Abstract
This study aimed to identify suitable siRNA delivery systems based on flexible generation 2–4 triazine dendrimers by correlating physico-chemical and biological in vitro and in vivo properties of the complexes with thermodynamic parameters calculated using molecular modeling. The siRNA binding properties of the dendrimers and PEI 25 kDa were simulated, binding and stability were measured in SYBR Gold assays, and hydrodynamic diameters, zeta potentials, and cytotoxicity were quantified. These parameters were compared with cellular uptake of the complexes and their ability to mediate RNAi. Radiolabeled complexes were administered intravenously, and pharmacokinetic profiles and biodistribution of these polyplexes were assessed both invasively and non-invasively. All flexible triazine dendrimers formed thermodynamically more stable complexes than PEI. While PEI and the generation 4 dendrimer interacted more superficially with siRNA, generation 2 and 3 virtually coalesced with siRNA, forming a tightly intertwined structure. These dendriplexes were therefore more efficiently charge-neutralized than PEI complexes, reducing agglomeration. This behavior was confirmed by results of hydrodynamic diameters (72.0 nm – 153.5 nm) and zeta potentials (4.9 mV – 21.8 mV in 10 mM HEPES) of the dendriplexes in comparison to PEI complexes (312.8 nm – 480.0 nm and 13.7 mV 17.4 mV in 10 mM HEPES). All dendrimers, even generation 3 and 4, were less toxic than PEI. All dendriplexes were efficiently endocytosed and showed significant and specific luciferase knockdown in HeLa/Luc cells. Scintillation counting confirmed that the generation 2 triazine complexes showed more than twofold prolonged circulation times as a result of their good thermodynamic stability. Conversely, generation 3 complexes dissociated in vivo, and generation 4 complexes were captured by the reticulo-endothelial system due to their increased surface charge. Molecular modeling proves very valuable for rationalizing experimental parameters based on the dendrimers’ structural properties. Non-invasive molecular imaging predicted the in vivo fate of the complexes. Therefore, both techniques effectively promote the rapid development of safe and efficient siRNA formulations that are stable in vivo.
Keywords: Triazine dendrimers, RNA interference, molecular modeling, structure function relationship, physico-chemical characterization, in vivo, biodistribution, pharmacokinetics, SPECT imaging
Introduction
Dendrimers are increasingly employed for siRNA delivery. While PAMAM and its derivatives are currently the most widely used [1], polypropylenimine (PPI) [2], dendritic poly-L-lysine (PLL) [3] and newer classes such as carbosilane dendrimers [4] and triazine dendrimers [5] are gaining more importance in the field of non-viral siRNA delivery. While more than 50 reports of dendrimer-mediated siRNA delivery can be found in the literature to date whereas only six studies describe in vivo results [2, 3, 5–8]. This discrepancy reflects the fact that dendrimers used for siRNA delivery still require further optimization in with respect to efficiency and toxicity to allow in vivo administration. For example, in several studies low generation dendrimers were unable to condense siRNA into uniformly small complexes [9–11]. This obstacle precludes in vivo administration. The use of higher generation materials (G6 and G7) [1, 3, 9, 11–14], however, is often accompanied by an increase in toxicity [15]. To counter this deficiency, many structural modifications have been made to enhance biocompatibility, including carboxylate-terminated [16], acetylated [17], internally cationic and hydroxyl-terminated PAMAM dendrimers [18]. However, these modifications decreased in vitro efficiency [17, 18]. The lack of in vitro knockdown efficiency of PAMAM has been attributed to incomplete endosomal release of the siRNA [19] and nuclear localization of oligonucleotides [20]. In an earlier study, endosomal release of siRNA was shown to be modulated by introduction of short, lipophilic C6-groups on the periphery of triazine dendrimers [5]. The alkylated G2 “rigid core” dendrimer efficiently mediatied in vitro gene silencing, however, accumulated strongly in the lung after intravenous injection although the hydrodynamic diameter of the siRNA dendriplexes was 103 nm as measured in buffer [5]. Since complex formation with the G3 rigid analogue did not lead to smaller dendriplexes (178 nm), but negatively affected cell viability to a significant extent [21], this study focuses on a new panel of “flexible” triazine dendrimers. Flexible triethanolamine core PAMAM dendriplexes of generation 7 were reported to exhibit almost no cytotoxicity in MTT and LDH assays [13], indicating that a flexible core may reduce the toxicity of higher generation dendrimers. Similarly, a flexible generation 2 triazine dendrimer F2-1 was previously shown to reduce hemolysis in comparison with related generation 1–3 rigid core analogues [21]. However, this flexible dendrimer F2-1 was found to present a “collapsed” topology, leading to less interaction with siRNA than observed for the dendrimers that were actually expected to be more rigid [22] and formation of loosely associated, large agglomerates 286 nm in size [5]. In an approach to enhance the interaction between flexible dendrimers and siRNA, new flexible dendrimers of generation 2, 3, and 4 were synthesized and characterized in this study concerning computed siRNA binding characteristics. The results obtained from in silico simulations were compared with experimental physico-chemical parameters such as siRNA complexation, complex stability, size, and zeta potentials. Since these properties are expected to determine siRNA packaging, dendriplex endocytosis, unpackaging, stability in the blood stream and thus the RNAi efficiency of siRNA formulations [23], the merit of using in silico results to predict the in vitro and in vivo performance of the dendriplexes was investigated. Furthermore, the ability to use radioactive in vivo imaging as a method to identify efficient siRNA delivery systems was probed.
Materials and Methods
Materials
Poly(ethylene imine) (Polymin™ 25 kDa, PEI 25k) was a gift from BASF (Ludwigshafen, Germany). Lipofectamine™ 2000 (LF) was bought from Invitrogen (Karlsruhe, Germany), Beetle Luciferin, and heparin sodium salt from Sigma-Aldrich Laborchemikalien GmbH (Seelze, Germany). 2′-O-Methylated 25/27mer DsiRNA targeting firefly luciferase, negative control sequence, TYE546- and 5′-sense strand C6-amine modified DsiRNA were obtained from Integrated DNA Technologies (IDT, Leuven, Belgium) [5]. Balb/c mice (6 weeks old) were bought from Harlan Laboratories (Horst, The Netherlands). The chelator 2-(4-isothiocyanatobenzyl)-diethylenetriaminepentaacetic acid (p-SCN-Bn-DTPA) was purchased from Macrocyclics (Dallas, TX, USA), SYBR® Gold from Invitrogen (Karlsruhe, Germany), and all chemicals used for synthesis were obtained from Sigma-Aldrich (St. Louis, MO).
Synthesis of new triazine dendrimers
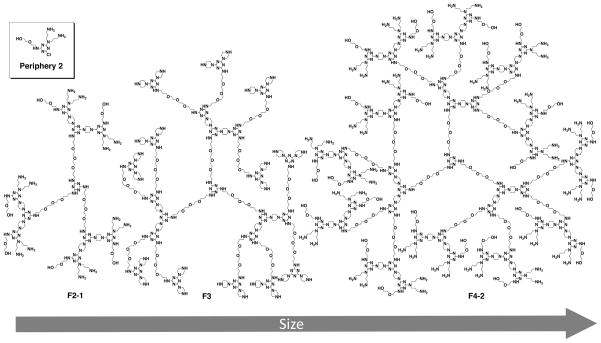
The flexible triazine dendrimers F2-2, F3, and F4–2 used in this study were synthesized following a previously described divergent approach [21, 24]. The final products and all intermediate structures were characterized by 1H and 13C NMR spectroscopy, and mass spectrometry, as shown in the Supplementary data. generations 2, 3, and 4 were synthesized of which generation 2 and 4 bear the same periphery (Figure 1), previously designated as periphery number 2 [24].
Figure 1.
Structures of flexible core triazine dendrimers (F) of generation number 2, 3 and 4. The monochlorotriazine from which the peripheral group (2) for generation 2 and 4 carries one hydroxyl group and two amines.
Molecular modeling
The model for Dicer Substrate siRNA was constructed with the NAB module within the AMBER 11 suite of programs [25]. To study the multivalent molecular recognition that characterizes the complexation, the binding between siRNA and dendrimers in a 1:1 ratio was modeled according to a validated strategy reported for the binding of dendrons and dendrimers with DNA [26, 27] and siRNA [28]. The structures of F2-2, F3 and F4–2 were composed of different residues according to previous studies on similar dendrimers [22]. The model of random branched PEI 25k was created and pre-optimized using the Materials Studio (MS) software package (Accelrys). For the simulations, the total charges of F2-2, F3, F4–2 and PEI were kept constant with +12, +24, +48 and +98, respectively, at pH 7.4 [29]. This simplification certainly does not reflect changes in response to variation of pH, ionic strength or other parameters and is therefore a limitation for the power of prediction at this point. Each of the nonstandard residues that compose the dendrimers was parameterized with the antechamber module of AMBER 11 consistently with the “general AMBER force field” (GAFF) following a well validated procedure previously adopted [30].
Molecular dynamic simulations
All simulations and data analyses were performed with the AMBER 11 suite of programs (sander.MPI and pmemd.cuda modules) [25]. The dendrimers were solvated in a TIP3P water box [31], minimized and then equilibrated by running 10 nanoseconds NPT molecular dynamics (MD) simulations, as described previously [22] to obtain a reliable configuration for PEI, F2-2, F3 and F4–2 in solution (Figure S1). The water molecules and counter ions were removed, and the polycations were placed in close proximity to the major groove of DsiRNA. The complexes were re-solvated, the resulting molecular systems were minimized [22] and equilibrated for 20 ns in NPT periodic boundary condition at 300 K and 1 atm using a time step of 2 fs, the Langevin thermostat, and a 10 Å cut-off. The particle mesh Ewald (PME) approach was adopted [32], and the SHAKE algorithm was used to constrain all bonds involving hydrogen atoms. Each of the molecular dynamics runs were carried out using parm99 all-atom force field [33] with NVIDIA Tesla 2050 GPU cards. In order to confirm that all of the systems were equilibrated, the root mean square deviation (RMSD) criterion was used. The RMSD data were obtained from the MD trajectories, which showed that all of the systems converged to the equilibrium with good stability. The free energies of binding (ΔGbind, ΔHbind and TΔSbind) were calculated according to the MM-PBSA approach [34] and the normal-mode analysis [35] on 100 MD frames taken from the equilibrated MD trajectories according to previous studies on the interactions between dendrimers and nucleic acids [22, 28].
Dendriplex formation
Dendriplexes were formed as previously described by adding 25 μl of a calculated concentration of dendrimer to an equal volume of 2 μM siRNA followed by vigorous pipetting [5]. Both siRNA and dendrimers were diluted with an isotonic solution of 5% glucose unless otherwise described. The appropriate amount and “protonable unit” of each dendrimer to afford a certain N/P ratio, which is the excess of polymer amines (N) over siRNA phosphates (P), was calculated as previously described [21]. Polyplexes with PEI 25 kDa were formed as described above, and lipoplexes with LF with 1 μl per 20 pmol were prepared as recommended by the manufacturer.
SYBR Gold® Assay
The ability of the three dendrimers to bind and protect siRNA was studied as previously reported and compared with PEI 25kDa [5]. Briefly, complexes of 1 μg siRNA were prepared at different N/P ratios, incubated for 20 minutes before 50 μl of a 1× SYBR® Gold solution was added and incubated for another 10 minutes in the dark. Free or accessible siRNA was quantified using a SAFIRE II fluorescence plate reader (Tecan Group Ltd, Männedorf, Switzerland) at 495 nm excitation and 537 nm emission wavelengths. The results are given as mean relative fluorescence intensity values (n=3) +/− the standard deviation (SD), where intercalation of free siRNA represents 100 % fluorescence, and non-intercalating SYBR® Gold in buffer represents 0 % remaining fluorescence.
Heparin Competition Assay
The stability of the dendriplexes against competing polyanions, such as the model molecule heparin, was studied as previously reported [5]. Briefly, dendriplexes were formed at N/P=5, incubated with 50 μl of a 1× SYBR® Gold solution, and treated with increasing amounts of heparin for 20 minutes. Fluorescence was quantified as described above. Results are given as mean relative fluorescence intensity values (n=3) +/− SD, where free siRNA represents 100 % fluorescence, and SYBR® Gold in buffer represents 0 % fluorescence.
Dynamic Light Scattering and Zeta Potential Analysis
Dendriplexes formed from each generation of flexible triazine dendrimers and siRNA were characterized concerning their hydrodynamic diameters and zeta potentials, whereas PEI 25kDa served as a control. Dendriplexes and PEI polyplexes were formed as described above in 5% glucose, HEPES-buffered glucose (HBG: 5% glucose, 10 mM HEPES, pH 7.4) or 10 mM HEPES buffer at increasing N/P ratios and measured as previously described in a disposable low volume UVette (Eppendorf, Wesseling-Berzdorf, Germany) using a Zetasizer Nano ZS (Malvern, Herrenberg, Germany) [5]. Zeta potentials were determined by laser Doppler anemometry (LDA) after diluting the samples with 750 μl of the equivalent solvent to a final volume of 800 μl and transferring the suspensions into a green zeta cuvette (Malvern, Herrenberg, Germany). Results are given as mean values (n=3) +/− SD.
Cell Culture
HeLa cells stably expressing luciferase (HeLa/Luc) [36] were maintained at 100 μg/ml hygromycin B in DMEM high glucose (PAA Laboratories, Cölbe, Germany) supplemented with 10 % fetal bovine serum (Cytogen, Sinn, Germany) in a humidified atmosphere with 5% CO2 at 37°C, and seeded for experiments in antibiotics-free medium. L929 cells were cultured in antibiotics-free DMEM high glucose (PAA Laboratories, Cölbe, Germany) supplemented with 10 % fetal bovine serum (Cytogen, Sinn, Germany).
Confocal laser scanning microscopy (CLSM)
As previously described, uptake and subcellular distribution of dendriplexes was investigated by confocal microscopy [5]. Briefly, HeLa/Luc cells were plated on 8-chamber slides and transfected 24 h later at N/P ratios of 10 and 20 with 25 pmol TYE-546-labeled siRNA per chamber in a total volume of 250 μl. Cells transfected with free siRNA or Lipofectamine™ 2000 were used as negative and positive controls, respectively. After 4 h of incubation, cells were washed, fixed with 4% paraformaldehyde, counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes, Invitrogen, Karlsruhe, Germany), and embedded with FluorSave (Calbiochem, Merck Biosciences, Darmstadt, Germany). For confocal microscopy on a Zeiss Axiovert 100 M microscope and a Zeiss LSM 510 scanning device (Zeiss, Oberkochen, Germany), lasers and filter settings were chosen as previously described [5].
Transfection efficiency
The ability of the different generation dendrimers to mediate RNA interference in vitro was quantified by luciferase knockdown in HeLa/Luc cells as previously reported [5]. Briefly, cells were seeded at a density of 15,000 cells per well in 48-well-plates and treated with dendriplexes of 50 pmol siRNA (siFLuc or siNegCon) and different N/P ratios 24 h after seeding. As positive controls, transfections were performed with Lipofectamine™ 2000. The medium was changed 4 h post transfection, cells were incubated for another 44 h before they were washed with PBS buffer, lysed with CCLR (Promega) and assayed for luciferase expression on a BMG luminometer plate reader (BMG Labtech, Offenburg, Germany) [5]. Results are given as mean values (n=4) +/− SD.
Radiolabeling and Purification
Pharmacokinetics and biodistribution after i.v. injection were investigated with radiolabeled siRNA administered either freely or as dendriplexes. Polyplexes formed with PEI 25kDa were administered as control. siRNA was labeled and purified as previously described [36, 37]. Briefly, amine-modified siRNA was reacted with p-Bn-SCN-DTPA at pH 8.5 for 3 h before it was precipitated in 10% sodium acetate and 70% ethanol overnight. After centrifugation for 5 min at 12,000 g, DTPA-coupled siRNA was dissolved in RNase free water, annealed in presence of 111InCl3 (Covidien Deutschland GmbH, Neustadt a.d. Donau, Germany) for 2 min at 94°C, incubated for 30 min at room temperature and purified from free 111InCl3 by size exclusion chromatography (SEC) on PD-10 Sephadex G25 (GE Healthcare, Freiburg, Germany) and RNeasy spin column purification as described earlier [36].
In vivo Imaging, Pharmacokinetics and Biodistribution
Circulation times and distribution within the body were determined as previously described [5]. Groups of 5 BALB/c mice were anesthetized intraperitoneally and injected with dendriplexes containing 35 μg of siRNA and the corresponding amount of dendrimer at N/P 5. Control animals received either free siRNA or PEI25kDa/siRNA polyplexes which were prepared and labeled as previously described [36]. Pharmacokinetics were assessed by retro-orbitally withdrawing blood samples, and biodistribution was recorded in three-dimensional SPECT and planar gamma camera images 2 h after injection using a Siemens e.cam gamma camera (Siemens AG, Erlangen, Germany) equipped with a custom built multiplexing multipinhole collimator and compared to results of scintillation counting of dissected organs measured using a Gamma Counter Packard 5005 (Packard Instruments, Meriden, CT) [36].
Statistics
All analytical assays were conducted in replicates of three or four, as indicated and in vivo experiments included 5 animals per group. Results are given as mean values +/− standard deviation (SD). Two way ANOVA and statistical evaluations were performed using Graph Pad Prism 4.03 (Graph Pad Software, La Jolla, USA).
Results and Discussion
Nomenclature, Synthesis and Design Criteria of the Synthesized Dendrimers
The nomenclature adopted for the new dendrimers described in this manuscript is consistent with previously described structures [21, 24] with respect to their generation, flexible core structure and surface group functionalities as shown in Figure 1. Previous investigations of gene delivery with triazine dendrimers showed that flexibility is a key factor for promoting pDNA transfection [21] whereas different triazine dendrimers had been most efficient for siRNA delivery [5]. This difference was explained by the rigidity of siRNA in comparison to the rather flexible behavior of pDNA [22]. Computation suggested that the “flexible” dendrimer F2-1 reported previously as an siRNA vector [5] adopted a collapsed structure [22]. The choice of peripheral group more strongly influenced siRNA delivery than flexibility of the dendrimer backbone. Therefore, a favored peripheral group, one with a diethylene glycol instead of two ethylene glycol chains, was incorporated into a flexible generation 2 and larger generation dendrimers. Details of synthesis and characterization including NMR and MS data for these new structures are provided in the Supporting Information. The atomic composition of the panel is summarized in Figure 1. For the physicochemical and biological assays, branched poly(ethylene imine) of 25 kDa (PEI 25kDa) and/or LF were used as controls.
Molecular dynamic simulations and energetic and structural analyses
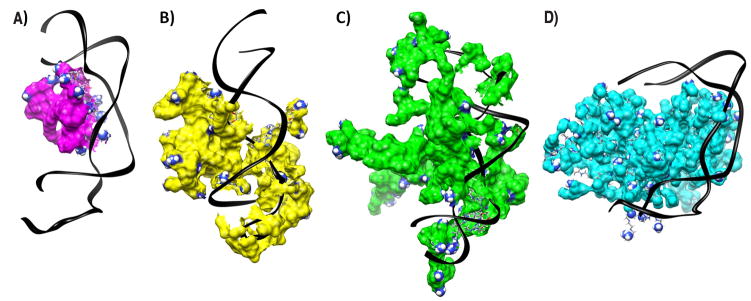
First, the solution phase structures were simulated as described above and shown in the Supplementary data (Figure S1). Table S1 shows the results of subsequent calculations of the binding affinity of the dendrimers and PEI towards partially 2′O-methylated siRNA. The binding energies were normalized per charge and expressed in kcal mol−1 to allow direct comparison between the different polycations with respect to the averaged interaction of each surface group (Table 1). High enthalpic gain accompanied by a lower and unfavorable entropic loss is typical of electrostatically driven molecular complexation with an overall gain in absolute free energy ΔG. Although the strength of RNA-dendrimer interactions is expected to increase with generation, the normalized binding energies showed a different trend. This observation arises from increased back folding of the peripheral groups with increasing generation. While the normalized ΔG value for the assembly of F2-1 with unmodified GL3 siRNA was only −4.5 kcal mol−1 [22], the binding of F2-2 to DsiRNA was comparably stronger with −9.1 kcal mol−1 per amine which indicates additional hydrophobic interactions between the dendrimers and partially 2′O-methylated siRNA. Interestingly, F3 was the dendrimer with the lowest affinity to DsiRNA (−5.8 kcal mol−1) reaching only reduced enthalpic attraction at the same entropic cost as F2-2. This behavior is attributed to the lack of primary amines in F3. Interestingly, all dendrimers showed similar enthalpic attraction towards DsiRNA that decreased slightly with generation (ΔH of −13.4, −10.2, and −9.7 kcal mol−1, respectively), while the entropic cost per charge in F4-2 was approximately half of that in F3 and F2-2. The binding with DsiRNA was therefore less entropically expensive for F4-2 than for F2-2 and F3, evidence that F4-2 maintains higher residual flexibility during the binding event, while F3 and F2-2 lost more degrees of freedom. The lower entropic cost of the complexation indicates that F4-2 interacts more superficially with DsiRNA, which is typically observed for PAMAM dendrimers [28] and was also shown here for the interaction of DsiRNA with PEI. The normalized ΔG value for PEI was the lowest among the tested panel. This indicates that PEI complexes are thermodynamically less stable than triazine dendrimer complexes, and that possibly not all charged amine groups are involved in a 1:1 complex with siRNA as a consequence of the sphere-like shape of solvated PEI. The entropic loss in all reactions was well compensated by the enthalpic gain leading to thermodynamically stable complexes, even in case of PEI. From the normalized free energies, it was hypothesized that the stability of the complexes decreases in the following order: F2-2>F4-2>F3>PEI. Additionally, the models in Figure 2 help to understand the differences between the siRNA binding modalities of a globular molecule like PEI (Figure 2D) and flexible molecules like F2-2 and F3 (Figures 2A-B), while F4-2 holds an intermediate position. For PEI, only on a limited part of the charged surface groups interacts actively with siRNA while a larger part of charged amines is back folded. This assembly leads to the formation of a complex with a distinct PEI domain next to a distinct siRNA domain, as shown in the upper panel of Scheme 1. The non-interacting interfaces shown in this 1:1 siRNA/dendrimer model could provide sites for further interactions with additional PEI or siRNA molecules or remain exposed to solvent. Experimental measurements in earlier reports led to the hypothesis that complexes of PEI 25k and pDNA contain both charge-neutralized regions as well as patches of uncomplexed, positively and negatively charged areas, leading to inter-particle electrostatic attractions [38]. The flexible dendrimers, however, appear to exert both electrostatic and hydrophobic interaction with DsiRNA, as hypothesized earlier [5]. This interaction leads to formation of coalesced complexes that appear to be a single neutralized or barely charged entity, as shown in the lower panel of Scheme 1. Interestingly, F4-2 deserves special attention since its intermediate binding behavior with DsiRNA is both affected by hydrophobic and electrostatic forces which are distributed over a larger and more rigid molecule.
Table 1.
ΔG energies and the contributing potentials of the binding between dendrimers or branched PEI 25 kDa and DsiRNA normalized to energy per charged surface amine expressed in kcal mol−1.
| F2-2 | F3 | F4-2 | PEI | |
|---|---|---|---|---|
| ΔH | −13.4 | −10.2 | −9.7 | −6.6 |
| −TΔS | 4.3 | 4.3 | 2.4 | 1.1 |
| ΔG | −9.1 | −5.8 | −7.3 | −5.5 |
Figure 2.
Equilibrated configurations (A) F2-2, (B) F3, (C) F4-2, and (D) PEI interacting with DsiRNA. Nucleic acids are represented as black ribbons and surface amines that carry a +1 charge are represented as spheres. Water molecules and counter ions are omitted for clarity.
Scheme 1.
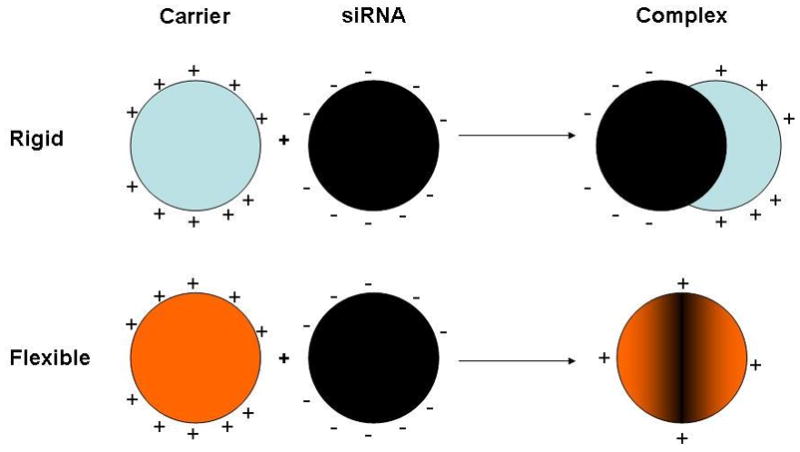
Hypothesized interaction of rigid polycations with siRNA leading to complexes of charged patches and flexible polycations yielding coalesced complexes of essentially neutralized charge.
These differences in the binding behavior are supported by the thermodynamic values calculated above. According to this hypothetical binding scheme, PEI complexes aggregate over time, which has been described earlier for pDNA [38]. If flexible triazine dendrimers coalesce with siRNA, leading to “neutralization” of their opposite charges, aggregation should be reduced. Single, distinct units of complexes between flexible triazine dendrimers and pDNA were previously shown by AFM [21]. After the aggregation tendency of DNA-polyelectrolyte complexes depending on the polymer structure was described in 1997 [39], the simulated results of dendrimer interactions with DsiRNA as a function of flexibility and generation described here will be compared with experimental data in this study.
Binding and protection efficiency and stability against competing polyanions
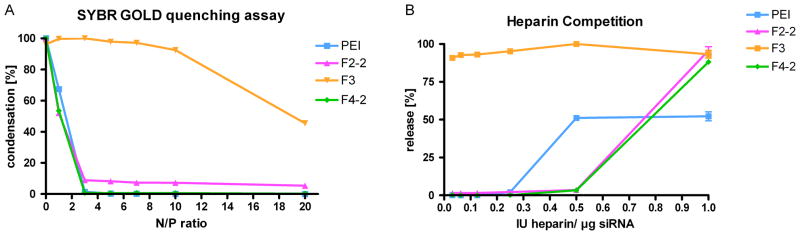
To compare the hypothesized order of complex stability with experimental data, the binding affinity, siRNA protection, and dendriplex stability were investigated in SYBR Gold assays [5]. SYBR Gold assays are related to ethidium bromide displacement assays which were found to yield DNA/polycation interaction profiles in agreement with thermodynamic microcalorimetry data [40] and allow quantification of siRNA available for intercalation of SYBR Gold. As shown in Figure 3A, the binding process was titrated by increasing the polycation concentration, specified as N/P ratio. The condensation of siRNA by PEI was very efficient and completely achieved at N/P 3, which is in line with previous reports [5, 41]. From the normalized binding enthalpy of a 1:1 PEI/siRNA complex, the complexation was expected to be less tight than the complexation of siRNA with flexible triazine dendrimers. However, the overall binding forces of PEI complexes were higher due to formation of multimolecular agglomerates. The condensation profile of F4-2 was comparable to that of PEI, while in F2-2 dendriplexes a fraction of 5% free siRNA remained accessible even at N/P 20. F3 exhibited only low affinity towards siRNA as expected from the simulations. F2-2 dendriplexes, which were hypothesized to be most stable, seemed to condense siRNA to a lesser extent than F4-2 and F2-1 [5]. Apparently, the energetic values obtained in the simulations described above can predict the affinity of macromolecules, but these numbers are not capable of predicting the spatial accessibility or the shielding and protection of siRNA. From the models in Figure 2, however, it can clearly be understood that a large molecule like F4-2 can mask siRNA more efficiently than a small dendrimer like F2-2. In the case of the flexible dendrimers, the increase in generation to F4-2 enhanced the shielding of siRNA, as previously seen for pDNA [21]. However, the increase in generation of the rigid dendrimer G3-1 improved its siRNA binding efficiency only marginally as compared to G2-1 [5]. The low affinity of F3 towards siRNA can be attributed to lack of the primary amines on the periphery. This hypothesis is in line with previous observations that siRNA condensation properties of triazine dendrimers are most importantly controlled by the end group modification rather than by the core structure [5]. The poor condensation is also reflected in a low enthalpic attraction and high entropic loss of the F3 complex. Stability of polyelectrolyte complexes is one of the main factors determining the efficacy of non-viral vectors, and is affected by the concentration of competing polyions [42], the presence of serum [37], and the interaction with negatively charged proteoglycans on the cell surface [43]. Therefore, the calculated thermodynamic stability of the complexes was compared with the experimental stability against the competing polyanionic model molecule heparin. As shown in Figure 3B, PEI complexes started to release siRNA at heparin concentrations of 0.25 IU per μg RNA. However, dendriplexes of F2-2 and F4-2 did not release siRNA up to 0.5 IU heparin per μg siRNA, which is comparable to previous observations for rigid, second generation dendriplexes, G2-1 [5]. The increased stability of these dendriplexes over PEI at intermediate heparin concentrations can be explained by the additional hydrophobic interactions of triazine dendrimers with amphiphilic 2′-O-methylated DsiRNA, previously assumed [5] and corroborated by simulation. These hydrophobic forces are not affected by competition with polyanions, but are weaker than electrostatic forces, and cannot promote assembly in the absence of electrostatic interactions. Therefore, both dendriplexes released about 90% of the load at 1 IU heparin per μg RNA, whereas PEI complexes seemed to be more stable due to the higher amount of positive charges in the periphery of PEI and the possibility of a multimolecular assembly. Even though F2-2 complexes were hypothesized to be thermodynamically more stable then F4-2 complexes, their profiles were comparable in terms of stability against competing polyanions. Dendrimer F3, however, was hypothesized to have only low affinity towards siRNA, according to the in silico data, and formed loose complexes with over 90% accessible siRNA at N/P 5, as shown in Figure 3A. Therefore, bound siRNA was easily released by low concentrations of heparin, as shown in Figure 3B, leading to a gain in entropy. This data reinforced the previous assumption that a low number of protonated amines in the periphery would result in a lack of stability [5]. Taken together, the hypothesized superior stability of F2-2 and F4-2 complexes over F3 based on the simulated energetic values was proven valid. Due to the fact that even the uncharged part of a dendrimer can shield encapsulated siRNA, the calculated differences in complex stability between F2-2 and F4-2 was not reproduced in these assays.
Figure 3.
Figure 3A: Complexation behavior of dendrimers as measured by SYBR Gold intercalation of residual free siRNA at increasing N/P ratios. 3B: Release profiles of siRNA from polyelectrolyte complexes at N/P 5 as function of the concentration of heparin.
Dendriplex size and zeta potential
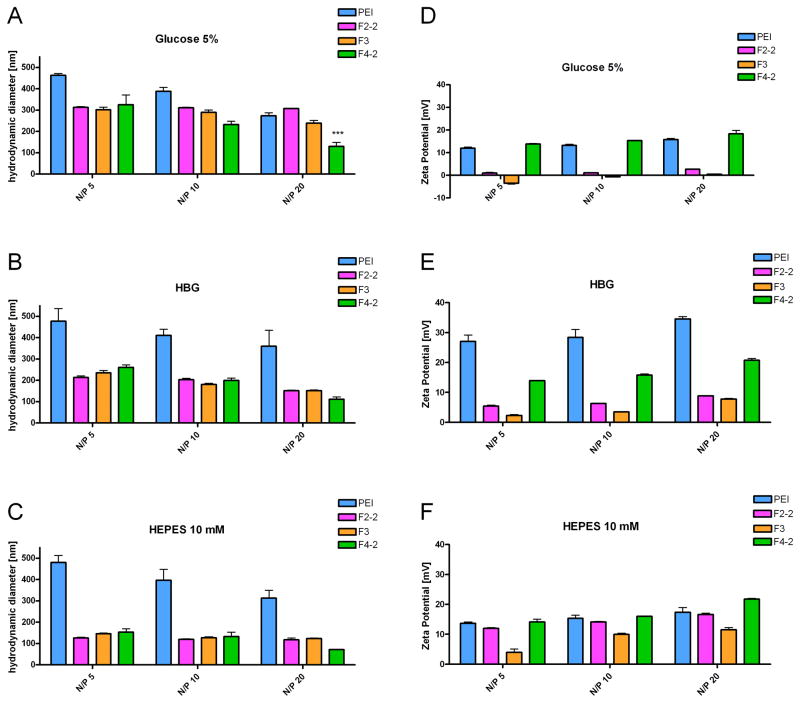
Previously, siRNA complexes with triazine dendrimers were formed in isotonic glucose solution [5]. To optimize the dendriplex formulation as a function of ionic strength and buffer capacity of the solvent [12], hydrodynamic diameters and zeta potentials were measured in glucose, HBG, and HEPES buffer as described above. PEI was highly efficient in condensing siRNA (Figure 3A), presumably due to the formation of multimolecular complexes. This behavior is consistent with the results of Figure 2D, showing that the a 1:1 polyplex of siRNA/PEI has regions that should promote aggregation [38], and was proven right for PEI/siRNA complexes by the measurement of hydrodynamic diameters. The size of the aggregates could be decreased if incubated at 0°C (data not shown), which is in line with PEI/DNA complexes [38]. Sizes of the dendriplexes decreased with increasing N/P ratio, as shown in Figure 4A–C. The decreased aggregation tendency at higher N/P ratios can be understood as a result of electrostatic repulsion of complexes with increased zeta potential (Figure 4D–F). Interestingly, the size of the dendriplexes formed at room temperature at a certain N/P ratio was comparable for all dendrimers despite their different charge densities, different peripheries, and quite different condensation profiles. Only at N/P 20 in 5% glucose, F4-2 formed significantly smaller complexes than F2-2 and F3 (Figure 4A). The polydispersity was low for F2-2 and F3 formulations (0.12<PDI<0.35) in contrast to F4-2 (0.19<PDI<0.44) and PEI complexes (0.49<PDI<0.67). Narrow size distribution is most important for biological activity and corresponds with coalesced complexes, which should be the more ideal structures for in vivo administration. This fact strengthens the previous hypothesis that differences in interactions between rigid or flexible polycations and siRNA leads to either the formation of multi-molecular agglomerates (in the case of PEI/siRNA) or coalesced dendriplexes (F2-2 and F3) as shown in Figure 2 and Scheme 1. As described above, F4-2 holds an intermediate position, which caused neither fully coalesced complexes nor agglomerates. Due to the higher flexibility of F4-2 in comparison to PEI, it can however be expected that no patches of negative charge were apparent on the surface of F4-2 complexes, leading to electrostatic repulsion instead of agglomeration. A 10 mM HEPES solution was chosen for the in vitro and in vivo evaluation of dendriplexes, since it both yielded the smallest particles (ca. 100 nm) and had the advantage of low ionic strength media, which has been described advantageous for PAMAM dendriplexes of low generation [12]. The sizes obtained in 5% glucose were comparable to the size of F2-1 dendriplexes (286 nm) previously reported [5]. The zeta potentials measured in 5% glucose were in agreement with the condensation behavior shown in Figure 3A and the differences in charge neutralization based on the simulations. While the siRNA was fully condensed into positively charged complexes by PEI and F4-2 at N/P 5, F2-2 complexes were almost neutral, and F3 complexes were slightly negatively charged. Within the dendrimer series, the cationic nature of dendriplexes derived from F4-2 is consistent with simulation, which shows a cationic surface within the 1:1 complex. Anionic surfaces derived from “unpackaged” siRNA are evident in simulated 1:1 dendriplexes comprising both F2-2 and F3. The rather neutral or slightly positive zeta potentials of F2-2 and F3 complexes can be explained by a higher molecular assembly than 1:1 and the influence of buffer capacity and ionic strength on the zeta potential of “charge-neutralized”, coalesced dendriplexes.
Figure 4.
Hydrodynamic diameters and zeta potentials of dendrimer/siRNA complexes in comparison to PEI complexes as a function of solvent and N/P ratio.
Subcellular Distribution of Dendriplexes
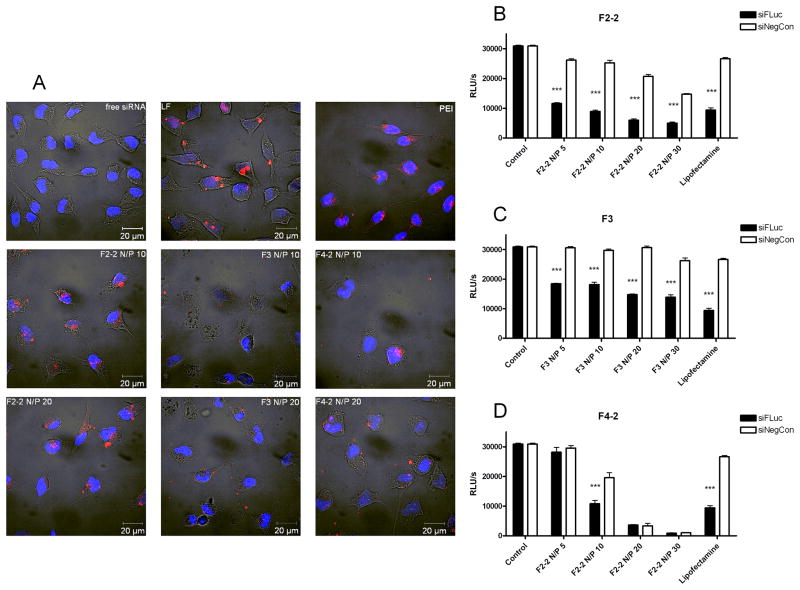
Cytotoxicity profiles (Supplementary data Figure S2) suggested that all of the dendrimers could be candidates for in vitro and in vivo siRNA delivery. Since physico-chemical parameters, such as complex size [44], surface charge [17], and stability [37], determine the intracellular delivery of siRNA, uptake efficiency of the dendriplexes was compared with their simulated and experimentally determined properties. For comparison, uptake of PEI complexes at N/P 10, lipoplexes made of Lipofectamine, and free siRNA was investigated as shown in Figure 5A. As expected, free siRNA was not taken up into HeLa cells. Lipoplexes showed very efficient uptake that was comparable to that of F2-2 complexes, both at N/P 10 and N/P 20. Previously described flexible generation 2 dendriplexes, F2-1, bound to the outer cell membrane after transfection [5], resulting in higher cytotoxicity of F2-1 compared to F2-2 [21]. The new periphery reported here may therefore be advantageous for siRNA delivery and endocytosis of the dendriplexes. Since it was previously shown that polymeric siRNA complexes easily release their load in presence of serum [37], the high efficiency of F2-2 complexes might be attributed to their thermodynamic stability, which was highest among the panel simulated. F3, which showed the lowest affinity (Figure 2B), lowest stability (Figure 3B), and was the least toxic due to the absence of primary amines (Figure S2), had the lowest zeta potential (Figure 4D–F), and poorly mediated uptake of siRNA. The inefficiency of F3 was consistent with in silico data and the previously reported reduced uptake of siRNA due to decreased surface charge and cytotoxicity of acetylated PAMAM derivatives [17]. The uptake of F4-2 complexes was reduced in comparison to F2-2. The difference may be explained by the lower thermodynamic stability of F4-2 complexes as compared to F2-2 complexes or by the higher cytotoxicity (Figure S2). Additionally, F4-2 complexes showed uptake into distinct subcellular locations, which may be an indication of incomplete endosomal release of the siRNA inside the cells as reported for siRNA complexes of PAMAM and Tat-conjugated PAMAM [19], SuperFect, and previously reported guanidinylated F2-1g triazine dendrimer complexes [5].
Figure 5.
Figure 5A. Confocal images showing the subcellular distribution of complexes made of Tye543-labeled siRNA (red) following cellular uptake in HeLa/Luc cells 4 hours after transfection. DAPI-stained nuclei are shown in blue. 5B-D. Knockdown of luciferase expression by dendrimer-siFLuc complexes in HeLa/Luc cells in comparison to dendriplexes with siNegCon (**p< 0.01, ***p< 0.001).
Transfection Efficiency
Since all dendriplexes were efficiently internalized into HeLa/Luc cells, their capacity to knockdown luciferase expression in the same cell line was assessed. All dendriplexes mediated RNAi depending on the N/P ratio, with some formulations achieving effects comparable to Lipofectamine (LF). While LF showed considerable off-target effects in cells treated with lipoplexes of the negative control sequence, as shown in Figures 5B–D, dendrimers F2-2 and F3 maintained strong transfection efficiency with comparably low cytotoxicity and off-target effects, although there appears to be a trend relating these effects to an increasing numbers of amines. The correlation between efficiency and toxicity has always been a major drawback of non-viral vectors [45] and was only recently reported to be overcome by disulfide cross-linked low molecular weight PEI for gene delivery [46] and conjugation of α-CD onto PAMAM for siRNA delivery [47]. Other modifications of dendrimers, however, such as internal quaternization in addition to a hydroxyl periphery of PAMAMs led to poor biological activity [18]. With these flexible triazine dendrimers, efficient luciferase knockdown was achieved even with the least toxic dendrimer F3, which supports the hypothesis of higher efficiency of flexible dendrimers at reduced toxicity, similar to what has been reported for G7 triethanolamine core PAMAM dendriplexes [13]. Although the efficiency of F3 complexes was lower than that of F2-2, its ability to elicit gene knockdown at all was surprising since F3 did not efficiently protect siRNA from intercalation of SYBR Gold. At N/P 20, the toxicity of F2-2 complexes was comparable to LF. However, stable complexes were also obtained at much lower N/P ratios, such as 5 and 10. These formulations seemed suitable for endocytosis concerning their hydrodynamic diameters, although this parameter provides no information on the morphology of the dendriplexes. Even so, these complexes were highly efficient in downregulating luciferase expression while causing no effect of the non-specific siRNA sequence. Due to its higher number of primary amines and lower IC50 value, F4-2 led to considerable cell death and off-target effects at N/P 20 and 30. At lower N/P ratios, F4-2 complexes were less efficient than F2-2, which was consistent with the simulated lower thermodynamic stability and reduced intracellular uptake, and may also be related to insufficient endosomal release. Stability [37], formation of larger aggregates with low generation dendrimers [9–11], nuclear localization [20], and incomplete endosomal release of the siRNA [19] are all major hurdles for polymeric and dendritic vectors. However, for the panel of triazine dendrimers investigated here, sufficient stability, which was predicted for F2-2 based on simulation of the thermodynamic binding profiles, and low cytotoxicity appeared to be the primary determining factors for successful gene knockdown.
Biodistribution and Pharmacokinetics
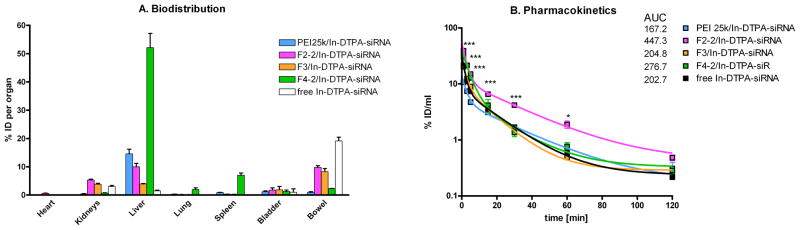
Since all dendrimer formulations successfully mediated RNAi in vitro with negligible toxicity at N/P 5, all were evaluated in vivo at this N/P ratio 5. SPECT imaging of in vivo administration of radiolabeled siRNA was adopted, because it can report on the pharmacokinetics and biodistribution with good correlation to results obtained by scintillation counting [36]. To date, information on biodistribution of dendritic siRNA carriers is limited to a study of surface-engineered PPI in which ex vivo fluorescence imaging and confocal laser scanning microscopy (CLSM) was employed to investigate the organ distribution of fluorescently labeled siRNA [2] and a one study that performed CLSM to trace the fluorescently labeled G4 cystamine-core PAMAM-based carrier [7]. The only report on pharmacokinetics of dendrimer-complexed siRNA used radioactively labeled siRNA and rigid triazine dendrimers to show strong lung accumulation of radioactively and fluorescently labeled siRNA [5]. As can be seen in Figure 6A and Movie 1 (Supporting Information), F2-2 dendriplexes did not accumulate in the lung, but rather accumulated in the liver, the kidneys, and to some extent in the bowel due to partial hepatobiliary excretion of amphiphilic DsiRNA [5]. Although F3 complexes were stable enough in 10% serum-containing medium to mediate RNAi in vitro, these complexes dissociated in vivo as previously described for PEI complexes [37], leading to quantitative excretion of free siRNA via the bowel and the bladder (Figure 6B and Movie 2). Interestingly, F4-2 complexes seemed to be most stable under in vivo conditions, with very strong uptake of radiolabeled siRNA into the liver, some excretion into the bladder, and no noticeable excretion of free siRNA via the bowel (Figure 6C and Movie 3).
Figure 6.
Three-dimensional biodistribution of A. F2-2-siRNA-dendriplexes, B. F3-siRNA-dendriplexes, and C. F4-2-siRNA-dendriplexes 2 hours after i.v. administration as registered by SPECT imaging.
These results were confirmed by scintillation counting of dissected organs, as shown in Figure 7A. Dendriplexes from F2-2 showed a comparably high signal in the heart 2 h after injection, which is consistent with significantly prolonged circulation times (Figure 7B). Their advantageous pharmacokinetics, reflected in a more than two-fold increased AUC versus free siRNA (Figure 7B), can be explained by reduced uptake into the reticulo-endothelial system (RES) which results from lower surface charge of F2-2 complexes compared to PEI or F4-2 complexes. Although F2-2 complexes were expected to be most stable, the SYBR Gold assay showed that some siRNA was still accessible for intercalation. This finding corroborates the considerable amount of 9.8% of the injected dose (ID) which was excreted as free siRNA via the bowel and 5.3% ID cleared through the kidneys. All other formulations led to rapid clearance from the blood pool, as shown by low AUC values (167.2–276.7 %ID*min/ml), as previously reported for native siRNA [48]. Both free siRNA and siRNA formulated with F3 were primarily cleared into the bowel and the bladder. The similarity of the pharmacokinetic profiles and the deposition of free siRNA and siRNA/F3 complexes into the bowel is a strong indication of instability, which was predicted by both simulated thermodynamic data and the results from the SYBR Gold assay. F4-2 complexes, however, were not rapidly cleared from the blood stream because of instability but rather because of extensive capture by the RES. In fact, the uptake of 52.1% of the injected siRNA is a sign of enhanced stability compared to PEI complexes, PEG-PEI complexes [37] and rigid triazine dendriplexes [5]. This strong uptake of siRNA into the liver was previously reported for surface-engineered PPI-based siRNA complexes [2] and was exploited for knockdown of ApoB in healthy C57BL/6 mice with poly-L-lysine-based vectors [3] or G3 tetra-oleoyl lysine dendrimers bound to the hydrophobic surface of single-walled carbon nanotubes (SWNT) [8]. While the spleen took up additional 7.0% ID of F4-2 formulated siRNA, only 0.7% ID accumulated in the kidneys and 2.3% ID were found in the bowel. As reported earlier, PEI complexes dissociated in the liver which was the only organ in which PEI-formulated siRNA accumulated. This is strongly in line with previous reports [37], and the differences of RES capture are in line with the different interaction of rigid and flexible polycations with siRNA as hypothesized based on the simulations. Radiolabeled siRNA complexed with PEI showed the same pharmacokinetic profile as free siRNA and siRNA complexed with F3, indicating the instability of both complexes as hypothesized from the thermodynamic values.
Figure 7.
Figure 7A. Biodistribution, 7B. Pharmacokinetics and AUC values in %ID*min/ml of siRNA-dendriplexes and polyplexes as measured by gamma scintillation counting of organ and blood samples.
Conclusions
Although dendrimers are increasingly used as non-viral vectors for siRNA delivery, the influence of dendrimer flexibility on in vitro and in vivo performance has not systematically been investigated. Molecular modeling approaches have been reported to explain the interactions of dendrimers with nucleic acids, but the predictive power of these simulations has not been challenged. In this study, simulated thermodynamic results were compared with experimental data. The results showed that the complexes that were predicted to be thermodynamically more stable and more “charge-neutralized” were indeed better vectors for siRNA delivery. The aggregation tendency of PEI complexes in comparison to dendriplexes could be predicted by molecular modeling. Importantly, internalization and transfection efficiency observed across the dendriplexes was consistent with the predicted order of calculated stability. However, as shown in the SYBR Gold assays, the simulated data does not account for differences in the shielding of siRNA by polycations. Although F2-2 was simulated to form the most stable complexes with siRNA, PEI and F4-2 most efficiently protected siRNA from intercalation with SYBR Gold. The lack of protection of siRNA by F2-2 and F3 complexes is reflected by the partial excretion of free siRNA via the bowel and kidneys upon intravenous administration. Also, despite the similarities between the binding of PEI and F4-2 with siRNA based on the modeling, dendriplexes of F4-2 were observed to be much more stable in vivo. Still, as predicted by the simulations, complexes of the more rigid PEI and F4-2 that contained large, non-charge neutralized patches, were subsequently captured by the RES to a significantly higher extent than F2-2 and F3 complexes. This accumulation may, however, be exploited for liver targeting or by local administration that circumvents the uptake into the liver. In this study, it became obvious that results from molecular modeling approaches can be used to help explain physico-chemical parameters and the in vitro behavior of dendriplexes. These attributes, in turn, determine in vivo behavior, such as stability in presence of serum or uptake into the RES. Since in vivo conditions, however, involve the complex interplay of dendriplexes with serum, cells, organs and metabolism, the prediction of the in vivo performance of dendriplexes can not solely be simulated, but must be evaluated by methods such as in vivo imaging. If combined, however, molecular modeling and in vivo imaging can be used as co-tools to predict the in vitro and in vivo performance of non-viral vectors for siRNA delivery.
Supplementary Material
Acknowledgments
We are grateful to Brian Sproat (IDT/Chemconsilium) for supplying the siRNA within MEDITRANS and to Eva Mohr (Dept. of Pharmaceutics and Biopharmacy) and Ulla Cramer (Dept. of Nuclear Medicine) for excellent technical support. MEDITRANS, an Integrated Project funded by the European Commission under the Sixth Framework (NMP4-CT-2006-026668), is gratefully acknowledged. EES and MAM thank the N.I.H. (R01 GM 65460). GMP and AD acknowledge respectively the Swiss State Secretariat for Education and Research (SER) and DECS-Canton Ticino for the support. MM acknowledges the Czech national COST project OC10053.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhou J, Wu J, Hafdi N, Behr JP, Erbacher P, Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem Commun (Cambridge, U K) 2006:2362–2364. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 2.Taratula O, Garbuzenko OB, Kirkpatrick P, Pandya I, Savla R, Pozharov VP, He H, Minko T. Surface-engineered targeted PPI dendrimer for efficient intracellular and intratumoral siRNA delivery. J Control Release. 2009;140:284–293. doi: 10.1016/j.jconrel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe K, Harada-Shiba M, Suzuki A, Gokuden R, Kurihara R, Sugao Y, Mori T, Katayama Y, Niidome T. In vivo siRNA delivery with dendritic poly(L-lysine) for the treatment of hypercholesterolemia. Mol Biosyst. 2009;5:1306–1310. doi: 10.1039/b900880b. [DOI] [PubMed] [Google Scholar]
- 4.Weber N, Ortega P, Clemente MI, Shcharbin D, Bryszewska M, de la Mata FJ, Gómez R, Muñoz-Fernández MA. Characterization of carbosilane dendrimers as effective carriers of siRNA to HIV-infected lymphocytes. J Control Release. 2008;132:55–64. doi: 10.1016/j.jconrel.2008.07.035. [DOI] [PubMed] [Google Scholar]
- 5.Merkel OM, Mintzer MA, Librizzi D, Samsonova O, Dicke T, Sproat B, Garn H, Barth PJ, Simanek EE, Kissel T. Triazine Dendrimers as Nonviral Vectors for in Vitro and in Vivo RNAi: The Effects of Peripheral Groups and Core Structure on Biological Activity. Mol Pharmaceutics. 2010;7:969–983. doi: 10.1021/mp100101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim ID, Lim CM, Kim JB, Nam HY, Nam K, Kim SW, Park JS, Lee JK. Neuroprotection by biodegradable PAMAM ester (e-PAM-R)-mediated HMGB1 siRNA delivery in primary cortical cultures and in the postischemic brain. J Control Release. 2010;142:422–430. doi: 10.1016/j.jconrel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal A, Min DH, Singh N, Zhu H, Birjiniuk A, von Maltzahn G, Harris TJ, Xing D, Woolfenden SD, Sharp PA, Charest A, Bhatia S. Functional delivery of siRNA in mice using dendriworms. ACS Nano. 2009;3:2495–2504. doi: 10.1021/nn900201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarroll J, Baigude H, Yang CS, Rana TM. Nanotubes functionalized with lipids and natural amino acid dendrimers: a new strategy to create nanomaterials for delivering systemic RNAi. Bioconjug Chem. 2010;21:56–63. doi: 10.1021/bc900296z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen XC, Zhou J, Liu X, Wu J, Qu F, Zhang ZL, Pang DW, Quelever G, Zhang CC, Peng L. Importance of size-to-charge ratio in construction of stable and uniform nanoscale RNA/dendrimer complexes. Org Biomol Chem. 2007;5:3674–3681. doi: 10.1039/b711242d. [DOI] [PubMed] [Google Scholar]
- 10.Juliano RL. Intracellular delivery of oligonucleotide conjugates and dendrimer complexes. Ann N Y Acad Sci. 2006;1082:18–26. doi: 10.1196/annals.1348.011. [DOI] [PubMed] [Google Scholar]
- 11.Inoue Y, Kurihara R, Tsuchida A, Hasegawa M, Nagashima T, Mori T, Niidome T, Katayama Y, Okitsu O. Efficient delivery of siRNA using dendritic poly(L-lysine) for loss-of-function analysis. J Control Release. 2008;126:59–66. doi: 10.1016/j.jconrel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 12.Perez AP, Romero EL, Morilla MJ. Ethylendiamine core PAMAM dendrimers/siRNA complexes as in vitro silencing agents. Int J Pharm. 2009;380:189–200. doi: 10.1016/j.ijpharm.2009.06.035. [DOI] [PubMed] [Google Scholar]
- 13.Liu XX, Rocchi P, Qu FQ, Zheng SQ, Liang ZC, Gleave M, Iovanna J, Peng L. PAMAM dendrimers mediate siRNA delivery to target Hsp27 and produce potent antiproliferative effects on prostate cancer cells. ChemMedChem. 2009;4:1302–1310. doi: 10.1002/cmdc.200900076. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Q, Lee E, Yeudall WA, Yang H. Dendrimer-triglycine-EGF nanoparticles for tumor imaging and targeted nucleic acid and drug delivery. Oral Oncol. 2010;46:698–704. doi: 10.1016/j.oraloncology.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svenson S. Dendrimers as versatile platform in drug delivery applications. Eur J Pharm and Biopharm. 2009;71:445–462. doi: 10.1016/j.ejpb.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Jevprasesphant R, Penny J, Jalal R, Attwood D, McKeown NB, D’Emanuele A. The influence of surface modification on the cytotoxicity of PAMAM dendrimers. Int J Pharm. 2003;252:263–266. doi: 10.1016/s0378-5173(02)00623-3. [DOI] [PubMed] [Google Scholar]
- 17.Waite CL, Sparks SM, Uhrich KE, Roth CM. Acetylation of PAMAM dendrimers for cellular delivery of siRNA. BMC Biotechnol. 2009;9 doi: 10.1186/1472-6750-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patil ML, Zhang M, Taratula O, Garbuzenko OB, He H, Minko T. Internally cationic polyamidoamine PAMAM-OH dendrimers for siRNA delivery: effect of the degree of quaternization and cancer targeting. Biomacromolecules. 2009;10:258–266. doi: 10.1021/bm8009973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang H, DeLong R, Fisher MH, Juliano RL. Tat-conjugated PAMAM dendrimers as delivery agents for antisense and siRNA oligonucleotides. Pharm Res. 2005;22:2099–2106. doi: 10.1007/s11095-005-8330-5. [DOI] [PubMed] [Google Scholar]
- 20.Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery. Bioconjug Chem. 2008;19:1396–1403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]
- 21.Merkel OM, Mintzer MA, Sitterberg J, Bakowsky U, Simanek EE, Kissel T. Triazine dendrimers as nonviral gene delivery systems: effects of molecular structure on biological activity. Bioconjug Chem. 2009;20:1799–1806. doi: 10.1021/bc900243r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavan GM, Mintzer MA, Simanek EE, Merkel OM, Kissel T, Danani A. Computational insights into the interactions between DNA and siRNA with “rigid” and “flexible” triazine dendrimers. Biomacromolecules. 2010;11:721–730. doi: 10.1021/bm901298t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanhai WR, Sakamoto JH, Canady R, Ferrari M. Seven challenges for nanomedicine. Nat Nanotechnol. 2008;3:242–244. doi: 10.1038/nnano.2008.114. [DOI] [PubMed] [Google Scholar]
- 24.Mintzer MA, Merkel OM, Kissel T, Simanek EE. Polycationic triazine-based dendrimers: effect of peripheral groups on transfection efficiency. New J Chem. 2009;33:1918–1925. doi: 10.1039/b908735d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Robertson B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell S, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sangui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 11. University of California; San Francisco: 2010. [Google Scholar]
- 26.Jones SP, Pavan GM, Danani A, Pricl S, Smith DK. Quantifying the Effect of Surface Ligands on Dendron-DNA Interactions: Insights into Multivalency through a Combined Experimental and Theoretical Approach. Chemistry. 2010;16:4519–4532. doi: 10.1002/chem.200902546. [DOI] [PubMed] [Google Scholar]
- 27.Pavan GM, Danani A, Pricl S, Smith DK. Modeling the multivalent recognition between dendritic molecules and DNA: understanding how ligand “sacrifice” and screening can enhance binding. J Am Chem Soc. 2009;131:9686–9694. doi: 10.1021/ja901174k. [DOI] [PubMed] [Google Scholar]
- 28.Pavan GM, Albertazzi L, Danani A. Ability to adapt: different generations of PAMAM dendrimers show different behaviors in binding siRNA. J Phys Chem B. 2010;114:2667–2675. doi: 10.1021/jp100271w. [DOI] [PubMed] [Google Scholar]
- 29.Suh J, Paik HJ, Hwang BK. Ionization of Poly(ethylenimine) and Poly(allylamine) at Various pH’s. Bioorg Chem. 1994;22:318–327. [Google Scholar]
- 30.Pavan GM, Kostiainen MA, Danani A. Computational approach for understanding the interactions of UV-degradable dendrons with DNA and siRNA. J Phys Chem B. 2010;114:5686–5693. doi: 10.1021/jp911439q. [DOI] [PubMed] [Google Scholar]
- 31.Jorgensen WK, Rice MJ. Morphology of a very extensible insect muscle. Tissue Cell. 1983;15:639–644. doi: 10.1016/0040-8166(83)90013-7. [DOI] [PubMed] [Google Scholar]
- 32.Darden T, York D, Pedersen L. Particle mesh Ewald: An N [center-dot] log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 33.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 34.Srinivasan J, Cheatham TE, Cieplak P, Kollman PA, Case DA. Continuum Solvent Studies of the Stability of DNA, RNA, and Phosphoramidate-DNA Helices. J Am Chem Soc. 1998;120:9401–9409. [Google Scholar]
- 35.Andricioaei I, Karplus M. On the calculation of entropy from covariance matrices of the atomic fluctuations. J Chem Phys. 2001;115:6289–6292. [Google Scholar]
- 36.Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Behe M, Kissel T. In vivo SPECT and real-time gamma camera imaging of biodistribution and pharmacokinetics of siRNA delivery using an optimized radiolabeling and purification procedure. Bioconjug Chem. 2009;20:174–182. doi: 10.1021/bc800408g. [DOI] [PubMed] [Google Scholar]
- 37.Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders NN, De Smedt SC, Behe M, Kissel T. Stability of siRNA polyplexes from poly(ethylenimine) and poly(ethylenimine)-g-poly(ethylene glycol) under in vivo conditions: effects on pharmacokinetics and biodistribution measured by Fluorescence Fluctuation Spectroscopy and Single Photon Emission Computed Tomography (SPECT) imaging. J Control Release. 2009;138:148–159. doi: 10.1016/j.jconrel.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 38.Sharma VK, Thomas M, Klibanov AM. Mechanistic studies on aggregation of polyethylenimine-DNA complexes and its prevention. Biotechnol Bioeng. 2005;90:614–620. doi: 10.1002/bit.20444. [DOI] [PubMed] [Google Scholar]
- 39.Tang MX, Szoka FC. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Ther. 1997;4:823–832. doi: 10.1038/sj.gt.3300454. [DOI] [PubMed] [Google Scholar]
- 40.Ehtezazi T, Rungsardthong U, Stolnik S. Thermodynamic analysis of polycation-DNA interaction applying titration microcalorimetry. Langmuir. 2003;19:9387–9394. [Google Scholar]
- 41.Merkel OM, Beyerle A, Librizzi D, Pfestroff A, Behr TM, Sproat B, Barth PJ, Kissel T. Nonviral siRNA delivery to the lung: investigation of PEG-PEI polyplexes and their in vivo performance. Mol Pharmaceutics. 2009;6:1246–1260. doi: 10.1021/mp900107v. [DOI] [PubMed] [Google Scholar]
- 42.Izumrudov VA, Bronich TK, Novikova MB, Zezin AB, Kabanov VA. Substitution reactions in ternary systems of macromolecules. Polym Sci USSR. 1982;24:367–378. [Google Scholar]
- 43.Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP. Sticky overhangs enhance siRNA-mediated gene silencing. Proc Natl Acad Sci U S A. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 45.Hong S, Leroueil PR, Janus EK, Peters JL, Kober MM, Islam MT, Orr BG, Baker JR, Jr, Banaszak Holl MM. Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjug Chem. 2006;17:728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 46.Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc Natl Acad Sci U S A. 2007;104:14454–14459. doi: 10.1073/pnas.0703882104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsutsumi T, Arima H, Hirayama F, Uekama K. Potential Use of Dendrimer/α-Cyclodextrin Conjugate as a Novel Carrier for Small Interfering RNA (siRNA) J Inclusion Phenom Macrocyclic Chem. 2006;56:81–84. [Google Scholar]
- 48.Dykxhoorn DM, Palliser D, Lieberman J. The silent treatment: siRNAs as small molecule drugs. Gene Ther. 2006;13:541–552. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.