Abstract
Amelogenin is the most abundant protein of the enamel organic matrix and is a structural protein indispensable for enamel formation. One of the amelogenin splicing isoforms, Leucine-rich Amelogenin Peptide (LRAP) induces osteogenesis in various cell types. Previously, we demonstrated that LRAP activates the canonical Wnt signaling pathway to induce osteogenic differentiation of mouse ES cells through the concerted regulation of Wnt agonists and antagonists. There is a reciprocal relationship between osteogenic and adipogenic differentiation in bone marrow mesenchymal stem cells (BMMSCs). Wnt10b-mediated activation of canonical Wnt signaling has been shown to regulate mesenchymal stem cell fate. Using the bipotential bone marrow stromal cell line ST2, we have demonstrated that LRAP activates the canonical Wnt/β-catenin signaling pathway. A specific Wnt inhibitor sFRP-1 abolishes the effect of LRAP on the stimulation of osteogenesis and the inhibition of adipogenesis of ST2 cells. LRAP treatment elevates the Wnt10b expression level whereas Wnt10b knockdown by siRNA abrogates the effect of LRAP. We show here that LRAP promotes osteogenesis of mesenchymal stem cells at the expense of adipogenesis through upregulating Wnt10b expression to activate Wnt signaling.
Introduction
Amelogenin is the most abundant protein of the enamel organic matrix and is a structural protein indispensable for enamel formation [1–6]. A serendipitous finding showed that amelogenin also can be used to induce the regeneration of periodontal tissues in monkeys and humans [7–9]. mdogain, a commercial product consisting largely of alternatively spliced and processed porcine amelogenins, can induce new bone, cementum and periodontal ligament formation in the jaws of dogs, monkeys and humans [7–11]. One naturally occurring amelogenin splicing isoform, Leucine-rich Amelogenin Peptide (LRAP), consisting of the N-terminal 33 and the C-terminal 26 residues of the full-length protein, has been shown to induce osteogenesis in various cell types [12–14]. We detected LRAP expression during osteogenesis of wild-type (WT) mouse embryonic stem (ES) cells and observed the absence of LRAP expression in amelogenin-null (KO) ES cells. LRAP treatment of WT and KO ES cells induces significant increases in mineral matrix formation, bone sialoprotein and osterix gene expression. In addition, the impaired osteogenesis of amelogenin-null ES cells is partially rescued by the addition of exogenous LRAP [15]. We also demonstrated that LRAP activates the canonical Wnt signaling pathway to induce osteogenic differentiation of mouse ES cells through the concerted regulation of Wnt agonists and antagonists [16].
Bone marrow mesenchymal stem cells (BMMSC) can differentiate into a number of cell types, including adipocytes and osteoblasts [17, 18]. Compelling evidence from both in vitro and in vivo experiments indicate a reciprocal relationship between these two cell lineages [19–21]. For example, bone marrow stromal cells and immortalized clonal lines (e.g. ST2) are capable of undergoing both osteogenic and adipogenic differentiation, depending upon culture conditions. Moreover, single cell clones from bone marrow can differentiate in vitro into either adipocytes or osteoblasts [22]. Activation of Wnt/β-catenin signaling inhibits adipogenesis and stimulates osteogenesis by a rapid suppression of the adipogenic transcription factors C/EBPα and PPARγ followed by an increase in osteoblastic transcription factors [23, 24]. The endogenous Wnt signal may be initiated by Wnt10b, which is expressed in preadipocytes and stromal vascular cells but is rapidly suppressed upon induction of adipogenesis [25, 26]. Although there is no evidence that Wnt10b deficiency in mice alters adipose tissue development, transgenic mice overexpressing Wnt10b in adipose tissues have ~50% less white adipose tissue and arrested development of brown fat [27, 28]. Furthermore, these mice resist expansion of adipose tissue under conditions of diet-induced and genetic obesity [27, 29]. Mice expressing the Wnt10b transgene also exhibit improved glucose homeostasis and increased insulin sensitivity [27, 29]. Mice expressing the Wnt10b transgene in bone marrow have increased bone mass and strength and resist the loss of bone that occurs with aging or estrogen deficiency. In addition, Wnt10b-null mice have decreased trabecular bone mass and serum osteocalcin levels, indicating that Wnt10b is an endogenous regulator of bone mass [23].
Previously, we have shown that LRAP stimulates osteogenic differentiation of murine ES cells through activating the canonical Wnt/β-catenin signaling pathway [16]. Given that Wnt10b-mediated activation of Wnt/β-catenin signaling stimulates osteogenesis and inhibits adipogenesis of bone marrow mesenchymal stem cells [23–26], we hypothesized that LRAP might affect fate determination (osteogenesis versus adipogenesis) of mesenchymal stem cells through Wnt/β-catenin signaling. In this study, we used the bipotential bone marrow stromal cells ST2 to characterize the effect of LRAP on mesenchymal stem cells and to delineate the underlying mechanism.
Materials and Methods
Reagents
LRAP was chemically synthesized and HPLC purified as described previously [16].
Cell Culture
ST2 cells were maintained in α-minimal essential medium containing 10% fetal bovine serum and 100 units/ml penicillin/streptomycin. For adipogenesis, cells that had been confluent for a day were treated with 10% fetal bovine serum, 1 µM dexamethasone, 0.5 mM methylisobutylxanthine, 1 µg/ml insulin, and 5 µM troglitazone (day 0). Cells were fed with 1 µg/ml insulin in 10% fetal bovine serum media (day 2), and refed with 10% fetal bovine serum media every 2 days subsequently. To induce osteogenesis, over-confluent cells were switched to mineralization media containing 25 µg/ml ascorbic acid and 10 mM β-glycerophosphate.
Analysis of Mineral Deposition or Lipid Deposition
Two-week post-osteogenic-induction, cells were stained with Alizarin Red. Quantification of calcium concentration was measured by spectrophotometry at 612 nm (QuantiChrom Calcium Assay kit; BioAssay Systems). The total amount of protein in each sample was used as a standard with which to normalize calcium concentration. Eight-day post-adipogenic-induction, cells were stained with Oil Red O (0.5 g Oil Red O in 100 ml isopropanolol) for analysis of triglyceride and lipid deposition. Oil Red O staining was extracted by isopropanol and measured by spectrophotometry at 490nm.
RNA Extraction and cDNA Synthesis
RNA was isolated using RNAqueous®-4PCR Kit (Ambion) following the manufacturer’s instructions. Synthesis of cDNA was performed using RETROscript® Kit (Ambion). For cDNA template preparation, 1 µg of total RNA was used in a 20 µL reaction.
Western Blot Analysis
Cell lysate was prepared by washing the cells with PBS twice followed by the addition of M-PER mammalian extraction reagent (Pierce). An aliquot of the protein was added to 2X SDS loading buffer consisting of 4% v/v SDS, 200 mM dithiothreitol, 100 mM Tris pH 6.8, 20% v/v glycerol, 0.2% w/v bromphenol blue for polyacrylamide gel electrophoresis (PAGE). Protein concentration was measured using the Bio-Rad protein assay on the lysate samples, with known amounts of bovine serum albumin to establish a standard curve. Approximately 10 µg of protein from each experimental sample group was loaded to a 4%–20% Tris-glycine SDS-polyacrylamide gel electropheoresis (PAGE) gel. The size-resolved proteins were transferred to Immobilon-P membranes (Millipore) for 1 hour at 100 mA. The membrane was blocked with 5% non-fat milk in TBST (1×TBS, 0.1% Tween-20) for 1 hour at room temperature. Mouse anti-β-catenin antibody (1:2000; BD Bioscience) was added to the TBST and the membrane was incubated at 4°C overnight. HRP-conjugated anti-mouse antibody (1:10000; Amersham Biosciences) was used as a secondary antibody and incubated with the membrane for 1h. The antigen-antibody signal was detected by ECL detection system and normalized to the amount of β-actin from the same sample. Quantification of the signal was described previously [16].
Detection of Wnt reporter activity
MC3T3 cells grown in a 12-well culture dish were transiently transfected with the Wnt responsive TOPFLASH construct (1.6 µg/well), which contains 16 copies of a TCF/LEF site, using Lipofectamine 2000 (Invitrogen). For control, parallel dishes were transfected with the FOPFLASH construct, which contains 16 copies of a mutant TCF/LEF site. In each case, CMV-lacZ (0.16 µg/well) was co-transfected. Sixteen-hour post-transfection, the media was replaced with the conditioned media from LRAP-treated ST2 cells. Luciferase activity was detected using the Dual-Light reporter gene assay system (Applied Biosystems) 24 hours later. Relative luciferase activity was calculated by normalizing the average luciferase activity to the β-galactosidase activity.
The siRNA knockdown assay
Knockdown of endogenous Wnt10b was accomplished using a commercially available siRNA (Ambion). The trypsinized ST2 cells were resuspended at 1 × 105 cells/mL in growth medium. Transfection agent siPORT NeoFX (3 µL) was diluted in OPTI-MEM I medium (47 µL). The siRNA was diluted in OPTI-MEM I medium by adding 1.5 µL of 20 µM siRNA to 48.5 µL of OPTI-MEM I medium, mixed with the transfection agent, and incubated at room temp for 10 min. The RNA/transfection agent complexes were dispensed into the empty wells of a 12-well culture plate and 900 µL of cells were transferred to each well of the culture plate containing the RNA/transfection agent complexes. The cells and RNA/transfection agent complexes were mixed gently and incubated at 37°C for 48 hours. An unrelated siRNA was included at the same concentration and conditions to serve as controls.
Quantitative RT-PCR
Quantitative PCR was performed according to the manufacturer's protocol. Briefly, a 25 µL reaction was prepared for each sample. Included in this reaction volume was 1 µL of the resulting cDNA, iQ SYBR green supermix (Bio-Rad) containing dNTP and iTaq DNA polymerase, and the appropriate primers. The resulting threshold cycle (CT) value from each primer pair was normalized with the CT value for 18S RNA, which serves as an internal control. After amplification, melting curve analysis was performed as described in the manufacturer's protocol, and samples with aberrant melting curves were excluded. The corresponding primer sequences are 18S RNA (forward, 5′-CGATGCTCTTAGCTGAGTGT-3′; reverse, 5′-GGTCCAAGAATTTCACCTCT-3′), Runx2 (forward 5’-CCGTGGCCTTCAAGGTTGT-3’, reverse 5’-TTCATAACAGCGGAGGCATTT-3’), osterix (forward 5’-CCCTTCTCAAGCACCAATGG-3’, reverse 5’-AAGGGTGGGTAGTCATTTGCATA-3’), Dlx5 (forward 5’-GTCCCAAGCATCCGATCCG-3’, reverse 5’-GCGATTCCTGAGACGGGTG-3’), collagen I (forward 5’-GCTCCTCTTAGGGGCCACT-3’, reverse 5’-CCACGTCTCACCATTGGGG-3’), C/EBPα (forward, 5′-TGAACAAGAACAGCAACGAG-3′; reverse, 5′-TCACTGGTCACCTCCAGCAC-3′), PPARγ (forward, 5′-GGAAAGACAACGGACAAATCAC-3′; reverse, 5′-TACGGATCGAAACTGGCAC-3′),Wnt10b (forward 5’-TTCTCTCGGGATTTCTTGGATTC-3’, reverse 5’-TGCACTTCCGCTTCAGGTTTTC-3’).
Statistical analysis
All experiments were performed in triplicate unless stated otherwise. Final values were reported as means±standard deviation (SD). Data were analyzed using Student's t-test and P<0.05 was considered statistically significant.
Results
Effect of LRAP on osteogenic and adipogenic differentiation of ST2 cells
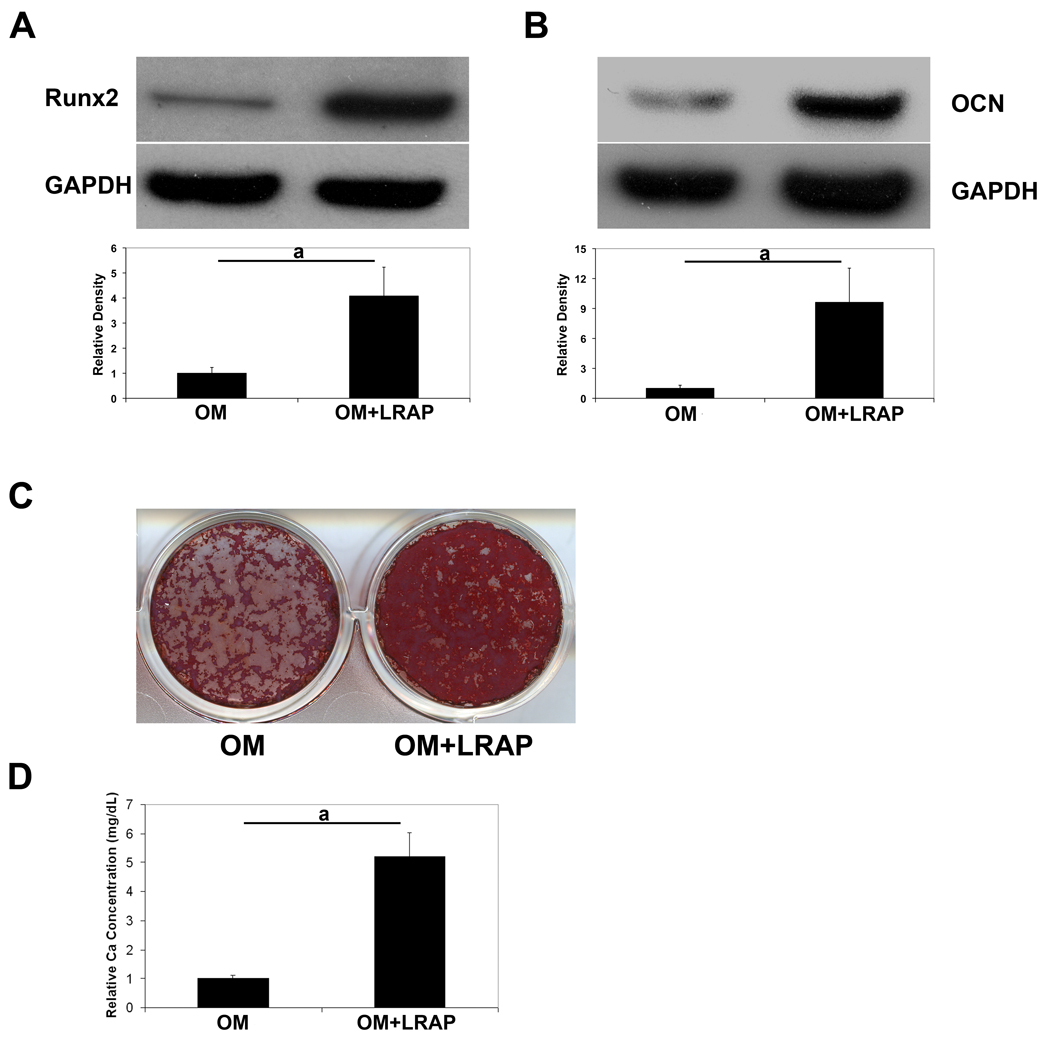
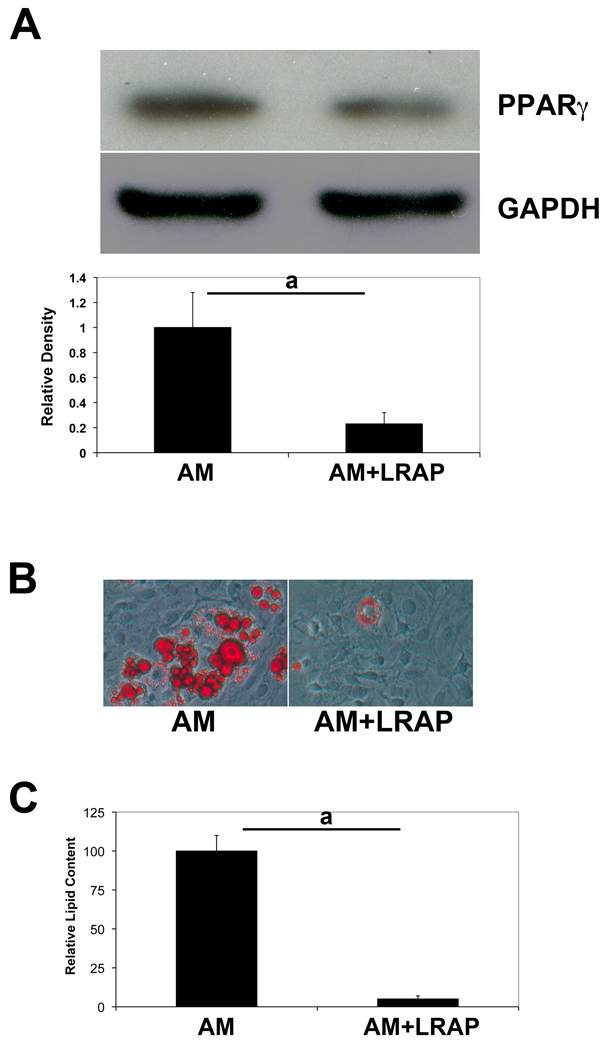
To decipher the mechanism underlying the signaling effect of LRAP on BMMSCs, we employed the bipotential stromal cell line ST2, which can be induced to both osteogenic and adipogenic differentiation. ST2 cells were cultured to over-confluency and induced to osteogenic differentiation for a week, and protein samples were extracted and analyzed for osteogenic markers. In the presence of LRAP (10 ng/ml) in the osteogenic medium, Runx2 protein levels were upregulated ~4 fold (Fig. 1A) and osteocalcin protein levels were increased by more than 9 fold (Fig. 1B). Two weeks after osteogenic induction, cells were subject to Alizarin Red staining (Fig. 1C). In the presence of LRAP, there was a 5.2-fold increase of mineral deposition (Fig. 1D). To examine whether LRAP alters the adipogenic differentiation potential, cells were cultured until over-confluent and induced to adipogenesis in the presence of LRAP (10 ng/ml). Five days after adipogenic induction, protein samples were collected for analyses of adipogenic markers. LRAP treatment resulted in a 3.5-fold decrease of the protein level of PPARγ, which is the master transcription factor of adipogenesis (Fig. 2A). Eight days after induction, the cells were subject to Oil Red O staining (Fig. 2B). Compared to cells cultured in adipogenic medium alone, LRAP-treated cells showed a marked decrease in the number of cells containing lipid droplets. Quantitative analyses revealed a statistically significant 3-fold decrease of triglyceride and lipid accumulation in LRAP-treated cells (Fig. 2C). We also tested LRAP on primary BMMSCs and observed that LRAP promoted osteogenesis whereas inhibited adipogenesis of BMMSCs (Fig. S1).
Figure 1. LRAP stimulates osteogenesis of ST2 cells.
ST2 cells were subject to osteogenic induction in the presence or absence of LRAP (10 ng/ml). One week after osteogenic induction, cells were collected for analysis of bone marker protein. (A) Western blot analysis of Runt-related transcription factor 2 (Runx2). “OM”: osteogenic medium alone; “OM+LRAP”: osteogenic medium with LRAP (10 ng/ml). (B) Western blot analysis of osteocalcin (OCN). “OM”: osteogenic medium alone; “OM+LRAP”: osteogenic medium with LRAP (10 ng/ml). GAPDH was used as an internal control. Band intensity ratio was calculated by normalizing the band intensity for each sample with the band intensity of GAPDH. (C) Mineral deposition assayed with Alizarin Red staining. (D) Quantification of mineral deposition. The graphs represent mean±SD (a: P<0.01).
Figure 2. LRAP inhibits adipogenesis of ST2 cells.
ST2 cells were induced to adipogenesis in the presence or absence of LRAP (10 ng/ml). (A) Western blot analysis of PPARγ. GAPDH was used as an internal control. Band intensity ratio was calculated by normalizing the band intensity for each sample with the band intensity of GAPDH. (B) Oil Red O staining of ST2 cells. “AM”: cells induced to adipogenic differentiation in the absence of LRAP; “AM+LRAP”: cells induced to adipogenic differentiation in the presence of LRAP. (C) Quantification of triglyceride and lipid deposition in cells induced to adipogenic differentiation. The graphs represent mean±SD (a: P<0.01).
To determine the effect of LRAP on cellular proliferation, ST2 cells were cultured in the presence of LRAP (10 ng/ml) and labeled with BrdU for 24 hours prior to cell sorting analyses. In comparison to the control culture, LRAP treatment had little effect on the proliferation of ST2 cells (Figure S2).
Effect of LRAP on the activation of Wnt/β-catenin signaling in ST2 cells
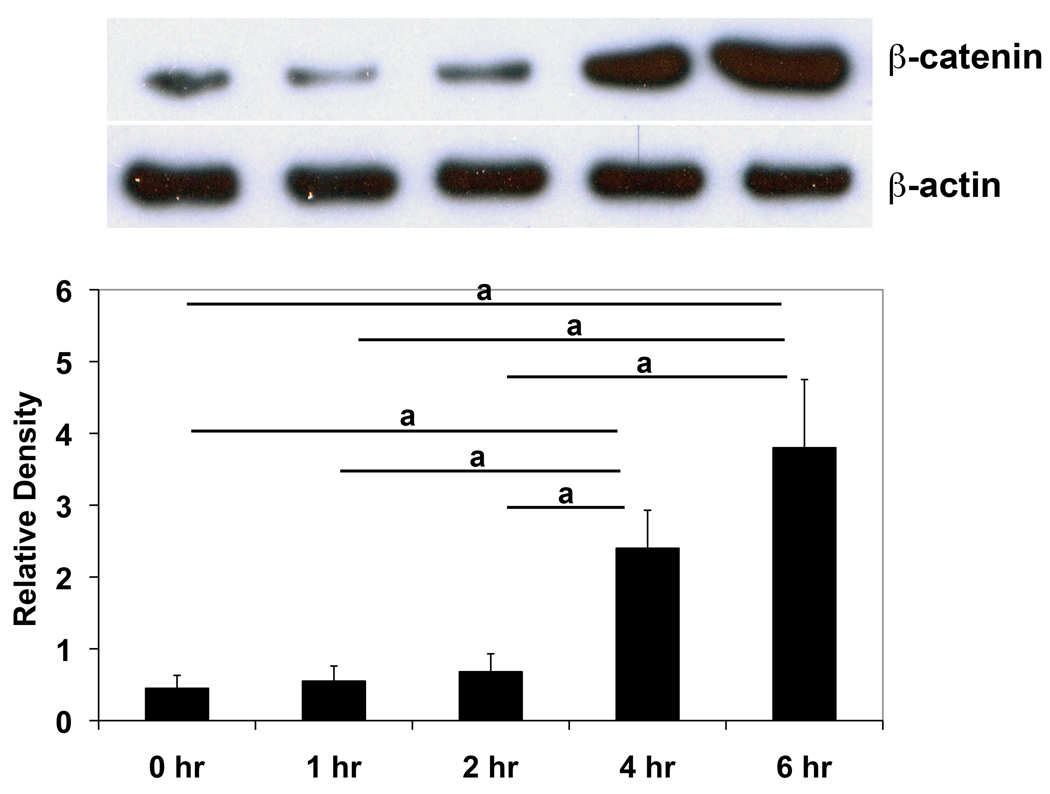
With active Wnt signaling, stabilized β-catenin accumulates in the cytosol and translocates to the nucleus, where it interacts with T cell factor/lymphoid enhancer binding factor (TCF/LEF) to mediate many of the effects of Wnts on gene transcription [30]. To determine whether canonical Wnt signaling pathway was activated in ST2 cells by LRAP, cytosolic β-catenin levels were measured with Western blot analysis at various time points post-LRAP treatment. At 1-hour and 2-hours post treatment, β-catenin levels remained at the baseline. However, marked increases in β-catenin levels were observed 4 hours and 6 hours after LRAP treatment (Fig. 3), indicating that the canonical Wnt/β-catenin signaling was activated.
Figure 3. LRAP treatment elevates β-catenin protein level in ST2 cells.
(A) Western blot analysis of cytosolic β-catenin in ST2 cells cultured in the presence of LRAP (10 ng/ml) for various time periods. β-actin was used as an internal control. (B) Band intensity ratio was calculated by normalizing the band intensity for each sample with the band intensity of β-actin. The graph represents mean±SD (a: P<0.01).
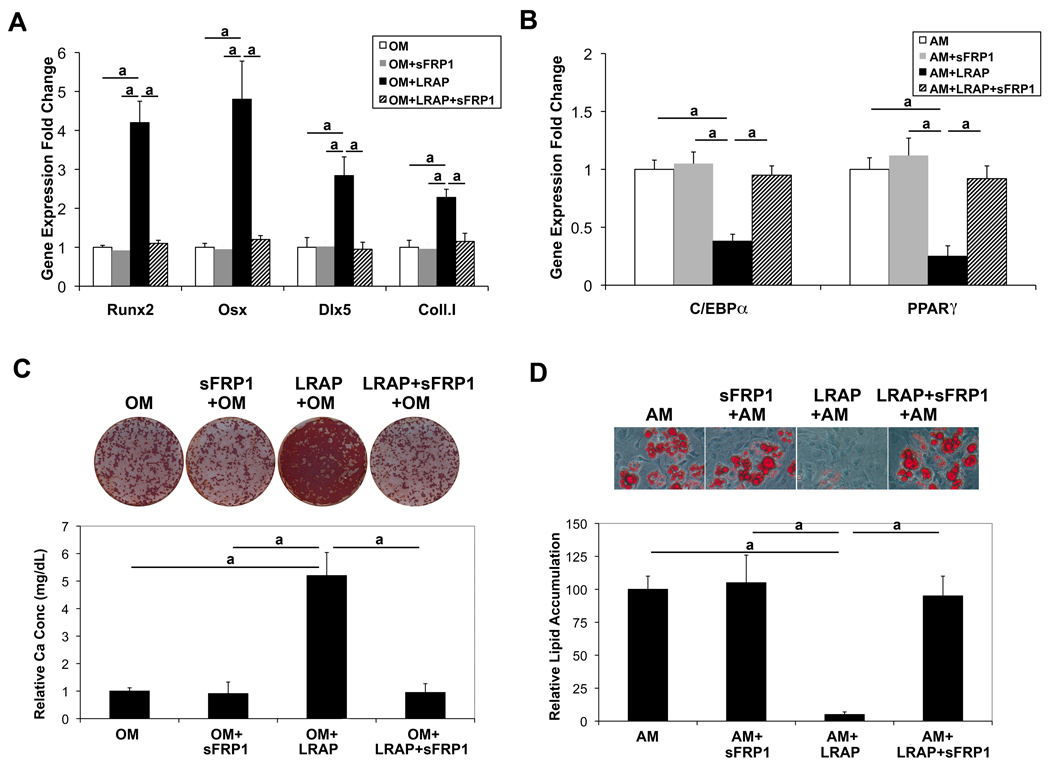
Wnt signaling is tightly regulated by members of several families of antagonists. Interactions between Wnts and frizzled receptors are inhibited by members of the secreted frizzled-related protein (sFRP) family [31]. To further address the involvement of Wnt signaling in LRAP-mediated regulation of osteogenic versus adipogenic lineage selection of ST2 cells, cells were treated with recombinant sFRP-1 (20 ng/ml, R&D Systems), along with LRAP (10 ng/ml), and induced to osteogenic and adipogenic differentiation, respectively. Two weeks after osteo-induction, cells were subject to Alizarin Red staining for analysis of mineral deposition. Eight days after adipo-induction, cells were subject to Oil Red O staining for analysis of triglyceride and lipid deposition. Treatment with sFRP-1 blocked LRAP-stimulated osteogenesis (Fig. 4A and 4C) and overcame the inhibitory effect of LRAP on adipogenesis (Fig. 4B and 4D), suggesting that the effect of LRAP was mediated by Wnt signaling.
Figure 4. Wnt antagonist sFRP-1 abolishes the effect of LRAP on the stimulation of osteogenesis and the inhibition of adipogenesis of ST2 cells.
ST2 cells were treated with recombinant sFRP-1 (20 ng/ml), along with LRAP (10 ng/ml), and induced to osteogenic and adipogenic differentiation, respectively. LRAP and/or sFRP-1 were maintained in the culture media throughout differentiation as indicated. (A) Two days after osteo-induction, RNA was isolated for quantitative Real-time RT-PCR analysis of osteoblast marker genes: Runx2, Osx, Dlx5 and type I collagen (coll. I). “OM”: osteo-induction in the absence of LRAP; “OM+sFRP-1”: osteo-induction in the presence of sFRP-1; “OM+LRAP”: osteo-induction in the presence of LRAP; “OM+LRAP+sFRP-1”: osteo-induction in the presence of LRAP and sFRP-1. (B) Two days after adipo-induction, RNA was isolated for quantitative Real-time RT-PCR analysis of adipocyte marker genes: C/EBPα and PPARγ “AM”: adipo-induction in the absence of LRAP; “AM+sFRP-1”: adipo-induction in the presence of sFRP-1; “AM+LRAP”: adipo-induction in the presence of LRAP; “AM+LRAP+sFRP-1”: adipo-induction in the presence of LRAP and sFRP-1. (C) Alizarin Red staining for analysis of mineral deposition 2 weeks after osteo-induction. (D) Oil Red O staining for analysis of triglyceride and lipid deposition 8 days after adipo-induction. The graphs represent mean±SD (a: P<0.01).
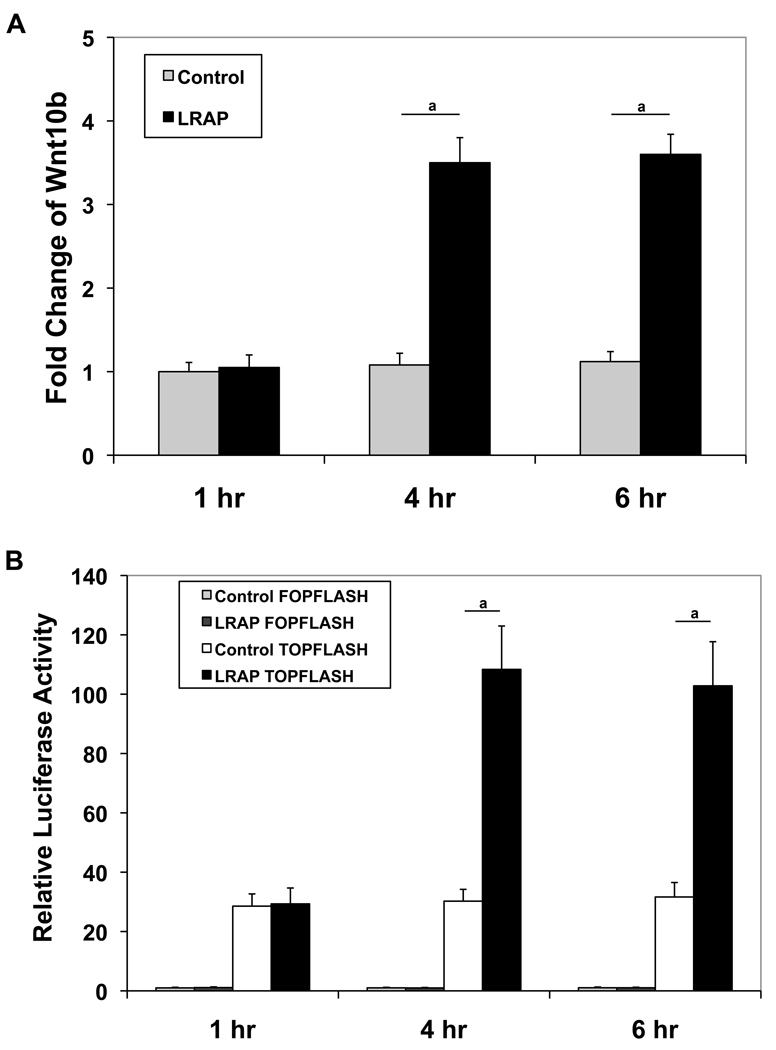
Wnt10b has been shown to stimulate osteogenesis and inhibit adipogenesis of BMMSC by suppression of the adipogenic transcription factors PPARγ and C/EBPα followed by induction of the osteoblastogenic transcription factors Runx2 and osterix [23–26]. Given that LRAP treatment gave rise to increased expression of Runx2 and decreased expression of PPARγ (Fig. 1 and 2), we postulated that the activation of canonical Wnt/β-catenin signaling by LRAP (Fig. 3) might be mediated by Wnt10b. To determine whether Wnt10b expression was upregulated, ST2 cells were treated with LRAP (10 ng/ml) for various time periods and the Wnt10b mRNA level was measured with quantitative real-time RT-PCR. In the absence of LRAP, there was no change in Wnt10b expression. Little change was observed at 1-hour post-treatment; however, a significant increase of Wnt10b expression was observed 4 hours post-LRAP treatment (Fig. 5A). To determine whether elevated Wnt activity appeared in the culture medium upon LRAP treatment and not simply an increase in mRNA levels, a TCF/LEF reporter assay was employed using a luciferase reporter construct that contains 16 TCF/LEF binding sites for β-catenin (TOPFLASH). Another construct containing the mutated TCF/LEF binding sites (FOPFLASH) upstream of the luciferase reporter was used as a control. TOPFLASH and FOPFLASH plasmids were transiently transfected into MC3T3 cells, respectively, along with pCMV-lacZ plasmid serving as an internal control for transfection. A transfection efficiency of approximately 40%, a level sufficient for this reporter assay, was achieved (data not shown). ST2 cells were treated with LRAP (10 ng/ml) for various time periods. The conditioned medium from each time point was then collected and transferred to the transiently transfected MC3T3 cells. At 1-hour post-treatment, there was little difference in luciferase activity between LRAP-treated and untreated samples. However, significant increases in luciferase activity were observed in LRAP-treated samples at 4 and 6 hours post-treatment intervals (Fig. 5B), suggesting that elevated Wnt activity was present in the conditioned medium.
Figure 5. LRAP treatment results in the upregulation of Wnt10b expression.
(A) ST2 cells were treated with LRAP (10 ng/ml) for various time periods and Wnt10b mRNA level was then measured with quantitative Real-time RT-PCR and normalized to GAPDH. “Control”: untreated; “LRAP”: treated with 10 ng/ml of LRAP. (B) ST2 cells were treated with LRAP (10 ng/ml) for various time periods. The conditioned medium from each time point was then collected and transferred to MC3T3 cells that had been transiently transfected with TOPFLASH or FOPFLASH reporter. Luciferase activity was measured using the Dual-Light reporter gene assay system (Applied Biosystems) 24 hours later. Relative luciferase activity was calculated by normalization of the average luciferase activity to the β-galactosidase activity. “Control FOPFLASH”: FOPFLASH-transfected MC3T3 cells with conditioned medium from untreated ST2 cells; “LRAP FOPFLASH”: FOPFLASH-transfected MC3T3 cells with conditioned medium from LRAP-treated ST2 cells; “Control TOPFLASH”: TOPFLASH-transfected MC3T3 cells with conditioned medium from untreated ST2 cells; “LRAP TOPFLASH”: TOPFLASH-transfected MC3T3 cells with conditioned medium from LRAP-treated ST2 cells. The graphs represent mean±SD (a: P<0.01).
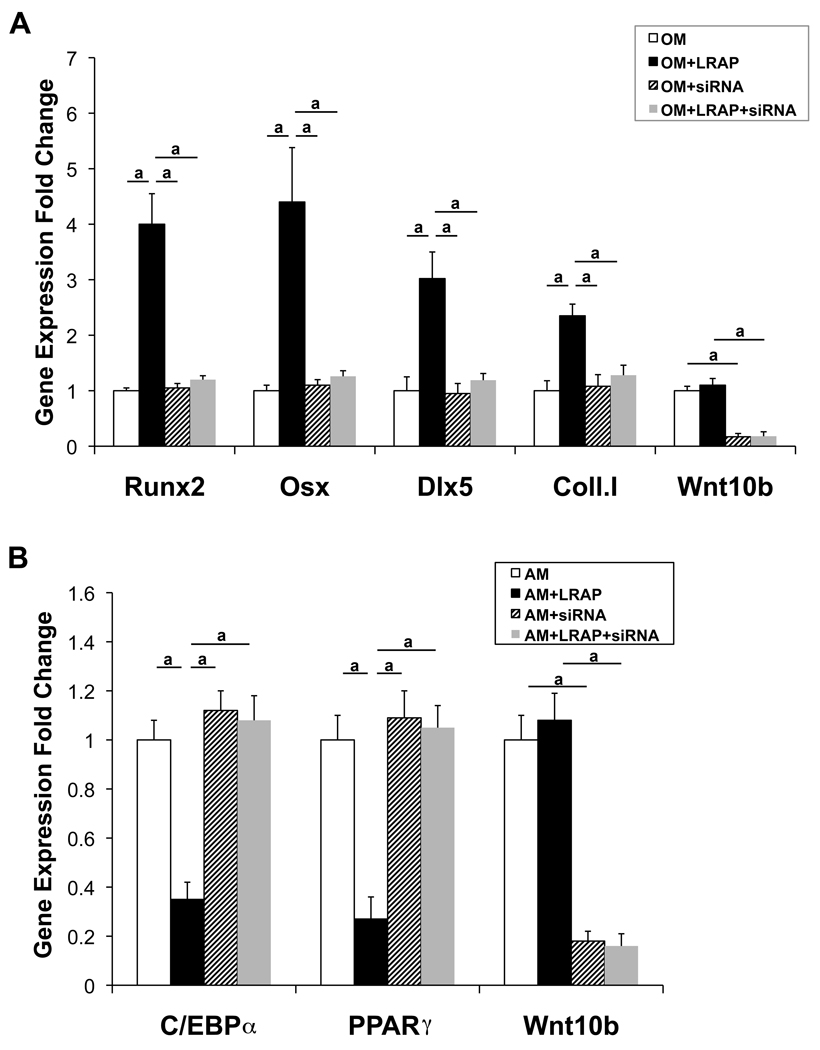
To determine whether the effect of LRAP was mediated by Wnt10b, siRNA was employed to specifically knockdown Wnt10b expression. ST2 cells were transfected with Wnt10b-specific siRNA for 48 hours and subsequently induced to osteogenic and adipogenic differentiation, respectively. Two days after osteogenic induction, RNA was isolated for quantitative Real-time RT-PCR analysis of osteoblast marker genes, Runx2, Osx, Dlx5 and type I collagen (Fig. 6A). Two days after adipogenic induction, RNA was isolated for quantitative real-time RT-PCR analysis of adipocyte marker genes, C/EBPα and PPARγ Fig. 6B). A knockdown efficiency of ~80% was achieved for Wnt10b (Fig. 6). This Wnt10b knockdown abolished the upregulation of osteoblast marker genes (Fig. 6A) and the downregulation of adipocyte marker genes (Fig. 6B) mediated by LRAP. These data demonstrated that the effect of LRAP on lineage selection between osteogenesis and adipogenesis of ST2 cells was mediated by Wnt10b.
Figure 6. Knockdown of Wnt10b expression by siRNA abolishes the effect of LRAP on the stimulation of osteogenesis and the inhibition of adipogenesis of ST2 cells.
ST2 cells were transfected with Wnt10b-specific siRNA for 48 hours and subsequently induced to osteogenic and adipogenic differentiation, respectively. (A) Two days after osteo-induction, RNA was isolated for quantitative Real-time RT-PCR analysis of osteoblast marker genes: Runx2, Osx, Dlx5 and type I collagen (coll. I). “OM”: osteo-induction in the absence of LRAP; “OM+LRAP”: osteo-induction in the presence of LRAP; “OM+siRNA”: osteo-induction in the presence of siRNA; “OM+LRAP+siRNA”: osteo-induction in the presence of LRAP and siRNA. (B) Two days after adipo-induction, RNA was isolated for quantitative Real-time RT-PCR analysis of adipocyte marker genes: C/EBPα and PPARγ “AM”: adipo-induction in the absence of LRAP; “AM+LRAP”: adipo-induction in the presence of LRAP; “AM+siRNA”: adipo-induction in the presence of siRNA; “AM+LRAP+siRNA”: adipo-induction in the presence of LRAP and siRNA. The graphs represent mean±SD (a: P<0.01).
Discussion
In previous studies, LRAP has been found to induce osteogenesis in various cell types, including rat muscle fibroblasts [12], mouse cementoblasts [13], and mouse oral mucosal cells [14]. We have shown that LRAP activates the canonical Wnt signaling pathway to induce osteogenesis of mouse ES cells [16]. In the present study, we demonstrated that LRAP stimulates osteogenic differentiation and inhibits adipogenic differentiation of ST2 cells through activating the canonical Wnt/β-catenin signaling pathway. Moreover, the effect of LRAP on lineage selection of mesenchymal stem cells was mediated by the upregulation of Wnt10b.
Wnt10b is expressed in bone marrow, and has been shown to function as an endogenous regulator of mesenchymal cell fate in bone marrow [23, 32, 33]. Amelogenin expression is detected in long bone, cartilage, epiphyseal growth plate, and bone marrow [34]. We have observed that LRAP influences the fate determination of primary mouse BMMSCs by stimulating osteogenic and inhibiting adipogenic differentiation (Fig. S1). Transient activation of Wnt/β-catenin signaling by Wnt10b rapidly suppresses the master adipogenic regulators C/EBPα and PPARγ in bipotential ST2 cells and this suppression precedes the Wnt-induced increase in osteogenic transcription factors [23]. Repression of C/EBPα and PPARγ appears to be a primary mechanism by which Wnt signaling controls mesenchymal cell fate between osteogenesis and adipogenesis. Mesenchymal precursors such as ST2 cells express low but biologically relevant levels of adipogenic transcription factors (C/EBPα and PPARγ) and osteoblast transcription factors (Runx2, Dlx5, and osterix) [23]. Expression of these 2 classes of transcription factors is maintained at low levels due to negative feedback, and imbalance between them results in changed cell fate leading to differentiation. Constitutive Wnt/β-catenin signaling favors expression of osteoblast genes at the expense of adipocyte genes [23]. Wnt signaling could regulate the fate of mesenchymal precursors by repressing adipocyte transcription factors, stimulating osteoblast transcription factors, or both. This is consistent with our observation that LRAP treatment upregulated bone marker genes and downregulated fat marker genes through the activation of Wnt/β-catenin signaling. Increased adipocyte and decreased osteoblast formation are often associated with aging and osteoporosis. Human genetic studies have implicated Wnt signaling in human bone and metabolic diseases [35–41].
A recent study has shown that Hedgehog signaling enhances the osteogenic differentiation of murine adipose-derived stromal cells, at the expense of adipogenesis [42, 43]. The Hedgehog signaling pathway is also involved in oxysterol-mediated pro-osteogenic and anti-adipogenic differentiation of bone marrow mesenchymal stem cells [43, 44]. However, LRAP treatment of ST2 cells did not activate Hedgehog signaling (unpublished data), suggesting that the Hedgehog pathway is not implicated in LRAP-mediated lineage selection of BMMSCs.
Although human recombinant full-length amelogenin protein has been shown to increase the proliferation of human bone marrow mesenchymal stem cells [45], we found that LRAP has little effect on the proliferation of ST2 cells, a murine bipotential stromal cell line. The difference could lie in the variations in cell type, species, and/or signaling motifs contained in amelogenin isoforms. For example, LRAP and full-length amelogenin have distinct conformations, with LRAP being highly hydrophilic and full-length amelogenin tending to aggregate.
It might not be surprising to find a bone-promoting effect for enamel matrix proteins. The development of teeth within the jaws induces bone around the dental primordia, suggesting a systems linkage between bone formation and tooth development so that teeth become better anchored into the jaw bones by supporting ligaments and tissues. Another dental enamel matrix protein ameloblastin, synthesized and secreted into developing enamel matrix by ameloblasts, has recently been shown to also express in mesenchymal stem cells, primary osteoblasts and chondrocytes [46]. Ameloblastin therefore is postulated to play an independent role in early bone formation and repair [46]. However, the molecular pathway by which ameloblastin achieves these outcomes has not been described. Here, we showed that the LRAP molecule promotes bone formation by inducing Wnt10b expression.
Conclusions
We have shown that LRAP activates Wnt signaling, through the upregulation of Wnt10b, to promote osteogenesis of mesenchymal stem cells at the expense of adipogenesis. The discovery of this signaling axis makes LRAP amenable to therapies and interventions to treat Wnt-related bone and metabolic disorders. LRAP has the potential to harvest the therapeutic value of Wnt signaling pathways without direct application of Wnt proteins, thereby mitigating the adverse effect on cells of direct Wnt protein exposure. Compared to the glycosylated Wnt proteins (over 40 KDa), LRAP is much smaller (~ 6 KDa), giving LRAP a distinct advantage as a small molecule for delivery as a therapeutic agent.
Supplementary Material
Acknowledgments
This work was supported by NIDCR Grants DE017362 (Y.Z.), DE13045 (M.L.S.), DE15920 (S.I.S and M.L.S), NIDDK Grants DK51563 (O.A.M) and DK62876 (O.A.M), the James H. Zumberge Faculty Research & Innovation Fund, University of Southern California (Y.Z.) and the Royal Commission for the Exhibition of 1851, United Kingdom (W.P.C).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aldred MJ, Crawford PJ, Roberts E, Thomas NS. Identification of a nonsense mutation in the amelogenin gene (AMELX) in a family with X-linked amelogenesis imperfecta (AIH1) Hum Genet. 1992;90:413–416. doi: 10.1007/BF00220469. [DOI] [PubMed] [Google Scholar]
- 2.Lagerstrom M, Dahl N, Nakahori Y, et al. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10:971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- 3.Lagerstrom-Fermer M, Nilsson M, Backman B, et al. Amelogenin signal peptide mutation: correlation between mutations in the amelogenin gene (AMGX) and manifestations of X-linked amelogenesis imperfecta. Genomics. 1995;26:159–162. doi: 10.1016/0888-7543(95)80097-6. [DOI] [PubMed] [Google Scholar]
- 4.Diekwisch T, David S, Bringas P, Jr., Santos V, Slavkin HC. Antisense inhibition of AMEL translation demonstrates supramolecular controls for enamel HAP crystal growth during embryonic mouse molar development. Development. 1993;117:471–482. doi: 10.1242/dev.117.2.471. [DOI] [PubMed] [Google Scholar]
- 5.Lyngstadaas SP, Risnes S, Sproat BS, Thrane PS, Prydz HP. A synthetic, chemically modified ribozyme eliminates amelogenin, the major translation product in developing mouse enamel in vivo. Embo J. 1995;14:5224–5229. doi: 10.1002/j.1460-2075.1995.tb00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson CW, Yuan ZA, Hall B, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 7.Hammarstrom L. Enamel matrix, cementum development and regeneration. J Clin Periodontol. 1997;24:658–668. doi: 10.1111/j.1600-051x.1997.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 8.Hammarstrom L. The role of enamel matrix proteins in the development of cementum and periodontal tissues. Ciba Found Symp. 1997;205:246–255. doi: 10.1002/9780470515303.ch17. discussion 55–60. [DOI] [PubMed] [Google Scholar]
- 9.Hammarstrom L, Heijl L, Gestrelius S. Periodontal regeneration in a buccal dehiscence model in monkeys after application of enamel matrix proteins. J Clin Periodontol. 1997;24:669–677. doi: 10.1111/j.1600-051x.1997.tb00248.x. [DOI] [PubMed] [Google Scholar]
- 10.Gestrelius S, Andersson C, Johansson AC, et al. Formulation of enamel matrix derivative for surface coating. Kinetics and cell colonization. J Clin Periodontol. 1997;24:678–684. doi: 10.1111/j.1600-051x.1997.tb00249.x. [DOI] [PubMed] [Google Scholar]
- 11.Gestrelius S, Andersson C, Lidstrom D, Hammarstrom L, Somerman M. In vitro studies on periodontal ligament cells and enamel matrix derivative. J Clin Periodontol. 1997;24:685–692. doi: 10.1111/j.1600-051x.1997.tb00250.x. [DOI] [PubMed] [Google Scholar]
- 12.Veis A, Tompkins K, Alvares K, et al. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J Biol Chem. 2000;275:41263–41272. doi: 10.1074/jbc.M002308200. [DOI] [PubMed] [Google Scholar]
- 13.Boabaid F, Gibson CW, Kuehl MA, et al. Leucine-rich amelogenin peptide: a candidate signaling molecule during cementogenesis. J Periodontol. 2004;75:1126–1136. doi: 10.1902/jop.2004.75.8.1126. [DOI] [PubMed] [Google Scholar]
- 14.Lacerda-Pinheiro S, Jegat N, Septier D, et al. Early in vivo and in vitro effects of amelogenin gene splice products on pulp cells. Eur J Oral Sci. 2006;114 Suppl 1:232–238. doi: 10.1111/j.1600-0722.2006.00320.x. discussion 54–6, 381–2. [DOI] [PubMed] [Google Scholar]
- 15.Warotayanont R, Zhu D, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis in mouse embryonic stem cells. Biochem Biophys Res Commun. 2008;367:1–6. doi: 10.1016/j.bbrc.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warotayanont R, Frenkel B, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis by activation of the Wnt pathway. Biochem Biophys Res Commun. 2009;387:558–563. doi: 10.1016/j.bbrc.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nuttall ME, Gimble JM. Controlling the balance between osteoblastogenesis and adipogenesis and the consequent therapeutic implications. Curr Opin Pharmacol. 2004;4:290–294. doi: 10.1016/j.coph.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Gimble JM, Zvonic S, Floyd ZE, Kassem M, Nuttall ME. Playing with bone and fat. J Cell Biochem. 2006;98:251–266. doi: 10.1002/jcb.20777. [DOI] [PubMed] [Google Scholar]
- 19.Sottile V, Halleux C, Bassilana F, Keller H, Seuwen K. Stem cell characteristics of human trabecular bone-derived cells. Bone. 2002;30:699–704. doi: 10.1016/s8756-3282(02)00674-9. [DOI] [PubMed] [Google Scholar]
- 20.Pereira RC, Delany AM, Canalis E. Effects of cortisol and bone morphogenetic protein-2 on stromal cell differentiation: correlation with CCAAT-enhancer binding protein expression. Bone. 2002;30:685–691. doi: 10.1016/s8756-3282(02)00687-7. [DOI] [PubMed] [Google Scholar]
- 21.Lecka-Czernik B, Moerman EJ, Grant DF, Lehmann JM, Manolagas SC, Jilka RL. Divergent effects of selective peroxisome proliferator-activated receptor-gamma 2 ligands on adipocyte versus osteoblast differentiation. Endocrinology. 2002;143:2376–2384. doi: 10.1210/endo.143.6.8834. [DOI] [PubMed] [Google Scholar]
- 22.Park SR, Oreffo RO, Triffitt JT. Interconversion potential of cloned human marrow adipocytes in vitro. Bone. 1999;24:549–554. doi: 10.1016/s8756-3282(99)00084-8. [DOI] [PubMed] [Google Scholar]
- 23.Bennett CN, Longo KA, Wright WS, et al. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci U S A. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282:14515–14524. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- 25.Ross SE, Hemati N, Longo KA, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 26.Bennett CN, Ross SE, Longo KA, et al. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277:30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- 27.Longo KA, Wright WS, Kang S, et al. Wnt10b inhibits development of white and brown adipose tissues. J Biol Chem. 2004;279:35503–35509. doi: 10.1074/jbc.M402937200. [DOI] [PubMed] [Google Scholar]
- 28.Kang S, Bajnok L, Longo KA, et al. Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1alpha. Mol Cell Biol. 2005;25:1272–1282. doi: 10.1128/MCB.25.4.1272-1282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright WS, Longo KA, Dolinsky VW, et al. Wnt10b inhibits obesity in ob/ob and agouti mice. Diabetes. 2007;56:295–303. doi: 10.2337/db06-1339. [DOI] [PubMed] [Google Scholar]
- 30.Molenaar M, van de Wetering M, Oosterwegel M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 31.Finch PW, He X, Kelley MJ, et al. Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci U S A. 1997;94:6770–6775. doi: 10.1073/pnas.94.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reya T, O'Riordan M, Okamura R, et al. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 2000;13:15–24. doi: 10.1016/s1074-7613(00)00004-2. [DOI] [PubMed] [Google Scholar]
- 33.Stevens JR, Miranda-Carboni GA, Singer MA, Brugger SM, Lyons KM, Lane TF. Wnt10b deficiency results in age-dependent loss of bone mass and progressive reduction of mesenchymal progenitor cells. J Bone Miner Res. 2010;25:2138–2147. doi: 10.1002/jbmr.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haze A, Taylor AL, Blumenfeld A, et al. Amelogenin expression in long bone and cartilage cells and in bone marrow progenitor cells. Anat Rec (Hoboken) 2007;290:455–460. doi: 10.1002/ar.20520. [DOI] [PubMed] [Google Scholar]
- 35.Gong Y, Slee RB, Fukai N, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 36.Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol. 2005;25:4946–4955. doi: 10.1128/MCB.25.12.4946-4955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyden LM, Mao J, Belsky J, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 38.Van Wesenbeeck L, Cleiren E, Gram J, et al. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet. 2003;72:763–771. doi: 10.1086/368277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christodoulides C, Scarda A, Granzotto M, et al. WNT10B mutations in human obesity. Diabetologia. 2006;49:678–684. doi: 10.1007/s00125-006-0144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo YF, Xiong DH, Shen H, et al. Polymorphisms of the low-density lipoprotein receptor-related protein 5 (LRP5) gene are associated with obesity phenotypes in a large family-based association study. J Med Genet. 2006;43:798–803. doi: 10.1136/jmg.2006.041715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mani A, Radhakrishnan J, Wang H, et al. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.James AW, Leucht P, Levi B, et al. Sonic Hedgehog influences the balance of osteogenesis and adipogenesis in mouse adipose-derived stromal cells. Tissue Eng Part A. 2010;16:2605–2616. doi: 10.1089/ten.tea.2010.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dwyer JR, Sever N, Carlson M, Nelson SF, Beachy PA, Parhami F. Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J Biol Chem. 2007;282:8959–8968. doi: 10.1074/jbc.M611741200. [DOI] [PubMed] [Google Scholar]
- 44.Kim WK, Meliton V, Amantea CM, Hahn TJ, Parhami F. 20(S)-hydroxycholesterol inhibits PPARgamma expression and adipogenic differentiation of bone marrow stromal cells through a hedgehog-dependent mechanism. J Bone Miner Res. 2007;22:1711–1719. doi: 10.1359/jbmr.070710. [DOI] [PubMed] [Google Scholar]
- 45.Huang YC, Tanimoto K, Tanne Y, et al. Effects of human full-length amelogenin on the proliferation of human mesenchymal stem cells derived from bone marrow. Cell Tissue Res. 2010;342:205–212. doi: 10.1007/s00441-010-1064-7. [DOI] [PubMed] [Google Scholar]
- 46.Tamburstuen MV, Reppe S, Spahr A, et al. Ameloblastin promotes bone growth by enhancing proliferation of progenitor cells and by stimulating immunoregulators. Eur J Oral Sci. 2010;118:451–459. doi: 10.1111/j.1600-0722.2010.00760.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.