P (Fig. 1B), and N (Fig. 1C) showed clear separations between HCC patients and healthy controls, with satisfactory modeling and predictive abilities (R2X = 0.39, R2Y = 0.944, Q2cum = 0.900 for GCT, R2X = 0.218, R2Y = 0.911, Q2cum = 0.742 for N mode, and R2X = 0.253, R2Y = 0.920, Q2cum = 0.783 for P mode, respectively). Forty-seven healthy controls and 55 HCC patients can be successfully differentiated by PC1 (the first principal component of the model) with statistical significance (Fig. 3). To further test the performance of this model, another group of 27 HCC patients with AFP values and 24 healthy controls were used as testing samples. Fig. 3 shows the prediction results of the 51 testing samples (green triangles and black stars) using the model established with the 102 training samples. All the test samples are correctly classified as HCC or healthy subjects, suggesting that these markers are of great potential value for HCC diagnosis. Moreover, HCC patients with AFP values lower than 20 ng/ml can be successfully differentiate from healthy controls with 100% accuracy (green triangles with labeled AFP values, see Fig. 3B). The permutation test (1000 times) of the PLS-DA model corresponding to PCA model including correlation coefficient between the original Y and the permuted Y versus the cumulative R2 and Q2, with the regression line was shown in supplemental Fig. S6. The intercept (R2 and Q2 when correlation coefficient is zero) which is correlated with the extent of overfitting is rather small (R2 = 0.21 and Q2 = −0.28) and the model is satisfactory.
Fig. 3.
The OPLS-DA prediction model of HCC. An OPLS-DA model was constructed using data from 47 healthy controls (blue squares) and 55 HCC patients (red dots) (the “training set”), this model was then used to predict HCC of a further 51 samples including 24 healthy controls (black stars) and 27 HCC patients (green triangles) that were not used in the construction of the model (the “testing set”). (A) scores plot; (B) magnified scores plot of HCC, the labled numbers are AFP values; (C) loadings plot with identified compounds.
Preliminary Analysis of HCC Stage
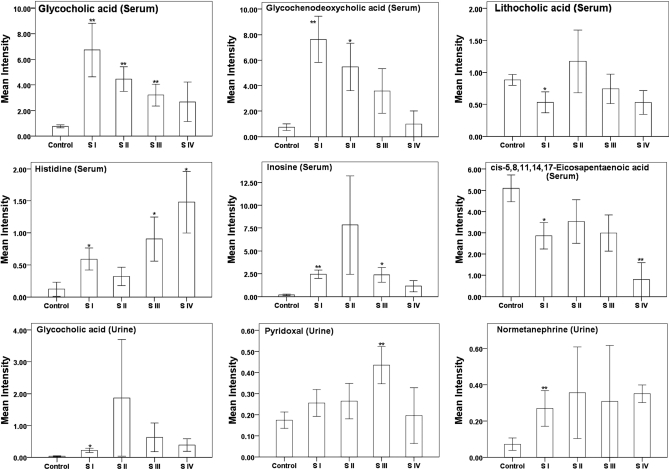
A total of 51, 48, and 49 most significantly altered serum metabolites (supplemental Table S5) in HCC group, HCC at stage I and II group, and HCC at stage III and IV group were identified from a three-component OPLS-DA model and determined based on the mass of molecular and fragment ions, respectively. These metabolites are listed in supplemental Table S5 along with the VIP and p values (Student's t test). There are several serum metabolites which showed a consistent trend of alteration (up- or down-regulation) from stage I to IV of HCC patients (Fig. 4). Glycochenodeoxycholic acid, and some metabolites including oleamide, aspartic acid, and 4-ketoglucose (not shown in Fig. 4) were consistently depleted at each of the four stages, whereas glycocholic acid and α-ketoglutaric acid fluctuated among different stages. Interestingly, some metabolites such as inosine and chenodeoxycholic acid altered differently at stage II (32), presumably because of the fact that stage II and III have drastically different pathological phenotypes, such as lymph node invasion. Therefore, inosine and chenodeoxycholic acid should be further investigated as potential markers for HCC stratification.
Fig. 4.
Bar charts showing fluctuations in integrated intensities of six representative serum differential metabolites and three urine metabolites among five phenotypic states; healthy, HCC stage I, II, III, and IV. (*, p < 0.05; **, p < 0.01, compared with healthy control).
DISCUSSION
Because the metabolomic data typically contains a large number of variables that are interrelated, multivariate statistical methods such as PCA and OPLS-DA coupled with univariate statistical methods such as Student's t test were used in this study. Therefore, feature selection from variables was performed using two parameters, a threshold of 1 and 0.05 by VIP and Student's t test P, respectively, to identify differential metabolites with biological significance as endpoints of altered interdependent biochemical pathways.
The cohort of patients with benign liver tumors (see Table I) collected in our study was highly heterogeneous, therefore, the analytical results of benign liver tumor patients are of little practical use. Because liver cirrhosis and hepatitis are chronic liver conditions that may have its own characteristic metabolomic markers, we identified a panel of markers specific for liver cirrhosis and hepatitis which were then excluded from the list of HCC markers. Altered bile acid levels associated with liver cirrhosis and hepatitis, HCC patients with and without cirrhosis and hepatitis were quantitatively determined and compared among the three conditions in Fig. 2.
Previous HCC metabolomic studies (19–21, 33) were not able to identify a sufficient number of metabolite markers because of the limitation in sample size and analytical platform(s) used. A recently published GC-MS based metabolomics study (20) identified eight serum metabolites using a library without validation of reference standards. Our combined use of two analytical platforms takes advantage of complementary analytical outcomes and therefore, broadens the “window” of important metabolic variations identified. Another advantage of using the LC-MS and GC-MS in combination is that we can cross-validate the metabolites mutually detected by these two analytical platforms. Twenty serum metabolites, including creatinine, lactic acid, nervonic acid, aspartic acid, citrulline, cysteine, cystine, serine, kynurenine, pyruvic acid, phenylalanine, oleamide, pyroglutamic acid, inosine, and ornithine, were identified in both analytical platforms with the same alteration direction (up- or down-regulation). The consistent results generated from these two platforms indicate the robustness of the metabolomic procedure used in this study. The OPLS-DA models derived from our current GC-TOFMS and UPLC-QTOFMS (both positive and negative ion mode) metabolic analysis showed good and similar separations between patients with HCC and healthy controls, highlighting the diagnostic potential of this noninvasive profiling approach.
The elevated level of conjugated bile acids appears to be associated with HCC at stage I, whereas levels of bile acids were elevated, to a lesser extent, in patients with more advanced HCC (stage II to IV) (Fig. 4). This was inconsistent with the quantitative result of bile acids. The reason for such an abrupt increase in conjugated bile acids at stage I of HCC and a gradual attenuation at stage II to IV is unknown, but is presumably associated with “acute” disruption of liver function at the early stage of tumorigenesis.
In the meantime, significantly lower levels of long-chain fatty acids and their derivates were observed in serum of HCC. Oleamide (cis-9, 10-octadecenoamide) was at a consistently low level from stage I to IV. Stearic acid, arachidonic acid, palmitic acid, myristic acid, etc. were down-regulated in the serum of HCC patients with great statistical significance. Apparently, the impaired liver function resulting from HCC impacts fatty acid metabolism and may be associated with a decreased consumption of conjugated bile acids, which in turn, resulted in a higher level of conjugated bile acids and a lower level of unconjugated bile acids in serum.
Blood samples provide an instant metabolome at the time of collection, whereas urine samples contain an average metabolomic change within a time course. GC-TOFMS based profiling identified a panel of 10 differential urinary metabolites, whereas UPLC-QTOFMS identified 28 differential metabolites in HCC patients. Among them, alanine, cysteine, cystine, cysteic acid, tyrosine, phenylalanine, and threonine were identified in both analytical platforms with the same alteration direction (up- or down-regulation).
Using either of the two panels of variables, 43 serum metabolites or 31 urine metabolites, HCC patients can be statistically separated from healthy controls by a three component PCA scores plot (supplemental Fig. S7), indicating that these metabolites hold the potential to be candidate diagnostic biomarkers.
Ornithine, citrulline, and arginine were detected significantly decreased in the serum of HCC patients as compared with healthy controls (34). Furthermore, ornithine decreased gradually from stage I to IV. Coincidentally, these three amino acids participate in the urea cycle (UC) (35), which allows for the disposal of excess nitrogen and takes place only in mammals' hepatocyte. A recent report on proteomic analysis of UC revealed the down-regulation of urea cycle enzymes (34), and the UC activity in HepG2 cells (a cell line of HCC) was also found decreased (36). As a result, aspartic acid, a nitrogen supplier in UC, was hence accumulated as the increased level of aspartic acid was detected in serum of HCC patients.
Similar to the metabolic observations in other cancers (25), higher levels of pyruvate and lactate were also discovered in the serum of HCC patients as compared with healthy controls, presumably because of higher energy consumption involving glycolysis in the solid tumor tissues.
A decreased level of serum creatinine in HCC patients was observed, because a reduced rate of creatinine production in patients with hepatic disease is likely expected because of the decreased hepatic conversion of creatine to creatinine (37). Concurrently, methionine and arginine, the amino acids involved in the synthesis of creatine (38), were also found significantly decreased in HCC serum. The decreased serum methionine was consistent with a low level of serine, one of the amino acid sources of one-carbon groups for tetrahydrofolic acid to synthesize methionine (THF, methionine cycle). A low level of cysteine and alanine in urine also suggests the down-regulation of methionine metabolism.
Alpha-fetoprotein (AFP), the most widely used tumor marker for detecting liver cancer, is affected by different pathophysiological conditions, including pregnancy, hepatitis, and the involvement of other types of cancer. Levels of AFP exceeding 50 ng/ml occur in only ∼40–60% of patients with HCC (39), and the false negative rate of diagnosis of HCC with AFP is usually 20–30%. Our metabolomic model was able to stratify between the HCC patients with AFP values higher than 20 ng/ml and healthy controls. Furthermore, HCC patients with AFP values lower than 20 ng/ml were successfully classified into the HCC group with an accuracy of 100%, using a panel of metabolite markers. Therefore, it is promising and also technically feasible to develop a panel of metabolite markers for the clinical diagnosis of the HCC patients, minimizing the false negative rate of diagnosis based on a single biomarker, such as AFP.
The aim of this study was to search for potential metabolite markers for human HCC. However, multiple phenotypes, such as cirrhosis and hepatitis, may complicate the biomarker selection for HCC and result in unique markers independent of HCC. We identified the markers resulting from cirrhosis and hepatitis conditions (see Table III) by comparing among benign liver tumor patients with liver cirrhosis and hepatitis, HCC with cirrhosis and hepatitis, and HCC without cirrhosis and hepatitis. But we were not able to investigate the metabolic influence by hepatitis or cirrhosis alone because the majority of HCC patients are accompanied with both hepatitis and cirrhosis. Therefore, the metabolic influence of hepatitis or cirrhosis is not investigated separately, which is a limitation of this study.
In summary, we have successfully applied a global metabolomic profiling approach to the study of HCC. We identified significant serum and urine metabolite markers relevant to the HCC and its different stages. These metabolite markers are involved in several key metabolic pathways such as bile acids, free fatty acids, urea cycle and methionine metabolism. Metabolites listed in Table II, such as bile acids, histidine, inosine, are of great statistical significance (high fold changes), and therefore, warrant further validation as a single biomarker or a panel of biomarkers for HCC. Several bile acids, cholic acid, glycocholic acid, deoxycholic acid and glycochenodeoxycholic acid, altered differently in concentration in the HCC patients with or without liver cirrhosis and hepatitis, which hold the potential as markers for the stratification of HCC subjects with and without cirrhosis and hepatitis (Fig. 2). The results of our study showed that the metabolomic profiling approach is a promising screening tool for the diagnosis and stratification of HCC.
Footnotes
* This work was financially supported by the National Basic Research Program of China (2007CB914700), the National Science and Technology Major Project (2009ZX10005-020) the Natural Science Foundation of Shanghai (10ZR1414800) and the National Science Foundation of China (20775048).
 This article contains supplemental Figs. S1 to S7 and Tables S1 to S8.
This article contains supplemental Figs. S1 to S7 and Tables S1 to S8.
1 The abbreviations used are:
- HCC
- Human hepatocellular carcinoma
- FDG-PET
- fluorodeoxyglucose-positron emission tomography
- AFP
- alpha fetoprotein
- HPLC
- high performance liquid chromatography
- LC-MS
- liquid chromatography-mass spectrometry
- GC-MS
- gas chromatography-mass spectrometry
- GC-TOFMS
- gas chromatography-time-of-flight mass spectrometry
- UPLC-QTOFMS
- ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry
- OPLS-DA
- orthogonal partial least squares-discriminant analysis
- PCA
- principal component analysis
- UC
- urea cycle
- VIP
- variable importance of the project
- ES+
- positive ion mode
- ES–
- negative ion mode
- TMCS
- trimethylchlorosilane
- BSTFA
- Bis(trimethylsilyl) trifluoroacetamide.
REFERENCES
- 1. El-Serag H. B., Rudolph K. L. (2007) Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576 [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization Mortality Database, WHO Statistical Information System. Availableat http://www.who.int/whosis/en/; accessed March 19, 2008
- 3. Kassahun W. T., Fangmann J., Harms J., Hauss J., Bartels M. (2006) Liver Resection and Transplantation in the Management of Hepatocellular Carcinoma: A Review. Exp. Clin. Transplant. 4, 549–558 [PubMed] [Google Scholar]
- 4.Fong T. L., Schoenfield L. J. Hepatocellular Carcinoma (Liver Cancer) available at http://www.medicinenet.com/liver-cancer/article.htm.
- 5. Anthony P. P. (2001) Hepatocellular carcinoma: an overview. Histopathology 39, 109–118 [DOI] [PubMed] [Google Scholar]
- 6. El-Serag H. B., Mason A. C. (1999) Rising incidence of hepatocellular carcinoma in the United States. N Eng J. Med. 340, 745–750 [DOI] [PubMed] [Google Scholar]
- 7. El-Serag H. B., Tran T., Everhart J. E. (2004) Diabetes increases the risk of chronic liver disease and hepatocellular carcinoma. Gastroenterology 126, 460–468 [DOI] [PubMed] [Google Scholar]
- 8. Onodera H., Ukai K., Minami Y. (1995) Hepatocellular-Carcinoma Cases with 5-Year Survival and Prognostic Factors Affecting the Survival-Time. Tohoku J. Exp. Med. 176, 203–211 [DOI] [PubMed] [Google Scholar]
- 9. Coston W. M., Loera S., Lau S. K., Ishizawa S., Jiang Z., Wu C. L., Yen Y., Weiss L. M., Chu P. G. (2008) Distinction of hepatocellular carcinoma from benign hepatic mimickers using glypican-3 and CD34 immunohistochemistry. Am. J. Surg. Pathol. 32, 433–444 [DOI] [PubMed] [Google Scholar]
- 10. Colli A., Fraquelli M., Casazza G., Massironi S., Colucci A., Conte D., Duca P. (2006) Accuracy of ultrasonography, spiral CT, magnetic resonance, and alpha-fetoprotein in diagnosing hepatocellular carcinoma: A systematic review. Am. J. Gastroenterol. 101, 513–523 [DOI] [PubMed] [Google Scholar]
- 11. Chan E. C., Koh P. K., Mal M., Cheah P. Y., Eu K. W., Backshall A., Cavill R., Nicholson J. K., Keun H. C. (2009) Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 8, 352–361 [DOI] [PubMed] [Google Scholar]
- 12. Nicholson J. K., Lindon J. C., Holmes E. (1999) ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 13. Kuo Y. T., Li C. W., Chen C. Y., Jao J., Wu D. K., Liu G. C. (2004) In Vivo Proton Magnetic Resonance Spectroscopy of Large Focal Hepatic Lesions and Metabolite hange of Hepatocellular Carcinoma Before and After Transcatheter Arterial Chemoembolization Using 3.0-T MR Scanner. J. Magn. Reson. Imaging 19, 598–604 [DOI] [PubMed] [Google Scholar]
- 14. Sreekumar A., Poisson L. M., Rajendiran T. M., Khan A. P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R. J., Li Y., Nyati M. K., Ahsan A., Kalyana-Sundaram S., Han B., Cao X., Byun J., Omenn G. S., Ghosh D., Pennathur S., Alexander D. C., Berger A., Shuster J. R., Wei J. T., Varambally S., Beecher C., Chinnaiyan A. M. (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 15. Woo H. M., Kim K. M., Choi M. H., Jung B. H., Lee J., Kong G., Nam S. J., Kim S., Bai S. W., Chung B. C. (2009) Mass spectrometry based metabolomic approaches in urinary biomarker study of women's cancers. Clin. Chim. Acta 400, 63–69 [DOI] [PubMed] [Google Scholar]
- 16. Denkert C., Budczies J., Kind T., Weichert W., Tablack P., Sehouli J., Niesporek S., Könsgen D., Dietel M., Fiehn O. (2006) Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 66, 10795–10804 [DOI] [PubMed] [Google Scholar]
- 17. Petrik V., Loosemore A., Howe F. A., Bell B. A., Papadopoulos M. C. (2006) OMICS and brain tumour biomarkers. Br. J. Neurosurg. 20, 275–280 [DOI] [PubMed] [Google Scholar]
- 18. Yan S. K., Wei B. J., Lin Z. Y., Yang Y., Zhou Z. T., Zhang W. D. (2008) A metabonomic approach to the diagnosis of oral squamous cell carcinoma, oral lichen planus and oral leukoplakia. Oral Oncol. 44, 477–483 [DOI] [PubMed] [Google Scholar]
- 19. Yang Y., Li C., Nie X., Feng X., Chen W., Yue Y., Tang H., Deng F. (2007) Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J. Proteome Res. 6, 2605–2614 [DOI] [PubMed] [Google Scholar]
- 20. Xue R., Lin Z., Deng C., Dong L., Liu T., Wang J., Shen X. (2008) A serum metabolomic investigation on hepatocellular carcinoma patients by chemical derivatization followed by gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 22, 3061–3068 [DOI] [PubMed] [Google Scholar]
- 21. Wu H., Xue R., Dong L., Liu T., Deng C., Zeng H., Shen X. (2009) Metabolomic profiling of human urine in hepatocellular carcinoma patients using gas chromatography/mass spectrometry. Anal. Chim. Acta 648, 98–104 [DOI] [PubMed] [Google Scholar]
- 22. Ni Y., Su M., Qiu Y., Chen M., Liu Y., Zhao A., Jia W. (2007) Metabolic profiling using combined GC-MS and LC-MS provides a systems understanding of aristolochic acid-induced nephrotoxicity in rat. FEBS Lett. 581, 707–711 [DOI] [PubMed] [Google Scholar]
- 23. Qiu Y., Cai G., Su M., Chen T., Zheng X., Xu Y., Ni Y., Zhao A., Xu L. X., Cai S., Jia W. (2009) Serum Metabolite Profiling of Human Colorectal Cancer Using GC-TOFMS and UPLC-QTOFMS. J. Proteome Res. 8, 4844–4850 [DOI] [PubMed] [Google Scholar]
- 24. Qiu Y., Su M., Liu Y., Chen M., Gu J., Zhang J., Jia W. (2007) Application of ethyl chloroformate derivatization for gas chromatography-mass spectrometry based metabonomic profiling. Anal. Chim. Acta 583, 277–283 [DOI] [PubMed] [Google Scholar]
- 25. Xie G. X., Ye M., Wang Y. G., Ni Y., Su M. M., Huang H., Qiu M. F., Zhao A. H., Zheng X. J., Chen T. L., Jia W. (2009) Characterization of Pu-erh Tea Using Chemical and Metabolic Profiling Approaches. J. Agri. Food Chem. 57, 3046–3054 [DOI] [PubMed] [Google Scholar]
- 26. Xie G., Plumb R., Su M., Xu Z., Zhao A., Qiu M., Long X., Liu Z., Jia W. (2008) Ultra-performance LC/TOF MS analysis of medicinal Panax herbs for metabolomic research. J. Sep. Sci. 31, 1015–1026 [DOI] [PubMed] [Google Scholar]
- 27. Jonsson P., Gullberg J., Nordström A., Kusano M., Kowalczyk M., Sjöström M., Moritz T. (2004) A Strategy for Identifying Differences in Large Series of Metabolomic Samples Analyzed by GC/MS. Anal. Chem. 76, 1738–1745 [DOI] [PubMed] [Google Scholar]
- 28. Jonsson P., Johansson A. I., Gullberg J., Trygg J. A. J., Grung B., Marklund S., Sjöström M., Antti H., Moritz T. (2005) High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal. Chem. 77, 5635–5642 [DOI] [PubMed] [Google Scholar]
- 29. Jansson J., Willing B., Lucio M., Fekete A., Dicksved J., Halfvarson J., Tysk C., Schmitt-Kopplin P. (2009) Metabolomics Reveals Metabolic Biomarkers of Crohn's Disease. Plos One 4, e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wiklund S., Johansson E., Sjöström L., Mellerowicz E. J., Edlund U., Shockcor J. P., Gottfries J., Moritz T., Trygg J. (2008) Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 80, 115–122 [DOI] [PubMed] [Google Scholar]
- 31. Ni Y., Su M., Lin J., Wang X., Qiu Y., Zhao A., Chen T., Jia W. (2008) Metabolic profiling reveals disorder of amino acid metabolism in four brain regions from a rat model of chronic unpredictable mild stress. FEBS Lett. 582, 2627–2636 [DOI] [PubMed] [Google Scholar]
- 32. Lewis G. D., Wei R., Liu E., Yang E., Shi X., Martinovic M., Farrell L., Asnani A., Cyrille M., Ramanathan A., Shaham O., Berriz G., Lowry P. A., Palacios I. F., Tasan M., Roth F. P., Min J., Baumgartner C., Keshishian H., Addona T., Mootha V. K., Rosenzweig A., Carr S. A., Fifer M. A., Sabatine M. S., Gerszten R. E. (2008) Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. J. Clin. Invest. 118, 3503–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao H., Lu Q., Liu X., Cong H., Zhao L., Wang H., Lin D. (2009) Application of H-1 NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 100, 782–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chaerkady R., Thuluvath P. J., Harsha H., Perumal V., Torbenson M., Pandey A. (2008) Quantitative proteomic approaches to identify biomarkers for hepatocellular carcinoma. HUGO's 13th Human Genome Meeting Hyderabad, India [Google Scholar]
- 35. Denkert C., Budczies J., Weichert W., Wohlgemuth G., Scholz M., Kind T., Niesporek S., Noske A., Buckendahl A., Dietel M., Fiehn O. (2008) Metabolite profiling of human colon carcinoma - Deregulation of TCA cycle and amino acid turnover. Molecular Cancer 7, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mavri-Damelin D., Eaton S., Damelin L. H., Rees M., Hodgson H. J., Selden C. (2007) Ornithine transcarbamylase and arginase I deficiency are responsible for diminished urea cycle function in the human hepatoblastoma cell line HepG2. Int. J. Biochem. Cell Biol. 39, 555–564 [DOI] [PubMed] [Google Scholar]
- 37. Cocchetto D. M., Tschanz C., Bjornsson T. D. (1983) Decreased rate of creatinine production in patients with hepatic disease: implications for estimation of creatinine clearance. Ther. Drug Monit. 5, 161–168 [DOI] [PubMed] [Google Scholar]
- 38. da Silva R. P., Nissim I., Brosnan M. E., Brosnan J. T. (2009) Creatine synthesis: hepatic metabolism of guanidinoacetate and creatine in the rat in vitro and in vivo. Am. J. Physiol. Endocrinol. Metab. 296, E256–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cottingham K. (2008) Candidate biomarkers for liver cancer. J. Proteome Res. 8, 428–428 [DOI] [PubMed] [Google Scholar]