Abstract
After the successful completion of the Human Genome Project, the Human Proteome Organization has recently officially launched a global Human Proteome Project (HPP), which is designed to map the entire human protein set. Given the lack of protein-level evidence for about 30% of the estimated 20,300 protein-coding genes, a systematic global effort will be necessary to achieve this goal with respect to protein abundance, distribution, subcellular localization, interaction with other biomolecules, and functions at specific time points. As a general experimental strategy, HPP research groups will use the three working pillars for HPP: mass spectrometry, antibody capture, and bioinformatics tools and knowledge bases. The HPP participants will take advantage of the output and cross-analyses from the ongoing Human Proteome Organization initiatives and a chromosome-centric protein mapping strategy, termed C-HPP, with which many national teams are currently engaged. In addition, numerous biologically driven and disease-oriented projects will be stimulated and facilitated by the HPP. Timely planning with proper governance of HPP will deliver a protein parts list, reagents, and tools for protein studies and analyses, and a stronger basis for personalized medicine. The Human Proteome Organization urges each national research funding agency and the scientific community at large to identify their preferred pathways to participate in aspects of this highly promising project in a HPP consortium of funders and investigators.
The success of the Human Genome Project (HGP)1 has provided a blueprint of genes encoding the entire human protein set potentially expressed in any of the ∼230 cell types that comprise the human body (the human proteome). At present, we have at least limited knowledge about the proteins of approximately two-thirds of the 20,300 protein-coding human genes mapped through the HGP. Based on the UniProtKB/Swiss-Prot database content, about 6000 (30%) of these genes currently lack any experimental evidence at the protein level; for many others, there is very little information related to protein abundance, distribution, subcellular localization, interactions, or cellular functions.
The Human Proteome Project (HPP) is designed to map the entire human proteome in a systematic effort using currently available and emerging techniques. Completion of this project will enhance understanding of human biology at the cellular level and lay a foundation for development of diagnostic, prognostic, therapeutic, and preventive medical applications. The proteomic space generated from these gene products is enormous, including up to an estimated one million different protein isoforms derived by DNA recombination, alternative splicing of primary transcripts, and numerous post-translational modifications of many types that vary with time, location, and physiologic, pathologic, and pharmacologic perturbations. These modifications expand the proteomic space by altering the primary products in a combinatorial manner. In early 2010, HUPO proposed a gene-centric approach to generate a human proteome map with an “information backbone” that would display the proteins expressed from each gene locus (1). A working groupq for an HPP was created in October 2009 by the HUPO Council to build an international consensus and a long-term plan for this project. We concluded that recent substantial advances in proteomic technologies including quantitative mass spectrometry, protein capture with antibodies, and bioinformatics for global exchange of large primary data sets and databases make the generation of such a human proteome map feasible (2, 3). As was done for the HGP, gene-centric human proteome mapping will be complemented with in-depth studies of protein variability in response to various physiologic and pathologic states. Supportive interest for the HPP has been expressed by the international scientific community, major scientific journals, industrial representatives, and funding agencies around the world. The overall plan for the HPP was launched at the 9th Annual HUPO World Congress in Sydney, Australia, on September 23, 2010. The presentation from that plenary session is available on the HPP web page at the HUPO website (http://hupo.org/research/hpp/).
The Three Pillars of the HPP
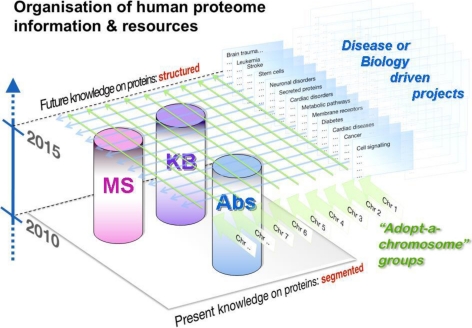
The HPP will deliver a comprehensive map of the human proteins in their biological context. It will generate publicly accessible data and informational resources supporting further exploration of the human proteome by basic and clinical scientists. The HPP will be built on the three major technological pillars of shotgun and targeted MS, polyclonal and monoclonal antibodies (Ab), and an integrated knowledge base (KB) (Fig. 1). The HPP will use the output and cross-analyses (see below) from the ongoing HUPO initiatives that have focused on tissue- and biofluid-based proteomes, as well as much other published work. The HPP will provide tools and reagents (e.g. cDNAs, peptides and their corresponding reference fragment ion spectra, recombinant proteins, antibodies, and annotation software) for the scientific community that will enable all researchers to design and perform hypothesis-driven and hypothesis-generating experiments to enhance existing knowledge, as has occurred with the HGP. The availability of reference samples and standardized informational resources will contribute to creating proteomic data with improved quality and comparability. Furthermore, the HPP will provide a blueprint for fast-track analogous projects in other species, again following the path of the HGP.
Fig. 1.
The HPP vision. The HPP is driven by two major scientific forces on top of the three pillars of MS, protein capture, and protein information databases: biology-powered projects and proteome activity mapped to a chromosomal orientation (see http://hupo.org/research/hpp/and Fig.). The HPP will deliver a comprehensive map of human proteins in their biological context.
Regarding the MS pillar, the HPP will track the development of and access to robust selected or multiple reaction monitoring (SRM/MRM)-based assays of proteotypic peptides, including stable isotope-labeled peptides, which enable high-throughput, targeted, quantitative protein measurements at a high degree of reproducibility. The short-term goal (within 2 years) is the completion of libraries of such peptides for at least one product of each gene, with critical knowledge of transitions in mass spectra based on fragment ion spectra derived from these peptides using different types of mass spectrometers (4). An intermediate-term goal (within 5 years) is the extension of the SRM library to include splice variants, single nucleotide polymorphisms, and possibly many established post-translational modifications. A specialized SRMAtlas database is currently being generated from fragment ion spectra for multiple proteotypic peptides of nearly every protein in the human proteome (5, 6), providing the information for the definitive identification and accurate quantification of each protein in biological samples (7) (www.peptideatlas.org; www.srmatlas.org). We expect that SRM-based data sets, with proper annotation and metadata, will be handled by the existing repositories (listed below). Comparison with SRM databases of other species will be facilitated (within 10 years). The entire research community will benefit from these resources.
Regarding the Ab pillar, the short-term HPP objective (within 3–5 years) is to focus on antibody-based identification and cellular and subcellular localization of protein expression. At least one protein product of each protein-coding gene will be used in the existing polyclonal antibody initiative, which has already reached 10,000 protein identifications in the Human Protein Atlas (8) (www.proteinatlas.org). Comprehensive libraries of monoclonal antibodies or renewable protein capture reagents will be established (within 5 years) (e.g. http://nihroadmap.nih.gov/proteincapture/); use will be encouraged. The long-term objectives are to construct a complete map or atlas of expression and subcellular localization of proteins in human cells, tissues, and organs during developmental stages and adult life and under various physiologic and pathologic conditions (within 10 years). Protein-specific antibodies will become widely available as reagents, and information on their specificity and usability in various assays such as Western blotting, enzyme-linked immunosorbent assays (ELISA), and other multiplexing systems will be provided through the web portal Antibodypedia (www.antibodypedia.org) with records of the use of antibodies in the PubMed literature (http://www.ncbi.nlm.nih.gov/pubmed). Comparisons of findings with MS and protein capture will be strongly encouraged and documented.
Regarding the KB pillar, the HPP working group has decided that the HPP would commit to draw upon UniProtKB/Swiss-Prot (9), PRIDE (10), PeptideAtlas (7), GPMDB (11), and Human Protein Atlas (8) databases and the ProteomeXchange (12) infrastructure for coordinating the proteomics databases through the global distributed data file-sharing system Tranche (13). Thus, the HPP will build upon resources that already exist and continue to be enhanced. It is anticipated that some of these resources will be extended to cope with specific needs of the HPP to integrate data and knowledge concerning human proteins. One of these resources, neXtProt, is currently under development specifically to become a human protein-centric knowledge platform that will address the needs of scientists aiming to connect the results of the HPP, particularly results from the MS and Ab pillars, with the full range of functional and structural knowledge about all human proteins. We recommend that participating laboratories use the emerging HUPO Proteomics Standards Initiative standardized exchange file formats. The plan calls for raw spectra for peptide and protein identification to be deposited with full annotation in Tranche. Data sets will be shared automatically with PeptideAtlas for appropriate combined analyses of the raw spectra. As noted, SRM data based on synthetic peptides will be stored in SRMAtlas, and antibodies will be referenced in Human Protein Atlas and Antibodypedia.
The HPP Web Portal
A web portal will be developed as the central focal point of the HPP for publicizing the goals, progress, and preliminary results, and for facilitating coordinated efforts. It will be composed of appropriate documents, roadmaps, and timelines, and be linked to the participating laboratories, funding agencies, major proteomic resources, and each group of HUPO initiatives. It will also provide direct links to the data sets and knowledge resources. To engage the participation of the broad scientific community, the design and development of the portal will be driven by user requirements. This web portal will also serve as a knowledge center for educational purposes, highlighting standardization of protocols employed in the HPP project.
HPP-Related Initiatives
Cross-Analyses of Specific Proteome Projects
Continuing efforts for cross-analyses of organ- and biofluid-based proteome data sets have established that data generated on different platforms can be compared after collective analysis from the primary spectra with uniform and rigorous criteria and bioinformatics tools. Such collective analyses of large data sets improve the quality of individual analyses via a cross-checking spectral library search. These PeptideAtlas analyses provide a baseline compendium of MS/MS observations, together with the Human Protein Atlas, to begin the HPP. These will be the foundation for an internal and external quality control program. In addition, we expect that the HPP will collaborate with human Protein Detection and Quantification (14).
Chromosome-centric HPP (C-HPP) Consortium
Many investigators or groups of investigators are considering ways to contribute to the HPP through projects of various types. One such approach, again in analogy to the HGP, is the C-HPP, which includes projects focused on all proteins that are coded by genes present on a particular chromosome (15). National initiatives have been announced as receiving interest and support from national funding agencies in Korea (chr 13), Russia (chr 18), and Iran (chr Y). Groups of scientists from Sweden, Spain, Japan, Canada, China, France, Switzerland, Norway, Germany, India and the United States have initiated discussions regarding the building of projects targeted on additional chromosomes (1, 2, 3, 6, 8, 14, 17, 19, 21, and X; not ordered by country). Thus, the C-HPP consortium will help map and annotate subsets of the human proteome. The C-HPP groups are expected to share emerging high-quality information on human proteins, as well as valuable reagents (SRM peptide information, antibodies, and other protein capture reagents). The overall HPP working group (and the future HPP consortium, see below) will encourage this “adopt-a-chromosome” strategy by implementing the same guidelines, operational approaches, data submission, and sharing of information in the portal. HUPO itself will not assign chromosome projects to specific countries or groups of scientists until the C-HPP guidelines are established.
Numerous Biologically Driven Projects
Many biology- or disease-driven projects will contribute to the HPP by feeding the HPP KB. Thus, the organization of the HPP and the population of the HPP KB will be derived from two major scientific realms: individually initiated and individually funded biology-driven projects, and the consortium mapping proteome subsets and their genes to specific chromosomal locations (see Fig. 1). The HPP will also interact with other international projects, such as the International Cancer Genomic Consortium, the Protein Data Bank, and the International Human Epigenomics Consortium.
Deliverables
The predicted deliverables of HPP are: (a) structured information on human proteins (protein parts list) and (b) well-described and available reagents and tools for protein studies. The investigators, through the HPP working group and Consortium (see below), will devise metrics to describe annually the extent of progress on identifying and characterizing the human proteome.
Within 3 years we expect to have SRM-based spectral libraries for multiple proteotypic peptides for at least one protein product of each of the 20,300 protein-coding genes, an SRMAtlas, and stably labeled peptide standards. We expect to have an expanded Human Protein Atlas with polyclonal antibodies to characterize the tissue expression and subcellular localization of more than 12,000 gene products. We will stimulate cross-comparisons of tissue expression by antibody and MS methods. The performance of antibodies will be characterized collectively by the proteomics community in Antibodypedia. Data sets and proteome atlases will be connected via Tranche and ProteomeXchange; the HUPO Proteomics Standards Initiative provides a standardized exchange file format that will be widely implemented. NeXtProt and UniProtKB/Swiss-Prot will be available as knowledge bases for human proteins and proteomes of other species.
Within 5 years we will extend SRM analyses and KBs to splice variants and post-translational modifications. We will have renewable protein capture methods and reagents for the characterization of protein tissue expression and modifications during physiologic and pathologic perturbations. We will have a greatly expanded ProteomeXchange comprising EBI/PRIDE, ISB/PeptideAtlas, Tranche, GPMDB, and any additional major data repositories. We will have substantial information comparing MS and protein capture findings for each organ-based or biofluid-based proteome.
Upon completion, HPP will inspire and encourage other stakeholders, beyond the basic research community, to use or target proteins for diagnosis, prognosis, prevention, therapy, and potentially cure of diseases, effects that will improve human health worldwide. Additional findings, currently not known or publicly available, will be catalyzed by the availability of an organized structure of the human proteome. Furthermore, the HPP will provide a blueprint for fast-track analogous projects in other species.
Costs and Funding
Each country will define its own process for HPP funding, with specific HPP calls for C-HPP projects and a great variety of biology- and disease-driven projects. Scientists will procure their own funding through their preferred national or regional funding agencies. Through its convening and educational functions, HUPO will seek to stimulate more funding for the HPP without being involved directly in the selection of projects to be funded.
Governance
A HPP Consortium will be created with a light organizational structure to promote interaction among funders (providing oversight), an international scientific steering committee (setting guidelines), and scientific groups and research institutions. This operation may or may not be directly associated with HUPO itself. In the immediate future, the HUPO HPP working group will continue to convene high-level discussion groups and reach out to the proteomics community and funding agencies. HUPO may be called upon to function as a supporting organization for certain aspects of the HPP, such as coordination of the C-HPP, coordination of KBs, and quality control and publication guidelines.
Institutions involved in a HPP labeled and funded project, research agencies involved in the funding of HPP labeled projects, and private or public institutions developing research projects on the human proteome and agreeing on the policies and guidelines of the HPP would be entitled to join or be represented in the HPP Consortium. Members of the HPP Consortium will be informed regularly about any decisions made regarding the structure of the HPP and on the progress of the HPP. They will be welcomed to all scientific events dealing with HPP results.
A governing body will consist of representatives of research institutions and funding agencies. It will encourage and oversee strategies to be followed by the HPP Consortium to ensure a match between national and international funding and HPP research plans to produce the expected deliverables on schedule.
An International Scientific Steering Committee will be constituted by recruiting the principal scientific leaders in the field of proteomics and some respected scientists involved in other biomedical research areas. The International Scientific Steering Committee will review and recommend guidelines that will be proposed by ad hoc working groups for the three pillars, the data set cross-analyses, and the C-HPP.
CONCLUSIONS
HUPO will continue to convene, encourage, facilitate, and coordinate human proteomics cross-analyses. It will stimulate dissemination of results, facilitate implementation of new technologies and methods, and organize education and training. HUPO is not seeking to claim responsibility for the specific HPP bodies for the governance of HPP. Nevertheless, for the present period, HUPO will continue to lead the creation of the HPP Consortium. The leaders of the C-HPP Consortium have been named: YK Paik (Korea) as chair and WS Hancock (USA) and G Marko-Varga (Sweden) as co-chairs. A renewed HPP working group will be charged with this objective and will help to establish the basic principles for action of this Consortium. The present HPP working group and the participants at the HUPO World Congress in Sydney have decided that now is the time to initiate the “Human Proteome Project.” The major technology platforms and data-sharing and database analytical capabilities are now in place. Major deliverables are within reach. Multiple national funding agencies are hosting or scheduling meetings to become part of this project. Thus, HUPO urges each national research funding agency and the scientific community at large to identify their preferred pathways to participate in aspects of this highly promising project in a HPP consortium of funders and investigators.
Footnotes
1 The abbreviations used are:
- HGP
- Human Genome Project
- HUPO
- Human Proteome Organization
- HPP
- Human Proteome Project
- KB
- knowledge base
- SRM
- selective reaction monitoring
- C-HPP
- chromosome-centric HPP.
REFERENCES
- 1. HUPO - The Human Proteome Organization (2010) A gene-centric human proteome project. Mol. Cell Proteomics 9, 427–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Editorial (2010) The call of the human proteome. Nat. Methods 7, 661. [DOI] [PubMed] [Google Scholar]
- 3. Nilsson T., Mann M., Aebersold R., Yates J. R., 3rd, Bairoch A., Bergeron J. J. (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods 7, 681–685 [DOI] [PubMed] [Google Scholar]
- 4. Anderson L., Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 5. Picotti P., Lam H., Campbell D., Deutsch E. W., Mirzaei H., Ranish J., Domon B., Aebersold R. (2008) A database of validated assays for the targeted mass spectrometric analysis of the S. cerevisiae proteome. Nat. Methods 5, 913–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. (2010) High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 [DOI] [PubMed] [Google Scholar]
- 7. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., Aebersold R. (2006) The PeptideAtlas project. Nucleic Acids Res. 34, D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berglund L., Björling E., Oksvold P., Fagerberg L., Asplund A., Szigyarto C. A., Persson A., Ottosson J., Wernérus H., Nilsson P., Lundberg E., Sivertsson A., Navani S., Wester K., Kampf C., Hober S., Pontén F., Uhlén M. (2008) A gene-centric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics 7, 2019–2027 [DOI] [PubMed] [Google Scholar]
- 9. The UniProt Consortium (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39, D214–D219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., Apweiler R. (2005) PRIDE: the proteomics identifications database. Proteomics 5, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 11. Fenyö D., Eriksson J., Beavis R. (2010) Mass spectrometric protein identification using the global proteome machine. Methods Mol. Biol. 673, 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Craig R., Cortens J. P., Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 13. Smith B. E., Hill J. A., Gjukich M. A., Andrews P. C. (2011) Tranche distributed repository and ProteomeCommons.org. Methods Mol. Biol. 696, 123–145 [DOI] [PubMed] [Google Scholar]
- 14. Anderson N. L., Anderson N. G., Pearson T. W., Borchers C. H., Paulovich A. G., Patterson S. D., Gillette M., Aebersold R., Carr S. A. (2009) A human proteome detection and quantitation project. Mol. Cell Proteomics 8, 883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hancock W., Omenn G., Legrain P., Paik Y. K. (2011) Proteomics, human proteome project, and chromosomes. J. Proteome Res. 10, 210. [DOI] [PubMed] [Google Scholar]