Abstract
Thin-sheet laser imaging microscopy (TSLIM) was used to serially section five whole cochleas from 4-wk-old CBA/JCr mice. Three-dimensional reconstructions of Rosenthal's canal (RC) were produced in order to measure canal length and volume, to generate orthogonal cross sections for area measurements, and to determine spiral ganglion neuron (SGN) number. RC length averaged 2.0 mm ± 0.04 (SEM) as measured along the centroid of the canal compared to an average basilar membrane (BM) length of 5.9 ± 0.05 (SEM). RC volume averaged 0.036 mm3 ± 0.009 (SEM). Significant increases in the radial area of RC were observed at the base (13%), middle (62%), and apex (90%) of its length. The total number of spiral ganglion neurons (SGNs) in RC in each of the five animals averaged 8,626 ± 96 (SEM). SGN number increased at the expanded regions of RC. Increased area and cell number at the base and apex are likely related to extensions of the organ of Corti past the length of RC in these areas. The increase in area and cell number in the middle of the RC appears to be related to the most sensitive frequency region of the organ of Corti. Volume imaging or tomography of the cochlea as provided by TSLIM has the potential to be an efficient and accurate semi-automated method for the quantitative assessment of the number of SGNs and hair cells of the organ of Corti.
Keywords: Spiral ganglion, Rosenthal's canal, cell count, TSLIM
1. Introduction
The advent of high resolution optical sectioning and 3D reconstruction of tissue structures has increased our understanding of complex three-dimensional (3D) structure and function from the level of organelles to embryos and whole tissues (Huang et al., 2008; Keller et al., 2008; Dodt et al., 2007). Tissues that have a complex 3D anatomy and whose function closely follows structure stand to gain the most from non-destructive methods that maintain and analyze 3D morphology. Such is the case with the mammalian cochlea where structural specializations help form its function/frequency specificity. Structures ranging from stereocilia and outer hair cell length to basilar membrane width and quantitative features of accessory structures have been shown to change significantly along the tonotopic axis of the mouse cochlea (Ehret, 1975; Ehret and Frankenreiter, 1977; Ehret 1979; and Ehret, 1983). Unfortunately, the ability to quantify these features is incomplete as sectioning the cochlea along its midmodiolar plane produces many tangential slices that are difficult to interpret. In addition, mechanical methods such as surface preparations of the organ of Corti are difficult to produce, especially in the mouse, and their reliability and accuracy depend upon the skill of the investigator.
In contrast, non-destructive imaging, such as Computed Tomography (CT), Magnetic Resonance Microscopy (MRI) and now TSLIM, allows researchers to visualize and digitize all of the cells and tissues contained within a cochlea at a resolution that allows for a quantitative assessment of cochlear elements relative to one another in 3D space. Digitization of the cochlea is consistent and automated by a computer program, which drives precise, closed feedback loop micropositioners. Virtual resectioning of the cochlea can be performed in any plane desired using a 3D reconstruction program such as Amira and structures can be isolated by segmentation to visualize their 3D anatomy and determine their morphometric characteristics. TSLIM optical serial sectioning of a cochlea clearly has advantages over other mechanical methods for comparing structural with functional characteristics of the normal and experimentally treated cochlea since the procedure is relatively rapid (30 min to produce a complete stack (~300 images) of 5 μm sections) and it preserves all of the cells and tissue in their 3D geometry in fixed, decalcified, and cleared cochleas.
While excellent descriptions of neuronal cell types and their projections exist in various species (Spoendlin 1972; Spoendlin, 1979; Liberman 1982a; Liberman 1982b; Liberman and Oliver, 1984; Berglund and Ryugo, 1987; Brown et al., 1988; Liberman et al., 1990), there is still some dispute over the total number of neurons in RC, especially in the mouse (Whitlon et al., 2006). This discrepancy seems to arise from the fact that each study implements a different method for counting cells and extrapolating that count over the entire volume of RC. While sampling a few sections of RC obtained from approximately the same regions of the cochlea may be acceptable for comparing groups within a single study, it lacks cross-study consistency (Luikart et al., 2003). Study methods for determining SGN number have been as diverse as their results even though many employ the well-accepted Abercrombie method for counting nuclei (1946). Table 1 summarizes some of the different methods and total number of SGN that have been estimated in the mouse by a number of different investigators. The total number of SGNs reported ranged from 4,421 ± 1325 (SEM) (Liebl et al., 1997) to 16,600 ± 600 (SD) (Camarero et al., 2001). While strain differences may account for some of this variability, it is unlikely that strain is a major factor and some of this variability is probably due to the different methods used to count SGNs. In the majority of these studies, cells were counted in mid-modiolar sections and extrapolated based upon their density over the entire volume, and few studies have accurately determined the volume or length of RC. Estimates of SGN number from relatively few sections can compound error from regions of variable density over the length of RC. Only one study (Whitlon et al., 2006), besides the present study, attempted to count every SGN in the cochlea, and to our knowledge, no previous study of SGN number has attempted to count every neuron and store its 3D position within RC. Much of the difficulty in studying the spiral ganglion/RC arises from its coiled shape, which when sectioned, leads to numerous tangential sections that sample highly variable regions of the cochlear spiral (Keithley and Feldman, 1983). Methods have been adopted to account for this (Keithley and Feldman, 1979) but may not preserve the ganglion-to-hair cell projection axis described by Liberman (1982b) and Keithley and Schreiber (1987).
Table 1.
| Investigator | Age Strain | Structure | Sample Size |
|---|---|---|---|
| Liebl et al., 1997 | P0.5 wild-type | nuclei | every 6th |
| Luikart et al., 2003 | P0 wild-type | cell bodies | every 10th |
| Postigo et al., 2002 | P7, P70 wild-type | nuclei | every 5th |
| Whitlon et al., 2006 | P1 CD-1 | nuclei | every cell |
| Farinas et al., 1994 | P0 wild-type | nucleoli | every 6th |
| Ehret, 1979 | P28 to P70 NMRI | afferents | tangential |
| Webster, 1985 | 7+ month deafness | cell bodies | every 10th |
| Camarero et al., 2001 | P5, P20 wild-type Igf-1 | nuclei | every 2nd |
Spiral Ganglion Neurons (SGN) counts
SD= standard deviation; SEM= standard error of the mean
The volume of RC, when reported, is also variable. Published values for the mouse range from 0.0375 mm3 (Webster, 1985) to 0.048 mm3 (Camarero et al., 2001). Few 3D anatomical reconstructions and descriptions of RC exist. 3D reconstruction of RC was reported for the gerbil, but the estimate was obtained from only five locations along the length of the canal (Echteler and Nofsinger, 2000). They reported that canal radial profile was reduced and cell number increased in the middle region of the gerbil cochlea. Length of spiral ganglion/RC is also underreported in the mouse, and in excised ganglia where it has obviously expanded without the surrounding bone; its length was reported at 4.5 mm ± 0.57 (SD) (Liu and Davis, 2007).
To remedy these shortcomings we imaged and reconstructed RC to produce accurate measurements of its length, volume and the total number SGNs in the mouse cochlea using thin-sheet laser imaging microscopy (TSLIM). TSLIM is a light-sheet fluorescence microscope (LSFM) with micrometer resolution that can image whole mouse cochleas and produce well-registered cross sections with 5 μm in thickness. TSLIM uses optically clear specimens and a focused light-sheet to optically section specimens with greater penetration depths than confocal and multiphoton systems. In TSLIM, a laser beam is focused into a planar light-sheet that induces a fluorescent plane (i.e., the optical section) in a cleared specimen. Images of the fluorescent plane are collected orthogonal to the illumination axis and only from the thinnest region of the light-sheet (see Santi et al., 2009). Cochleas were first studied using light-sheet microscopy by Voie et al. (1993). TSLIM is an enhanced version of OPFOS that is capable of producing higher resolution images, with full-width specimen focus, and with rapid imaging speed which drastically reduces photobleaching of the specimen. Further details of TSLIM and other light-sheet microscopes can be found in Santi et al., (2009); Schacht et al., (2010), and Santi (2011).
2. Materials and methods
2.1 Tissue preparation
Five female 4-wk-old CBA/JCr mice were anesthetized by an I.P. ketamine/xylazine injection followed by a 2 ml injection of 4% paraformaldehyde into the left ventricle of the heart after cutting the right atrium. Cardiac injection of fixative without a saline rinse (Eichenbaum et al., 2005) was found to be suitable and consistent for good fixation of SGNs. Cochleas were removed from the skull and bulla and post-fixed by perilymphatic perfusion and immersion in 4% paraformaldehyde overnight at 4 °C. After fixation, specimens were rinsed in phosphate buffered saline (PBS) and decalcified for four days in a 10% solution of disodium salt of ethylaminediamine tetraacetic acid (EDTA). After rinsing in PBS, cochleas were dehydrated in an ascending ethanol series (50, 75, 100%) and stained in Rhodamine B isothiocyanate (5 μg/mL in 100% ethanol) overnight. After rinsing in 100% ethanol, cochleas were cleared to transparency in Spalteholz solution (5:3 methyl salicylate:benzyl benzoate) and mounted on a plastic rod for TSLIM imaging.
2.2 Imaging
TSLIM optically sections the whole cochlea with a thin light-light sheet that is moved in the x- and z- axes. The light-sheet is produced by passing a 1 mm diameter 532 nm laser beam through a 10× Galilean beam expander and a cylindrical lens to produce a light-sheet with a minimum thickness of 3.5 μm. Due to the optical characteristics of the light-sheet the tissue was scanned in the x-axis to produced a well-focused image across the full width of the cochlea. A z-stack of well-aligned images was produced by moving the specimen through the light-sheet and the images are collected using either a Retiga 2000R CCD camera (QImaging) or a Piranha line scan camera (Dalsa) (Schacht et al., 2010). At 5 μm section thickness, the whole mouse cochlea was contained in ~300 images that take ~ 30 min to produce. Images were adjusted for brightness, contrast and either unsharp masking or deconvolution using ImageJ (NIH) and the stacks were loaded into Amira 5 (Visage Imaging) for placement within a Cartesian coordinate system, structure segmentation, 3D reconstruction of RC, and morphometry (Fig. 1A).
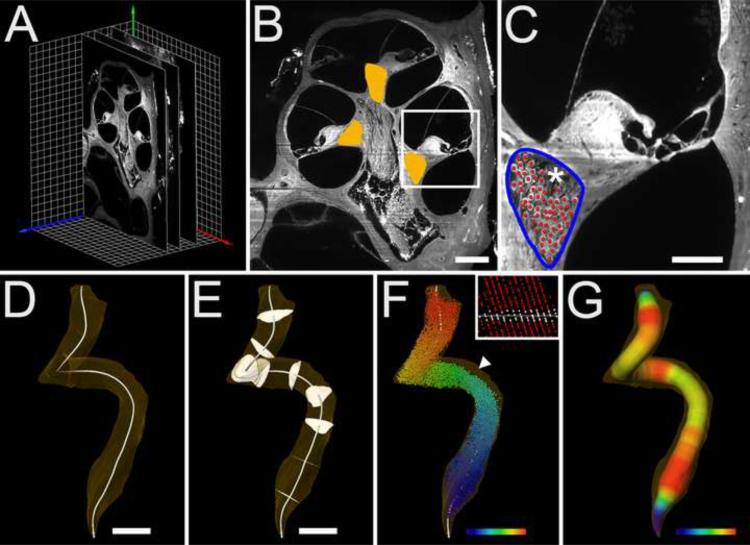
Fig. 1.
Data collection and analysis. Panel A shows three of the ~300 serial sections of a z-stack placed within a Cartesian coordinate system (red, green and blue arrows indicate x, y and z axes respectively). Each grid square is 100 × 100 μm. Panel B shows an example of Rosenthal's canal segmentation (orange overlay) in a representative mid-modiolar section. Bar indicates 200 μm. Panel C shows a lower turn from a cross section in B with the border of the canal outlined in blue and SGNs marked by manually placed red spheres using Amira. The * indicates a region of the canal that contains nerve fibers of the intraganglionic spiral bundle. It was this type of cross section that was used to determine the reproducibility of RC area by two observers. Bar indicates 100 μm. Panel D shows a 3D rendering of Rosenthal's canal (transparent orange) with B-spline generated centroid (white line). Bar indicates 200 μm. Panel E shows the orientation of orthogonal sections distributed every 10% distance from base to apex (10–90%). Bar indicates 200 μm. Panel F shows spheres representing SGN cell bodies color mapped by base-apex location along the centroid. Color gradient indicates the base (purple), middle (green, yellow-green) and apex (red). The clear area of RC (arrowhead) indicates the position of the intraganglionic spiral bundle within the canal. SGNs were counted in serial sections as shown in C. Panel F (insert) indicates the plane of an orthogonal section which in this case bisects 14 original imaging planes through the canal. Orthogonal plane at this percent distance location is represented by a white line. Panel G shows a representation of the canal's radius measured as the Euclidian distance from the exterior of the canal to its centroid. A color gradient map indicates radii between 20 μm (purple) and 65 μm (red).
2.3 Data collection
RC was segmented in each image by tracing its border in each serial section using a Wacom tablet (Fig. 1BC). Segmentation was partially automated in Amira by manually segmenting every other section, and then automatically segmenting the skipped section by linear interpolation. 3D reconstruction of RC was accomplished by outlining the bony wall containing SGNs in every profile in every section (Fig. 1B,C). This channel included the relatively acellular region of the intraganglionic spiral bundle (Fig. 1C) but did not include the nerves traveling to/from the osseous spiral lamina or to/from the modiolus. To determine the reliability in segmenting RC, two observers outlined the RC in a basal cross section in each of the five animals. Table 2 shows the results of their segmentations, which showed a strong correlation (r=0.999) in the areas determined by both observers. In addition, these observers also counted SGNs in each of the cross sections of RC and a strong correlation (r=0.995) between their results was also obtained. For ease of segmentation, the 5 μm z-step stacks were sub-sampled to 20 μm z-steps which Amira used to produce isosurface volume renderings. Other structures were segmented and morphometrically assessed in these cochleas, but only the RC, basilar membrane (BM), and the scala tympani (ST) are described in the present report.
Table 2.
| Animal | RC Area (mm2) | SGN # | |
|---|---|---|---|
| Observer: | SJ | HS | SJ |
| 1 | 0.019 | 0.019 | 71 |
| 2 | 0.012 | 0.012 | 104 |
| 3 | 0.024 | 0.023 | 72 |
| 4 | 0.051 | 0.05 | 112 |
| 5 | 0.046 | 0.046 | 90 |
|
| |||
| Correlation | r=0.999 | ||
Repeated measures of RC area and SG # by two observers (SJ and HS) in a basal cross
To produce average centroid positions and well-aligned orthogonal cross-sections, Amira's ScanConvertSurface module was used to generate 2 μm isometric voxel stacks of RC from the surface data. Amira's CenterlineTree module generated rough centroids from the isometric stacks using the TEASAR algorithm (Sato et al., 2000). The algorithm was assisted by manually placing a root point at the most apical point of the isosurface. Subsampling the rough centroid points to 32 B-spline control points produced a smoother centroid as well as equally-spaced points for percent distance measurements (Fig. 1D). A similar procedure was used to measure the length of the BM and its centroid was mathematically determined to extend from base to apex along a line below the third row of outer hair cells.
Using this point-by-point equidistant centroid, virtual orthogonal area measurements were produced using custom Tcl scripting of Amira's Trajectory module (Fig. 1E). The module computed planes orthogonal to point vectors representing 1–99% of the distance from the base of the RC with 0% and 100% being the absolute base and apex, respectively (Fig. 1F). Depending upon the position within RC and the imaging sectioning plane, the virtual orthogonal sections at determined locations along the length of RC may contain SGNs from a number of different imaging planes, which were not originally orthogonal to the axis of RC. Fig. 1F (insert) shows that SGNs from 14 different imaging planes comprise a single orthogonal section at this location along the length of RC. Figure 1F also shows a clear (i.e., without SGNs) region (arrowhead) near the organ of Corti side of RC containing fibers of the intraganglionic spiral bundle (see also Fig. 1C). A skeletonized representation of the canal's radius was also produced by computing the Euclidian distance from the exterior of the canal to its centroid (Fig. 1G). Skeletonization enhances the radial differences in the RC as a function of its length to show its difference in size and as a color map as well. Significant changes in mean orthogonal area were determined by Student's paired two-tailed t-test.
2.4 Cell counting
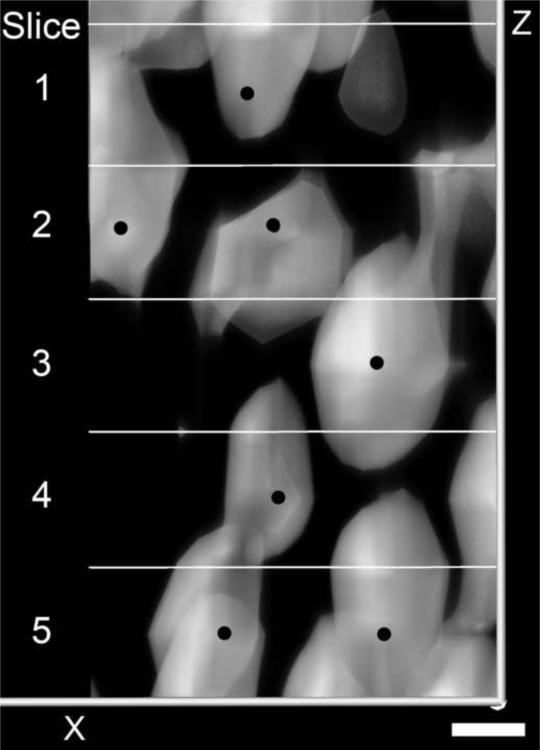
The total number of SGNs in all five animals was counted in RC by one observer (S.J.). Each of the approximately 42,500 cells were counted manually; however, this task was facilitated using Amira's ability to mark cells and travel through the segmented volume of RC. SGNs were distinguished from other cells by their large diameter (~10 μm) and morphology. To reduce the chances of double counting cells, 5 μm z-stacks were subsampled to 10 μm, which is approximately the diameter of the SGNs (Berglund and Ryugo, 1987). We could not reliably differentiate between type I and II SGNs and counted all of the cells we considered as SGNs. Cell counts were performed by placing points on the center of each SGN in the subsampled 10 μm stack (Fig. 1C). Previously selected points were observed on top of the next section to check for previously counted cells (Fig. 2). The observer moved forward and back in the z-stack to determine the extent of the cell and verify that it was counted only once. To identify cells based on where they occur along the length of the RC, a Tcl script was written to calculate the distance from each SGN to the nearest centroid point. To control for the fact that identified cells might occur anywhere within a certain cell-sized radius of the identified point, the z-dimension of each cell location in one animal was randomized. Randomized data were then analyzed for percent distance location and compared to the original dataset to determine the quantitative effect of 10 μm z-steps between serial sections. Z-axis randomized cells were identified with a different percent distance location less than 10% of the time and always within 1% of the original distance point.
Fig. 2.
This is direct volume rendering of a portion of the RC containing at least seven SGNs (black dot) that were counted in five, 10 μm sections (panels labeled 1–5). Notice that although the cells spanned across different optical sections they could be marked and identified to insure that they were only counted once. Bar indicates 5 μm.
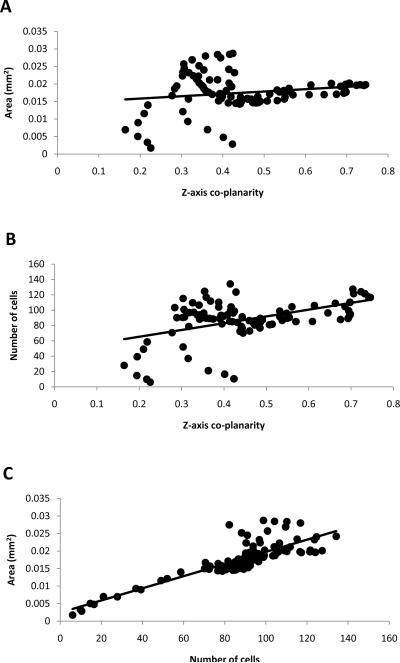
Since specimens were imaged in the mid-modiolar plane it was necessary to determine whether this biased the data. For example, mid-modiolar sections are inherently more orthogonal in the middle region of the cochlea than in the base or apex. Each specimen's centroid was assessed for how orthogonal it was to the imaging plane (z-axis) by calculating the z-component magnitude of the orthogonal plane unit vector at each point 1–99%. Greater unit vector z-components indicated whether the centroid-orthogonal planes were co-planar with the serial sections of the stack. Z-axis coplanarity of orthogonal centroid planes was plotted against normalized area and cell count data at the corresponding points 1–99% and analyzed by linear regression. Z-axis co-planarity of orthogonal centroid planes correlated poorly with orthogonal area (r=0.152) (Fig. 3A). Z-axis co-planarity also correlated weakly with cell number (r=0.448) (Fig. 3B). In contrast, orthogonal area correlated well with cell number (r=0.843) (Fig. 3C).
Fig. 3.
Panel A is a scatter plot of area versus z-axis co-planarity at each percent distance point. Panel B is a plot of cell number versus z-axis co-planarity. Panel C is a scatter plot showing area versus number of cells at each percent distance. Points falling below and above the trend line are areas with lesser or greater cell density respectively.
3. Results
In order to capture a complete cross section of the whole mouse cochlea in a single image we used a final Olympus microscope magnification of 5×. Since the numerical aperture of the 2× objective of the microscope is 0.50, its expected resolving capability is 0.5 μm. However, due to other optical factors (e.g., refractive index mismatch, specimen chamber thickness, and light scatter) we could resolve lines on a grid that were spaces 1 μm apart, thus our resolution is less than 2 μm in the center of the fluid-filled chamber. The line scan camera produces an image 4096×>3000) pixels and with our setup each pixel on the monitor represents 1.5 μm of tissue with the CCD camera and 0.7 μm of tissue with the line scan camera. Fig. 4 is an enlarged view (from a low power midmodiolar section) of the scala media where SGNs, hair cells and other cellular components are clearly recognized.
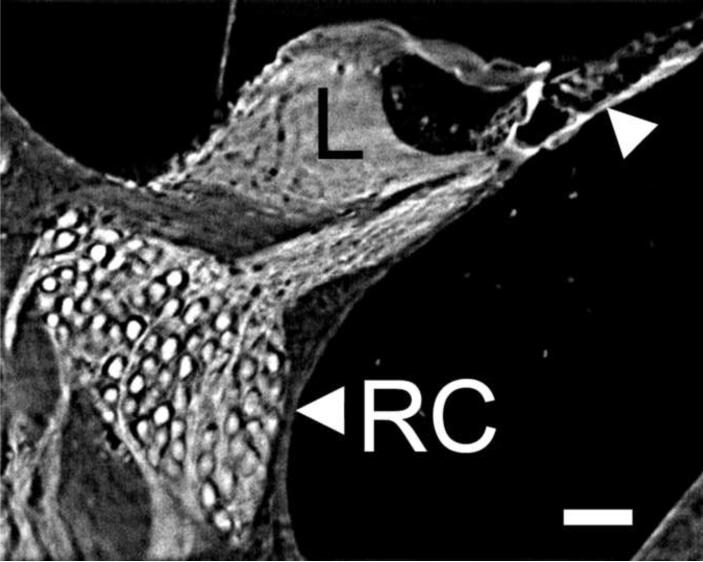
Fig. 4.
This is a higher magnification view (scala media cross section) of a midmodiolar cross section through the whole cochlea imaged by TSLIM. Note that image can be further enlarged within Amira and on a computer screen to view details of the cells and tissues of the scala media. Even at this magnification one can easily resolve Rosenthal's canal (RC) containing SGNs, the spiral limbus (L), and the outer hair cells (arrowhead). Bar indicates 50 μm.
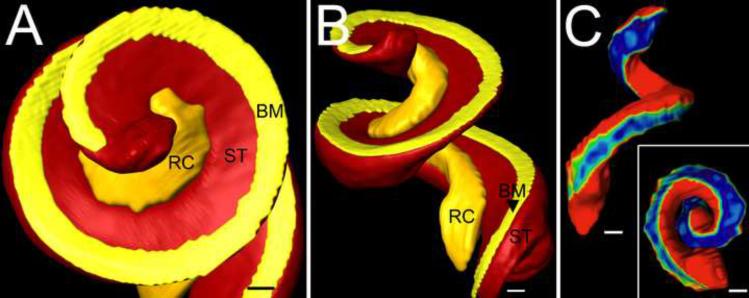
In three dimensions RC is a coiled, centrally located channel containing SGNs and nerve fibers. It has noticeable enlargements at its base and apex and openings in its bony walls to allow nerve fibers to travel to/from the osseous spiral lamina and the modiolus (Fig. 5). When viewed orthogonal to its centroid axis, RC can be described as a rounded triangle. One face is formed by its bony interface with the modiolus, which is perforated by several parallel rows of nerve bundles. At the base and apex large bundles of nerve fibers pierce the bony wall of the modiolar side of RC (Fig. 5). RC ends in the apex at the notch where scala tympani turns to join scala vestibuli at the helicotrema (Fig. 6A). Another face of RC is formed by the bone separating it from scala tympani (Fig. 6B.). This shelf of bone appears to decrease in thickness from base to apex (Fig. 6C). The third face of the canal is formed by the spiral limbus and osseous spiral lamina. Scala vestibuli is located above the point formed by the modiolar and limbal faces and nerve fibers travel to/from the organ of Corti, which exit the canal at the point formed by the tympanic and limbal faces.
Fig. 5.
3D isosurface reconstruction of Rosenthal's canal (RC) also showing the nerve fiber bundles traveling to/from the modiolus and the osseous spiral lamina. Large nerve bundles (arrowheads) entering the modiolus were present at the base and apex. Color map indicates relative thickness of nerve bundles (blue=5 μm and red=40 μm diameter). Bar indicates 100 μm.
Fig. 6.
3D relationships of RC. Figure shows a 3D isosurface reconstruction of basilar membrane (BM), scala tympani (ST) and Rosenthal's canal (RC) in apical (panel A) and side (panel B) views. Panel C shows the width of the bone between ST and RC with a color map (blue and red indicate 5 μm and 30+ μm respectively). Bars indicate 100 μm in A, B and C.
Whole-structure morphometry of RC was consistent across all five animals and is summarized in Table 3. Average canal length was 2.0 mm ± 0.04 (SEM) and ranged from 1.9 to 2.1 mm. For comparison, the length of the BM averaged 5.9 ± 0.05 (SEM) and ranged from 5.8 to 6.0 mm. Average RC surface area was 1.1 mm2 ± 0.02 (SEM), and ranged from 1.0 mm2 to 1.1 mm2. Average volume was 0.036 mm3 ± 0.009 (SEM) and ranged from 0.034 to 0.039 mm3. Average SGN total was 8,626 ± 96 (SEM). The minimum number of cells was 8,408 and the maximum was 8,912. Density was calculated for 99 planes from the base to the apex (1–99%). Individual cell totals correlated well with corresponding values for total volume (r=0.91) but more poorly with RC centroid length (r=0.65), suggesting that RC may expand radially rather than longitudinally as its cellular content increases. Individual BM lengths correlated well with cell totals (r=0.99), total volume (r=0.86) and to a lesser degree with RC length (r=0.748).
Table 3.
| Specimen ID | RC Length (mm) | BM length (mm) | RC Surface Area (mm2) |
|---|---|---|---|
| 9002 | 1.9 | 5.8 | 1.0 |
| 9003 | 2.1 | 6.0 | 1.1 |
| 9004 | 2.1 | 5.8 | 1.1 |
| 1004 | 2.0 | 6.0 | 1.1 |
| 1021 | 1.9 | 5.8 | 1.0 |
|
| |||
| Mean ± SEM | 2.0 ± 0.04 | 5.9 ± 0.05 | 1.1 ± 0.02 |
Whole structure morphometry in five animals
3.1 Area
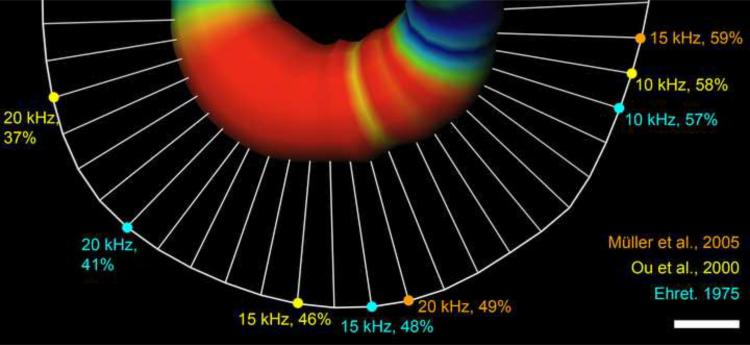
The orthogonal, cross-sectional area of RC as a function of percent distance from base to apex was non-uniform (Fig.7). The most significant minima and maxima are summarized in Table 4. Mean canal area increased rapidly to 0.029 mm2 ± 0.001 (SEM) at 13% of the base-to-apex distance, then decreased to 0.015 mm2 ± 0.001 (SEM) at 38% (13 vs 38% p<0.001). The area increased in the middle region, reaching significant peaks at 56% (0.020 mm2 ± 0.001 SEM) and 62% (0.020 mm2 ± 0.001 SEM) (38 vs 56% and 62%, p<0.01). Area decreased again after the middle turn to 0.014 mm2 ± 0.001 (SEM) at 76% (56 vs 76% p<0.01) and increased to 0.024 mm2 at 90% (76 to 90% p<0.001). The peak at 13% was significant compared to the other maxima at 56%, 62% and 90% (p<0.001). Maxima 56% and 90% were not significantly different from each other, nor were the minima at 38% and 76%. The expansion at the middle turn, shown as the thick red region in Fig. 8, seems to correspond well with the region of the lowest hearing thresholds in place-frequency maps reported by both Ehret (1975; 1983) and Ou et al. (2000), and less well with Müller (2005).
Fig. 7.
Mean Rosenthal's canal orthogonal area (black line) as a function of percent distance from cochlear base with standard error (grey shading). Circles indicate statistically significant peaks at 13, 56, 62 and 90% and valleys at 38 and 76%.
Fig. 8.
Skeletonized view of RC expansion in one specimen. Orthogonal area is represented on the centroid as both thickness and color (blue indicates 0.014 mm2 on either side of the middle expansion and red indicates expansions over 0.018 mm2). The centroid of BM is shown as a curved white line connected to the RC centroid by straight lines representing the shortest distance between the respective centroids. The region with the lowest hearing thresholds (10-kHz – 20kHz) is marked on the BM according to place-frequency equations by Ehret (1975; 1983), Ou et al., (2000) and Müller (2005). White bar indicates 100μm.
3.2 Cell number by percent distance from base
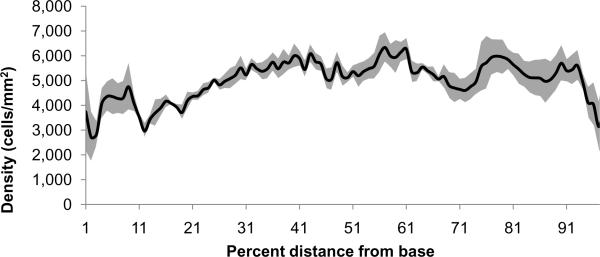
By identifying with the nearest percent point SGNs generally organized into groups orthogonal to the percent point (Fig. 1F insert). Percent distance cell data was more variable than orthogonal area but followed a similar pattern (Fig. 9). More SGNs were found in the base at 16% (117 cells ± 9 SEM); in the middle at 56% (122 cells ± 14 SEM) and 61% (127 cells ± 15 SEM); and the apex at 90% (134 cells ± 11 SEM). The fewest cells occurred at 37% (81 cells ± 7 SEM) and 73% (70 cells ± 6 SEM). The maxima at 16, 56, 61, and 90% were all statistically significant compared to the minima (p<0.05).
Fig. 9.
Graph showing number of cells as a function of percent distance from base to apex and standard error represented with grey shading. More cells were found at the base, middle and apex of Rosenthal's canal, reflecting orthogonal area peaks found in Fig.7.
Percent distance locations of maxima and minima for orthogonal area and cell number measurements were well correlated by a linear relationship (r=0.99) (values in Tables 4 and 5). Orthogonal area correlated with cell number as a function of distance (r=0.842) (Fig. 2C). This correlation, taken together with the strong relationship between cell total and RC total volume between animals (r=0.91), indicates that changes in cell number along the base-apex axis are not due sampling bias.
3.3 Cell density
Compared to area and cell number as a function of percent distance, cell density expressed as cells/mm2, was relatively constant, but with greater variability at each percent (Fig. 10). Cell density was lowest at the extreme base and apex and increased near the areas with restricted orthogonal area at 38% and 76% as well as the middle region at 56–62%. Average spiral ganglion neuron density was 5,000 cells per mm2 ± 227 (SEM) per section. Since cells occurred over a range of locations around the orthogonal plane (Fig. 1F insert), section thickness was defined as the distance between centroid points or 20 μm.
Fig. 10.
Graph showing SGN density (cells/mm2) as a function of percent distance from base to apex in five animals with standard error shown as grey shading. Since the basal orthogonal area peak (Fig. 7) was greater than the corresponding cell number peak (Fig. 9), there is a resulting decrease in density. Though not statistically significant, it appears that there are areas of greater cell density in the middle region from 56 to 62% and in the area-constricted regions near 38% and 76%.
4. Discussion
This study determined the 3D structure of RC and total SGN number using TSLIM. The canal is a coiled, spiral structure that contains the SGNs and nerve fibers and wraps itself around the modiolus of the cochlea. RC has three bony sides or faces that form its triangular structure. The most accessible, i.e. thinnest, face is the one it shares with scala tympani which grows thinner from base to apex. The combination of its 3D morphology and tonotopic organization means RC functional properties are packed into a relatively small region relative to distal cochlear structures like the organ of Corti/basilar membrane complex. This combination of properties also makes it difficult to study with mechanical sectioning methods that disrupt its organization. Previous studies have attempted to quantify and convey some anatomical features of the ganglion, but in many cases, these reports have been based upon limited samples along its length.
TSLIM's optical sectioning allowed us to quantify the entire ganglion without misaligned or lost sections. We found that average RC volume is 0.036 mm3 and it contains an average number 8,626 ± 96 (SEM) SGNs in the CBA/JCr mouse. RC volume was somewhat lower compared to previous estimates of 0.0375 mm3 and 0.048 mm3 but these previous studies were based upon few samples and they also reported greater SGN totals (Webster, 1985; Camarero et al., 2001). Our value for total SGN number is within the range of the largest number of published estimates. While many previous reports of SGN totals were accompanied by larger standard errors or deviation, our data has a narrower range. This may be due to the fact that we counted all of the cells as well as TSLIM's ability to produce well-aligned, loss-free sections. Average orthogonal section cell density data was in the range of data from previous reports (Willott et al., 1987; Dazert et al., 1996).
TSLIM's optical sectioning allowed us to study morphometric features along new axes. RC has orthogonal area expansions at the base, middle and apex of its centroid axis. These expansions are reflected in the number of SGNs along the centroid. At first, it seems that the increased number of cells at the base and apex conflicts with previous reports. In tangential sections taken at various locations of the mouse cochlea, Ehret (1979) found a significant increase in the number of nerve fibers in the middle region but not in the base or apex. Given that RC is roughly one-third the length of the basilar membrane (2.0 vs. 5.9 mm) it is likely that the expansions at its base and apex form to accommodate the greater number of neurons required to innervate longer basal and apical extensions of organ of Corti on the basilar membrane. This, however, does not explain why the middle region of the RC is expanded and shows an increase in the number of nerve fibers found there by Ehret (1979), afferent contacts at inner hair cells (Liberman et al., 1990) and an increased number of SGNs in the present study. In the human cochlea, there is evidence of a similar increase in the number of SGNs in middle region of RC (Pollak et al., 1987).
Studies have found that the middle region of the mouse cochlea has anatomical and neurophysiological features that favor lower hearing thresholds (Liu and Davis, 2007). Nerve fiber numbers and hair cell density all increase in and around the 15 kHz region of the basilar membrane, which is also the frequency region with the lowest hearing thresholds (Ehret, 1975; Ehret and Frankenreiter, 1977; Müller et al., 2005). SGNs in this region also been shown to have lower electrophysiological thresholds (Liu and Davis, 2007). While the mouse basilar membrane devotes nearly equal portions of its length to its complete frequency range (Müller et al., 2005), there appear to be anatomical (i.e. cell number) and physiological adaptations that favor this middle region.
SGN density was relatively constant compared to RC orthogonal area and cell number. This relative consistency may be the result of a compensating relationship between SGNs and the shape of Rosenthal's canal. Despite this consistency, density changed enough that extrapolations from a few sections may have yielded an inaccurate estimate of the total number of SGNs (Richter et al., 2011 [this issue]). SGN density was lowest in the base of the cochlea and may result from a combination of larger cell size and innervation demands of the organ of Corti which does not extend as far past RC as it does in the apex.
Cochlear implant studies could also stand to gain from the insights provided by TSLIM imaging methods. OPFOS was used to image a human temporal bone and that data was used to assist cochlear implant surgery by registering it with clinical CT data from live patients (Skinner et al., 2007). Again, data sets produced by light-sheet microscopes produce well-aligned sections and allow investigators to digitally analyze data in any plane. This flexibility and preserved morphology will also be important for computer models that seek to determine the functional contributions of anatomical features in the cochlea.
TSLIM also has the potential to facilitate studies of protein expression gradients in the developing and adult ganglia, which continue to be an important area of research (Rubel and Fritzsch, 2002, Whitlon et al., 2006). Using TSLIM it should be feasible to automatically count every neuron that is positive for a protein of interest in an intact cochlea by co-localizing neuron-specific antibodies and fluorescent nuclear stains. The challenge will be to label whole cochleas with antibodies. We have successfully labeled hair cells with myosin VII and prestin antibodies and all of the nuclei in a cochlea with a fluorescent DNA label (data not shown). Data from such experiments could potentially be combined into a 3D probabilistic atlas. Such data repositories are becoming increasingly common in brain research but are notably missing in the auditory field. A database and coordinate system that stores location-specific anatomical and functional data for the cochlea would greatly benefit the field.
Acknowledgements
Funding for this study was provided by the National Institute for Deafness and Communication Disorders (NIDCD) grants RO1DC007588 and DC007588-03S1 to PAS.
Abbreviations
- TSLIM
Thin-sheet laser imaging microscopy
- RC
Rosenthal's canal
- SGN
Spiral ganglion neuron
- 3D
Three-dimensional
- SEM
Standard error of the mean
- SD
Standard deviation
- IP
Intraperitoneal
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abercrombie M. Estimation of nuclear population from microtome sections. Anat. Rec. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
- Berglund AM, Ryugo DK. Hair cell innervation by spiral ganglion neurons in the mouse. J. Comp. Neur. 1987;255:560–670. doi: 10.1002/cne.902550408. [DOI] [PubMed] [Google Scholar]
- Brown MC, Berglund AM, Kiang NYS, Ryugo DK. Central trajectories of type II spiral ganglion neurons. J. Comp. Neur. 1988;278:581–590. doi: 10.1002/cne.902780409. [DOI] [PubMed] [Google Scholar]
- Camarero G, Avendaño Fernández-Moreno, Villar A, Contreras J, de Pablo F, Pichel JG, Varela-Nieto I. Delayed inner ear maturation and neuronal loss in postnatal igf-1-deficient mice. J. Neurosci. 2001;21:7630–7641. doi: 10.1523/JNEUROSCI.21-19-07630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazert S, Feldman ML, Keithley EM. Cochlear spiral ganglion cell degeneration in wild-caught mice as a function of age. Hearing Res. 1996;100:101–106. doi: 10.1016/0378-5955(96)00100-1. [DOI] [PubMed] [Google Scholar]
- Dodt HU, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgänsberger Becker, K. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Echteler SM, Nofsinger YC. Development of ganglion cell topography in the postnatal cochlea. J. Comp. Neurol. 2000;425:436–446. [PubMed] [Google Scholar]
- Ehret G. Masked auditory thresholds, critical ratios, and scales of the basilar membrane of the house mouse. J. Comp. Physiol. 1975;103:329–341. [Google Scholar]
- Ehret G, Frankenreiter M. Quantitative analysis of cochlear structures in the house mouse in relation to mechanisms of acoustical information processing. J. Comp. Neur. 1977;122:65–85. [Google Scholar]
- Ehret G. Quantitative Analysis of Nerve Fibre Densities in the Cochlea of the House Mouse (Mus Musculus) J. Comp. Neur. 1979;183:73–88. doi: 10.1002/cne.901830107. [DOI] [PubMed] [Google Scholar]
- Ehret G. Peripheral anatomy and physiology II. In: Willott JF, editor. The Auditory Psychobiology of the Mouse. Charles C. Thomas; Springfield, IL: 1983. pp. 169–200. [Google Scholar]
- Eichenbaum JW, Fadiel A, Miller DC, Demir M, Naftolin F, Stern A, Pevsner PH. Minimally invasive method for murine brain fixation. BioTechniques. 2005;39:487–490. doi: 10.2144/000112003. [DOI] [PubMed] [Google Scholar]
- Fariñas I, Jones KR, Backus C, Wang X,Y, Reichardt LF. Severe sensory and sympathetic deficits in mice lacking neurotrophin-3. Nature. 1994;369:658–661. doi: 10.1038/369658a0. [DOI] [PubMed] [Google Scholar]
- Huang B, Jones SA, Brandenburg B, Zhuang X. Whole-cell 3D STORM reveals interactions with nanometer-scale resolution. Nat. Methods. 2008;5:1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keithley EM, Feldman ML. Spiral ganglion cell counts in an age-grade series of rat cochleas. J. Comp. Neur. 1979;188:429–442. doi: 10.1002/cne.901880306. [DOI] [PubMed] [Google Scholar]
- Keithley EM, Feldman ML. The spiral ganglion and hair cells of Bronx waltzer mice. Hearing Res. 1983;12:381–391. doi: 10.1016/0378-5955(83)90007-2. [DOI] [PubMed] [Google Scholar]
- Keithley EM, Schreiber RC. Frequency map of the spiral ganglion in the cat. J. Acoust. Soc. Am. 1987;81:1036–1042. doi: 10.1121/1.394675. [DOI] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- Liberman MC. Single-neuron labeling in the cat auditory nerve. Science. 1982;216:1239–1241. doi: 10.1126/science.7079757. [DOI] [PubMed] [Google Scholar]
- Liberman MC. The cochlear frequency map for the cat: labeling auditory-nerve fibers of known characteristic frequency. J. Acoust. Soc. Am. 1982;72:1441–1449. doi: 10.1121/1.388677. [DOI] [PubMed] [Google Scholar]
- Liberman MC, Oliver ME. Morphometry of intracellularly labeled neurons of the auditory nerve: correlations with functional properties. J. Comp. Neur. 1984;223:163–176. doi: 10.1002/cne.902230203. [DOI] [PubMed] [Google Scholar]
- Liberman MC, Dodds LW, Pierce S. Afferent and efferent innervation of the cat cochlea: quantitative analysis with light and electron microscopy. J. Comp. Neur. 1990;301:443–460. doi: 10.1002/cne.903010309. [DOI] [PubMed] [Google Scholar]
- Liebl DJ, Tessarollo L, Palko ME, Parada LF. Absence of sensory neurons before target innervation in brain-derived neurotrophic factor-, neurotrophin 3-, and TrkC-deficient embryonic mice. J. Neurosci. 1997;17:9113–9121. doi: 10.1523/JNEUROSCI.17-23-09113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Davis RL. Regional specification of threshold sensitivity and response time in CBA/caJ mouse spiral ganglion neurons. J. Neurophysiol. 2007;98:2215–2222. doi: 10.1152/jn.00284.2007. [DOI] [PubMed] [Google Scholar]
- Luikart BW, Nef S, Shipman T, Parada LF. In vivo role of truncated TrkB receptors during sensory ganglion neurogenesis. 2003;117:847–858. doi: 10.1016/s0306-4522(02)00719-4. [DOI] [PubMed] [Google Scholar]
- Müller M, von Hünerbein K, Hoidis S, Smolders JWT. A physiological place-frequency map of the cochlea in the CBA/J mouse. Hearing Res. 2005;202:63–73. doi: 10.1016/j.heares.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Ou HC, Harding GW, Bohne BA. An anatomically based frequency-place map for the mouse cochlea. Hearing Res. 2000;145:123–129. doi: 10.1016/s0378-5955(00)00082-4. [DOI] [PubMed] [Google Scholar]
- Pollak A, Felix H, Schrott A. Methodological aspects of quantitative study of spiral ganglion cells. Acta Otolaryngol. 1987;436:37–42. doi: 10.3109/00016488709124974. [DOI] [PubMed] [Google Scholar]
- Postigo A, Calella AM, Fritzsch B. Distinct requirements for TrkB and TrkC signaling in target innervation by sensory neurons. Genes Dev. 2002;16:633–645. doi: 10.1101/gad.217902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CP, Kumar G, Webster E, Banas SK, Whitlon DS. Unbiased counting of neurons in the cochlea of developing gerbils. Hearing Res. 2011;(this issue) doi: 10.1016/j.heares.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Rubel EW, Fritzsch B. Auditory system development: primary auditory neurons and their targets. Ann. Rev. Neurosci. 2002;25:51–101. doi: 10.1146/annurev.neuro.25.112701.142849. [DOI] [PubMed] [Google Scholar]
- Santi PA, Johnson SB, Hillenbrand M, GrandPre PZ, Glass TJ, Leger JR. Thin-sheet laser imaging microscopy for optical sectioning of thick tissues. BioTechniques. 2009;46:287–294. doi: 10.2144/000113087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi PA. Light-Sheet Fluorescence Microscopy: A Review. J. Histochem. Cytochem. 2011;59:129–138. doi: 10.1369/0022155410394857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Bitter I, Bender MA, Kaufman AE, Nakajima M. TEASAR: tree-structure extraction algorithm for accurate and robust skeletons. Proc. Pacific Graphics. 2000;2000:281–289. [Google Scholar]
- Schacht P, Johnson SB, Santi PA. Implementation of a continuous scanning procedure for thin-sheet laser imaging microscopy. Biomed. Opt. Express. 2010;1:598–609. doi: 10.1364/BOE.1.000598/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MW, Holden TA, Whiting BR, Voie AH, Brunsden B, Neely JG, Saxon EA, Hullar TE, Finley CC. In vivo estimates of the position of advanced bionics electrode arrays in the human cochlea. Ann. Otol. Rhinol. Laryngol. 2007;116:1–24. [PubMed] [Google Scholar]
- Spoendlin H. Innervation densities of the cochlea. Acta Otolaryngol. 1972;73:235–248. doi: 10.3109/00016487209138937. [DOI] [PubMed] [Google Scholar]
- Spoendlin H. Neural connections of the outer hair cell system. Acta Otolaryngol. 1979;87:381–387. doi: 10.3109/00016487909126437. [DOI] [PubMed] [Google Scholar]
- Voie AH, Burns DH, Spelman FA. Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J. Microsc. 1993;170:229–236. doi: 10.1111/j.1365-2818.1993.tb03346.x. [DOI] [PubMed] [Google Scholar]
- Webster DB. The spiral ganglion and cochlear nuclei of deafness mice. Hearing Res. 1985;18:19–27. doi: 10.1016/0378-5955(85)90107-8. [DOI] [PubMed] [Google Scholar]
- Whitlon DS, Ketels KV, Coulson MT, Williams T, Grover M, Edpao W, Richter CP. Survival and morphology of auditory neurons in dissociated cultures of newborn mouse spiral ganglion. Neuroscience. 2006;138:653–662. doi: 10.1016/j.neuroscience.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Willott JF, Jackson LM, Hunter KP. Morphometric study of the anteroventral cochlear nucleus of two mouse models of presbycusis. J. Comp. Neur. 1987;260:472–480. doi: 10.1002/cne.902600312. [DOI] [PubMed] [Google Scholar]