Abstract
The heterochromatin-like structure formed by the yeast silent information regulator complex (SIR) represses transcription at the silent mating type loci and telomeres. Here, we report that tight protein–DNA complexes induce ectopic recruitment of the SIR complex, promoting gene silencing and changes in subnuclear localization when cis-acting elements are nearby. Importantly, lack of the replication fork-associated helicase Rrm3 enhances this induced gene repression. Additionally, Sir3 and Sir4 are enriched genome-wide at natural replication pause sites, including tRNA genes. Consistently, inserting a tRNA gene promotes SIR-mediated silencing of a nearby gene. These results reveal that replication stress arising from tight DNA–protein interactions favors heterochromatin formation.
Keywords: heterochromatin, replication, SIR, Rrm3, nuclear organization, lacO
In budding yeast, repressed chromatin is generated at telomeres and cryptic mating type loci (HM) by the recruitment of a complex of silent information regulators; namely, Sir2, Sir3, and Sir4 (Rusche et al. 2003; Moazed et al. 2004). These silent loci are preferentially located at the nuclear envelope, forming foci that sequester SIR factors (Gotta et al. 1996) in a manner comparable with heterochromatic chromocenters, which sequester HP1 in higher eukaryotes (Maison and Almouzni 2004). At HM loci, repression is nucleated by short silencer elements that flank the target genes. These elements contain combinations of binding sites for Rap1, Abf1, Sum1, and the origin recognition complex (ORC) (Rusche et al. 2003; Irlbacher et al. 2005). Notably, all of these factors also have functions outside silencing: Rap1 and Abf1 act as activators at many gene promoters, Sum1 is a specific repressor of meiotic genes also involved in the control of replication initiation, and ORC is the replication initiator that binds both active and inactive origins of replication.
One issue is how the combination of binding sites for factors with independent roles in the cell can create a silencer able to nucleate gene silencing. The current view is that the property of silencers emerges from the close juxtaposition of these factors, three of which have been shown to interact with one or more Sir proteins (Rusche et al. 2003). No single binding site for any one of these factors by itself can create a sufficiently high local concentration of Sir proteins to sustain silencing, but in combination, these factors can.
Here we show that tight DNA–protein interactions can contribute to the formation of silent chromatin. Importantly, this effect is increased in the absence of the Rrm3 helicase, known to facilitate replication crossing nonhistone protein–DNA complexes (Ivessa et al. 2003), thus revealing a novel mechanism linking replication stress with gene repression.
Results and Discussion
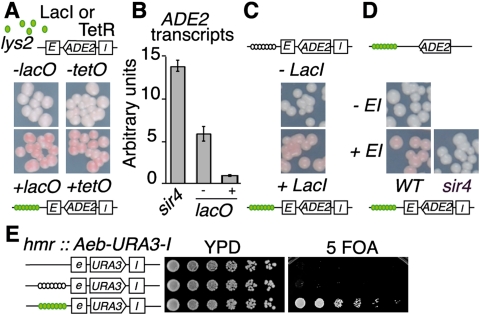
The efficiency of HM silencers is sensitive to the local SIR concentration and thus to the distance to telomeres, which are located in compartments of the nucleoplasm enriched in silencing factors (Taddei et al. 2010). Accordingly, the ADE2 reporter gene flanked by two functional silencers of the left HM (HML; referred to as lys2:E-ADE-I) is weakly repressed by the SIR complex when integrated at the LYS2 locus, located 339 kb away from a telomere (Fig. 1A,B; Maillet et al. 2001). Unexpectedly, we found that integration of an array of 256 lac operators (lacO) (Robinett et al. 1996) 1.5 kb upstream of this construct was sufficient to reduce ADE2 expression, as seen by both a colony color assay and quantitative RT–PCR (Fig. 1A,B). This transcriptional repression was observed only in strains expressing the LacI (lacO-binding protein) fused or not to GFP (Fig. 1C; Supplemental Fig. S1A). A similar ADE2 repression was observed when a tet operator (tetO) array was integrated at this locus in a strain expressing the tetO-binding protein TetR-GFP (Fig. 1A). Thus, arrays of sequences bound by either the LacI or the TetR proteins induce the repression of a reporter gene flanked by silencer elements away from a “heterochromatic environment.”
Figure 1.
Arrays of DNA–protein complexes cooperate with cis-acting elements to induce SIR-dependent silencing in budding yeast. (A) Yeast cells expressing GFP-LacI (yAT1023) or GFP-TetR (yAT525) fusions bear an ADE2 gene flanked by the E and I HML silencers integrated at LYS2 (lys2:E-ADE2-I) to monitor ectopic silencing. These strains form white colonies when ADE2 is expressed. Integration of 256 lacO (yAT58) or 2x112 tetO (yAT557) repeats 1.5 kb upstream of the reporter construct in strains expressing the cognate fusion proteins gives rise to pink colonies, indicative of ADE2 repression. (B) ADE2 mRNA levels were determined by quantitative real-time PCR on mRNA isolated from strains bearing the E-ADE2-I reporter system and expressing GFP-LacI in sir4 mutant (yAT1394) and wild-type context without (yAT1023) or with (yAT58) a lacO array integrated at the LYS2 locus. Shown are ADE2 transcript levels normalized to ACT1 mRNA. Error bars denote the SEM of three independent experiments. (C) Gene repression induced by lacO array insertion requires the expression of the GFP-LacI fusion. ADE2 repression is monitored as in A in strains expressing (yAT752) or not (yAT751) the GFP-LacI fusion. (D) ADE2 expression is monitored as in A in strains expressing GFP-LacI fusions with (yAT415) or without (yAT414) silencers flanking the ADE2 reporter gene in the wild type or sir4 mutant (yAT502). (E) lacO array integration at HMR induces silencing of URA3 adjacent to a modified E silencer (hmr:Aeb-URA3), in which the Rap1 and Abf1 elements are replaced by four lexA-binding sites (yAT1504). Serial fivefold dilutions of strains, without (yAT1504) or with (yAT1505) a lacO array inserted 970 pb upstream of the hmr:Aeb silencer and expressing the GFP-LacI fusion (yAT1506) as indicated, are grown on YPD or 5-FOA plates. Growth on 5-FOA reflects URA3 silencing.
Importantly, this repression depended on an intact SIR complex and required the presence of the E and I silencers flanking the reporter gene (Fig. 1B,D; Supplemental Fig. S1B). Thus, protein-bound arrays act as protosilencers, defined in Saccharomyces cerevisiae as DNA elements incapable of establishing silencing on their own but able to cooperate with a silencer to locally promote the formation and maintenance of a heterochromatin-like structure (Fourel et al. 2002). Consistent with this notion, lacO or tetO arrays bound by their cognate proteins induced silencing when integrated as far as 8 kb away from the silencer, although the silencing decreased with increasing distance between the lacO and silencer (Supplemental Fig. S1C). Furthermore, this protosilencer effect correlated directly with the number of lacO units in the array (Supplemental Fig. S1D).
In order to test whether LacI-bound lacO arrays can act as protosilencers in a different context, we introduced a lacO array 1 kb upstream of a mutated version of HMR missing the Rap1- and Abf1-binding sites of the E silencer element. This crippled version of HMR is unable to nucleate repression of the URA3 reporter gene introduced at this locus (Chien et al. 1993), as shown by the absence of growth on 5-Fluoroorotic acid (5-FOA) plates. Again, integration of the lacO array in a strain expressing the LacI protein induced the silencing of this locus (Fig. 1E). Therefore, LacI-bound lacO arrays are not only able to favor silencing mediated by the ectopic HML silencers, but are also able to compensate for the absence of cis elements essential for silencing at HMR. Moreover, a LacI/lacO array integrated 8.3 kb from the telomere VI-R was also able to cooperate with the telomeric TG repeats to induce gene silencing of the URA3 reporter gene located 6 kb downstream (Supplemental Fig. S1E). Thus, the protosilencer activity of the LacI-bound lacO array appears to be a general phenomenon that can affect different reporter genes at different loci.
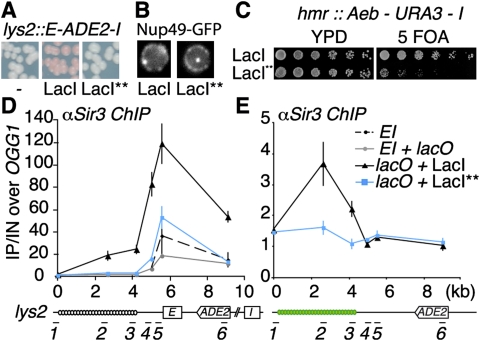
Since the combinations of either lacO/LacI or tetO/TetR showed similar protosilencer activities despite the absence of any homology between these operators or their binding proteins, we reasoned that the common feature responsible for this effect could be the strong affinity of these proteins for their binding sites (Lewis 2005; Ramos et al. 2005). We tested this hypothesis by reducing this binding affinity. First, we used tetracycline to compete with the TetR/tetO interaction and found that 1μg/mL was sufficient to suppress the protosilencer effect (Supplemental Fig. S2A). We next tested a set of LacI variants expected to have weak affinity for their binding sequences based on previous studies (Falcon and Matthews 1999). Notably, one of these variants, LacI** (see the Materials and Methods), enabled the detection of a lacO array in vivo without interfering with the expression of a neighboring silencer-flanked reporter gene (Fig. 2A,B; Supplemental Fig. S2B,C), both at the ectopic HML silencers and at the crippled HMR locus (Fig. 2C). These findings strongly argue that the effect of the lacO/LacI or tetO/TetR arrays on silencing is due to the high affinity of these proteins for their DNA-binding sequence.
Figure 2.
Arrays of tight DNA–protein complexes recruit the SIR complex. (A) lacO array bound by a LacI variant with low affinity does not induce silencing of a silencer-flanked reporter gene. Expression of the ADE2 reporter gene at LYS2 is monitored as in Figure 1A, in strains expressing (yAT752) or not (yAT751) the commonly used GFP-LacI or its variant, GFP-LacI** (yAT755). (B) lacO arrays integrated at the LYS2 locus are detected with GFP-LacI (yAT752) or its LacI** variant (yAT755) in strains expressing the GFP-Nup49 fusion. (C) Silencing is monitored at the hmr:Aeb-URA3 reporter as in Figure 1E in strains expressing either version of the GFP-LacI as indicated (yAT1506 and yAT1507). (D,E) Sir3 association with chromatin assessed by ChIP using antibodies against Sir3. Primers are described in Supplemental Table S2. Primer pair (1) was used as a reference (0 kb) to plot ChIP results according to the distance to this primer pair. (D) LacI-bound array increases Sir3 recruitment at the LYS2 locus bearing the silencer-flanked ADE2 reporter gene (yAT826). In contrast, unbound (yAT825) or LacI**-bound (yAT827) lacO arrays show Sir3 recruitment similar to that in wild-type strain lys2:E-ADE2-I (yAT1023). (E) Sir3 recruitment is detected by ChIP at a lacO array in a strain expressing the LacI protein (yAT995) but not the LacI** variant (yAT996) in the absence of silencer at the LYS2 locus. Error bars denote the SEM of three independent experiments.
To check whether the repression induced by the LacI-bound lacO array reflects an increase in Sir protein recruitment, we performed chromatin immunoprecipitation (ChIP) experiments to monitor the association of Sir3 and Sir4 at this locus. We observed a basal recruitment of Sir3 and Sir4 at the E silencer and at the ADE2 reporter gene flanked by the HML silencers with or without an unbound lacO array located 1.5 kb upstream of the silencers (Fig. 2D; Supplemental Fig. S2D). This basal recruitment is consistent with the 2.3-fold repression of the reporter gene by the SIR complex (Fig. 1B). Strikingly, both Sir3 and Sir4 recruitment increased dramatically when the lacO array was bound by the commonly used LacI but not when bound by the LacI** variant (Fig. 2D; Supplemental Fig. S2D). Thus, the close proximity of a tightly bound lacO array favors the recruitment of silencing factors at a reporter gene flanked by silencer elements.
Intriguingly, we observed that Sir3 and Sir4 were significantly enriched along the LacI-bound lacO array. We thus tested whether tightly bound arrays had the ability to recruit the SIR complex in the absence of a neighboring silencer. Indeed, we found that Sir3 was enriched 3.5-fold above the background level at lacO arrays in cells expressing the common LacI but not in cells expressing the LacI** variant (Fig. 2E). Although this recruitment alone was not sufficient to increase the silencing of the reporter gene (Supplemental Fig. S1B), it could cooperate with silencer elements to reach the threshold of SIR recruitment necessary for silencing.
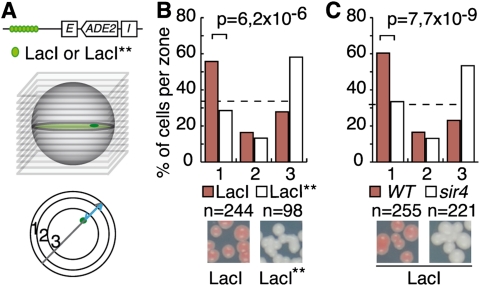
Since silent chromatin is often found at the nuclear periphery, we wondered whether SIR recruitment at LacI-bound lacO arrays affected the localization of the tagged locus. We thus compared the localization of the HML silencers flanking ADE2 when introduced at the lacO-tagged LYS2 locus in strains expressing either LacI or LacI** fused to GFP. The locus was found enriched at the nuclear periphery (zone 1) in strains expressing the GFP-LacI fusion, while it was found mostly in the nuclear interior (zone 3) in strains expressing the LacI** variant (Fig. 3A,B). Thus, arrays of protein tightly bound to DNA can affect both the transcription and the localization of the tagged locus.
Figure 3.
Tightly bound lacO arrays cooperate with cis-acting elements to induce Sir4-dependent perinuclear localization. (A) Position of the LYS2-tagged locus relative to the nuclear envelope. Z-stacks were acquired from strains bearing a lacO array inserted at the LYS2 locus and expressing Nup49-GFP and either the GFP-LacI or the GFP-LacI** mutant. The localization of the tagged locus in one of the three equal concentric zones was scored on the corresponding focal plane (Hediger et al. 2002). (B) Tightly bound lacO arrays induce the perinuclear association of the lys2:E-ADE2-I locus. This locus is localized either mostly at the nuclear periphery or mostly in the nuclear interior in wild-type cells expressing the common GFP-LacI (yAT752) or its variant, GFP-LacI** (yAT755), respectively. The colony color of the same strains is shown at the bottom of the graph. (C) The perinuclear localization of the LacI-tagged lys2:E-ADE2-I locus depends on SIR4. (Bottom) Subnuclear localization of the lys2:E-ADE2-I and the colony color as in B for the wild type (yAT58) and its sir4 derivative (yAT502) are shown.
Previous studies have shown that silencing can be either a cause or a consequence of perinuclear anchoring (Andrulis et al. 1998; Taddei et al. 2004). We first asked whether the silencing induced by lacO/LacI arrays could stem from the perinuclear anchoring of the locus by bringing it to an area of high Sir protein concentration. Following this hypothesis, lacO/LacI arrays should not induce silencing in a yku70 mutant, in which the SIR complex is not concentrated at the nuclear periphery (Laroche et al. 1998). However, the lacO/LacI-induced silencing was still efficient in this mutant, arguing that the concentration of SIR at the nuclear periphery was not necessary to induce silencing in this context and that the silencing is not a direct consequence of perinuclear localization in this case (Supplemental Fig. S3A).
Reciprocally, we tested whether the formation of silent chromatin induced by the lacO/LacI array could be the cause of its perinuclear anchoring. Indeed, we showed previously that recruiting Sir4 to an internal locus is sufficient to induce its perinuclear localization (Taddei et al. 2004). Accordingly, we found that SIR4 deletion restored the internal localization of the lacO/LacI-tagged locus (Fig. 3C), while deleting SIR3 or SIR2 had a weaker effect (Supplemental Fig. S3B). These results suggested that the recruitment of Sir4 is responsible for the perinuclear localization of the locus.
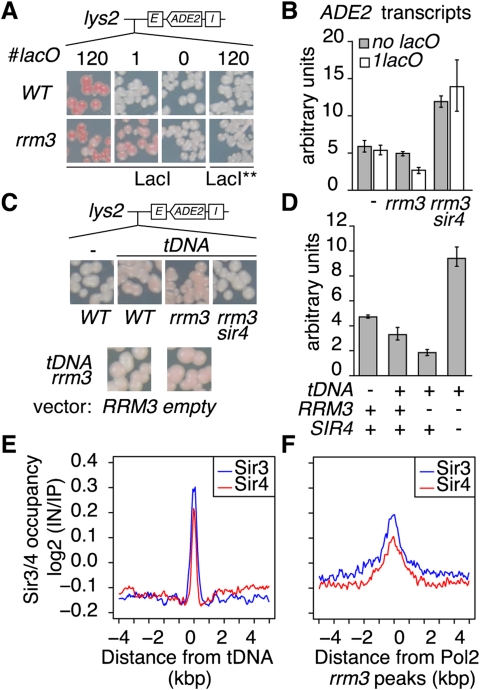
We next sought the mechanism responsible for the recruitment of the SIR complex at arrays of tight DNA–protein complexes. Interestingly, LacI-bound arrays have been shown to induce replication fork stalling in bacteria (Possoz et al. 2006). We thus explored the possibility that replication stress was responsible for this recruitment. Indeed, previous reports showed that sites of DNA stress such as double-strand breaks or chromatin stretching recruit SIR proteins (Martin et al. 1999; Mills et al. 1999; Thrower and Bloom 2001). Strains bearing a LacI-bound lacO array exhibited no cell cycle defect, indicating that, if these arrays induce replicative stress, it is likely to be transient and to not be detected by the checkpoint machinery, as shown for a single replication block in Schizosaccharomyces pombe (Lambert et al. 2005). Consistent with the notion that LacI-bound arrays create a replicative stress, we found that the stability of these arrays were compromised in strains deleted for components of the checkpoint machinery (Supplemental Fig. S4A), indicating that the cells have difficulty in replicating LacI-bound arrays. Similarly, lacO arrays became highly unstable in the absence of the Rrm3 DNA helicase (Fig. 4A), which helps replication forks move across protein–DNA complexes (Ivessa et al. 2003).
Figure 4.
rrm3-sensitive replication pause sites induce SIR recruitment. Expression of the ADE2 reporter gene at LYS2 was monitored as in Figure 1. (A,B) A single LacI-bound lac operator is sufficient to promote silencing in the absence of the Rrm3 DNA helicase. (A) Colony color assay performed in the wild-type (yAT826, yAT528, yAT1023, and yAT827) or rrm3 mutant (yAT945, yAT944, yAT1024, and yAT886) strains expressing either LacI or the LacI** variant and bearing 120, one, or no lacO sites inserted 1.5 kb upstream of the E-ADE2-I reporter gene at the LYS2 locus. (B) ADE2 expression monitored in the wild-type strain with (yAT528) or without (yAT1023) a single lacO/LacI complex integrated upstream of the lys2:E-ADE2-I construct in rrm3 (yAT944 and yAT1024, respectively) or rrm3 sir4 mutants (yAT1425 and yAT1384, respectively) as indicated. (C,D) Integration of a tRNA gene (tDNA) favors SIR4-dependent silencing in the absence of Rrm3. (C, top panel) ADE2 expression monitored in strains with (yAT1294) or without (yAT1023) a tDNA integrated 5.7 kb upstream of the lys2:E-ADE2-I construct in wild-type strains or in rrm3 (yAT1295) or rrm3 sir4 (yAT1300) mutants as indicated. Expression of a wild-type copy of RRM3 rescues the increased silencing observed in the rrm3 mutant strain. (D) ADE2 mRNA levels were determined by quantitative real-time PCR on mRNA isolated from the strains shown in C. (E) Sir3 and Sir4 are enriched at tDNAs. Mean profiles of Sir3/Sir4 occupancy (Sperling and Grunstein 2009) around tDNAs. The enrichment profiles were computed as the log2 score of immunoprecipitate versus input, on both sides of each tDNA center (annotations from the Saccharomyces Genome Database), in 50-base-pair adjacent windows as in Sperling and Grunstein (2009). (F) Sir3 and Sir4 are enriched at rrm3-sensitive replication pause sites. Mean profiles of Sir3 and Sir4 occupancy (Sperling and Grunstein 2009) were computed as in E around DNA Pol2 peak centers detected in RRM3-deleted strains (Azvolinsky et al. 2009).
Importantly, we found that, contrary to the situation in wild-type strains, in an RRM3-deleted strain, a single LacI-bound lacO was sufficient to induce SIR-dependent silencing of the neighboring silencer-flanked reporter gene inserted at the LYS2 locus (Fig. 4A–B). Moreover, this repression was dependent on SIR4 and the presence of the flanking silencers (Supplemental Fig. S4B,C). In contrast, no detectable silencing occurred in the absence of the lacO sequence in an rrm3 mutant strain expressing LacI, or in the presence of 120 lacO bound by the LacI** variant, ruling out an indirect effect of RRM3 deletion on silencing (Fig. 4A,B). Thus, a single lacO, tightly bound by a LacI dimer, is sufficient to induce silencing in the absence of the Rrm3 DNA helicase, strongly indicating that a replicative stress is at the origin of SIR complex recruitment. To further test this hypothesis, we monitored the effect of replacing the LacI-bound lacO array by a tRNA gene [tS(AGA)E, referred to as tDNA], known to induce replication pausing that is increased in the absence of Rrm3 (Ivessa et al. 2003). Strikingly, we found that this tRNA gene induced a weak but reproducible decrease in the expression of the silencer-flanked ADE2 gene whose promoter was located 5.7 kb downstream (Fig. 4C,D). Supporting the hypothesis that this transcriptional silencing stems from a replicative stress, ADE2 repression further increased in the absence of Rrm3 in a SIR4-dependent manner. This demonstrates that natural replication stress can also cooperate with silencer elements to establish SIR-dependent silencing.
Consequently, we checked whether Sir proteins were enriched at tDNA by analyzing the high-resolution genome-wide binding site maps of Sir3 and Sir4 published by Sperling and Grunstein (2009). Averaging Sir3 and Sir4 occupancy at tRNA genes revealed a narrow peak of both Sir3 and Sir4 (Fig. 4E). More generally, we tested whether natural replication stress sites were associated with the SIR complex at a genome-wide scale. This was achieved by averaging Sir3 and Sir4 occupancy (Sperling and Grunstein 2009) at replication pause sites detected by genome-wide mapping of loci enriched for the catalytic subunit of the DNA replication polymerase ɛ, Pol2 (Azvolinsky et al. 2009). Interestingly, Sir4 but not Sir3 showed a weak enrichment around the Pol2 peaks detected in wild-type cells (Supplemental Fig. S4D). In contrast, both Sir3 and Sir4 were clearly enriched in wild-type cells at Pol2 peaks detected in an rrm3 mutant (Fig. 4F), in which pause sites corresponding to stable protein–DNA complexes are exacerbated (Azvolinsky et al. 2009). Thus, Sir3 and Sir4 enrichments appear as general features of replication pause sites.
Together, our data indicate that arrays of protein tightly bound to DNA, such as LacI and TetR, or the stable association of the RNA polymerase III transcription initiation factor complex (TFIIIC) at tRNA genes induce the recruitment of silencing factors. Although weak, this SIR recruitment can cooperate with silencers or telomeric repeats if located in the vicinity (<8 kb), leading to silencing and association of the locus with the nuclear periphery (Supplemental Fig. S5).
Stable DNA–protein complexes are known to induce replication fork pausing and possibly replication stress. Intriguingly, several previous studies have shown that damaged chromatin is recruited to the nuclear periphery (Gartenberg 2009). However, the lacO/LacI-induced relocalization to the nuclear periphery does not appear to be the consequence of persistent DNA damage, but rather to result from Sir4 recruitment (Fig. 3; Supplemental Fig. S3).
Our data suggest that SIR proteins are recruited to stable DNA–protein complexes as a consequence of the replication stress occurring at these sites. Supporting the role of DNA replication in SIR recruitment, a tightly bound lacO array, integrated at an internal locus bearing an ectopic copy of the HML silencers, is recruited to the nuclear periphery only in S phase in a cac1 mutant in which silent chromatin is unstable (Supplemental Fig. S6A,B).
Interestingly, replication has been proposed to be required to establish silencing in S. cerevisiae (Miller and Nasmyth 1984). Although this notion has been challenged (Kirchmaier and Rine 2001; Li et al. 2001), it is noteworthy that Sir proteins are recruited at HM silencers, telomeric regions, and tRNA genes that are sites of transient replication fork pausing (Ivessa et al. 2003). At these sites, Rrm3 helps the replication fork to pass through the DNA–protein complexes formed by Rap1, Orc1, and Abf1, even in the absence of Sir proteins (Ivessa et al. 2003). Importantly, we showed that a tRNA gene, which represents a natural replication pause site, also promotes SIR-mediated silencing when inserted close to an ectopic copy of the HML silencers. However, in normal cells, massive and stable SIR recruitment occurs only at sites harboring combinations of DNA–protein complexes that have affinity for components of the SIR complex (i.e., Rap1, Abf1, and Orc1). It is thus possible that replication fork pausing imposed by these complexes contributes to SIR complex recruitment at these sites, where they are then maintained by the affinity of these proteins for Sir3 or Sir4. Consistent with our hypothesis, SIR-dependent silencing of cryptic mating type loci in Kluyveromyces lactis, another yeast closely related to S. cerevisiae, requires a different set of DNA-binding proteins, including a yet unknown protein, Reb1 and Ume6 (Sjostrand et al. 2002; Barsoum et al. 2010). Interestingly, direct interaction between these two latter proteins and the SIR complex could not be shown so far (Barsoum et al. 2010), suggesting that any protein tightly bound to DNA can help recruit the SIR complex.
Although the detailed mechanism leading to the recruitment of silencing factors remains to be deciphered, it may well be conserved in other species. Indeed, previous work in Arabidopsis thaliana showed that transgenic repeats frequently associate with pericentric heterochromatin, and that this frequency is further increased for LacI-bound lacO arrays compared with unbound arrays (Pecinka et al. 2005). Furthermore, in a human cell line, similar to the situation in yeast, HP1α localization at lacO arrays is triggered by tight binding of LacI but not LacI** (P Beuzer, M Dubarry, A Taddei, and G Almouzni, unpubl.). Thus, the recruitment of silencing factors at arrays of protein tightly bound to DNA likely reflects a widespread phenomenon. Importantly, LacI-bound arrays were also shown to induce replication fork stalling in human cells as in bacteria (Possoz et al. 2006; Jegou et al. 2009). Links between replication and transcriptional repression have been reported recurrently in many different species. However, it remained unclear how general replication factors could trigger silencing factors at specific sites. Here we propose that replication stress contributes to the establishment of silencing at natural sites in eukaryotic genomes, possibly contributing to genome integrity by preventing collision between the replication and transcription machineries.
Our results also have important technical bearings. Operator-based gene-tagging systems have been used broadly over the last decade, and have proven very useful for following the dynamics of individual loci or for purifying associated complexes. Our data uncover that, in some instances, using such systems could introduce a significant bias on both the localization and the expression status of the tagged locus. Furthermore, we characterized a LacI variant that does not introduce this bias. This variant will be helpful to both revisit previous observations and study chromatin dynamics in the future.
Materials and methods
Media and growth conditions
Yeast cells were grown in either YPD-rich medium (yeast extract–peptone–dextrose; Difco and Carlo Erba Reagent) or synthetic medium (YNBAS from MP Biomedicals), supplemented with 2% glucose (w/v) and the appropriate complete supplement mixture (CSM from BIO101) as in Ruault et al. (2011). The selection for operator arrays was maintained by growing cells on synthetic medium lacking tryptophan.
Strains and plasmids
Genotypes of the strains used in this study are described in Supplemental Table S1. Details of strains and plasmid constructions are provided in the Supplemental Material.
Silencing assays
For the ADE2 color colony assay, cells were plated on synthetic medium lacking tryptophan (SC-TRP) for 3 d at 30°C and then shifted for roughly 3 d to 4°C to allow the pink color to develop. Strains presented in a given figure panel were always cropped from a picture of the same plate to allow proper comparison.
The expression of the URA3 reporter at HMR and telomere VI-R was monitored by a spot assay (Gottschling et al. 1990) on YPD-rich medium or 5-FOA medium as in Ruault et al. (2011).
Microscopy
For in vivo position analysis, a Metamorph-driven inverted Nikon TE2000 microscope was used to capture 21 image stacks of 0.2-μm step size. The zonal position of the GFP spot was determined on one focal section as described in Hediger et al. (2002) on cells grown on SC-TRP plate for 20 h.
Quantitative transcript analysis
Total RNA was isolated from yeast cells using RNeasy kit (Qiagen) and then DNase-treated (Qiagen) to remove contaminating DNA. First strand cDNA was prepared from 0.5 μg of RNA and random hexamers using SuperScript III (Invitrogen) for 1 h at 42°C in 50 μL. Primer sequences are provided in Supplemental Table S2. Detailed procedure of qPCR analyses can be found in the Supplemental Material. Values were normalized by ACT1 expression levels.
ChIP
ChIP was adapted from Ruault et al. (2011) using a polyclonal antibody anti-Sir3 (Ruault et al. 2011) or monoclonal anti-GFP (Roche). The signal from a region was normalized to that from the OGG1 control locus in immunoprecipitated and input DNA samples. Detailed procedures of ChIP and qPCR analyses can be found in the Supplemental Material.
ChIP-on-chip analyses
The position of DNA Pol2 peaks in wild-type and rrm3Δ strains were retrieved from Azvolinsky et al. (2009). Data of Sir3- and Sir4-binding enrichment for the wild-type strain were retrieved from Sperling and Grunstein (2009). The enrichment profiles were computed on both sides of each DNA Pol2 peak center in 50-base-pair adjacent windows.
Acknowledgments
We thank the members of the Taddei laboratory for helpful discussions; A. Cook, V. Borde, and G. Almouzni for critical reading of the manuscript; Fabrizio Martino for providing recombinant Sir3 protein; and D. Sherrat, F.X. Barre, and E. Fabre for providing reagents. M.D. was funded by the Ministère de l'Enseignement Supérieur et de la Recherche and the Association pour la Recherche sur le Cancer (ARC). The research leading to these results has received funding from the European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement number 210508 and the ANR “Jeune chercheur.”
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.611011.
References
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595 [DOI] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum E, Sjostrand JO, Astrom SU 2010. Ume6 is required for the MATa/MATα cellular identity and transcriptional silencing in Kluyveromyces lactis. Genetics 184: 999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Buck S, Sternglanz R, Shore D 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75: 531–541 [DOI] [PubMed] [Google Scholar]
- Falcon CM, Matthews KS 1999. Glycine insertion in the hinge region of lactose repressor protein alters DNA binding. J Biol Chem 274: 30849–30857 [DOI] [PubMed] [Google Scholar]
- Fourel G, Lebrun E, Gilson E 2002. Protosilencers as building blocks for heterochromatin. Bioessays 24: 828–835 [DOI] [PubMed] [Google Scholar]
- Gartenberg MR 2009. Life on the edge: telomeres and persistent DNA breaks converge at the nuclear periphery. Genes Dev 23: 1027–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol 134: 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM 2002. Live imaging of telomeres. yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol 12: 2076–2089 [DOI] [PubMed] [Google Scholar]
- Irlbacher H, Franke J, Manke T, Vingron M, Ehrenhofer-Murray AE 2005. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes Dev 19: 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Jegou T, Chung I, Heuvelman G, Wachsmuth M, Gorisch SM, Greulich-Bode KM, Boukamp P, Lichter P, Rippe K 2009. Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase-negative human cell line. Mol Biol Cell 20: 2070–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier AL, Rine J 2001. DNA replication-independent silencing in S. cerevisiae. Science 291: 646–650 [DOI] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM 2005. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702 [DOI] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol 8: 653–656 [DOI] [PubMed] [Google Scholar]
- Lewis M 2005. The lac repressor. C R Biol 328: 521–548 [DOI] [PubMed] [Google Scholar]
- Li YC, Cheng TH, Gartenberg MR 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291: 650–653 [DOI] [PubMed] [Google Scholar]
- Maillet L, Gaden F, Brevet V, Fourel G, Martin SG, Dubrana K, Gasser SM, Gilson E 2001. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep 2: 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Almouzni G 2004. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5: 296–304 [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621–633 [DOI] [PubMed] [Google Scholar]
- Miller AM, Nasmyth KA 1984. Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312: 247–251 [DOI] [PubMed] [Google Scholar]
- Mills K, Sinclair D, Guarente L 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97: 609–620 [DOI] [PubMed] [Google Scholar]
- Moazed D, Rudner AD, Huang J, Hoppe GJ, Tanny JC 2004. A model for step-wise assembly of heterochromatin in yeast. Novartis Found Symp 259: 48–56 [PubMed] [Google Scholar]
- Pecinka A, Kato N, Meister A, Probst AV, Schubert I, Lam E 2005. Tandem repetitive transgenes and fluorescent chromatin tags alter local interphase chromosome arrangement in Arabidopsis thaliana. J Cell Sci 118: 3751–3758 [DOI] [PubMed] [Google Scholar]
- Possoz C, Filipe SR, Grainge I, Sherratt DJ 2006. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J 25: 2596–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69: 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C, Straight A, Li G, Wilhelm C, Sudlow G, Murray A, Belmont A 1996. In vivo localization of DNA sequences and visualization of large scale chromatin organization using lac-operator/repressor recognition. J Cell Biol 135: 1685–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruault M, De Meyer A, Loiodice I, Taddei A 2011. Clustering heterochromatin: Sir3 promotes telomere clustering independently of silencing in yeast. J Cell Biol 192: 417–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Sjostrand JO, Kegel A, Astrom SU 2002. Functional diversity of silencers in budding yeasts. Eukaryot Cell 1: 548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling AS, Grunstein M 2009. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc Natl Acad Sci 106: 13153–13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J 23: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Schober H, Gasser SM 2010. The budding yeast nucleus. Cold Spring Harb Perspect Biol 2: a000612 doi: 10.1101/cshperspect.a000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower DA, Bloom K 2001. Dicentric chromosome stretching during anaphase reveals roles of Sir2/Ku in chromatin compaction in budding yeast. Mol Biol Cell 12: 2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]