Abstract
Amyloid fibrils of Alzheimer’s β-amyloid peptide (Aβ) are a primary component of amyloid plaques, a hallmark of Alzheimer’s disease (AD). Enormous attention has been given to the structural features and functions of Aβ in amyloid fibrils and other type of aggregates in associated with development of AD. This report describes an efficient protocol to express and purify high-quality 40-residue Aβ(1–40), the most abundant Aβ in brains, for structural studies by NMR spectroscopy. Over-expression of Aβ(1–40) with glutathione S-transferase (GST) tag connected by a Factor Xa recognition site (IEGR▼) in E. Coli resulted in the formation of insoluble inclusion bodies even with the soluble GST tag. This problem was resolved by efficient recovery of the GST-Aβ fusion protein from the inclusion bodies using 0.5% (w/v) sodium lauroyl sarcosinate as solubilizing agent and subsequent purification by affinity chromatography using a glutathione agarose column. The removal of the GST tag by Factor Xa enzymatic cleavage and purification by HPLC yielded as much as ~7 mg and ~1.5 mg of unlabeled Aβ(1–40) and uniformly 15N- and/or 13C-protein Aβ(1–40) from 1 L of the cell culture, respectively. Mass spectroscopy of unlabeled and labeled Aβ and 1H/15N HSQC solution NMR spectrum of the obtained 15N-labeled Aβ in the monomeric form confirmed the expression of native Aβ(1–40). It was also confirmed by electron micrography and solid-state NMR analysis that the purified Aβ(1–40) self-assembles into β-sheet rich amyloid fibrils. To the best of our knowledge, our protocol offers the highest yields among published protocols for production of recombinant Aβ(1–40) samples that are amendable for an NMR-based structural analysis. The protocol may be applied to efficient preparation of other amyloid-forming proteins and peptides that are 13C- and 15N-labeled for NMR experiments.
Keywords: Amyloid β, GST fusion protein, sodium lauroyl sarcosinate, NMR
Introduction
Amyloid β (Aβ) peptides (39–43 residues) are the primary components of amyloid plaques in Alzheimer’s disease (AD). The 40-residue Aβ peptide Aβ(1–40) is the most abundant species in brain [1, 2]. Due to their intrinsic hydrophobicity, Aβ peptides misfold into amyloid fibrils and other diffusible aggregates [3–7]. Amyloid fibrils and some of these diffusible aggregates for Aβ have been reported to be neurotoxic [4, 5, 8–11]; thus, it is widely believed that Aβ plays a central role in neural dysfunctions in AD [2]. Consequently, extensive efforts have been made to understand the detailed structural features of fibrils and diffusible aggregates of Aβ. Since high quality crystals are not available for the full-length Aβ peptides, X-ray crystallography has not been an option for the structure elucidation of Aβ in these aggregates. Hence, solid-state NMR (SSNMR) and solution NMR have been widely used for this purpose [6, 7, 10, 12–23]. These NMR analyses typically require relatively large amounts (ca. mg-scale) of isotope-labeled Aβ peptides. Although Aβ peptides have been produced by solid-phase peptide synthesis (SPPS) [10, 13, 14], it is not cost-effective to chemically synthesize a large quantity of uniformly labeled or highly isotope-labeled Aβ peptides by SPPS because of the high costs for isotope-labeled amino acids for synthesis reagents. Biologically expressed uniformly isotope-labeled Aβ samples are commercially available. However, besides their high costs, the types of commercially available isotope-labeled samples are limited, further restricting applications that require more sophisticated labeling schemes [24, 25].
Thus, intensive efforts have been made for biological expression of Aβ peptides using E. Coli and other expression systems [16, 26–32]. Despite these studies, because of the strong intrinsic aggregation propensity of Aβ peptides, it is difficult to express and purify Aβ peptides from bacterial or insect cells efficiently. Also, modifications of the amino acid sequence or addition of extra residues in the N-terminal have been shown to alleviate the problems associated with the expression and purification of the Aβ peptide; however, this can cause significant alteration of its properties [16, 26, 28, 31, 32]. To overcome these problems, we developed a new protocol that involves the high-efficiency solubilization of bacterially expressed, glutathione S-transferase (GST)-fused Aβ(1–40) from the inclusion bodies using sodium lauroyl sarcosinate. After the cleavage of the GST-tag and the purification, this convenient and cost-effective procedure allows for the high-yield preparation of uniformly 15N and/or 13C-labeled Aβ(1–40) for NMR measurements without the complex unfolding-refolding process.
Materials and Methods
Materials
The expression vector pGEX-2T was purchased from GE Healthcare (Piscataway, NJ). Host cell BL21-CodonPlus (DE3) was purchased from Stratagene (La Jolla, CA). Restriction endonucleases BamHI and EcoRI were obtained from New England BioLabs (Ipswich, MA). Glutathione agarose, 15NH4Cl and 13C-glucose were purchased from Sigma (Saint Louis, MO). Isopropyl β-D-thiogalactopyranoside (IPTG) and reduced glutathione (GSH) were obtained from Fisher Scientific (Pittsburgh, PA). The DNA template encoding Aβ(1–40) and primers were synthesized by IDT (Coralville, IA). Bovine Factor Xa was purchased from Haematologic Technologies Inc. (Essex Junction, Vermont). Fmoc-protected amino acids and Wang resins were purchased from Peptide International (Louisville, KY). Other reagents for peptide synthesis were purchased from Applied Biosystems (ABI, Foster City, CA).
Cloning and Construction
Construction of the pGEX-amyloid beta expression vector
The expression vector pGEX-2T-Aβ(1–40) was constructed in the laboratory of Prof. Wonhwa Cho in the University of Illinois at Chicago. The DNA template ‘5- TA GGA TCC ATT GAA GGT CGT GAT GCG GAA TTT CGT CAT GAT AGC GGC TAC GAA GTT CAT CAC CAG AAA CTG GTG TTC TTT GCG GAA GAC GTT GGT AGC AAC AAA GGC GCA ATT ATC GGC CTG ATG GTT GGT GGT GTG GTT TAG GGA ATT CA-3’ was purchased from IDT. After PCR amplification, the DNA duplex was digested by BamHI and EcoRI and inserted into pGEX-2T vector. It encoded 26-kDa glutathione S-transferase (GST) and the amino acid sequences of Aβ(1–40) connected by the site-specific recognition sequences for both thrombin (LVPR▼GS) and Factor Xa (IEGR▼) [33]. The expression vectors were verified by DNA sequencing, and transformed to E. coli strain BL21-CodonPlus (DE3) competent cells.
Expression of unlabeled GST-Amyloid beta fusion protein
For the expression of the unlabeled Aβ, BL21-CodonPlus (DE3) competent cells with expression vector were grown at 37°C on a LB agar plate containing 100 μg/mL ampicillin for ~16 h. A single colony was picked and grown at 27°C for overnight in 100 mL of a LB medium containing 100 μg/mL ampicillin. The bacteria were diluted (1:100) into a TB medium and grown at 37 °C until OD600 was ~2.0. Protein expression was induced with 0.8 mM IPTG, and then the cells were harvested after 6~8 h of the incubation at 27 °C.
Expression of isotope labeled GST-Amyloid beta fusion protein
For the expression of uniformly 15N- or/and 13C-isotope labeled Aβ(1–40), a single colony was picked and grown in a LB medium at 27°C for overnight, as described for the expression of unlabeled Aβ. To change the a LB medium to a M9 minimal medium, the cells were pelleted at 5000 g for 10 min, then washed by using 20 mL of a 1X M9 salt solution and pelleted again. The cell pellet was resuspended in a 1000-mL M9 media containing 1g/L NH4Cl, 2g/L glucose, 2 mM MgSO4, 0.05 mM CaCl2, 10 mg/L thiamine, 10 mg/L biotin, and 100 mg/L ampicillin [34]. When OD600 was about 0.8, protein expression was induced by adding 0.8 mM IPTG at 27°C to the culture. The cells were harvested after 16 h of the incubation.
Purification of GST-Aβ
After centrifugation, the harvested cells were suspended in a cold STE buffer (20 mM Tris, 100 mM NaCl, 3 mM EDTA, pH8.0) containing 5 mM DTT. The cells were sonicated 6–8 times for 15 s by using a Branson Sonifier150 (Branson Ultrasonics Corporation, CT) on ice. It was reported that the heat caused by the sonication may permanently denature some of the GST [35, 36]; we have tested other cell lysis method such as the Avestin system, but only marginal or no improvement was observed in our preliminary analysis. 10% (w/v) sodium lauroyl sarcosinate was added to the lysate until the final concentration of sodium lauroyl sarcosinate became 0.5% (w/v). The lysate was stirred for 1 min, and then it was ultra-centrifuged at 40,000 g for 15 min for the removal of cell debris and other particles. Triton X-100 was added to the supernatant to a final concentration of 0.8% (v/v). The GST-Aβ fusion protein from the clear supernatant was affinity purified using a glutathione agarose column, equilibrated in an ice-cold Tris-HCl buffer (20 mM Tris, 100 mM NaCl, pH 8.0). The supernatant was loaded to the glutathione agarose column (~ 20 ml) at a flow rate of 1.0 mL/min. The column was washed with 5–10 column volume of Tris-HCl buffer to remove unbound proteins. GST-Aβ was eluted from the column with elution buffer (25 mM Tris, pH 9.0, 5 mM reduced GSH). The eluted solution containing GST-Aβ had A280 of 1 to 2. The purity of GST-amyloid beta was evaluated by using 15% SDS-PAGE.
Purification of Aβ(1–40) from GST-Aβ fusion protein
The GST tag was cleaved by adding Factor Xa to the eluted solution (pH was adjusted to 9.0) from glutathione agarose column for overnight at 16 °C. For 1 mg of the fusion protein, 50 U of Factor Xa was added. The cleaved protein solution was filtered through an Amicon Ultra-15 centrifugal filter unit (30-kDa-MWCO), and the pH was adjusted to ~ 3.0 before the injection to a C18 column (Grace Vydac, Hesperia, CA) column, which was equilibrated with a solvent with 26% (v/v) acetonitrile, 74% water (v/v), and 0.1% tri-fluoro aceticacid (TFA). The acetonitrile concentration in an elution solvent was ranged from 26% (v/v) to 90% (v/v) with constant TFA concentration, and the flow rate was kept at 3 ml/min. The Aβ(1–40) was eluted approximately when the acetonitrile concentration reached 40 % (v/v) [10]. The collected sample was immediately frozen in liquid nitrogen and lyophilized. The molecular mass and purity were analyzed by MALDI-TOF mass spectroscopy.
Synthesis and purification of synthetic Aβ(1–40) peptide
The 40-residue Aβ peptide (Aβ(1–40)) was synthesized on an ABI 433A peptide synthesizer by using solid-phase synthesis with standard FMOC synthesis protocols (Applied Biosystems). After cleavage from the Wang resin, the crude peptides were lyophilized and further purified by HPLC as described previously [6].
Gel electrophoresis
Proteins were analyzed by SDS-PAGE using 15% polyacrylamide gels. The experiments were performed with a mini Protein 3 system (Bio-Rad, Hercules, CA). Protein samples of 8 μL were mixed with 2 μL of a 4× SDS-PAGE sample buffer, and loaded on the gel. The samples were run for 1.5 h at 125 V. Then, the resulting gels were stained with Coomassie Brilliant Blue R-250.
Solution NMR spectroscopy
The sample used for the NMR experiment was prepared based on the protocol suggested in the previous studies [17, 37]. The lyophilized Aβ(1–40) peptide of 0.3 mg was first dissolved in 80 μL of 10 mM NaOH solution and followed by 1 min sonication in a ice-cold water bath. The mixture was diluted with of an ice-cold mixture of 160 μL H2O and 60 μL D2O to half of the final volume followed by another 1 min sonication. Then 40 mM sodium phosphate buffer (pH 6.9) of 300 μL was added to adjust the pH, and the final pH of the solution was 7.1. The solution was filtered with a 50-kDa-MWCO filter to remove any preformed aggregates before the NMR experiment. The concentration of Aβ(1–40) was 82 μM from the UV-Vis measurement. The 2D 1H/15N HSQC experiment was performed on a Bruker 900 MHz spectrometer at 10°C The spectrum was processed by using NMRPipe [38].
Solid state NMR spectroscopy
A solution of 0.5 mM Aβ(1–40) was prepared by dissolving 1.8 mg of lyophilized Aβ peptide in 80 μL of 50 mM NaOH, and then diluted about 10 times with water containing 0.02% NaN3. The pH was adjusted to 7.4 by adding 100 mM HCl. The solution was sonicated in an ice-cold water bath and filtered with a 50-kDa-MWCO filter to remove any preformed aggregates. The final concentration is 350 μM. The sample was incubated at room temperature for 7 days with agitation. The formation of amyloid fibrils was confirmed by thioflavin T (ThT) fluorescence assay following the protocol in the previous studies [6]. The amyloid fibrils were collected as a pellet after the centrifugation at 16.1 × 103 g for 20 min. Then the gel-like pellet was transferred into a 1.8-mm MAS rotor. Cu-EDTA was introduced to the sample for sensitivity enhancement of SSNMR using the PACC method [39] before the experiments (final concentration of Cu-EDTA was ~ 200 mM).
All the SSNMR experiments were performed at Ishii’s group at UIC with a Varian InfinityPlus SSNMR spectrometer at a 1H NMR frequency of 400.2 MHz and a 1.8-mm triple-resonance MAS probe. The MAS probe was constructed by Dr. Ago Samoson’s group at National Institute of Chemical Physics and Biophysics at Estonia. The sample temperature was ~ 15°C at a spinning speed of 40 kHz. In the 2D 13C/13C correlation SSNMR measurement for Aβ(1–40), we used a fpRFDR 13C/13C dipolar recoupling sequence [40] with a mixing time 2.0 ms and a 13C β-pulse width of 13 μs. For each t1 point, 1024 scans were accumulated with an acquisition period of 10.24 ms with recycle delays of 0.225 s. The signals during the t1 and t2 periods were collected under 1H low-power TPPM decoupling at 10 kHz [39, 41]. A total of 126 complex t1 points were recorded with a t1 increment 48 μs. The overall experimental time was 35 h.
Electron micrograph of Aβ amyloid fibrils
Morphologies of Aβ(1–40) fibrils were analyzed by a JEOL JEM-1220 transmission electron microscope (TEM) at the UIC Research Resource Center using an accelerating voltage of 120 kV. A 10-βL solution containing Aβ fibrils was placed onto a carbon-coated Formvar 200-mesh copper grid (Electron Microscopy Sciences, Hatfield, PA) for 1 min. An excess solution on the grid was removed with a piece of filter paper. Then, the sample was negatively stained with 10 βL of 2 % uranyl acetate solution for 1 min. An excess solution was subsequently removed; the grid was allowed to air dry and used for the analysis.
Results
Cloning and construction of the recombinant protein expression system in E. Coli
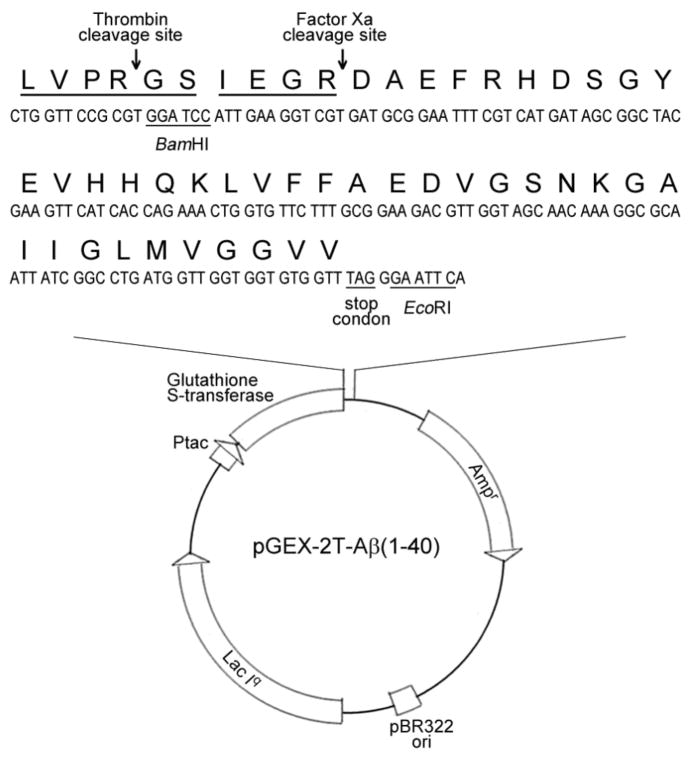
The pGEX-2T-Aβ vector we constructed was shown in Fig. 1. The vector was designed to express amyloid beta peptide fused to the C-terminus of GST in E. Coli. Between the GST and Aβ(1–40), there were two site-specific recognition sequences for thrombin (LVPR▼GS) and Factor Xa (IEGR▼), respectively. The vector pGEX-2T contains a DNA sequence encoding a thrombin recognition site (LVPR▼GS). If thrombin is utilized for the cleavage, there would be two extra residues (GS) in the N-terminal of the Aβ(1–40) peptide. The presence of the Gly and Ser may lead to unwanted changes in chemical and biological properties. To avoid this, a Factor Xa recognition site (IEGR▼) was inserted between thrombin recognition site and Aβ(1–40) peptide. The orientation and the sequence of DNA for Factor Xa cleavage site and Aβ(1–40) peptide were verified by DNA sequencing.
Figure 1.
Schematic diagram of the expression vector pGEX-2T-Aβ(1–40). Factor Xa recognition site (IEGR) and Aβ(1–40) sequence were inserted into the BamHI and EcoRI sites of pGEX-2T downstream from GST gene. The vector also had the tac promotor (Ptac), ampicillin resistance gene (Ampr), pBR322 origin of replication (pBR322 ori) and lac Iq gene.
Expression of GST-Aβ
Sequence-verified pGEX-2T vectors containing Aβ(1–40) peptides gene were used for the expression in E. Coli as described in Materials and Methods. In this system, the target protein was expressed as a fusion partner with GST and the GST tag (26-kDa) allows rapid purification of the protein by affinity chromatography using a glutathione-agarose resin (GSH-agarose). When this fusion protein was expressed in E. coli, however, less than 10% of GST-Aβ (31 kDa) was recovered in the supernatant even with a mild detergent 1% Triton X-100, compared with the protein recovered from the total lysate with a stronger detergent 1% SDS (see the SDS-PAGE result in Fig. 2 (Lane 1 and 2)). As shown in Lane 3 in Fig. 2, a much greater amount of the induced fusion protein was recovered from the pellet, which suggests that a majority of the fusion proteins form insoluble inclusion bodies in the cultured cells. To prevent the aggregation, we tried alternative expression conditions, such as lower culture temperature, induction with lower IPTG concentration, and variable induction times, but none of them improved the yield of soluble proteins. The expressed fusion protein was consistently found in the pellet fraction in all cases.
Figure 2.
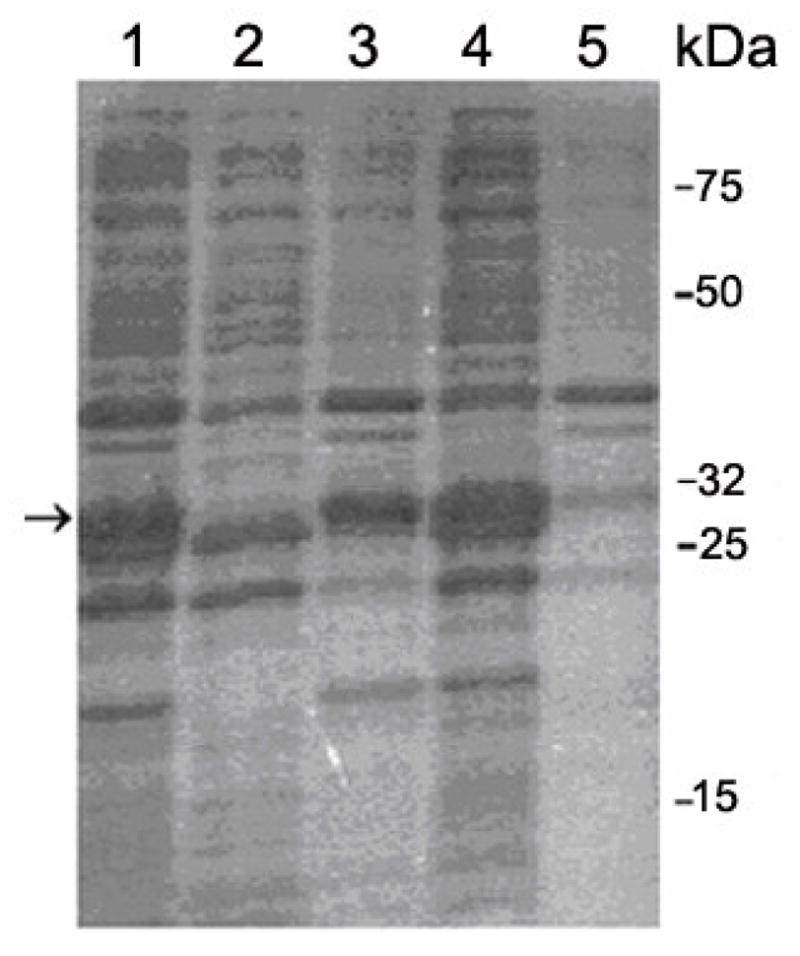
15% SDS-PAGE analysis of the cells extracts. Lane 1: total lysate treated with 1% (w/v) SDS. Lane 2: soluble supernatant from cells treated with 1% Triton X-100 (v/v). Lane 3: insoluble pellet from cells treated with 1% Triton X-100 (v/v). Lane 4: soluble supernatant from cells treated with 0.5% sodium lauroyl sarcosinate and 0.8% Triton X-100 (v/v). Lane 5: insoluble pellet from cells treated with 0.5% sodium lauroyl sarcosinate and 0.8% Triton X-100 (v/v). It is clear from the data in Lane 4 and Lane 5 that most of the fusion protein was successfully solubilized. The arrow indicates the position of GST-Aβ having the molecular mass of ~ 31 kD
Purification of GST- Aβ(1–40)
We therefore optimized the conditions to dissolve the inclusion body and recover GST-Aβ. Urea and guanidine hydrochloride are the most common solubilizing agents for insoluble proteins. However, the use of the agents typically requires subsequent slow and low-yield refolding processes. As an alternative, a strong anionic surfactant, sodium lauryl sarcosinate (sarkosyl) has been successfully used for refolding of insoluble GST fusion proteins, such as GST-tagged chicken muscle pyruvate kinase (residue 17–426) [36]. We thus employed sodium lauroyl sarcosinate for solubilization of GST-Aβ inclusion bodies. As shown in Lane 4 in Fig. 2, the GST-Aβ fusion protein was recovered from the supernatant fraction when the cells were treated with 0.5% (w/v) sodium lauroyl sarcosinate in a lysis buffer. The band corresponding to GST-Aβ in Lane 4 is much stronger than that of the Triton X-100-treated cells (Lane 1–3). In contrast, nearly no GST-Aβ was found in the pellet fraction after the treatment (Lane 5, in Fig. 2). These results indicate that most of the fusion proteins were solubilized by sodium lauroyl sarcosinate.
We then established a protocol to purify GST-Aβ in sodium lauroyl sarcosinate. Nonionic detergents were reported to form mixed micelles with sodium lauroyl sarcosinate [36] and thus help the refolding of the partially denatured GST-Aβ. We thus added Triton X-100 to the sodium lauroyl sarcosinate solution to a final concentration of 0.8% (v/v) and purified GST-Aβ by affinity chromatography using GSH-agarose. At least 80% of GST-Aβ bound to the resin under these conditions and the bound fusion protein was successfully eluted by 5 mM reduced glutathione (GSH). The SDS-PAGE showed a strong band of ~31 kDa, which corresponds to GST-Aβ(1–40) fusion protein (see Lane 3 of Fig. 3). Thus, this protocol allowed purification of the GST-Aβ fusion protein to homogeneity.
Figure 3.
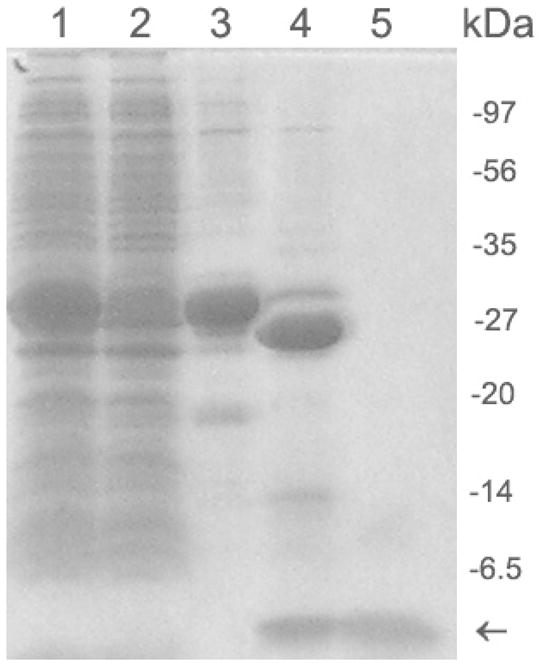
15% SDS-PAGE analysis of cells exacts and cleavage of GST-Aβ(1–40) fusion protein. Lane 1, soluble supernatant from cell treated with 0.5% (w/v) sodium lauroyl sarcosinate and 0.8% Triton X-100 (v/v). Lane 2, soluble supernatant treated with sodium lauroyl sarcosinate and TX-100, and flowed through glutathione agarose column. Lane 3, GST-Aβ(1–40) fusion protein eluted from glutathione agarose. Lane 4: a cleavage mixture after incubation of GST-Aβ(1–40) fusion protein with Factor Xa for 16 h. Lane 5: the cleaved protein solution was filtered through an Amicon Ultra-15 centrifugal filter unit (30-kDa-MWCO). The arrow indicates the position of Aβ(1–40) having a molecular mass of 4–6 kDa. The cleavage of the fusion protein was performed by 1U of Factor Xa per 20 μg of the fusion protein at 16°C and pH 9.0. The molecular mass of GST tag is ~26 kDa.
Purification of Aβ(1–40) peptide
To separate Aβ(1–40) peptide from GST fusion protein, Factor Xa cleavage was carried out as described in Materials and Methods. Before the cleavage, the pH of the eluted solution containing the fusion protein was adjusted to 9.0 by adding 0.2 M NaOH. At lower pH (< 8.5), the cleaved Aβ(1–40) showed tendency to aggregate, while at higher pH (> 9.5), the Factor Xa cleavage efficiency was low. Thus, the careful adjustment of the pH is critical. Under our conditions, more than 90% of GST was cleaved from the fusion protein after 16 h incubation with Factor Xa (Lane 4 in Fig. 3). Lane 4 in Fig. 3 also indicates a band for a protein with molecular mass of less than 6.5 kDa, which is consistent with the successful production of Aβ as shown below.
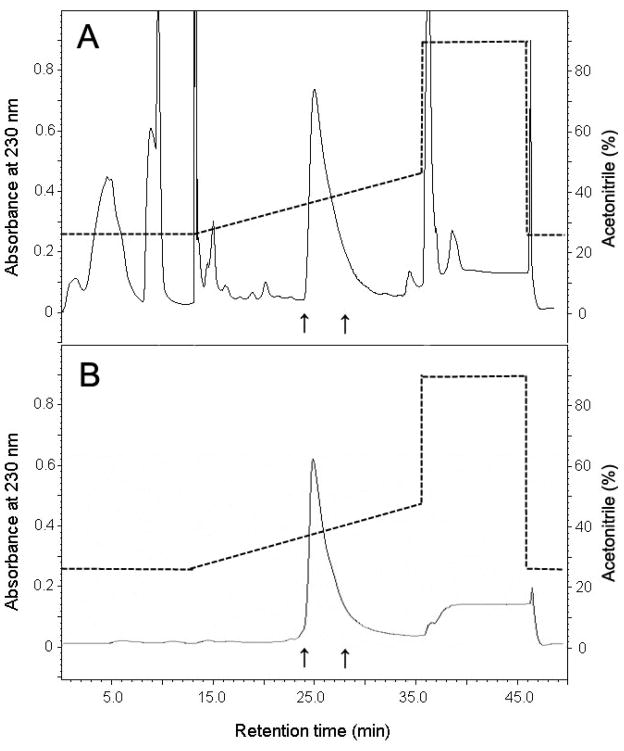
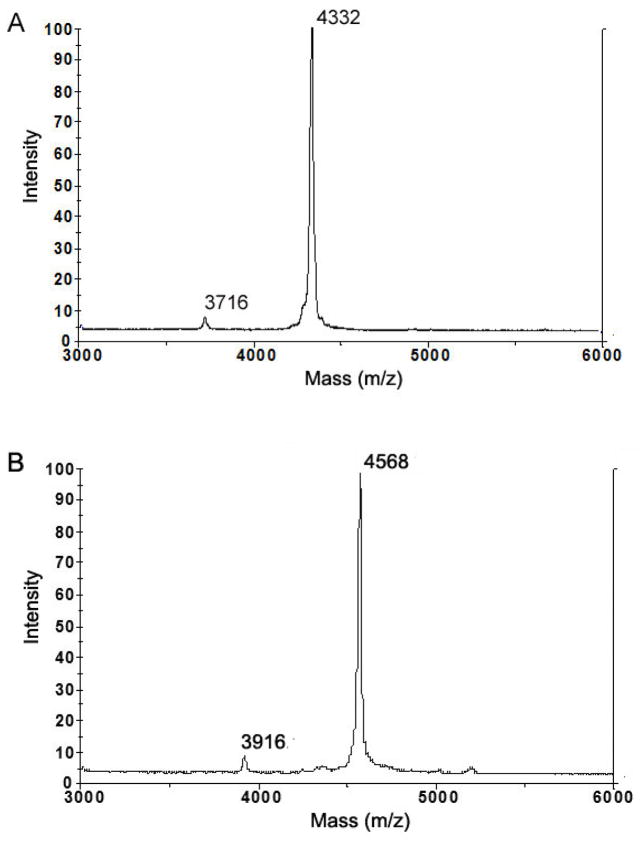
After the cleavage, GST and the uncleaved GST-Aβ(1–40) was removed by filtering the cleavage mixture through an Amicon Ultra-15 centrifugal filter unit (30-kDa-MWCO), and subsequently collecting monomeric Aβ from the filtrate. Because GST is known to form a dimer (~52 kDa) at a neutoral pH without detergent [42], it is quite possible that GST-Aβ also forms a dimer (~61 kDa), which can be also removed by the ultrafiltration. Lane 5 in Fig. 3 clearly shows that the filtrate displays only a band of the proteins with low MW (< 6.5 kDa). Our method of separating the cleaved Aβ by filtering is advantageous over the previously reported protocol employing dialysis and purification on a glutathione agarose column [43] in that it is much faster and does not cause sample dilution while allowing high yield recovery of the peptide. Then, purification of Aβ(1–40) was conducted by reversed-phase HPLC using a Vydac C18 column as described previously [6]. The fraction eluted between 24–28 min was collected and lyophilized (Fig. 4). The identity of the purified Aβ(1–40) peptide was confirmed by MALDI-TOF mass spectroscopy (Fig. 5A). The measured molecular mass of 4332 ± 1 Da agrees well with the theoretical value (4329.8 Da). The protein recovery of unlabeled and isotope labeled Aβ(1–40) peptides at different purification steps is summarized in Table 1 and Table 2, respectively. The purity of the final product is estimated to be more than 90% from the mass spectrum. The minor peak at the mass per charge of 3716 Da is likely to be assigned to Aβ(6–40), whose theoretical molecular mass is 3711.2 Da. Since the 2–5 residues at the N-terminal of Aβ(1–40) is AEFR, that is similar to Factor Xa recognition site IEGR, the first 5 residues may also be cleaved by the protease. About 7 mg of the purified Aβ(1–40) was successfully obtained from a 1 L of the E. coli cell culture. As summarized in Table 3, this yield for Aβ(1–40) expression is notably higher than any previously reported value from similar expression systems [30].
Figure 4.
(A), Purification of recombinant Aβ(1–40) by reverse-phase HPLC on a Vydac C18 column. The elution solution from 24 min to 28 min (arrows) was collected and lyophilized. (B), Chromatographic profile of a chemically synthesized purified Aβ(1–40) on the same Vydac C18 column using the same elution gradient for comparison. The dashed line indicated the percentage (v/v) of acetonitrile in the elution solution.
Figure 5.
Mass spectrometry of the purified peptides for (A) unlabeled Aβ(1–40) and (B) uniformly 13C- and 15N-labeled Aβ(1–40). Theoretical molecular masses for Aβ(1–40) are (A) 4329.8 Da and (B) 4576.8 Da. The error of the mass measurement is ±1 Da. The estimated purity from the mass spectroscopy is more than 90%. The impurity is likely to be Aβ(6–40) as a result of the non-specific cleavage by Factor Xa (see text). The total labeling ratio is ~ 98%.
Table 1.
Yield of Aβ(1–40) from E. coli cell (~10 g) in 1 L of TB media at each purification step
| V (ml) | GST-Aβ(1–40) (mg) | Aβ(1–40) (mg) | Recovery (%) | |
|---|---|---|---|---|
| Supernatant | 60 | ND | ND | ND |
| GSH column | 100 | 95 | 13.7a | 100 |
| Factor Xa cleavage | 50 | ND | 13.7 a | ND |
| HPLC | 50 | ND | 7.0 | 51 |
the estimated amount from the amount of fusion protein.
Table 2.
Yield of Aβ(1–40) from E. coli cell (~3 g) in 1 L of M9 media for expression of an isotope labeled sample at each purification step
| V (ml) | GST-Ab1–40 (mg) | Aβ(1–40) (mg) | Recovery (%) | |
|---|---|---|---|---|
| Supernatant | 20 | ND | ND | ND |
| GSH column | 25 | 24 | 3.4 a | 100 |
| Factor Xa cleavage | 10 | ND | 3.4 a | ND |
| HPLC | 10 | ND | 1.5 | 44 |
the estimated amount from the amount of fusion protein.
Table 3.
Yield of the full-length Aβ peptides Aβ(1–40) or Aβ(1–42) from 1 L of culture of E. coli in previous works
| Expression system | Aβ peptide | Fusion protein system | Yield of Aβ (mg) | Recovery (%) | Reference |
|---|---|---|---|---|---|
| E. coli BL21(DE3) | Aβ(1–42) | GST-Aβ | 0.36 | ~5 | [29] |
| E. coli MG1655 | Aβ(1–40) | IFABP-Aβ | 4 | ~50 | [30] |
| Aβ(1–42) | IFABP-Aβ | 3 | ~35 | [30] | |
| E. coli BL21 (DE3) | Aβ(1–42) | H6-Ubq-Aβ | 4 | ~50 | [27] |
| E. coli BL21(DE3) | Aβ(1–40) | GST-Aβ | 7.1 | ~50 | This work |
| E. coli BL21(DE3) | 13C- and 15N-labeled Aβ(1–40) | GST-Aβ | 1.5 | ~45 | This work |
Expression and Purification of Isotope labeled Aβ(1–40) peptide
We expressed 13C- and 15N-labeled GST-Aβ(1–40) in E. coli cells grown in M9 media containing 13C-labaled glucose and 15N-labeled NH4Cl, as described in Materials and Methods. Since the activity of E. Coli was lower in M9 media than in TB media, a longer induction time (16 h) was used. Purification was performed in the same manner as described for the unlabeled Aβ. A mass spectrum for this sample in Fig 5B shows a strong peak at m/z of 4568 Da, which is consistent with the molecular mass for uniformly 13C- and 15N-labeled Aβ(1–40) (4574 Da). The isotope labeling ratio calculated from MS was ~ 98%. The minor peak at m/z of 3916 Da suggests the presence of a minor impurity due to uniformly 13C- and 15N-labeled Aβ(6–40), which has molecular mass of 3921 Da. Although the impurity is detectable, the high labeling ratio and purity are sufficient for structural studies of Aβ by NMR and other methods.
Morphologies of Aggregation profile of Aβ and electron micrograph analysis
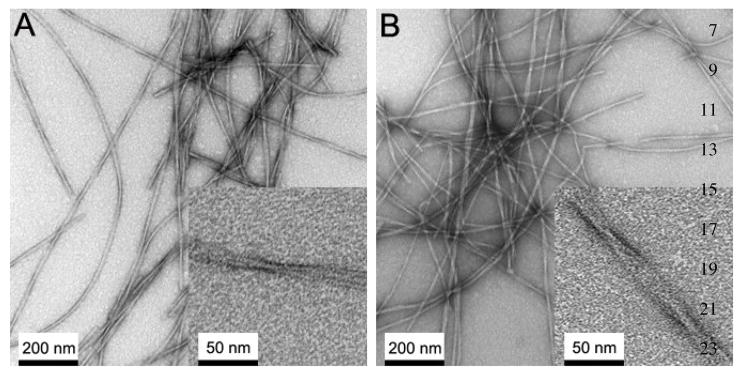
Next, we examined the misfolding capabilities of the biologically expressed Aβ. To confirm that the expressed Aβ misfolds into amyloid fibrils that have similar morphology and physical properties to those formed by chemically synthesized peptides, we prepared amyloid fibrils with the purified Aβ(1–40) peptides as described in Materials and Methods. The formation of Aβ(1–40) fibrils was confirmed by ThT fluorescence assay after 7 days of the incubation. Then, we inspected the morphologies of Aβ fibrils by electron microscopy. The morphology of the fibrils formed by the Aβ peptide was examined in negatively stained TEM images. The TEM image (Fig. 6) showed fibrils of the diameter of average 12–20 nm and the length of > 1 μm. The observed morphologies are consistent with those reported for chemically synthesized peptides [44].
Figure 6.
Negatively stained transmission electron micrograph (TEM) images of Aβ(1–40) fibrils after 1 week of the incubation at room temperature with agitation for (A) recombinant Aβ(1–40) and (B) synthetic Aβ(1–40). The insets show magnified images. The observed morphologies and the width are similar between synthetic and recombinant Aβ peptides.
HSQC solution NMR analysis
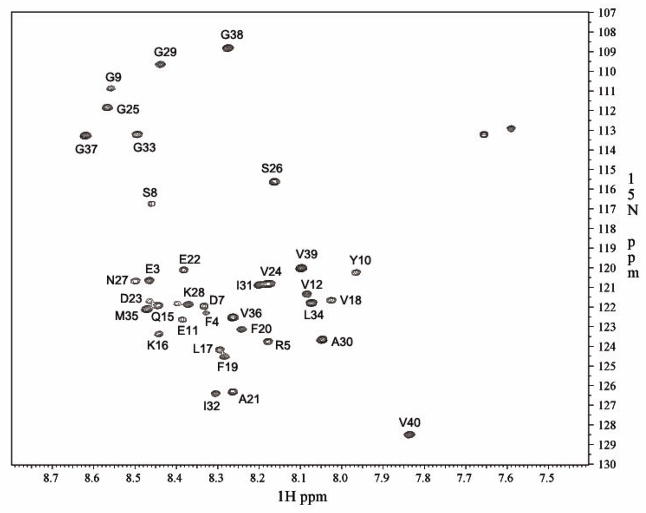
The 2D 15N/1H HSQC experiment of uniformly 15N-labeled Aβ(1–40) was carried out in a 20-mM sodium phosphate buffer at pH 7.1 and 10°C at a Bruker Avance 900 MHz spectrometer with a 5mm inverse TCI cryoprobe at the UIC Center for Structural Biology (CSB). The monomeric Aβ(1–40) showed a well dispersed HSQC spectrum (Fig. 7), which is largely consistent with the spectra reported previously for 15N-labeled Aβ(1–40) at 4–5 °C. The signal assignments were based on the previous studies [17, 37]. The chemical shifts of Met-35 and Val-36 indicated that the monomeric Aβ has the reduced state [37]. Presumably because of the high sensitivity at 900 MHz spectrometer, a previously unsigned peak was found adjacent to Lys-28, which can be assigned to Ala-2. Although the cross peaks for Asp-1, His-6, His-13, and His-14 are missing as previously reported, this confirms that the obtained sample is most likely to be intact Aβ(1–40) in the reduced form.
Figure 7.
2D 1H/15N HSQC spectrum of Aβ(1–40). The experiment was performed at 10°C at a Bruker Avance II 900 MHz NMR spectrometer with a 5mm inverse TCI cryoprobe. The assignments of the residues are based on previous publications [17, 37].
Solid-state NMR analysis
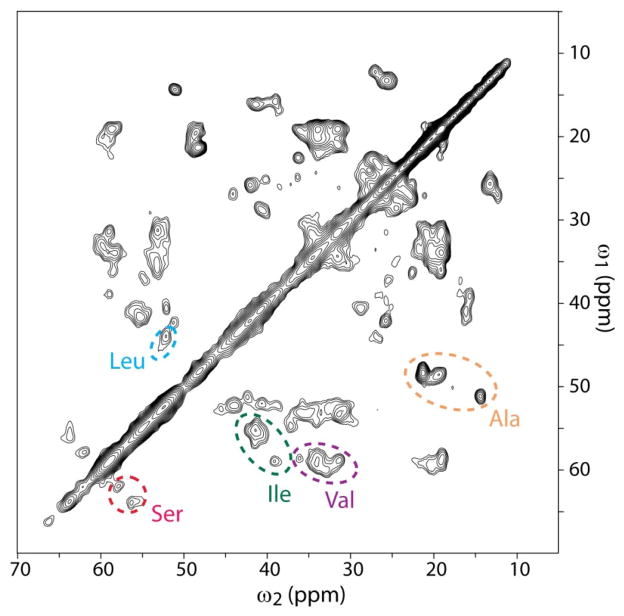
Figure 8 shows a 2D 13C/13C chemical-shift correlation SSNMR spectrum of hydrated uniformly 13C- and 15N-labeled Aβ(1–40) fibrils, together with preliminary assignments based on amino acid types for cross peaks between 13Cα and 13Cβ. With the aid of the PACC method,[39] we could characterize the amyloid fibril sample of Aβ(1–40) by SSNMR at minimal sample requirements (~1 mg). Although many cross peaks are overlapping, line widths of the cross peaks are about 1.0–1.5 ppm in the full width at the half height. The narrow line widths suggest the presence of well ordered structures. The preliminary analysis of 13Cα and 13Cβ chemical shifts shows that a majority of the assigned residues are likely to have β-sheet structure. The high resolution in the SSNMR spectrum provides excellent prospects of structural analysis of Aβ(1–40) fibrils with uniformly 13C-labeled samples.
Figure 8.
The aliphatic region of 2D 13C-13C correlation SSNMR spectra for uniformly 13C- and 15N-labeled Aβ(1–40) fibrils. The signals were collected with the PACC method with the pulse delays of 0.225 s with 1024 scans for each real or imaginary t1 point in a total of 126 complex t1 points. The overall experiment time was 35 h. The experiments were performed at ~15°C under magic angle spinning at 40 kHz.
Discussion
The objective of this work is to establish an efficient protocol for mg-scale preparation of uniformly 15N- and/or 13C- labeled Aβ peptides for NMR studies. Because the chemical synthesis of isotope-labeled Aβ peptides for NMR studies is prohibitively expensive, bacterial expression and purification of the peptides has been tried as alternative. However, the highly hydrophobic nature of the Aβ peptides has made the expression and purification of Aβ by standard recombinant expression methods considerably difficult. Frieden and co-workers [30] reported that inclusion body of the fusion protein of Aβ with a IFABP tag could be successfully dissolved with guanidine hydrochloride, and that the subsequent purification yields moderately high recovery (3–4 mg/L of unlabeled Aβ) [30]. However, it was not demonstrated that the sample obtained by this protocol was pure enough for structural analysis by NMR. It should be noted that the sample must be free from contamination of aggregated Aβ for NMR structural analysis, since a trace of “seed” aggregates can promote rapid fibrillization of Aβ, making NMR analysis of a monomer or soluble oligomers impossible. Ueda and co-workers [28] reported that monomeric Aβ(1–40) was expressed and purified as a fusion protein with hen egg white lysozyme. Although the 1H/15N HSQC spectrum for their Aβ(1–40) sample is consistent with previously reported data [37], the procedure involves complex reduction and alkylation steps and its overall yield was not reported.
When compared with these methods, our protocol allows faster and higher-yield preparation of the pure Aβ(1–40) peptide ready for NMR analysis. A key new element in our method is the use of sodium lauroyl sarcosinate as solubilizing agent for the inclusion bodies that not only improves the overall protein yields but also makes the purification simpler and more straightforward. Our new procedure allows for the preparation of about 7 mg of the unlabeled Aβ(1–40) peptide from 1 L of TB media and about 1.5 mg of the purified isotope-labeled Aβ(1–40) peptide from 1 L of M9 media supplemented with 15NH4Cl and/or 13C-labeled glucose. The morphology of the Aβ(1–40) fibrils was confirmed by EM; no difference was observed between the recombinant and synthetic peptide. Most importantly, multi-dimensional NMR and SSNMR analysis proved that the protocol offers a high quality 15N- and/or 13C-labeled sample amenable for NMR-based structural analysis of Aβ peptides and possibly of other amyloid-forming proteins. Applications of this method to other Aβ such as Aβ(1–42) and other pathogenic mutants of Aβ(1–40) are currently underway in our laboratory. This protocol is also likely to be effective for other relatively small amyloid forming proteins and peptides. It is also likely that more sophisticated labeling schemes such as amino acid selective uniformly 13C- and 15N-labeling [24, 45, 46] is possible based on this protocol with minor modifications.
A possible disadvantage of choosing GST fusion system is that GST is a relatively large tag compared with Aβ; thus, much of a molar yield for the fusion protein may be “wasted” in regard to the overall yield of Aβ. It is possible that yields of the expression level are improved by use of other smaller tag such as Protein G B1 domain (GB1). On the other hand, it is likely that a fusion protein for Aβ with a smaller tag aggregates irreversibly in a manner that such an aggregate is difficult to resolubilize. Also, GB1 is not an affinity tag, and we normally need to add another affinity tag such as His6-tag for further Aβ purification. The imidazole needed for elution of His6-tagged proteins may cause some unwanted consequences such as protein aggregation [47]. Another possible disadvantage is that imidazole can inhibit Factor Xa protease activity [48], possibly making the GB1 cleavage process less straight forward. But the screen of proper fusion tags is very worth performing in order to improve the final yield of isotope labeled proteins. Although further optimization of a proper fusion tag may improve the final yield of isotope labeled proteins, this is likely to require the further optimization in the recovery process. As the present protocol has provided the best yield among published protocols, we will examine the yield of the solubilization and recovery for different fusion tags in our future study.
Acknowledgments
We thank Dr. B. E. Ramirez at the UIC CSB and Dr. L. Casabianca in our group for the assistance with the solution NMR experiment. We are also grateful to L. Juarez for assistance with TEM studies. This work was supported in part primarily by the NIH RO1 program (AG028490, GM098033) and Alzheimer’s Association IIRG grant (08-91256), and also in part by the NSF (CHE-0957793) and the Dreyfus Foundation Teacher-Scholar Award program.
Abbreviations
- Aβ
Alzheimer’s β-amyloid peptide
- NMR
nuclear magnetic resonance
- SSNMR
solid-state NMR
- HSQC
heteronuclear single quantum correlation
- AD
Alzheimer’s disease
- EM
electron microscopy
- IPTG
Isopropyl β-D-thiogalactopyranoside
- GSH
reduced glutathione
- GST
Glutathione S-Transferase
- LB
Lysogeny broth
- TB
Terrific broth
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Naslund J, Schierhorn A, Hellman U, Lannfelt L, Roses AD, Tjernberg LO, Silberring J, Gandy SE, Winblad B, Greengard P, Nordstedt C, Terenius L. Relative abundance of Alzheimer A-beta amyloid peptide variants in Alzheimer-disease and normal aging. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:8378–8382. doi: 10.1073/pnas.91.18.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Cell biology of protein misfolding: The examples of Alzheimer’s and Parkinson’s diseases. Nature Cell Biology. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 3.Walsh DM, Hartley DM, Kusumoto Y, Fezoui Y, Condron MM, Lomakin A, Benedek GB, Selkoe DJ, Teplow DB. Amyloid beta-protein fibrillogenesis - Structure and biological activity of protofibrillar intermediates. Journal of Biological Chemistry. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 4.Gong YS, Chang L, Viola KL, Lacor PN, Lambert MP, Finch CE, Krafft GA, Klein WL. Alzheimer’s disease-affected brain: Presence of oligomeric A beta ligands (ADDLs) suggests a molecular basis for reversible memory loss. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10417–10422. doi: 10.1073/pnas.1834302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoshi M, Sato M, Matsumoto S, Noguchi A, Yasutake K, Yoshida N, Sato K. Spherical aggregates of beta-amyloid (amylospheroid) show high neurotoxicity and activate tau protein kinase I/glycogen synthase kinase-3 beta. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6370–6375. doi: 10.1073/pnas.1237107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chimon S, Ishii Y. Capturing intermediate structures of Alzheimer’s beta-amyloid, A beta(1–40), by solid-state NMR spectroscopy. Journal of the American Chemical Society. 2005;127:13472–13473. doi: 10.1021/ja054039l. [DOI] [PubMed] [Google Scholar]
- 7.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Quarterly Reviews of Biophysics. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 8.Lorenzo A, Yankner BA. Beta-amyloid neurotoxicity requires fibril formation and is inhibited by congo red. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:12243–12247. doi: 10.1073/pnas.91.25.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Natural oligomers of the amyloid-protein specifically disrupt cognitive function. Nature Neuroscience. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 10.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Evidence of fibril-like beta-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s beta-amyloid. Nature Structural & Molecular Biology. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 11.Yankner BA, Duffy LK, Kirschner DA. Neurotrophic and Neurotoxic Effects of Amyloid Beta-Protein - Reversal by Tachykinin Neuropeptides. Science. 1990;250:279–282. doi: 10.1126/science.2218531. [DOI] [PubMed] [Google Scholar]
- 12.Lansbury PT, Costa PR, Griffiths JM, Simon EJ, Auger M, Halverson KJ, KD A, Hendsch ZS, Ashburn TT, Spencer RGS, Tidor B, Griffin RG. Structural model for the beta-amyloid fibril based on interstrand alignment of an antiparellel-sheet comprising a c-terminal peptide. Nat Struct Biol. 1995;2:990–998. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- 13.Antzutkin ON, Balbach JJ, Leapman RD, Rizzo NW, Reed J, Tycko R. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of beta-sheets in Alzheimer’s beta-amyloid fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO. Structural conversion of neurotoxic amyloid-beta(1–42) oligomers to fibrils. Nature Structural & Molecular Biology. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou LM, Shao HY, Zhang YB, Li H, Menon NK, Neuhaus EB, Brewer JM, Byeon IJL, Ray DG, Vitek MP, Iwashita T, Makula RA, Przybyla AB, Zagorski MG. Solution NMR studies of the A beta(1–40) and A beta(1–42) peptides establish that the met35 oxidation state affects the mechanism of amyloid formation. Journal of the American Chemical Society. 2004;126:1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 18.Whittemore NA, Mishra R, Kheterpal I, Williams AD, Wetzel R, Serpersu EH. Hydrogen-deuterium (H/D) exchange mapping of A ss(1–40) amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 19.Yan YL, Wang CY. A beta 42 is more rigid than A beta 40 at the C terminus: Implications for A beta aggregation and toxicity. Journal of Molecular Biology. 2006;364:853–862. doi: 10.1016/j.jmb.2006.09.046. [DOI] [PubMed] [Google Scholar]
- 20.Hou LM, Zagorski MG. NMR reveals anomalous copper(II) binding to the amyloid A beta peptide of Alzheimer’s disease. Journal of the American Chemical Society. 2006;128:9260–9261. doi: 10.1021/ja046032u. [DOI] [PubMed] [Google Scholar]
- 21.Yan YL, Wang CY. A beta 40 protects non-toxic A beta 42 monomer from aggregation. Journal of Molecular Biology. 2007;369:909–916. doi: 10.1016/j.jmb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 22.Fawzi NL, Ying JF, Torchia DA, Clore GM. Kinetics of Amyloid beta Monomer-to-Oligomer Exchange by NMR Relaxation. Journal of the American Chemical Society. 2010;132:9948–9951. doi: 10.1021/ja1048253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lian LY, Middleton DA. Labelling approaches for protein structural studies by solution-state and solid-state NMR. Progress in Nuclear Magnetic Resonance Spectroscopy. 2001;39:171–190. [Google Scholar]
- 25.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nature Protocols. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 26.Sharpe S, Yau WM, Tycko R. Expression and purification of a recombinant peptide from the Alzheimer’s beta-amyloid protein for solid-state NMR. Protein Expression and Purification. 2005;42:200–210. doi: 10.1016/j.pep.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 27.Lee EK, Hwang JH, Shin DY, Kim DI, Yoo YJ. Production of recombinant amyloid-beta peptide 42 as an ubiquitin extension. Protein Expression and Purification. 2005;40:183–189. doi: 10.1016/j.pep.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 28.Nagata-Uchiyama M, Yaguchi M, Hirano Y, Ueda T. Expression and purification of uniformly N-15-Labeled amyloid p peptide 1–40 in Escherichia coli. Protein and Peptide Letters. 2007;14:788–792. doi: 10.2174/092986607781483741. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Yu HX, Song CC, Lin XF, Chen B, Tan C, Cao GX, Wang ZW. Expression, purification, and characterization of recombinant human beta-amyloid42 peptide in Escherichia coli. Protein Expression and Purification. 2009;64:55–62. doi: 10.1016/j.pep.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Garai K, Crick SL, Mustafi SM, Frieden C. Expression and purification of amyloid-beta peptides from Escherichia coli. Protein Expression and Purification. 2009;66:107–112. doi: 10.1016/j.pep.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh DM, Thulin E, Minogue AM, Gustavsson N, Pang E, Teplow DB, Linse S. A facile method for expression and purification of the Alzheimer’s disease-associated amyloid beta-peptide. Febs Journal. 2009;276:1266–1281. doi: 10.1111/j.1742-4658.2008.06862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macao B, Hoyer W, Sandberg A, Brorsson AC, Dobson CM, Hard T. Recombinant amyloid beta-peptide production by coexpression with an affibody ligand. Bmc Biotechnology. 2008;8 doi: 10.1186/1472-6750-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenny RJ, Mann KG, Lundblad RL. A critical review of the methods for cleavage of fusion proteins with thrombin and factor Xa. Protein Expression and Purification. 2003;31:1–11. doi: 10.1016/s1046-5928(03)00168-2. [DOI] [PubMed] [Google Scholar]
- 34.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. Journal of Biomolecular Nmr. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 35.Yoon SO, Hirata RDC, da Silva ACR, Nguyen NY, Hirata MH. Cloning and expression of soluble recombinant protein comprising the extracellular domain of the human type I interferon receptor 2c subunit (IFNAR-2c) in E-coli. Biotechnology Letters. 2002;24:1443–1448. [Google Scholar]
- 36.Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione-s-transferase (PGEX) fusion proteins. Analytical Biochemistry. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 37.Danielsson J, Andersson A, Jarvet J, Graslund A. N-15 relaxation study of the amyloid beta-peptide: structural propensities and persistence length. Magnetic Resonance in Chemistry. 2006;44:S114–S121. doi: 10.1002/mrc.1814. [DOI] [PubMed] [Google Scholar]
- 38.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe - A multidimensional spectral processing system based on Unix pipes. Journal of Biomolecular Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 39.Wickramasinghe NP, Parthasarathy S, Jones CR, Bhardwaj C, Long F, Kotecha M, Mehboob S, Fung LWM, Past J, Samoson A, Ishii Y. Nanomole-scale protein solid-state NMR by breaking intrinsic H-1 T-1 boundaries. Nature Methods. 2009;6:215–218. doi: 10.1038/nmeth.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishii Y. 13C-13C dipolar recoupling under very fast magic angle spinning in solid-state NMR: Applications to distance measurements, spectral assignments, and high-throughput secondary-structure elucidation. J Chem Phys. 2001;114:8473–8483. [Google Scholar]
- 41.Wickramasinghe NP, Shaibat M, Casabianca LB, de Dios AC, Harwood JS, Ishii Y. Progress in 13C and 1H solid-state NMR for paramagnetic systems under very fast MAS. J Chem Phys. 2008;128:52210. doi: 10.1063/1.2833574. [DOI] [PubMed] [Google Scholar]
- 42.Kaplan K, Hüsler P, Klump H, Erhardt J, Sluis-Cremer N, Dirr H. Conformational stability of pGEX-expressed Schistosoma japonicum glutathione S-transferase: a detoxification enzyme and fusion-protein affinity tag. Protein Science. 1997;6:399–406. doi: 10.1002/pro.5560060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan F, Wu YM, Liu JX. Expression and purification of two different antimicrobial peptides, PR-39 and Protegrin-1 in Escherichia coli. Protein Expression and Purification. 2010;73:147–151. doi: 10.1016/j.pep.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 45.Atreya HS, Chary KVR. Amino acid selective ‘unlabelling’ for residue-specific NMR assignments in proteins. Current Science. 2000;79:504–507. [Google Scholar]
- 46.Shi LC, Ahmed MAM, Zhang WR, Whited G, Brown LS, Ladizhansky V. Three-Dimensional Solid-State NMR Study of a Seven-Helical Integral Membrane Proton Pump-Structural Insights. Journal of Molecular Biology. 2009;386:1078–1093. doi: 10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Hefti MH, Caroline JG, der Toorn VV, Dixon R, Vervoort J. A novel purification method for histidine-tagged proteins containing a thrombin cleavage site. Analytical Biochemistry. 2001;295:180–185. doi: 10.1006/abio.2001.5214. [DOI] [PubMed] [Google Scholar]
- 48.He W, Hanney B, Myers MR, Spada AP, Brown K, Colussib D, Chub V. Nonbenzamidine Compounds as Selective Factor Xa Inhibitors. Bioorganic & Medicinal Chemistry Letters. 2000;10:1737–1739. doi: 10.1016/s0960-894x(00)00316-4. [DOI] [PubMed] [Google Scholar]