Abstract
We report a reagentless, electrochemical sensor for the detection of double-stranded DNA targets that employs triplex-forming oligonucleotides (TFOs) as its recognition element. These sensors are based on redox-tagged TFO probes strongly chemisorbed onto an interrogating gold electrode. Upon the addition of the relevant double-stranded DNA target, the probe forms a rigid triplex structure via reverse Hoogsteen base pairing in the major groove. The formation of the triplex impedes contact between the probe’s redox moiety and the interrogating electrode, thus signaling the presence of the target. We first demonstrated the proof of principle of this approach by using a well-characterized 22-base polypurine TFO sequence that readily detects a synthetic, double-stranded DNA target. We then confirmed the generalizability of our platform with a second probe, a 19-base polypyrimidine TFO sequence that targets a polypurine tract (PPT) sequence conserved in all HIV-1 strains. Both sensors rapidly and specifically detect their double-stranded DNA targets at concentrations as low as ~10 nM and are selective enough to be employed directly in complex sample matrices such as blood serum. Moreover, to demonstrate real-world applicability of this new sensor platform, we have successfully detected unpurified, double-stranded PCR amplicons containing the relevant conserved HIV-1 sequence.
Many DNA detection methods are based on hybridization and thus require the generation of single-stranded DNA prior to analysis. For example, to properly generate a signal, methods such as DNA microarrays, Southern blotting, and in situ hybridization require denaturation of the double-stranded DNA of interest into single strands, followed by their subsequent renaturation with specific probes. In response, a number of approaches have been developed in which double-stranded targets are detected directly, such as the use of intercalating dyes.1–3 As these approaches lack sequence specificity, however, they are prone to false positives arising due to, for example, spurious amplification.4 The development of assays that are both sequence specific and avoid the cumbersome need to generate single-stranded DNA targets would thus significantly simplify DNA detection.
Several approaches have been recently proposed for the direct, sequence-specific detection of double-stranded DNA. Most of these employ non-DNA recognition probes and optical read-outs. Such probes include low molecular weight polyamides, which bind to specific minor groove sequences,5,6 or DNA binding proteins, which target specific duplex sequences.7–9 However, while several of these methods have been reduced to practice in the laboratory, their use in “real-world” settings is beset by drawbacks. Protein-based methods, for example, showed promising features of selectivity and sensitivity toward double-stranded targets,10 but they require cumbersome selection processes for the production of the relevant affinity reagents, and the use of proteins as recognition elements reduces the stability of the platform. Polyamide probes are likewise limited by the length of the targeted DNA sequence, which is generally between four to six base pairs, although, with recognition footprints of 10–12 base pairs, head-to-head bis-hairpin minor groove binders (MGBs) are a recent exception.11,12 The utility of these approaches to the direct, sequence-specific detection of double-stranded DNA has thus proven limited to date.
A potential route toward the direct, sequence-specific detection of double-stranded DNA is the observation that many such sequences support the formation of triplex structures in which a third strand of DNA runs along the major groove of the double helix, where it forms Hoogsteen base pairs.13–15 Specifically, triplex-forming oligonucleotides (TFOs), which are homopurine or homopyrimidine oligonucleotides that bind in the major groove of homopurine–homopyrimidine duplexes,16 exhibit high affinity and specificity, suggesting that they might serve as viable probes for the detection of double-stranded DNA targets without invoking the prior generation of single-stranded DNA. Motivated by these arguments, several groups have reported the use of TFOs in the optical detection of double-stranded DNA.17–20 Here, we expand this approach into a reagentless, electrochemical platform for the direct detection of double-stranded DNA targets.
MATERIALS AND METHODS
Probe and Target DNA Sequences
Polypurine TFO Probes and Targets
The modified polypurine 22-base TFO was obtained from Biosearch Technologies (Novato, CA) and employed as the probe DNA without further purification.
The polypurine probe sequence is as follows: 5′-HS-(CH2)6-CGTTC-GAAGG-AGGAA-GGAGG-GA-(CH2)τNH2-MB-3′.
The probe is modified at the 5′-end with a mercaptohexanol moiety and at the 3′-end with a methylene blue (MB) redox label. The MB redox moiety conjugation has been performed at the 3′-end of the oligonucleotide via succinimide ester coupling to a 3′-amino modification. The 15 internal bases of this sequence (underlined above) target the duplex obtained by hybridizing (before the injection in the working solution) oligos T1-R and T2-R. The complementary 15-base single-stranded target was also tested. Sequences of these oligonucleotides are reported below:
T1-R (15 bases, 5′-GGAGG-AAGGA-GGAAG-3′).
T2-R (15 bases, 5′-CTTCC-TCCTT-CCTCC-3′).
ss-DNA target complementary to the polypurine TFO probe (15 bases, 5′-CCTCC-TTCCT-CCTTC-3′).
Polypyrimidine TFO Probes and Targets
The modified polypyrimidine 19-base TFO was obtained from Biosearch Technologies (Novato, CA) and employed as the probe DNA without further purification. The polypyrimidine probe sequence is as follows:
5′-HS-(CH2)6-TATTT-TTCTT-TTCCC-CCCT-(CH2)τ-NH2-MB-3′.
The probe is modified at the 5′-end with a mercaptohexanol moiety and at the 3′-end with a methylene blue (MB) redox label. The 15 internal bases of this sequence (underlined above) target the duplex obtained by hybridizing (before the injection in the working solution) oligos T1-Y and T2-Y or by directly injecting the hairpin self-complementary target (T3-Y) whose sequences are reported below:
T1-Y, oligopurine target strand (15 bases, 5′-AAAAG-AAAAG-GGGGG-3′).
T2-Y, oligopyrimidine target strand (15 bases, 5′-CCCCC-CTTTT-CTTTT-3′).
T3-Y, oligopurine–oligopyrimidine hairpin target (34 bases, 5′-AAAAG-AAAAG-GGGGG-TTTT-CCCCC-CTTTT-CTTTT-3′).
The polypyrimidine TFO probe was also tested for specificity by challenging with a hairpin target sequence mutated at four positions. The hairpin is still able to form fully complementary double-stranded DNA, but four base pairs (underlined in the sequence below) are not complementary to the TFO probe:
Mutated oligopurine–oligopyrimidine hairpin for specificity tests (34 bases, 5′-AAACG-CAAAG-GTGGT-TTTT-ACCAC-CTTTG-CGTTT-3′).
The complementary 15-base single-stranded target forming a double-strand DNA with the TFO was also tested. Sequences of this oligonucleotide are reported below:
ss-DNA target complementary to the polypyrimidine TFO probe (15 bases, 5′-GGGGG-GAAAA-GAAAA-3′).
The polypyrimidine TFO was also tested with PCR amplification products. In order to demonstrate the feasibility of detecting the PCR products with this sensor, a synthetic 63 base-pair PCR duplex sequence, which mimics the actual HIV1 double-stranded PCR product, was preliminarily tested (target region is underlined):
plus strand, 5′-GTAGATCTTAGCCACTTTTTAAAAGAAAA-GGGGGGACTGGAAGGGCTAATTCACTCCCAAAGAA-3′.
minus strand, 5′-TCTTTGGGAGTGAATTAGCCCTTCCAGTC-CCCCCTTTTCTTTTAAAAAGTGGCTAAGATCTAC-3′.
Reagents
Reagent grade chemicals, including 6-mercapto-1-hexanol (C6-OH), sulfuric acid, potassium phosphate monobasic and dibasic, and sodium chloride (all from Sigma-Aldrich, St. Louis, MO), were used without further purification.
Sensor Fabrication
The sensors were fabricated by depositing the relevant TFO probe on gold rod electrodes (3.0 mm diameter) as previously described.21 Prior to use, the electrodes were cleaned using a series of oxidation and reduction cycles in 0.5 M H2SO4, 0.01 M KCl/0.1 M H2SO4, and 0.05 M H2SO4.21 The thiol-containing oligonucleotide we have employed is supplied as a mixed disulfide of 6-mercaptohexanol in order to minimize the risk of oxidation. The first step in sensor fabrication is the reduction of the DNA probes (100 μM) for 1 h in a solution of 0.4 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in 1 M NaCl/10 mM potassium phosphate, pH 7. This solution was then diluted to 50 nM with 1 M NaCl/10 mM potassium phosphate buffer, pH 7.
Electrodes (thoroughly rinsed with DI water) were incubated in 250 μL of this DNA probe solution for 60 min. Electrodes were rinsed with DI water and incubated in 2 mM mercaptohexanol in 1 M NaCl/10 mM potassium phosphate buffer (pH 7) for 2 h to displace nonspecifically adsorbed DNA and passivate the remaining electrode area. After thoroughly rinsing with DI water, electrodes were stored in the working buffer solution for 30 min before use. Polypurine TFO probes were tested in 0.1 M pH 7 Tris buffer containing 10 mM MgCl2, while polypyrimidine TFO probes were tested in 0.1 M pH 6.5 Tris buffer solution also containing 10 mM MgCl2. For the PCR experiments, the PCR product solution was diluted 1:5 prior to measurement using a highly acidic, pH 2.3 Tris buffer (0.2 M) containing 11 mM of MgCl2 to achieve a final, postdilution pH of 6.5.
PCR of HIV-1 RNA
HIV-1 RNA was obtained by extraction from inactivated, intact viral particles (Optiqual HIV-1 RNA Positive Controls, Acrometrix, Benecia, CA) using a QIAamp Viral RNA kit (Qiagen, Valencia, CA). To improve the yield of target DNA for electrochemical detection, we amplified this starting material using a nested, reverse transcriptase-PCR (RT-PCR) protocol. This protocol requires two sets of primers, here termed outer and inner primers. All primers were obtained from IDT (Coralville, IA).
The outer primers, which flank the targeted duplex region, contain the following sequences:
Outer_F, 5′-CACAAGAGGAGGAGGAGGTG-3′.
Outer_R, 5′-TGGCCCTGGTGTGTAGTTCT-3′.
The sequences for the inner primers are as follows:
Inner_F, 5′-GTAGATCTTAGCCACTTTTTAAAAG-3′.
Inner_R, 5′-TCTTTGGGAGTGAATTAGCCCTTCCA-3′.
The following procedure was adopted for the nested RT-PCR. One microliter of a 1 μg/μL solution of extracted viral RNA was added to a RT-PCR tube containing the following reaction mixture (all from Qiagen, Valencia, CA): 1× OneStep RT-PCR buffer mix [Tris-Cl/KCl/(NH4)2SO4/1.25 mM MgCl2/DTT/pH 8.7]/2.5 mM MgCl2/400 μM of each dNTP/0.6 μM of each outer primer/2 units of OneStep RT-PCR enzyme mix. The reaction tubes were then placed in a standard thermal cycler programmed with the following settings: reverse transcription at 50 °C for 30 min; polymerase activation at 95 °C for 15 min; then 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 72 °C for 1 min. Finally, a 10 min elongation step at 72 °C was used to complete the polymerization of any less than full length products. Immediately following RT-PCR, 1 μL of the amplified products was added to a new tube containing a standard PCR mixture (all products from Qiagen, Valencia, CA) composed of the following: 1× Taq polymerase buffer [Tris-Cl/KCl/(NH4)2SO4/1.5 mM MgCl2/pH 8.7]/2.5 mM of MgCl2/400 μM of each dNTP/0.5 μM of each inner primer/2.5 units of HotStar Taq polymerase. The reaction tubes were placed in a standard thermal cycler programmed with the following settings: initial polymerase activation step at 95 °C for 15 min; 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 72 °C for 1 min. A final elongation step of 10 min at 72 °C was applied to complete polymerization of any less than full-length products. Visualization of the products was performed using gel analysis with 4–20% TBE polyacrylimide gels and stained with SYBR gold (Invitrogen, Carlsbad, CA).
Electrochemical Measurements
The sensor response was measured by incubating the electrodes in a solution containing the appropriate target DNA. The sensors were interrogated at different intervals in the same target solution until a stable current peak was obtained (typically after 20 min). The ratio between the stabilized current peak in the presence of target DNA and the current peak in the absence of target DNA gives the measure of the signal suppression caused by the target.
Between target detection experiments, the electrodes were rinsed with an 8 M guanidine hydrochloride and subsequently interrogated in target-free buffer. This method provides a measure of the extent to which each sensor can be regenerated. All measurements were performed at room temperature using an Autolab potentiostat (EcoChemie, Utrecht, The Netherlands). Square wave voltammetry (SWV) was recorded at 60 Hz, 50 mV amplitude, and with an increment potential of 1 mV over a potential range from −0.1 to −0.45 V in a standard cell with a platinum counter electrode and a Ag/AgCl (3 M NaCl) reference electrode.21
RESULTS
We have employed TFOs as recognition elements for the direct detection of specific double-stranded DNA targets in an electrochemical sensor format analogous to the previously described E-DNA platform.21 Our sensors are comprised of a redox-tagged TFO probe that is strongly chemisorbed to an interrogating gold electrode (Figure 1). In the absence of its double-stranded DNA target, the probe is flexible, allowing an attached methylene blue to approach the electrode and exchange electrons. Upon the addition of its specific double-stranded target, the probe forms a rigid triplex structure that impedes contact between the methylene blue and the interrogating electrode. Thus, the TFO probe serves as an E-DNA sensor23 and maintains the various positive features of this class of devices.21,23–29 For example, we show below that this new class of sensor is reagentless, reusable, and suitable for deployment directly in complex matrices.
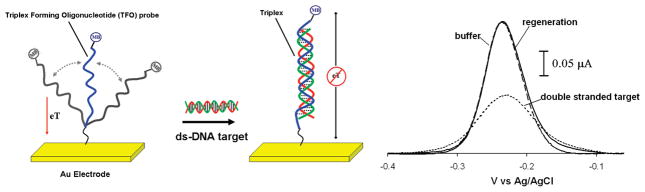
Figure 1.
An E-DNA sensor employing a triplex-forming oligonucleotide (TFO) probe readily detects double-stranded DNA targets. (Left) The sensor consists of a polypurine or polypyrimidine TFO probe modified at its 3′-terminus with a methylene blue redox tag and at its 5′-terminus with a mercaptohexanol moiety for attachment on a gold electrode. (Right) The Faradaic current arising from the flexible TFO probe is significantly reduced in the presence of the double-stranded DNA target, presumably because triplex formation reduces the efficiency with which the terminal redox tag collides with the electrode surface and transfers electrons.
To test the principle of a triplex-forming E-DNA platform, we first fabricated a sensor using a well-characterized 22-base polypurine TFO probe designed to bind a specific double-stranded target DNA via reverse Hoogsteen base pairing in the major groove18 (Figure 1). The sensor was fabricated by the formation of a mixed self-assembled monolayer (SAM) comprised of the TFO probe and 6-mercaptohexanol on the gold electrode, which gives rise to a sharp, well-defined peak at ~260 mV (vs Ag/AgCl), consistent with the formal potential of the methylene blue redox moiety (Figure 1, right). This TFO probe readily binds to its specific double-stranded DNA target (formed by the previous hybridization of oligonucleotides T1-R and T2-R) via triplex formation,18 producing a readily measurable decrease in Faradaic current (Figure 1, right, Figure 2, left). Support for the proposed formation of triplex is provided by the observation that the 65% signal suppression observed in the presence of the double-stranded target is somewhat greater than the 55% suppression obtained when the TFO probe is instead hybridized to its fully complementary, single-stranded DNA target to form a simple duplex (Figure 2, left). This presumably occurs because the greater bulk and charge of triplex DNA is more effective at reducing the efficiency with which the reporting redox tag approaches the electrode. As control experiments we also investigated the effects of adding the two single-stranded oligonucleotides (T1-R) employed in the double-stranded target in isolation. We find that the first of these, T1-R, does not lead to any detectable change in current, even at high concentrations (data not shown). In contrast the addition of the other, T2-R, leads to a nontrivial signal change, presumably because an 11-base portion of this strand is fully complementary to the TFO probe. Despite this, the signal suppression observed in the presence of this strand is much less than that obtained in presence of the duplex target (data not shown), further suggesting that the signal change produced by the duplex target arises due to triplex formation and not due to the dissociation of the duplex target and the consequent formation of a duplex with the probe DNA.
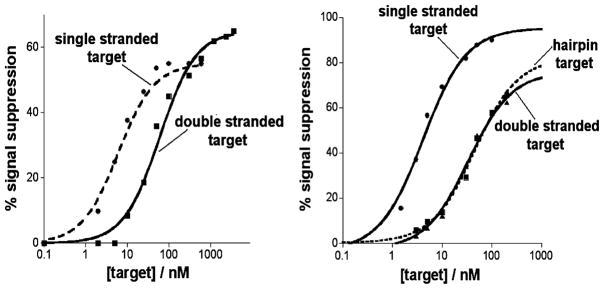
Figure 2.
The polypurine (left) and polypyrimidine (right) TFO based E-DNA sensors respond well to their specific double-stranded DNA targets (triplex formation). As expected, they also respond to fully complementary, single-stranded targets (via the formation of duplex DNA). As previously reported, the affinity of TFO probes for their double-stranded DNA targets is approximately an order of magnitude poorer than that for their single-stranded targets.30,31
The polypurine TFO-based sensor rapidly and specifically detects its double-stranded DNA target at concentrations as low as ~10 nM. The ~60 nM dissociation constant observed when the TFO probe is challenged with double-stranded target is, as expected,30,31 about an order of magnitude poorer than the 6 nM dissociation constant produced with fully complementary single-stranded target (Figure 2, left). Triplex formation is relatively rapid, exhibiting an equilibration half-life of ~5 min (Figure 3) that, perhaps because of the greater negative charge and increased hydrodynamic radius of the duplex target, is slightly slower than the rate of simple duplex formation (half-life ~3 min). The sensor is also specific: random sequences of single- and double-stranded DNA oligonucleotides ranging from 15 to 25 bases do not produce any detectable signal change at concentrations as high as 1 mM (data not shown).
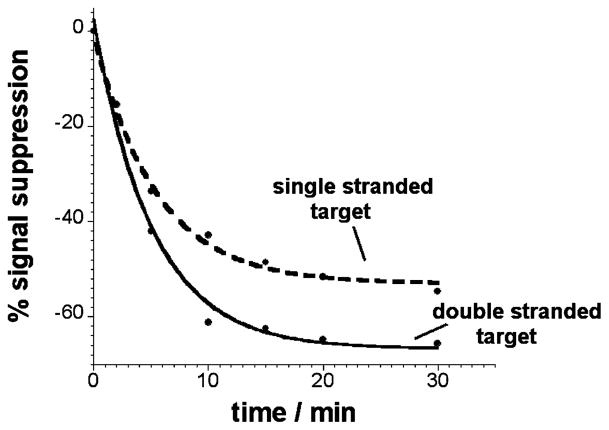
Figure 3.
The TFO-based E-DNA sensor is rapid. We observe an equilibration half-life (time required for half of the total signal change to occur) of ~5 min for the double-stranded (triplex-forming) DNA target and ~3 min for a fully complementary single-stranded target. The results shown here were obtained with the polypurine TFO probe.
In order to determine the generality of our approach to the sequence-specific detection of double-stranded DNA, we fabricated a second sensor employing a TFO probe targeting the 15 base pair polypurine tract (PPT) sequence conserved in all HIV-1 strains and present twice in HIV-1 proviral DNA.32 In contrast to our first sensor, which is a polypurine tract, the TFO probe in this sensor is a polypyrimidine sequence. We find that this second sensor achieves a dissociation constant of 33 nM (again, only an order of magnitude higher that that obtained for the single-stranded target) and supports the detection of its double-stranded target at concentrations as low as 10 nM (Figure 2, right). In order to rule out a strand-exchange mechanism in which the sensor is instead detecting dissociated, single-stranded target, we also tested this system using a self-complementary hairpin target containing a poly-T linker between the two strands. Because the hairpin target is self-complementary across 15 bases, duplex dissociation is thermodynamically unfavorable and simple duplex formation with the TFO probe is unlikely. The observation that the 37 nM affinity of this hairpin target is effectively indistinguishable from that of a double-stranded target comprised of separate strands (Figure 2, right) further supports the proposed triplex-based detection mechanism.
Observations regarding the effects of divalent cations on the response of our sensors provide still more support of the triplex-formation mechanism. Specifically, while triple-helix formation requires the presence of magnesium or other divalent cations,17,33 duplex formation does not. Consistent with this, we do not observe any signal change in the presence of even very high concentrations of duplex target if magnesium and other divalent cations are absent (Figure 4, left). In contrast, when the sensor is challenged with single-stranded target (and thus signaling is linked to the formation of a double helix), we observe similarly large signal changes in both the presence (10 mM) and the absence of magnesium (Figure 4, right).
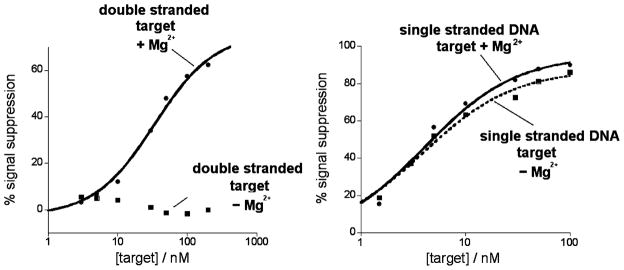
Figure 4.
The proposed triplex formation mechanism, shown here with our polypyrimidine TFO probe, is supported by experiments in which the concentration of magnesium ions is altered. As triplex formation is dependent on magnesium ion concentration, the absence of magnesium produces no signal, even at high concentrations of double-stranded target (left). A duplex-forming target, in contrast, is readily detected in both the presence and absence of magnesium ions (right). The magnesium ion concentration was 10 mM in both experiments.
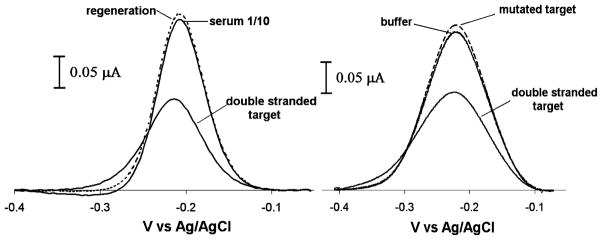
This new electrochemical sensor architecture is convenient and selective. For example, because the TFO probe is strongly adsorbed to the sensor electrode, the sensor is reagentless and reusable: the original current is completely recovered via a 30 s wash with room temperature 8 M guanidine hydrochloride, allowing us to perform repetitive measurements with a single device (Figure 1, right and Figure 5, left). Likewise, because the signal change upon triplex formation is solely due to a binding-specific change in DNA flexibility, and not simply to the adsorption of charge or mass on the sensor surface, it performs well even when deployed directly in complex, multicomponent samples. For example, the sensor detects its double-stranded DNA target directly in blood serum that is diluted 1:10 with buffer with a signal change similar to that observed in buffer alone (Figure 5, left). The specificity of the TFO sensor was tested by using a synthetic hairpin target mutated at four positions.46 The mutated target is similar to the hairpin target (T3-Y) in that it contains the poly-T linker that holds the complementary strands together, creating a stable duplex DNA molecule. However, while both strands are complementary to one another, four base pairs are mutated such that reverse Hoogsteen interactions with the TFO probe are compromised. When challenging the sensor with high concentrations (200 nM) of this mutated duplex, the sensor does not produce any measurable signal change, thus demonstrating the high specificity of the TFO E-DNA platform (Figure 5, right).
Figure 5.
The TFO-based E-DNA sensors are selective, reusable, and specific. Because signal change upon triplex formation is solely due to a DNA binding-specific event, the TFO-based E-DNA sensor performs well, even when deployed directly in complex, multicomponent samples, including 1:10 diluted blood serum (left). The original current is completely recovered via a 30 s rinse in room temperature 8 M guanidine hydrochloride solution, thus allowing us to perform repetitive measurements with a single sensor (left). Moreover, the TFO E-DNA probe displays specificity when challenged with mutated double-stranded DNA (right). Here are shown voltammograms obtained when challenging the polypyrimidine TFO probe with 200 nM of double-stranded target and 200 nM of mutated target (right).
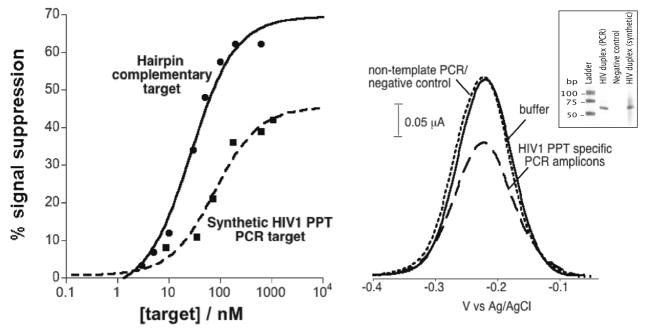
The above proof-of-principle studies employed fully synthetic targets that would be of little if any interest in a clinical setting. Thus motivated, we have also explored the utility of employing this new sensor architecture in the detection of authentic, unpurified PCR amplicons generated from HIV-1 samples. We amplified a region of the HIV-1 genomic RNA containing the PPT sequence recognized by the polypyrimidine TFO probe employed above. As a first step toward this goal, we determined the ease with which we can use our sensor to quantify synthetic, 63-base pair oligonucleotides equivalent to the PCR amplicon (Figure 6, left). We find that these produce signal changes similar to the large, easily measurable signal changes associated with the short hairpin targets described above. The dissociation constants for these larger targets, however, are slightly poorer than those obtained for the short hairpin target. This is presumably due to steric and/or electrostatic effects associated with the larger targets.
Figure 6.
TFO sensor responds to unpurified PCR amplicons containing a triplex-forming element in the HIV-1 genome. (left) Specifically, test results using a synthetic, duplex oligonucleotide equivalent to the relevant HIV-1 sequence binds to the probe, albeit with a poorer dissociation constant and somewhat poorer signaling than those seen with a short hairpin target containing the same recognition footprint. These effects are presumably due to the larger size of the amplicon. (right) The sensor also responds rapidly and robustly to authentic PCR amplified target molecules (at 100 nM), while producing no signal change in response to negative control PCR samples. Polyacrylimide gel (right, inset) containing amplicons from HIV-specific PCR, a negative control (no template added to the PCR), and synthetic HIV duplex show the specificity of the PCR reaction. The size of our HIV target DNA is 63 bp.
Moving forward, we find that our new sensor platform readily detects double-stranded DNA produced via reverse transcriptase polymerase chain reaction amplification of HIV-1 genomic RNA (Figure 6, right). To demonstrate this, we employed a conventional nested, amplification protocol to produce a high concentration of amplified products. To inhibit any potential hybridization of the primers–or primer-dimer or other primer-sequence-containing amplification artifacts–to the TFO probes, we placed the sensor’s recognition footprint 20 bases internal to the end of the amplicon. Starting from 1 μg of HIV-1 genomic RNA, we obtain about 100 nM of the appropriate, double-stranded DNA amplification product (nucleic acid concentrations obtained using Nanodrop instrument). After incubation of the E-DNA sensors with the PCR amplicon solution, we observe about a 40% drop in the sensor current signal, which is indicative of the presence of the expected target sequence (Figure 6, right). Negative controls performed exclusively with the primers sets and the TFO probe produced no change in signal (data not shown), and a control experiment using nontemplate negative control RT-PCR did not show any significant signal change over the same time period (Figure 6, right).
DISCUSSION
Among the numerous methods recently proposed for the sequence-specific detection of DNA, E-DNA sensors, the electrochemical analog of optical molecular beacons,22,29,34–38 present several advantages over other optical or electrochemical hybridization detection methods.21 E-DNA sensors are based on the hybridization-induced folding of an electrode-bound, redox-tagged DNA probe. The E-DNA platform is reagentless, electronic (electrochemical), and highly selective (they perform well even when challenged directly in complex, multicomponent samples such as blood serum or soil) and can discriminate between a perfect match and a single mismatch target.21,23 For all these reasons, they appear to be a promising and appealing approach for the sequence-specific detection of DNA and RNA.39,40 But like other hybridization-based methods, traditional E-DNA sensors require the generation of single-stranded DNA targets prior to detection. Here, in contrast, we describe an E-DNA sensor that overcomes this limitation by hybridizing directly to double-stranded DNA targets. And while this approach is limited to the detection of homopurine or homopyrmidine tracks, these triplex-forming sequences are nevertheless common enough that it is straightforward to generate 16–20 base probes with sufficient specificity to target unique sites in human or pathogen genomes.41,42
Our approach is not the first to employ triplex formation in the detection of double-stranded DNA. Several groups, for example, have previously reported the use of TFOs in the optical detection of double-stranded DNA.16–19 Similarly, two electrochemical methods have been proposed to date where triplex formation is monitored via a direct electrochemical signal from guanine43 or via HPLC separation of the triplex coupled with electrochemical detection.44 While these latter approaches are interesting and achieve remarkable detection limits, their detection principles are limited by several drawbacks: the direct oxidation of guanine appears impractical for realistic applications due to the high overpotential required, the possibility of electrochemical interferences in complex samples (from, for example, exogenous, nontarget DNA), and a high background currents.45 Likewise, the use with HPLC renders the second platform complicated and less suitable for PCR coupling or rapid measurements. For these reasons, the E-DNA sensor described here would appear to offer potentially important advantages for the rapid, electrochemical detection of specific double-stranded DNA sequences.
Like earlier E-DNA counterparts,21 sensors based on triplex-forming oligonucleotides are label-free, reusable, and selective enough to employ directly in complex sample matrices such as blood serum. Directly measuring double-stranded DNA targets makes these TFO sensors an optimal candidate for use with the PCR amplification process. We have demonstrated this coupling here by performing a nested, reverse-transcriptase PCR of HIV-1 genomic RNA followed by the measurement of the amplified target with the TFO-based E-DNA sensors. As E-DNA sensors have proved suitable for low-cost sensors and portable instrumentations,26 our TFO sensors provide many optimal features that demonstrate the possibility of adopting these sensors in real-world applications. All these attributes suggest that E-DNA sensors may be better suited for clinical applications than the previous, mostly optical methods supporting the direct detection of double-stranded DNA, including approaches based on non-DNA probes.
References
- 1.Persil O, Hud NV. Trends Biotechnol. 2007;25(10):433–436. doi: 10.1016/j.tibtech.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Fang TH, Ramalingam N, Xian-Dui D, Ngin TS, Xianting Z, Kuan ATL, Huat EYP, Hai-Qing Gong. Biosens Bioelectron. 2009;24(7):2131–2136. doi: 10.1016/j.bios.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Ihmels H, Otto D. Top Curr Chem. 2005;258:161–204. [Google Scholar]
- 4.Markoulatos P, Saifakas N, Moncany M. J Clin Lab Anal. 2002;16(1):47–51. doi: 10.1002/jcla.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh I, Stains CI, Ooi AT, Segal D. J Mol Biosyst. 2006;2(11):551–560. doi: 10.1039/b611169f. [DOI] [PubMed] [Google Scholar]
- 6.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 7.Ooi AT, Stains CI, Ghosh I, Segal DJ. Biochemistry. 2006;45:3620–3625. doi: 10.1021/bi0517032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stains CI, Furman JL, Segal DJ, Ghosh I. J Am Chem Soc. 2006;128:9761–9765. doi: 10.1021/ja060681j. [DOI] [PubMed] [Google Scholar]
- 9.Stains CI, Porter JR, Ooi AT, Segal DJ, Ghosh I. J Am Chem Soc. 2005;127:10782–10783. doi: 10.1021/ja051969w. [DOI] [PubMed] [Google Scholar]
- 10.Paleček E, Masařiḱ M, Kizek R, Kuhlmeier D, Hassmann J, Schülein J. Anal Chem. 2004;76(19):5930–5936. doi: 10.1021/ac049474x. [DOI] [PubMed] [Google Scholar]
- 11.Halby L, Ryabinin VA, Sinyakov AN, Boutorine AS. Bioorg Med Chem Lett. 2005;15:3720–3724. doi: 10.1016/j.bmcl.2005.05.106. [DOI] [PubMed] [Google Scholar]
- 12.Buchmann W, Boutorine A, Halby L, Tortajada J, De Pauw E. J Mass Spectrom. 2009;44(8):1171–81. doi: 10.1002/jms.1592. [DOI] [PubMed] [Google Scholar]
- 13.Thuong TN, Hélène C. Angew Chem, Int Ed Engl. 1993;32:666–690. [Google Scholar]
- 14.Helene C. Anti-Cancer Drug Des. 1991;6(6):569–584. [PubMed] [Google Scholar]
- 15.Helene C, Toulme JJ. Biochim Biophys Acta Gene Struct Expression. 1990;1049(2):99–125. doi: 10.1016/0167-4781(90)90031-v. [DOI] [PubMed] [Google Scholar]
- 16.Dervan PB. Bioorg Med Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 17.Johnson MD. Chromosoma. 1999;108:181–189. doi: 10.1007/s004120050367. [DOI] [PubMed] [Google Scholar]
- 18.Escude C, Garestier T, Helene C. Proc Natl Acad Sci USA. 1999;96:10603–10607. doi: 10.1073/pnas.96.19.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geron-Landre B, Roulon T, Desbiolles P, Escude C. Nucleic Acids Res. 2003;31:e125. doi: 10.1093/nar/gng125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geron-Landre B, Roulon T, Escude C. FEBS J. 2005;272:5343–5352. doi: 10.1111/j.1742-4658.2005.04932.x. [DOI] [PubMed] [Google Scholar]
- 21.Xiao Y, Lai RY, Plaxco KW. Nature Prot. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 22.Ricci F, Plaxco KW. Mikrochim Acta. 2008;163(3–4):149–155. [Google Scholar]
- 23.Lubin AA, Plaxco KW. Acc Chem Res. 2010;43(4):496–505. doi: 10.1021/ar900165x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai RY, Lagally ET, Lee SH, Soh HT, Plaxco KW, Heeger AJ. Proc Natl Acad Sci USA. 2006;103(11):4017–4021. doi: 10.1073/pnas.0511325103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavlovic E, Lai RY, Wu TT, Ferguson BS, Sun R, Plaxco KW, et al. Langmuir. 2008;24:1102–1107. doi: 10.1021/la702681c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ricci F, Zari N, Caprio F, Recine S, Amine A, Moscone D, et al. Bioelectrochem. 2009;76(1–2):208–213. doi: 10.1016/j.bioelechem.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricci F, Lai RY, Heeger AJ, Plaxco KW, Sumner JJ. Langmuir. 2007;23(12):6827–6834. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci F, Lai RY, Plaxco KW. Chem Commun. 2007;36:3768–3770. doi: 10.1039/b708882e. [DOI] [PubMed] [Google Scholar]
- 29.Xiao Y, Lubin AA, Baker BR, Plaxco KW, Heeger AJ. Proc Natl Acad Sci USA. 2006;103(45):16677–16680. doi: 10.1073/pnas.0607693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shafer RH. Prog Nucleic Acid Res Mol Biol. 1998;59:55–94. doi: 10.1016/s0079-6603(08)61029-6. [DOI] [PubMed] [Google Scholar]
- 31.Antony T, Thomas T, Sigal LH, Shirahata A, Thomas TJ. Biochemistry. 2001;40(31):9387–9395. doi: 10.1021/bi010397z. [DOI] [PubMed] [Google Scholar]
- 32.Giovannangeli C, Perrouault L, Escude C, Thuong N, Helene C. Biochemistry. 1996;35:10539–10548. doi: 10.1021/bi952993x. [DOI] [PubMed] [Google Scholar]
- 33.Malkov VA, Voloshin ON, Soyfer VN, Frank-Kamenetskii MD. Nucleic Acids Res. 1993;21:585–591. doi: 10.1093/nar/21.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Immoos CE, Lee SJ, Grinstaff MW. Chem Bio Chem. 2004;5:1100–1104. doi: 10.1002/cbic.200400045. [DOI] [PubMed] [Google Scholar]
- 35.Immoos CE, Lee SJ, Grinstaff MW. J Am Chem Soc. 2004;126:10814–10815. doi: 10.1021/ja046634d. [DOI] [PubMed] [Google Scholar]
- 36.Mao T, Luo C, Ouyang Q. Nucleic Acids Res. 2003;31:108–172. doi: 10.1093/nar/gng108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phares N, White RJ, Plaxco KW. Anal Chem. 2009;81:1095–1100. doi: 10.1021/ac8021983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lubin AA, Vander Stoep Hunt B, White RJ, Plaxco KW. Anal Chem. 2009;81:2150–2158. doi: 10.1021/ac802317k. [DOI] [PubMed] [Google Scholar]
- 39.Paleček E. Trends Biotechnol. 2004;22(2):55–58. doi: 10.1016/j.tibtech.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 40.Thorp HH. Trends Biotechnol. 2003;21:522–524. doi: 10.1016/j.tibtech.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Goñi JR, de la Cruz X, Orozco M. Nucleic Acids Res. 2004;32(1):354–360. doi: 10.1093/nar/gkh188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. Nucleic Acids Res. 2008;36(16):5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai X, Rivas G, Shirashi H, Farias P, Wang J, Tomschik M, Jelen F, Palecek E. Anal Chim Acta. 1997;344:65–76. [Google Scholar]
- 44.Ihara T, Maruo Y, Takenaka S, Takagi M. Nucleic Acids Res. 1996;24:4273–4280. doi: 10.1093/nar/24.21.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 46.Giovannangeli C, Diviacco S, Labrousse V, Gryaznov S, Charneau P, Helene C. Proc Natl Acad Sci USA. 1997;94(1):79–84. doi: 10.1073/pnas.94.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]