Summary
Alginate overproduction by P. aeruginosa strains, also known as mucoidy, is associated with chronic lung infections in cystic fibrosis (CF). It is not clear how alginate induction occurs in the wild type (wt) mucA strains. When grown on Pseudomonas isolation agar (PIA), P. aeruginosa strains PAO1 and PA14 are nonmucoid producing minimal amounts of alginate. Here we report the addition of ammonium metavanadate (AMV), a phosphatase inhibitor, to PIA (PIA-AMV) induced mucoidy in both these laboratory strains and early lung colonizing nonmucoid isolates with a wt mucA. This phenotypic switch was reversible depending on the availability of vanadate salts and triclosan, a component of PIA. Alginate induction in PAO1 on PIA-AMV was correlated with increased proteolytic degradation of MucA, and required envelope proteases AlgW or MucP, and a two-component phosphate regulator, PhoP. Other changes included the addition of palmitate to lipid A, a phenotype also observed in chronic CF isolates. Proteomic analysis revealed the upregulation of stress chaperones, which was confirmed by increased expression of the chaperone/protease MucD. Altogether, these findings suggest a model of alginate induction and the PIA-AMV medium may be suitable for examining early lung colonization phenotypes in CF before the selection of the mucA mutants.
Keywords: Alginate induction, Vanadate, AlgU/T, AlgW, LPS, lipid A
Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium capable of thriving in various niches in the environment. P. aeruginosa is also an opportunistic pathogen that can colonize the airways of patients with cystic fibrosis (CF) (Lyczak et al., 2002). Once in the lung, P. aeruginosa is subjected to selective pressures that result in the emergence of strains which produce copious amounts of an exopolysaccharide known as alginate (Govan & Deretic, 1996). Alginate overproduction is advantageous for P. aeruginosa as it promotes the formation of an extracellular matrix that protects the bacterium from host defenses (Govan & Deretic, 1996, Lyczak et al., 2002).
Alginate overproducing strains of P. aeruginosa generally harbor mutations in a gene known as mucA. The mucA gene is located within the algUmucABCD cluster in the genome of P. aeruginosa. For the prototypic strain PAO1, these genes correspond to the loci of PA0762-PA0766 within the genome (www.pseudomonas.com). AlgU (also known as AlgT, σ22, or σE) (DeVries & Ohman, 1994, Martin et al., 1993a) is an alternative stress-related sigma factor, which belongs to the extracytoplasmic function (ECF) sigma factor superfamily. One of the major functions controlled by AlgU is to promote alginate production. Transcriptome analysis has revealed that AlgU can activate a vast number of genes (Firoved et al., 2002, Firoved & Deretic, 2003, Firoved et al., 2004a, Firoved et al., 2004b, Tart et al., 2005, Wood et al., 2006, Wood & Ohman, 2009b), some of which are beyond alginate regulation. AlgU is required for the oxidative stress and heat shock responses (Yu et al., 1996). Regulation of AlgU occurs at transcriptional and post-translational levels. At the level of transcription, the algU operon has five promoters, and two of them are dependent upon AlgU (DeVries & Ohman, 1994, Schurr et al., 1995). At post-translational level, AlgU is negatively regulated by MucA, the cognate anti-sigma factor. MucA is a transmembrane protein whose N-terminus sequesters AlgU to the cytoplasmic membrane. Release of AlgU from MucA causes increased AlgU activity, and the subsequent expression of the alginate biosynthetic operon via the algD promoter. The alginate biosynthetic operon (PA3540-PA3551) encodes most all of the genes required to synthesize and secrete alginate. However, the gene encoding the AlgC enzyme, which is involved both lipopolysaccharide (LPS) (Coyne et al., 1994) and alginate synthesis (Zielinski et al., 1991), is located outside of the biosynthetic operon (PA5322). AlgU also activates expression of transcription factors, encoded by algB (Wozniak & Ohman, 1991), algR (Mohr et al., 1991), and amrZ (Baynham et al., 2006), which participate in transcription from the algD promoter.
The emergence of mucoid isolates in the CF lung is thought to be a selective process. Environmental isolates of P. aeruginosa are generally nonmucoid. The CF patients probably acquire the initial colonizing strains from the environment. Emergence of the mucA mutants signifies the onset of chronic infection because overproduction of alginate facilitates the formation of a mucoid biofilm (Govan & Deretic, 1996). But before these mutants become dominant in the CF lung, it is possible that there is a time point that P. aeruginosa displays a transitional mucoid phenotype without mucA mutations. This probably occurs during the initial colonization to a time before the establishment of chronic infection. But this transitional mucoid phenotype is not well studied due to lack of an in vitro growth condition that mimics the induction. Analysis of this phenotype is important because it will help to understand the early colonizing phenotypes and the associated stress conducive in the conversion to the mucoid phenotype. Previous studies have shown the phosphate availability (Terry et al., 1991) and hydrogen peroxide (Mathee et al., 1999) can cause conversion to the stable mucoid phenotype. Furthermore, anaerobiosis can induce alginate expression in vivo (Bragonzi et al., 2005). Recently, two studies have shown that cell wall stress agents can activate proteolysis of MucA and alginate gene expression in vitro (Wood et al., 2006, Wood & Ohman, 2009b). However, such activation is not enough to result in mucoid conversion (Wood et al., 2006, Wood & Ohman, 2009b). To achieve alginate overproduction in vitro by wt mucA strains, genetic manipulations are required. For example, overexpression of the envelope protein MucE (Qiu et al., 2007), or inactivation of a sensor kinase KinB (Damron et al., 2009b) can both cause AlgW-dependent proteolysis of MucA and alginate overproduction (Fig. S1). Within the algUmucABCD cluster, inactivation of mucB (Martin et al., 1993b) or mucD also results in alginate overproduction in the wt mucA strains (Boucher et al., 1996). MucB protects the C-terminus of MucA (Cezairliyan & Sauer, 2009); without MucB, increased degradation of MucA likely occurs. When the chaperone protease MucD is absent, MucA degradation is dependent upon the MucP protease (Damron & Yu, 2011). However, each of these models lacks description of the events that occur upstream of regulated proteolysis of MucA by AlgW and MucP. Conditions that cause the envelope proteins to accumulate and/or misfold thereby activating AlgW and the resulting mucoidy have not been characterized.
P. aeruginosa also expresses other CF specific phenotypes upon adaptation to the CF airway. These phenotypes include increased antibiotic tolerance, biofilm formation, loss of flagellar motility, decreased secretion of virulence factors, and changes to the O-antigen and lipid A components of LPS. LPS is the major component of the outer leaflet of the Gram-negative outer membrane and is comprised of three distinct regions: O-antigen, core, and a hydrophobic anchor known as lipid A. O-antigen and core are made up of polysaccharide chains. CF isolates commonly display a rough phenotype due to the loss of the O antigen (Hancock et al., 1983). Lipid A consists of β-(1′,6)-linked diglucosamine backbone with phosphates at the 1 and 4′ positions, amide-linked fatty acids at the 2 and 2′ positions, and ester-linked fatty acids at the 3 and 3′ positions. The chain lengths of the fatty acids linked to lipid A are C12 or C10. In P. aeruginosa, penta-acylated lipid A is the most common form, however, hexa- and hepta- forms are also found (Moskowitz & Ernst, 2010). Addition of a C16 fatty acid, or palmitate, results in the synthesis of a hexa-acylated lipid A. In a study, 100% of CF isolates have palmitate on lipid A (n=86) (Ernst et al., 2007). It has been suggested that acylation alters the recognition of lipid A by the host and leads to chronic colonization of the CF airway (Moskowitz & Ernst, 2010). These lipid A modifications also change the bacterial cell surface and outer leaflet integrity.
In Escherichia coli, modifications to lipid A structure can induce σE activity (σE is the E. coli homologue of AlgU) (Tam & Missiakas, 2005). When palmitate is added to lipid A by the palmitoyltransferase PagP, increased σE activity is observed (Tam & Missiakas, 2005). Palmitoylation of lipid A and increased σE expression has relevance to P. aeruginosa and CF because palmitoylation of lipid A is observed in P. aeruginosa strains isolated from the CF airway (Ernst et al., 1999). In E. coli, addition of palmitate is regulated by specific growth conditions, such as low divalent cation concentration or by the addition of ammonium metavanadate (NH4VO3, AMV) to the growth medium (Tam & Missiakas, 2005). AMV is a general phosphatase inhibitor that causes increased σE activity without a major impact on the growth of E. coli (Tam & Missiakas, 2005). We wondered if AMV would induce a similar effect on the activation of σE homolog AlgU in P. aeruginosa.
The goals of this work were to develop an in vitro growth medium for the analysis of alginate induction in P. aeruginosa and to characterize the nature of environmental sensing which activates alginate overproduction. Here, we report Pseudomonas isolation agar (PIA) supplemented with AMV (PIA-AMV) can induce mucoidy in strains of P. aeruginosa with a wt mucA. We found that AMV and triclosan can synergistically activate alginate overproduction. To our knowledge, this is the first example that mucoidy can be artificially induced in P. aeruginosa in vitro, independent of mutations. We also show that alginate induction in PAO1 is due to the increased MucA degradation leading to activation of AlgU. Furthermore, growth on PIA-AMV induces synthesis of palmitoylated lipid A. It also seems that growth of PAO1 on PIA-AMV may cause protein misfolding in the envelope because there is an increase in the expression of general stress chaperones including the chaperone protease MucD. We propose that the PIA-AMV medium simulates environmental conditions which can activate alginate overproduction in P. aeruginosa.
Results
P. aeruginosa PAO1 cultivated on PIA-AMV is mucoid
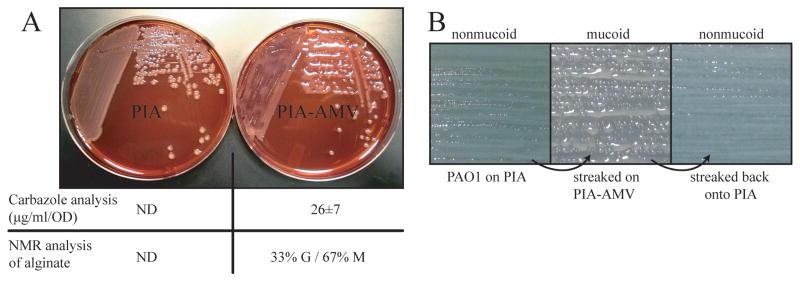
In E. coli, modifications to lipid A structure have been shown to induce a σE-dependent response (Tam & Missiakas, 2005). To determine if AMV could induce activation of the P. aeruginosa σE homologue, AlgU, the wt nonmucoid strain PAO1 was cultivated on the PIA-AMV medium. Interestingly, mucoid colony morphology of PAO1 was observed (Fig. 1A). PIA is a complex growth medium for isolation and selection of pseudomonads. PAO1 on PIA is always nonmucoid. The AMV concentration was then optimized for this phenotype. We determined PIA supplemented with 0.27 mM AMV resulted in the highest visible mucoid phenotype. Other salts of metavanadate, such as cesium, sodium and potassium could also induce this phenotype similar to AMV (data not shown), suggesting that the vanadate component of the salt is responsible for this phenotype. Carbozole analysis for uronic acid content was performed on cell free suspensions. The uronic acid content of PAO1 cultured on PIA-AMV was 26±7 μg/mL/OD600, but PAO1 on PIA was not detectable as determined by the method of collection and carbazole analysis as described in this study. For comparison, mucA25 mucoid strain PAO581 (Qiu et al., 2008b) on PIA produced the same amount of alginate upon 24 hr growth. To determine if the exopolysaccharide was alginate, standard nuclear magnetic resonance (NMR) analysis of precipitated polysaccharide was performed. As expected, the precipitated material of PAO1 on PIA-AMV was alginate and composed of 33% guluronic acid and 67% mannuronic acid (Fig. 1A). The composition of PAO581 alginate (Qiu et al., 2008b) was 31% guluronic acid and 69% mannuronic acid.
Figure 1. PIA-AMV induces alginate production in strain PAO1.
A. Shown is strain PAO1 cultured on PIA and PIA supplemented with 0.27 mM AMV for 24 hr at 37°. Both PIA and PIA-AMV plates in the picture contained Congo Red for imaging contrast. The average uronic acid content and the ratio of guluronate to mannuronate residues of the alginate produced (G/M) are indicated. NMR analysis was used to determine the percentage of G and M residues in the alginate produced by PAO1 on PIA-AMV. ND indicates the alginate concentration was below the detection limit. B. PAO1 was cultured on PIA and then streaked on PIA-AMV where alginate overproduction occurs. When mucoid PAO1 was transferred back to PIA, alginate production ceased and the strain was nonmucoid. These passages were performed daily for two weeks during which PAO1 was mucoid on PIA-AMV and nonmucoid on PIA.
To establish that the AMV-mediated mucoidy in PAO1 was not due to the mutation of mucA, the colonies on PIA-AMV were then transferred to PIA lacking AMV. This returned the bacterium to wt nonmucoid morphology (Fig. 1B). So AMV on PIA has an alginate induction effect. We wondered whether AMV was solely responsible for this phenotype or components of PIA were also required. When PAO1 was cultivated on LB supplemented with 0.27 mM AMV, the inducible mucoid phenotype was not evident (data not shown). This suggests PIA has some ingredient(s) that facilitate the induction. PIA contains tricolsan (also known as irgasan), a cell wall stress agent that has been shown to activate algD expression in P. aeruginosa (Wood et al., 2006). When triclosan was added to LB containing 0.27 mM AMV, the inducible mucoid phenotype was observed (data not shown). To determine if AMV or triclosan alone could activate alginate overproduction, AMV or triclosan from 0-10 mM and 0-2000 μg ml-1 respectively, in L agar was used to grow PAO1. PAO1 showed a reduced growth on high concentrations of AMV, but was nonmucoid (data not shown). PAO1 is capable of growing on high amounts of triclosan (Chuanchuen et al., 2003, Zhu et al., 2010). On LB agar, PAO1 was nonmucoid for all concentration of triclosan tested (data not shown). PIA also contains 15 mM magnesium chloride to stimulate pyocyanin production for identification of P. aeruginosa. When the magnesium chloride concentration was lowered to 1.5 mM in PIA-AMV, alginate overproduction ceased (data not shown), suggesting that a high divalent cation concentration is needed for alginate overproduction by PAO1 on PIA-AMV. Collectively, these data indicate the combination of triclosan, AMV and high magnesium are required for strain PAO1 to overproduce alginate.
PIA-AMV induces mucoid conversion in strains with wt alleles of algU mucA
Since PIA-AMV activated alginate production in strain PAO1 with a wt mucA, we wondered if this inducible phenotype can be reproduced in other laboratory strains. Commonly used P. aeruginosa strains PA14 and PAK were both mucoid on PIA-AMV (Table 1). However, strain PA103 did not overproduce alginate, suggesting that PA103 may have a mutation that impairs alginate synthesis. We hypothesized that the most likely place for a mutation in PA103, which blocks alginate overproduction, would be algU. So we sequenced algU from PA103 but found no mutations in PA103 in algU (Table 1). Therefore, it seems PA103 may have an unknown mutation(s) compared to PAO1. We next tested CF P. aeruginosa isolates for alginate induction (Table 1). Early colonizing CF isolates with a wt mucA (C01232, C3715, C7406) were nonmucoid on PIA, but they all became mucoid on PIA-AMV. Therefore, AMV has an alginate induction effect in strains with a wt mucA. We then examined whether AMV could also have an alginate inhibitory effect in strains that were already mucoid such as the mucA mutants. Mucoid CF isolate, FRD1 (algT+mucA22) was tested on PIA-AMV and remained mucoid, indicating the absence of such an effect. However, nonmucoid CF strain FRD2 and CF149 were not mucoid on PIA-AMV. FRD2 and CF149 have suppressor mutations in algT/U (Damron et al., 2009a, DeVries & Ohman, 1994, Ohman & Chakrabarty, 1981), which would block alginate production. These results suggest that PIA-AMV will induce mucoid conversion in nonmucoid laboratory and CF strains with the genotype of algU+mucA+. The strains with algU suppressor mutations will not respond to induction on PIA-AMV.
Table 1. Alginate induction in P. aeruginosa strains on PIA-AMV.
| Phenotypeb | |||
|---|---|---|---|
| Strain or mutantsa | Genotype of algU mucA, inactivated genes or description of strain | PIA | PIA-AMV |
| Laboratory strains | |||
| PAO1 | algU+mucA+ | NM | M |
| PA14 | algU+mucA+ | NM | M |
| PAK | algU+mucA+ | NM | M |
| PA103 | algU+mucA+ | NM | NM |
| CF clinical strains | |||
| C0132 | Early colonizing CF isolate, algU+mucA+ | NM | M |
| C3715 | Early colonizing CF isolate, algU+mucA+ | NM | M |
| C7406 | Early colonizing CF isolate, algU+mucA+ | NM | M |
| FRD1 | Mucoid CF isolate, algT(U)+mucA22 | M | M |
| FRD2 | Spontaneous nonmucoid CF isolate, mucA22with algT(U) suppressor mutation | NM | NM |
| CF149 | Nonmucoid CF isolate, mucA mutant with algU suppressor mutation | NM | NM |
| Sigma factors | |||
| PAO1ΔalgU | In-frame deletion of alginate master regulator/sigma factor AlgU (σ22) | NM | NM |
| PAO1rpoH | Knockout of the heat shock sigma factor RpoH (σ32) | NM | M |
| PAO1ΔrpoN | In-frame deletion of the alternative sigma factor RpoN (σ54) | NM | NM |
| PAO1rpoS∷tet | Knockout of the alternative sigma factor RpoS (σ38) | NM | M |
| Alginate regulators | |||
| PAO581 | mucA25 encoding the N-terminal portion of anti-sigma factor MucA | M | M |
| PAO1mucB | Knockout of mucB encoding a protein that the C-terminus of MucA | M | M |
| PAO1mucC | Knockout of mucC encoding a protein with unknown function | NM | M |
| PAO1mucD | Knockout of mucD encoding a periplasmic chaperone / protease | M | M |
| PAO1kinB | Knockout of kinB encoding a two-component histidine kinase | M | M |
| Alginate biosynthesis | |||
| PAO1algD | Knockout of algD encoding initial enzyme in alginate biosynthesis | NM | NM |
| PAO1algE | Knockout of algE encoding alginate biosynthesis porin pump | NM | NM |
| Transcription factor | |||
| PAO1ΔalgR | In-frame deletion of transcriptional activator AlgR of PalgD | NM | NM |
| PAO1algZ | Knockout of a two-component histidine kinase | NM | M |
| PAO1ΔalgB | In-frame deletion of transcriptional activator AlgB of PalgD | NM | NM |
| Alginate proteases | |||
| PAO1ΔalgW | In-frame deletion of algW encoding an envelope protease for MucA | NM | NM |
| PAO1mucP | Knockout of mucP encoding an envelope protease for MucA | NM | NM |
| PAO1prc | Knockout of mucP encoding a tail-specific protease for MucA | NM | M |
| Lipid A enzyme | |||
| PAO1ΔphoP | In-frame deletion of phoP encoding a regulator of palmitoylation of lipid A | NM | NM |
| PAO1pmrA | Knockout of pmrA encoding a regulator of aminoarabinose of lipid A | NM | M |
| PAO1pmrB | Knockout of pmrB encoding a histidine kinase/regulator of aminoarabinose of lipid A | NM | M |
| Envelope proteins | |||
| PAO1oprH | Knockout of oprH encoding PhoP-regulated OMP | NM | M |
| PAO1mucE | Knockout of mucE encoding AlgW activating envelope protein | NM | M |
The origin of the strains and references are indicated in Supplemental Table 1
The phenotype is indicated as NM or M for nonmucoid or mucoid respectively. Mucoid phenotype was assigned to strains that were similar to PAO1 cultured on PIA-AMV. This phenotype was further characterized by the standard observation of exopolysaccharide strings upon touch of a colony with wooden toothpick.
ND stands for not determined.
Alginate induction in PAO1 requires two sigma factors, AlgU and RpoN
We tested a group of sigma factor mutants for alginate induction. As expected, we observed the algU and rpoN genes in PAO1 were both required for alginate induction on PIA-AMV medium (Table 1). Heat shock sigma factor RpoH and stationary phase growth sigma factor RpoS have also been previously associated with alginate production. AlgU can drive expression of RpoH (Schurr & Deretic, 1997). Here we observed rpoH was not required for mucoidy on PIA-AMV (Table 1). This is somewhat expected because rpoH expression is downstream of AlgU (Schurr and Deretic, 1997). Previous work has shown that inactivation of rpoS causes decreased alginate production in FRD1 (Suh et al., 1999). But rpoS was not required for alginate induction in PAO1 on PIA-AMV.
Alginate induction in alginate regulator and biosynthetic gene mutants
Mutation of the negative regulators of AlgU such as mucA (Martin et al., 1993c), mucB (Goldberg et al., 1993, Martin et al., 1993b), mucD (Boucher et al., 1996), kinB (Damron et al., 2009b) cause alginate overproduction. Mutants in the negative regulators of AlgU all remained mucoid on PIA-AMV (Table 1). The gene mucC has been associated with alginate overproduction, but has not been fully characterized (Boucher et al., 1997). Here, we observed that alginate overproduction occurred in the mucC mutant (Table 1). We next tested the requirement of alginate biosynthetic genes using this system. As anticipated, alginate biosynthetic genes algD and algE, encoding GDP-mannose dehydrogenase and the alginate porin pump, respectively, were both required for mucoidy on PIA-AMV (Table 1). Several two-component regulatory systems are involved in regulation of alginate production. Alginate overproduction requires algR, which encodes a transcription factor involved in algD expression (Deretic et al., 1989). PAO1algR was not mucoid on PIA-AMV (Table 1). However, the cognate histidine kinase of algR, encoded by algZ, was not required for alginate overproduction on PIA-AMV. The NtrC-family transcription factor, AlgB, has been shown to be required for algD expression (Leech et al., 2008) as well as mucoidy in a wt mucA strains (Damron et al., 2009b). As predicted, PAO1algB remained nonmucoid on PIA-AMV (Table 1).
Alginate induction in PAO1 requires MucA proteases AlgW and MucP
Since PAO1 has a wt mucA gene, alginate induction would relieve suppression on the proteolytic degradation of MucA. Two proteases, AlgW and MucP, are critical for the degradation of wt MucA. Therefore, we tested PAO1 strains lacking each of the two characterized protease genes, algW and mucP. Both algW and mucP were required for mucoidy on PIA-AMV (Table 1). This suggested that alginate induction on PIA-AMV was occurring due to the activation of these two proteases. Prc protease is a periplasmic protease needed for mucoidy in mucA mutants (Reiling et al., 2005). However, prc is dispensible for mucoidy under conditions in which AlgW is activated (Qiu et al., 2007). Without prc, mucoidy occurred on PIA-AMV, which indicated Prc had a minimal role in regulated proteolysis of wt MucA in PAO1 (Table 1). MucE is the only identified envelope protein that activates regulated proteolysis of MucA. However, many P. aeruginosa outer membrane proteins have C-termini peptide sequences that could activate AlgW (Qiu et al., 2007). With mucE inactivated in PAO1, alginate induction still occurred. This suggests that mucE is not involved in alginate induction in PAO1 on PIA-AMV.
AMV increases PalgU and PalgD promoter activity in PAO1
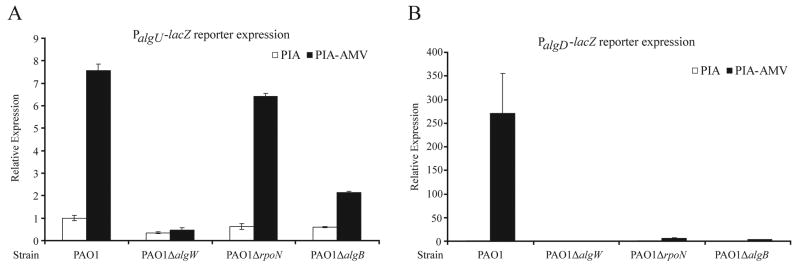
As mentioned, without the proteases AlgW or MucP, alginate induction was not observed (Table 1). This suggests MucA degradation may be occurring in PAO1 on PIA-AMV. The proteolysis of MucA results in increased promoter activity of algU (Damron et al., 2009b, Qiu et al., 2007). This effect is due to the fact that two of the algU promoters are dependent upon AlgU (DeVries & Ohman, 1994, Schurr et al., 1995). To observe the effect of PIA-AMV on the alginate-related promoters PalgU and PalgD, a single copy of the algU or algD promoters fused with lacZ (Damron et al., 2009b), were integrated onto the chromosomes of PAO1, PAO1ΔalgW, PAO1ΔrpoN, PAO1ΔalgB. The β-galactosidase activities were measured on PIA and PIA-AMV (Fig. 2AB). Compared to the activities on PIA, PAO1 on PIA-AMV had high PalgU and PalgD activities, which were dependent upon algW (Fig. 2AB). This result suggests that AlgW-dependent regulated proteolysis is the mechanism for alginate induction on PIA-AMV. In the absence of rpoN, high PalgU but low PalgD activities were observed (Fig. 2AB). This is consistent with a model that RpoN activates transcription at PalgD (Boucher et al., 2000). In the absence of algB, there was decreased PalgU expression associated with minimal PalgD expression. Previously we have noted in a kinB mutant that PalgU activity can be dependent on algB (Damron et al., 2009b).
Figure 2. PalgU and PalgD expression on PIA-AMV.
PalgU-lacZ and PalgD-lacZ reporter constructs were integrated into the chromosome of the strains as indicated via the miniCTX1 chromosomal shuttle vector (Hoang et al., 2000). β-galactosidase activities were determined after 24 hr growth on PIA or PIA-AMV. Values were normalized to PAO1 on PIA and indicated as mean ± SD from three independent experiments. A. The PalgU activity of P. aeruginosa strains on PIA and PIA-AMV. B. The PalgD activity of P. aeruginosa strains on PIA and PIA-AMV. No PalgD activity was detected for strain PAO1 on PIA or strain PAO1ΔalgW on PIA or PIA-AMV.
PIA-AMV induced mucoidy of PAO1 causes decreased stability of MucA and an increase in MucD expression
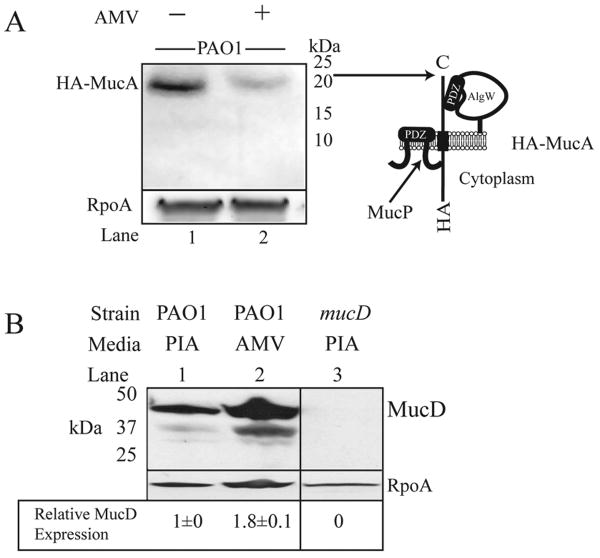
Since algW was required for alginate induction on PIA-AMV, we hypothesized that MucA was being degraded under this condition. Previously, we have reported degradation of HA-epitope tagged MucA expressed from a multicopy plasmid in kinB mutant (Damron et al., 2009b), mucD mutant (Damron & Yu, 2011), and mucE overexpressing strains (Qiu et al., 2007). To improve the resolution for measuring HA-MucA degradation in vivo, we constructed a single copy of HA-mucA (Fig. S2) on the chromosome shuttled via miniCTX1 (Hoang et al., 2000). Western blot analysis revealed that when PAO1 was grown on PIA-AMV, there was less HA-MucA detected (Fig. 3A lanes 1 and 2). Since the PalgU promoters were used to drive expression of HA-mucA, it should be noted that PIA-AMV caused 7-fold increase in PalgU activity (Fig. 2A). Therefore, PIA-AMV caused high expression of HA-MucA, but HA-MucA in PAO1 on PIA-AMV was detected in a low quantity (Fig. 3A). Our interpretation of this result is that the growth of PAO1 on PIA-AMV caused increased MucA degradation which leads to AlgU activation and alginate overproduction.
Figure 3. Strain PAO1 shows a decreased level of HA-MucA and increased MucD expression on PIA-AMV.
A. Western blot analysis was performed as previously described (Damron et al., 2009b). Cell lysates were prepared from cells after 24 hr growth on PIA and PIA-AMV. The membranes were probed with anti-HA (Roche) and anti-alpha subunit of RNAP antibodies (Neoclone) as a loading control. Shown are representative panels of blots from three independent experiments with 50μg of total lysate. Positions of apparent molecular masses are indicated aligned with a schematic of HA-tagged MucA C. Total protein lysates of PAO1 cultured on PIA and PIA-AMV as well as a mucD mutant strain, were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were probed sequentially with anti-MucD (Wood & Ohman, 2006) and anti-alpha RNA polymerase (Neoclone). MucD signal was normalized to 1 for PAO1 on PIA for comparison to PAO1 on PIA-AMV. Shown is a representative of three independent experiments with standard deviation indicated for biological replicates.
When the chaperone protease mucD is inactivated, alginate overproduction occurs (Boucher et al., 1996). Inactivation of mucD causes instability of MucA (Damron & Yu, 2011, Wood & Ohman, 2009a). Since MucD appears to control proteins that activate regulated proteolysis of MucA, we used MucD expression as an indicator of protein quality in the periplasm. Western blot analysis with cell lysates of PAO1 on PIA and PIA-AMV revealed that MucD expression increased on PIA-AMV (Fig. 3B). This suggests a possibility that the protein quality of the periplasm was affected by PAO1 growth on PIA-AMV compared to PIA. However, the MucD upregulation on PIA-AMV was not sufficient to suppress mucoidy.
PhoP, but not PmrAB, is required for induced mucoidy in PAO1
Previously Zhou et al. (Zhou et al., 1999) showed that one effect of cultivating E. coli in the presence of AMV was synthesis of alternative forms of lipid A. The transcription factor PhoP is required for expression of the genes that encode the enzymes which modify lipid A (Ernst et al., 1999). We reasoned that lipid A modifications may induce alginate overproduction on PIA-AMV. PAO1 growing on PIA-AMV required phoP for alginate overproduction (Table 1), which suggests a link between modified lipid A and activation of alginate production. In E. coli, PagP is the enzyme that adds palmitate to lipid A; however a homologue in P. aeruginosa has not yet been identified (Bishop et al., 2005). In E. coli, it has been shown that overexpression of the PagP enzyme caused increased σE activity (Tam & Missiakas, 2005). Zhou et al. (Zhou et al., 1999) showed that the addition of aminoarabinose to lipid A occurs due to AMV in the culture media. In light of this, we thought it logical to test pmrA and pmrB mutants that control aminoarabinose modification of lipid A in P. aeruginosa (McPhee et al., 2003). Neither pmrA nor pmrB were required for mucoidy on PIA-AMV (Table 1). Together, this suggests the composition of lipid A in the cell may influence the conserved σE system in P. aeruginosa.
PAO1 lipid A is modified with palmitate when cultivated on PIA-AMV
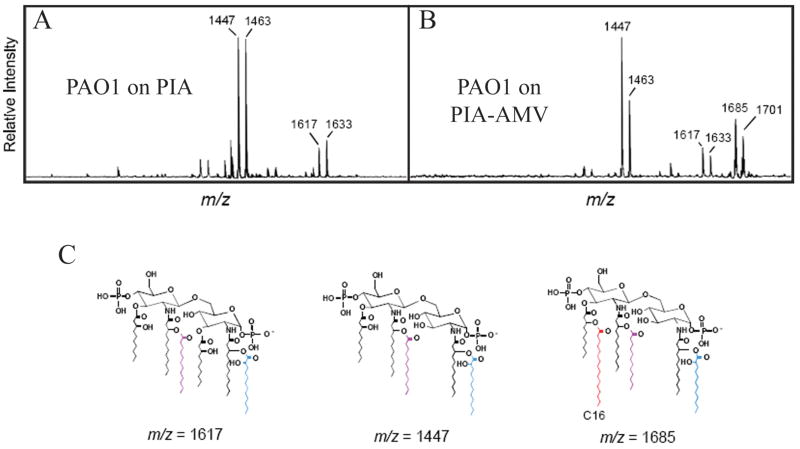
To determine if vanadate played a role in altering the structure of lipid A, mass spectrometry analysis was performed on lipid A extracted after growth on PIA-AMV (Fig. 4ABC). Consistent with previous reports (Ernst et al., 2006, Ernst et al., 2007), lipid A structures of PAO1 were a blend of penta- and hexa-acylated molecules, identified on spectra by mass to charge ratios (m/z) of 1447 and 1463 (penta) and 1617 and 1633 (hexa) (Fig. 4C). The difference of 16 mass units in Fig. 4 spectra represents the presence of an additional hydroxyl residue in the lipid A structure. Hexa-acylated lipid A (m/z 1617) was the major type of lipid A from PAO1 on PIA though penta-acylated lipid A (m/z 1447) was also observed. Interestingly, when PAO1 was cultured on PIA-AMV, a third species of lipid A was observed that represents a second hexa-acylated lipid A (m/z 1685). This ion species results from the addition of palmitate (C16) to the penta-acylated species (m/z 1447) (Fig. 4B and C). Interestingly, and different to previous results using E. coli, the addition of aminoarabinose was not detected (Fig. 4ABC).
Figure 4. PIA-AMV regulation of P. aeruginosa lipid A modification.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) in the negative-reflection ion mode of lipid A isolated from P. aeruginosa PAO1 grown in the absence or presence of vanadate. (A) PAO1 grown on PIA. (B) PAO1 grown on PIA-AMV. (C) Indicates the three structures of the lipid A molecules identified.
Since PhoP was required for mucoidy on PIA-AMV, it was imperative to test an oprH mutant because the genes phoPQ and oprH are genetically linked and are autoregulated via phoP (Macfarlane et al., 1999). Here, we observed that oprH was not required for mucoidy of PAO1 on PIA-AMV. Recently it has been shown that the absence of phoQ upregulates alginate genes (Gooderham et al., 2009). Collectively, these data implicate a role of phoP in the induction of the alginate pathway, but oprH is not essential for alginate induction (Table 1).
Proteomic comparison of PAO1 grown on PIA vs. PIA-AMV
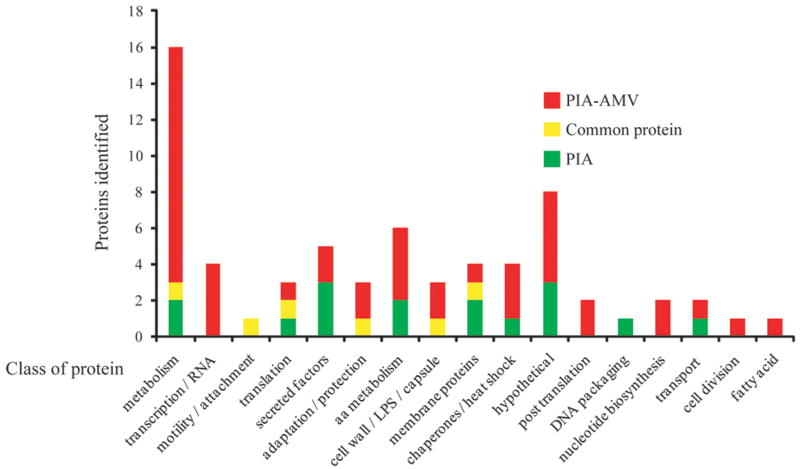
Growth of PAO1 mutants on PIA-AMV was used to directly test the importance of alginate related genes in the overproduction of alginate (Table 1). However, we expected other effects might have occurred that were overlooked by our direct analysis. To determine effects of PIA-AMV on the proteome of PAO1, a shotgun proteomics approach was utilized. Total protein extracts were submitted to Multidimensional Protein Identification Technology (MudPIT). MudPIT analysis involves two liquid-column chromatographic separations followed by MALDI-TOF MS (Matrix-assisted laser desorption-time of flight mass spectrometry) peptide fingerprinting to identify peptides with a complex sample. PAO1 was cultured on PIA and PIA-AMV. Proteins were extracted by lysis using an acid-labile surfactant and MS was performed. 1845 peptides were identified with a cutoff level of 99 % confidence corresponding to 69 unique proteins in the two samples. Fig. 5 shows the various classes of proteins identified and Table 2 shows selected proteins identified in the two samples. Table S2 provides all of the peptides identified in the two samples. Of interest many proteins associated with metabolism were identified in the PIA-AMV sample (Fig. 5). Several potential explanations exist for this observation. It is likely that since PAO1 is mucoid on PIA-AMV, then increased metabolism is necessary for alginate production. Interestingly, a two-component histidine kinase referred to as AruS (PA4982), which has been shown to be associated with the arginine transaminase pathway (Yang & Lu, 2007), has been identified with a high number of peptides in PAO1 cultured on PIA-AMV (Table 2 and Table S2). Observation of this high number of peptides corresponding to AruS, suggests that AMV does alter histidine kinases. AruS has been previously identified as important in early infection of CF airway in a phage display library (Beckmann et al., 2005). It is possible AruS may be a direct target of AMV, but we cannot exclude the possibility that AruS was upregulated as part of the metabolic scheme for increased alginate production.
Figure 5. Distribution of proteins as determined by MudPIT mass spectrometry of PAO1 on PIA versus PIA-AMV.
Proteins were grouped into classes (Winsor et al., 2009) based on the proteins identified by MudPIT MS analysis. Proteins found on PIA and PIA-AMV are indicated in green and red, respectively. Proteins that were common for both samples are indicated in yellow.
Table 2. Selected proteins identified by MudPIT MS from strain PAO1 cultured on PIA and PIA-AMV.
| PA loci | Class | Protein | Product Name | PAO1 on PIA | PAO1 on PIA-AMV | ||
|---|---|---|---|---|---|---|---|
| Peptides | relative % of peptides | Peptides | relative % of peptides | ||||
| PA3540 | cell wall / LPS / capsule | AlgD | GDP-mannose 6-dehydrogenase | 24 | 2.0 | ||
| PA4982 | Metabolism | AruS | two-component sensor kinase | 80 | 6.5 | ||
| PA2976 | transcription / RNA | Rne | ribonuclease E | 72 | 5.9 | ||
| PA1092 | motility / attachment | FliC | flagellin type B | 133 | 21.5 | 62 | 5.1 |
| PA5369 | secreted factors | PstS | periplasmic phosphate-binding protein | 72 | 11.6 | ||
| PA3319 | secreted factors | PlcN | non-hemolytic phospholipase C | 51 | 4.2 | ||
| PA4236 | adaptation / protection | KatA | Catalase | 50 | 8.1 | 36 | 2.9 |
| PA4366 | adaptation / protection | SodB | superoxide dismutase | 11 | 0.9 | ||
| PA2532 | adaptation / protection | Tpx | thiol peroxidase | 10 | 0.8 | ||
| PA1777 | membrane proteins | OprF | structural outer membrane porin | 16 | 2.6 | 35 | 2.9 |
| PA0976 | membrane proteins | OprL | peptidoglycan associated lipoprotein | 13 | 2.1 | ||
| PA1053 | membrane proteins | conserved hypothetical protein | 4 | 0.6 | |||
| PA3280 | membrane proteins | OprO | pyrophosphate-specific outer membrane porin | 6 | 0.5 | ||
| PA4385 | chaperones / heat shock | GroEL | GroEL protein | 29 | 2.4 | ||
| PA4761 | chaperones / heat shock | DnaK | DnaK protein | 19 | 1.5 | ||
| PA4762 | chaperones / heat shock | GrpE | heat shock protein | 17 | 1.4 | ||
| PA1793 | chaperones / heat shock | PpiB | peptidyl-prolyl cis-trans isomerase B | 4 | 0.6 | ||
Since PAO1 growing on PIA-AMV produces alginate, we would predict that AlgD would be identified in the proteome (Table 2). Furthermore, because PAO1 has high AlgU activity on PIA-AMV (Fig. 2A), we would expect flagella would be down regulated since AlgU represses flagella synthesis (Garrett et al., 1999). FliC or flagelin B was observed with less peptides in PAO1 on PIA-AMV than PIA (Table 2).
There were more proteins corresponding to secreted factors observed after growth on PIA than on PIA-AMV (Fig. 5 and Table 2). Interestingly, PstS which is an extracellular phosphate binding protein that is secreted out of the cell as finger like appendages (Zaborina et al., 2008) was only observed in PAO1 grown on PIA and not on PIA-AMV (Table 2 and Table S2). Inversely, phospholipase C was observed only in the PIA-AMV sample and not in the PIA sample (Table 2). Catalase, which is involved in adaptation and protection, was observed on both PIA and PIA-AMV. However, adaptation and protection proteins such as superoxide dismutase and thiol peroxidase were only detected in PAO1 grown on PIA-AMV. SodB or superoxide dismutase has been previously shown to be responsible for resistance to vanadium (Baysse et al., 2000). Growth on PIA-AMV likely caused increased SodB to deal with the antibacterial properties of vanadium.
It is known in E. coli that the DegS protease is activated by misfolded proteins (Ades et al., 1999). Previously when E. coli was cultured in AMV, there was no apparent change in the profiles of outer membrane proteins (OMPs) (Tam & Missiakas, 2005). Our MudPIT proteome data also suggested little change in the total amount of OMPs identified since major outer membrane porin OprF has an equal percent of peptides identified on both PIA and PIA-AMV (Table 2). However, we have previously shown that overexpression of OprF alone does not activate alginate production (Qiu et al., 2008a). Also, OprO was identified in the PIA-AMV sample (Table 2). OprO has a C-terminal sequence (YVF) which can activate alginate production (Qiu et al., 2007). It is possible that increased OprO could activate AlgW resulting in MucA degradation. If PIA-AMV causes general protein misfolding, identification of chaperone proteases would occur in MudPIT MS analysis. A greater amount of the chaperones GroEL, DnaK and GrpE were identified in the PIA-AMV sample than the PIA sample (Table 2). This suggests PIA-AMV may cause protein misfolding which would cause upregulation of chaperones to maintain the cellular integrity. Previously it has been observed that chronically infected CF patients have a high antibody titre against GroEL (Ulanova et al., 1997). Since PIA-AMV seems to upregulate GroEL (Table 2), this further suggests PIA-AMV mimics the CF lung condition. In terms of the other chaperones, DnaK has been shown to have a role in regulated accumulation of heat shock proteins in Pseudomonas putida (Kobayashi et al., 2010). It has not been demonstrated if inactivation of any of these chaperones would activate alginate overproduction. MudPIT analysis revealed that PIA-AMV affects the proteome mainly in the classes of metabolism and chaperones.
Discussion
Alginate overproduction by P. aeruginosa has been extensively examined because this phenotype marks the onset of chronic infection that will ultimately lead to the decline in health of a CF patient. Further complicating this phenotype is the fact that the CF lung environment selects for mutation(s) in P. aeruginosa that alter antibiotic resistance, motility, virulence factors, and many other phenotypes. Little information is known about the early infecting P. aeruginosa strains other than they are nonmucoid and similar to environmental strains. Previously, we suggested that the biodiversity of CF isolates may be a cause for the variation of morbidity and mortality in CF (Head & Yu, 2004). Since most agree the CF lung drives evolution of the initial infecting strains, it could be speculated the best chance for therapeutic intervention is before the emergence of resistant mutants. Therefore, the study of environmental strains and their response to CF conditions is necessary.
It has been shown that alginate production genes are induced before any mutations in mucA have occurred (Bragonzi et al., 2005). This suggests that alginate production may be protective to the colonizing strains in the CF lung. Furthermore, the alginate biosynthetic enzyme gene algD has been shown to be necessary for the development of chronic infections using initially nonmucoid strains (Coleman et al., 2003). Our recent work, as well as the work of others (Cezairliyan & Sauer, 2009, Damron et al., 2009b, Damron & Yu, 2011, Qiu et al., 2008b, Qiu et al., 2007, Wood et al., 2006, Wood & Ohman, 2009a), has detailed that proteolytic cleavage of MucA by AlgW and MucP as a mechanism for alginate production independent of mutation in mucA. Interestingly, in a study which utilized signature tagged mutagenesis, algU and algW were required for chronic infection in a rat model (Potvin et al., 2003). These findings preceded the characterization of AlgW as the regulated protease of MucA and now further support the notion that AlgW and the proteolysis of MucA are important in pathogenesis. Collectively, these studies suggest alginate production may contribute to early infection.
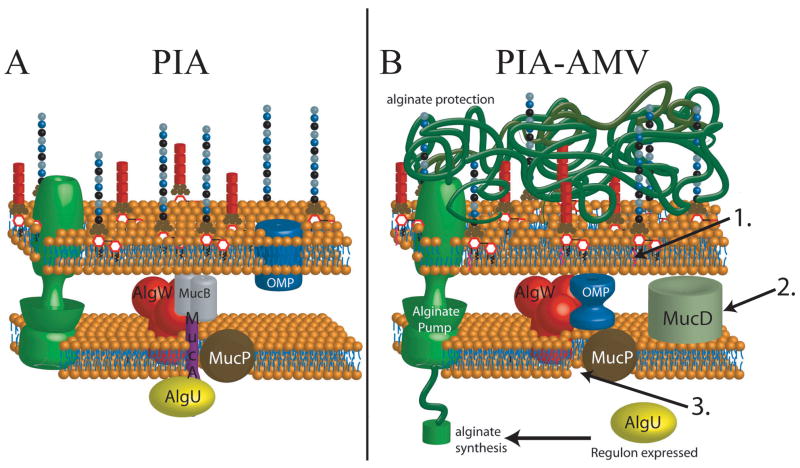
In this study, we have defined a medium (PIA-AMV) which induces mucoidy in PAO1. Cell wall synthesis inhibitors such as triclosan in PIA have been shown to induce expression of alginate genes, but not to a level high enough to cause mucoidy (Wood et al., 2006). Here, we show that PIA-AMV causes alginate overproduction through the AlgW-dependent pathway and utilized most all of the previously described alginate genes as well as other genes not previously associated in alginate overproduction. The PIA-AMV model suggests that modifications to the cellular envelope can activate the AlgW-dependent pathway for alginate production (Fig. 6). However, is PIA-AMV a relevant condition or an artificial lab condition? PIA-AMV medium caused palmitoylation of lipid A (Fig. 4) and hexa-acylated lipid A, which we observed here in this study, has also been previously observed in chronic P. aeruginosa isolates from the airways of CF patients (Ernst et al., 1999). It has been suggested that palmitoylation of lipid A could create a situation where β-barrel OMPs are not inserted into the outer leaflet (Bishop, 2008, Tam & Missiakas, 2005), resulting in the activation of the σE system. It has been proposed that the E. coli σE system functions as the ‘canary in a coal mine’(Alba & Gross, 2004). It is possible that the AlgU system functions the same way downstream as σE, but responds to different environmental cues by overproducing alginate to remedy the situation and maintain homeostasis. It is also possible that this response is for protection from stress agents (Fig. 6). Since AlgU is upregulated and proteolysis of MucA is activated, periplasmic housekeeping chaperones should be increased in expression on PIA-AMV. If OMPs are not being properly inserted or folded, then we would expect chaperones would be upregulated to compensate. MudPIT MS analysis of the proteome of PAO1 cultured on PIA-AMV suggests that chaperones are upregulated on PIA-AMV (Table 2). From these data, we may also consider the increased expression of chaperone/protease MucD indicates that the periplasmic protein quality may be affected by growth on PIA-AMV. Our data suggest a model where outer leaflet changes can activate the AlgW-dependent pathway (Fig. 6). As mentioned above, we observed lipid A modification when PAO1 is cultured on PIA-AMV (Fig. 4). In spite of this data, we do not know if palmitoylation of lipid A is directly responsible for the activation of alginate production. Regulation of the lipid A modification transcription factor phoP is required for mucoid phenotype of PIA-AMV (Table 1). Interestingly, the same lipid A modifications observed on PIA-AMV also occur in CF isolates (Ernst et al., 1999). Here we also looked at O-antigen expression and did not observe changes in A- or B-band O antigen due to PIA-AMV (Fig. S3)
Figure 6. Model for PIA-AMV induction of alginate production in P. aeruginosa with wt MucA.
A. On PIA, PAO1 is nonmucoid due to AlgU being sequestered by anti-sigma factor MucA. B. PIA-AMV employs ammonium metavanadate, which induces synthesis of pamitoylated lipid A (1.). The combination of AMV and triclosan (a cell wall stress agent) induces alginate overproduction. It may be possible pamitoylated lipid A is a side effect of AMV induction. However, without phoP, which controls these modifications, alginate production does not occur (Table 1). This condition induces increased expression of chaperone protease MucD (2.) and AlgW-regulated proteolysis of MucA (3.). Based on the data presented here, we propose that it is likely that MucD is upregulated along with other chaperones because OMPs are misfolded due to the modified lipid A and the cell wall stress caused by triclosan. Regulated proteolysis of MucA leads to activation of AlgU (3.) and alginate production for protection due to the perturbations in the envelope.
Our data suggest the PIA-AMV medium induces clinically relevant phenotypes, but is there vanadate in the CF lung environment? More specifically, is there enough vanadate and stress to activate alginate overproduction? Vanadium is commonly found in minerals such as sandstone, coal, shale, crude oil, and tar. It is therefore conceivable that all humans come in contact with this element. Vanadium is the least abundant element in the human body and in a healthy 70 kg person there is about 0.11 mg (Emsley, 1988). However, vanadium accumulates in the human lung with age and can reach a level of 6.5 μg g-1 in elderly people due to decreased alveolar and mucociliary clearance (Waters, 1977). To our knowledge, it is not known how much vanadium is in the CF lung. However, it is known that the CF lung has decreased mucociliary clearance. Assuming the CF lung has the same amount of vanadium as an elderly person, how does that compare to the amount of vanadium in PIA-AMV? Fortuitously, PIA-AMV medium contains 6.1 μg g-1 vanadium, which is nearly equivalent to the amount found in elderly humans. However, it is not clear if compounds such as triclosan exist in the lung that would inhibit fatty acid synthesis, but there is little to argue that the CF lung condition applies stress to P. aeruginosa. Of additional interest, Ascidians, otherwise known as sea squirts, are creatures that accumulate massive amounts of vanadium in vanadocytes (vanadium-containing blood cells). Recently, a chloride channel has been identified that participates in transport of vanadium from seawater into vanadocytes. The chloride channel of vanadocytes is similar to the mammalian CIC3/4/5-type chloride channels (Ueki et al., 2003). This is interesting because the defect in CF is the cystic fibrosis transmembrane conductance regulator which is also a chloride ion channel. Collectively these data from the literature, combined with our data presented here, suggest that PIA-AMV may mimic a clinically relevant phenotype of P. aeruginosa.
CF patients are typically colonized with nonmucoid P. aeruginosa (Govan & Deretic, 1996) that would theoretically resemble the laboratory strain PAO1, producing minimal amounts of alginate. However, the pathophysiological significance associated with the phenotypic changes from initial colonizing nonmucoid, to hypothetical temporary mucoid, to the constitutively mucoid is not fully understood. Because PIA-AMV induces alginate production in nonmucoid strains with algU+ mucA+, this medium will allow examination of the dynamics and distribution of the infecting population of P. aeruginosa in CF. It is not clear at what stage of the infection the algU mutant emerges, and whether the emergence and/or dominance of this mutant correlates with an acute exacerbation in CF. Since a wt algU, encoding a functional AlgU is required for alginate induction on PIA-AMV (Table 1), it may be possible to quickly screen isolates for mutations in algU. This is of importance because mutation of algU results in increased virulence (Yu et al., 1996). Surveillance of CF patient sputa for the emergence of algU mutants via the cultivation on PIA-AMV may provide vital information about the P. aeruginosa infecting population to the clinician. The PIA-AMV medium creates a condition that causes alginate overproduction and lipid A modification that have been observed in CF isolates. We are currently testing P. aeruginosa non-redundant mutant libraries on PIA-AMV to determine all of the genes required for alginate production. It will also be interesting to see the effect of PIA-AMV on the transcriptome of P. aeruginosa. In conclusion, we suggest PIA-AMV medium can serve as an in vitro model to address questions regarding the initial interaction of P. aeruginosa and the CF airway.
Experimental procedures
Bacterial strains, media, growth conditions, and oligonucleotides
Bacterial strains and plasmids used in this study are indicated in Table S1. To clone miniCTX1-PalgU-algU-HAmucA as shown in Fig. S2, splicing by overlap PCR (SOE-PCR) was used to generate the DNA fragment that was ligated into miniCTX1 to be shuttled to the PAO1 chromosome. A meridiploid strain was generated by integrating the HA-tagged mucA into the attB attachment site of the PAO1 chromosome. PCR and sequencing were used to show that the HA-mucA construct was properly integrated into the attB site (data not shown). Since the HA-mucA was expressed from a copy of the algU promoters, HA-MucA expression is physiologically similar to wt MucA of that strain. This system allows for better detection of changes in MucA concentration since HA-MucA is not expressed from a multicopy plasmid. P. aeruginosa strains were grown at 37° C in lysogeny broth Lennox formulation (10g tryptone, 5 g yeast extract, 5 g NaCl) (LB), on LB agar or Pseudomonas isolation agar plates (PIA; Difco) or on PIA-AMV. PIA plates were prepared containing 20 mL of glycerol per liter as recommended by the manufacturer. PIA-AMV plates were prepared as follows. One liter of PIA medium with 20 mL of glycerol was autoclaved and cooled. 20 mL of distilled water was boiled with a stir bar to which 50 mg of ammonium metavanadate (Sigma-Aldrich) was added. After several minutes the AMV dissolved and tinted the liquid yellow. 12.5 mL of the 2.5 mg mL-1 AMV solution was then added to the molten PIA to make PIA-AMV with AMV at a concentration of 0.27 mM. For contrast in digital images shown in Fig. 1A, Congo Red (Sigma-Aldrich) was added at 20 mg L-1. Oligonucleotides used in this study are indicated in Table S1.
Carbozole determination of uronic acid content
P. aeruginosa strains were grown at 37°C on PIA or PIA-AMV for 24hr. Bacteria and alginate were removed from plates with PBS and suspended in 100 mL of PBS per plate. The optical density at OD600 of bacterial suspension in PBS was measured and adjusted. Cells were then cleared from suspension by centrifugation. The supernatants containing bacterial alginates combined with 3 volumes of cold ethanol (99%), mixed thoroughly to precipitate the alginate fibers that were removed through filtration by 10 μM titanium dutch weave filters (Dorstener Wire Tech). The collected alginate was used for carbozole assay for the amounts of the uronic acid using a standard curve made with D-mannuronic acid lactone (Sigma-Aldrich) in the range of 0-100 μg mL-1 as described (Knutson & Jeanes, 1968).
Analysis of bacterial alginate via nuclear magnetic resonance (NMR)
Cells and alginate were harvested by scraping the colonies off PIA-AMV plates and re-suspended in phosphate buffered saline (PBS). Cell pellets were removed by centrifugation, and the supernatants were combined with 3 volumes of cold ethanol (99%), mixed thoroughly to precipitate the alginate fibers that were removed through filtration by 10 μM titanium Dutch weave filters (Dorstener Wire Tech). The alginate fibers were then dried under a vacuum. The 1H–NMR analysis of the seaweed and bacterial alginates were performed according to the protocol F2259-03 of the American Society for Testing and Materials (ASTM) International. Alginic acid, from brown seaweed, (Sigma-Aldrich Catalog A7003) was used as a standard. To determine the composition and sequence of alginates by 1H-NMR, the alginate samples (∼10 mg) were dissolved in 10 mL distilled water with pH adjusted to 5.6 with 1 M HCl and incubated at 100°C for 1 h. Next the pH was reduced to 3.8 followed by incubation at 80°C for 30 min. After the two-step acid hydrolysis, the pH was elevated to 7.5 with 1 M NaOH before the samples were subject to freeze-drying. The dried samples were re-dissolved in 1 mL 99.9 % D2O, and underwent 3 additional cycles of freeze-drying were performed to remove the excess exchangeable protons. The samples were solubilized in 0.5 mL 99.96% D2O, transferred to a NMR tube and analyzed by high-field 1H-NMR spectroscopy at 80°C using a JEOL JNM-ECP600 (600-MHz) spectrometer at 378K. Acetone-d6 (2.04 ppm) was used as an internal standard in the samples. The composition, expressed as percent of monomers G (FG) and M (FM) was determined from the integration of the relevant 1H-NMR spectra. The area of each peak, which is proportional to the amount of residues giving rise to the signal, was used to calculate the above parameters.
β-galactosidase activity assay
The promoter fusion constructs miniCTX-PalgU-lacZ and miniCTX-PalgD-lacZ were integrated onto the P. aeruginosa chromosome at the CTX phage att site (Hoang et al., 2000). β-galactosidase activity assay was based on the method as originally described by Miller (Miller, 1972) with the following modifications. The cells were grown on PIA and PIA-AMV plates in triplicate for 24hr at 37° C and harvested in PBS. The β-galactosidase activity was assayed after toluene permeabilization of the cells. The reported values represent the average in triplicate of three independent experiments. The values displayed are normalized to PAO1 cultured on PIA medium.
Western blot analysis of proteins
Total cell lysates were prepared with Protea Preps (Protea Biosciences) by manufacturer's protocol. Protein lysates were quantified by DC assay (BIO-RAD). 50 μg of protein was boiled in SDS-loading buffer. The samples were electrophoresed on 15% ProteaGel (Protea Biosciences) SDS-PAGE polyacrylamide and then electro blotted (Trans-Blott Cell, BIO-RAD) on to 0.45μm nitrocellulose. The membrane was blocked with 3% non-fat dry milk in PBS (pH 7.4). Primary antibodies were diluted 1:5000 in 3% non-fat dry milk in PBS. The membranes were probed with mouse monoclonal antibodies against alpha RNA subunit polymerase subunit (Neoclone) and rat monoclonal antibody against HA (Roche) or anti-MucD (Wood & Ohman, 2006) overnight at 4°C with shaking. HRP-labeled goat anti-mouse IgG or HRP-labeled anti-rabbit IgG were diluted 1:10000 in 3% non-fat dry milk in PBS and used as the secondary antibodies. Advanced ECL or ECL chemiluminescence (Amersham Biosciences) was used for detecting HRP-labeled goat anti-mouse IgG or anti-rabbit IgG (Roche) by the manufacturer's protocol. The signals were detected with an EC3 Imaging System (UVP) by capturing with a BioChemi HR camera (UVP) or by exposure to Amersham Hyperfilm ECL (Amersham Biosciences). For re-probing, membranes were stripped with 62.5 mM Tris-HCl pH 6.8, 2% SDS, 100 mM β-mercaptoethanol for 15 min at 50°C and then washed in PBS.
Isolation of LPS
LPS was isolated using a rapid small-scale isolation method (Yi & Hackett, 2000). In brief, 1.0 mL of Tri-reagent (Molecular Research Center) was added to cell culture pellets resuspended in 10 mL LB and incubated at room temperature for 15 minutes. 200 μL of chloroform was added, vortexed, and incubated for an additional 15 minutes. Samples were centrifuged for 10 min at 13,500 × g and the aqueous layer was removed. Five hundred μL of water was added to the lower organic layer and vortexed. After 15 to 30 minutes, samples were centrifuged again and the aqueous layers were combined. This process was repeated two more times. The combined aqueous layers were frozen and lyophilized overnight.
In order to hydrolyze the lipid A from the LPS, 500 μL of 1% (wt/vol) SDS in 10 mM Na-acetate at pH 4.5 was added to lyophilized sample. Samples were incubated at 100°C for 1 hour, frozen, and lyophilized. The samples were then washed with 100 μL water and 1 mL of acidified ethanol (100 μL 4M HCl in 20 mL 95% ethanol). Samples were centrifuged at 2,350 × g for 5 minutes and ethanol removed. The pellet was washed three times in 1 mL of 95% ethanol and the entire series of washes repeated twice. Samples were resuspended in 500 μL of water, frozen and lyophilized.
Mass spectrometry procedures for lipid A
Negative-ion matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) experiments were performed as previously described with the following modifications (Ernst et al., 1999, Guo et al., 1998). Lyophilized lipid A was dissolved in 10 μL 5-chloro-2-mercaptobenzothiazole (CMBT) MALDI matrix in chloroform/methanol 1:1 (v/v), and then 1 μL applied onto the sample plate. All MALDI-TOF experiments were performed using a Bruker Autoflex II MALDI-TOF mass spectrometer (Bruker Daltonics Inc.). Each spectrum was an average of 300 shots. ES tuning mix (Agilent) was used to calibrate the MALDI-TOF MS.
Total protein preparation for MS
Total protein preparations of PAO1 cultured on PIA and PIA-AMV were obtained by processing protein lysates with ProteaPrep MS Cell Lysis kit (Protea Biosciences), per manufacturer's instructions. Protea MS Preps use Progenta anionic acid surfactant II to disrupt membranes. The surfactant is then inactivated by acidification with trifluoroacetic acid as instructed by the manufacturer. The lack of SDS or other detergents increases the sensitivity of downstream mass spectrometry analysis. Protein concentrations were determined using Bio-Rad DC Protein Assay. SDS-PAGE (14% polyacrylamide) with Coomassie staining was performed to determine the complexity of the samples and to insure protease degradation had not occurred to the samples.
Multidimensional Protein Identification Technology (MudPIT) mass spectrometric analysis
To identify the proteins present in P. aeruginosa total cell lysates, MudPIT with tandem MALDI TOF-TOF mass spectrometry was employed. Total protein samples (50μg) were desalted using C4 ProteaTip SpinTips (Protea Biosciences) as per manufacturer's protocol. The collected samples were lyophilized and dissolved in 100μl of 50mM ammonium bicarbonate in 20% acetonitrile for tryptic digestion. The samples were then reduced and alkylated with 10μL of 250mM DTT (60min/55°C), and 10μl of 625mM iodoacetamide (60min/room temperature/in the dark). Proteolytic digestion was performed in 50mM ammonium bicarbonate buffer using a trypsin to protein ratio of 1:100. The digestion was carried out overnight at 37°C. The digests were cleaned by repeated lyophilizing and reconstituting in a 0.1M acetic acid solution. After final lyophilization, the digests were reconstituted in a strong cation exchange loading buffer (5mM ammonium formate in 20% acetonitrile, pH 3.0) to be fractionated with ProteaTip spin tips as per manufacturer's protocol. The SpinTip was transferred to a new centrifuge tube to collect the sample during elution with 200μL of elution solution. Eight different elution solutions were used fractionate the peptides (20, 60, 100, 150, 200, 250, 400, 500 mM ammonium formate in 20% acetonitrile) in a step-wise manner. The collected fractions were cleaned by repeated lyophilization and reconstitution in a 0.1M acetic acid solution. After the final lyophilization, the digests were reconstituted in LC run buffer. The fractions were then submitted to LC MALDI spotting and MALDI TOF/TOF spectral analysis by Protea Biosciences as detailed previously (Damron et al., 2009a) to survey the proteome of the P. aeruginosa samples. Peptides corresponding to proteins were counted and reported as the relative percent of peptides identified in the sample. The classifications of each proteins indicated were derived from the Pseudomonas Genome Database (Winsor et al., 2009).
Supplementary Material
Acknowledgments
F.H.D. was supported by grants from the NASA Graduate Student Researchers Program (NNX06AH20H), NASA West Virginia Space Grant Consortium, and a post-doctoral fellowship from the Cystic Fibrosis Foundation (DAMRON10F0). M.R.D was partially supported by NIH through the University of Virginia Infectious Disease training grant AI07406. T.R.W. was supported by the NASA West Virginia Space Grant Consortium. R.K.R was supported by NIH R01 AI047938. J.B.G was supported by NIH R01 AI068112. G. Y. was supported by NSFC (31070724), PCSIRT (IRT0944), and Special Fund for Marine Scientific Research in the Public Interest of China (201005024). H.D.Y. was supported by research grants from the NASA West Virginia Space Grant Consortium, NIH P20 RR016477 to the West Virginia IDeA Network for Biomedical Research Excellence and the Cystic Fibrosis Foundation (YU11G0). This work was also partially funded by Progenesis Technologies, LLC.
We thank Dongru Qiu for generation of strains, Megan Luciano for technical assistance, Erica Kintz for conjugation of the A-band antibody, and Matt Powell of Protea Biosciences for assistance with the proteomic analysis. We also would like to thank Dennis Ohman of Virginia Commonwealth University for the MucD antibodies. We also posthumously acknowledge Jin Lei of Zhejiang University, China for her help with the promoter activity assays. We would also like to thank the anonymous reviewers for their helpful suggestions and critiques.
References
- Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J Bacteriol. 2006;188:132–140. doi: 10.1128/JB.188.1.132-140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysse C, De Vos D, Naudet Y, Vandermonde A, Ochsner U, Meyer JM, Budzikiewicz H, Schafer M, Fuchs R, Cornelis P. Vanadium interferes with siderophore-mediated iron uptake in Pseudomonas aeruginosa. Microbiology. 2000;146:2425–2434. doi: 10.1099/00221287-146-10-2425. [DOI] [PubMed] [Google Scholar]
- Beckmann C, Brittnacher M, Ernst R, Mayer-Hamblett N, Miller SI, Burns JL. Use of phage display to identify potential Pseudomonas aeruginosa gene products relevant to early cystic fibrosis airway infections. Infect Immun. 2005;73:444–452. doi: 10.1128/IAI.73.1.444-452.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop RE. Structural biology of membrane-intrinsic beta-barrel enzymes: sentinels of the bacterial outer membrane. Biochim Biophys Acta. 2008;1778:1881–1896. doi: 10.1016/j.bbamem.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop RE, Kim SH, El Zoeiby A. Role of lipid A palmitoylation in bacterial pathogenesis. J Endotoxin Res. 2005;11:174–180. doi: 10.1179/096805105X35242. [DOI] [PubMed] [Google Scholar]
- Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JC, Schurr MJ, Deretic V. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol Microbiol. 2000;36:341–351. doi: 10.1046/j.1365-2958.2000.01846.x. [DOI] [PubMed] [Google Scholar]
- Boucher JC, Schurr MJ, Yu H, Rowen DW, Deretic V. Pseudomonas aeruginosa in cystic fibrosis: role of mucC in the regulation of alginate production and stress sensitivity. Microbiology. 1997;143:3473–3480. doi: 10.1099/00221287-143-11-3473. [DOI] [PubMed] [Google Scholar]
- Bragonzi A, Worlitzsch D, Pier GB, Timpert P, Ulrich M, Hentzer M, Andersen JB, Givskov M, Conese M, Doring G. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J Infect Dis. 2005;192:410–419. doi: 10.1086/431516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezairliyan BO, Sauer RT. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol. 2009;72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuanchuen R, Karkhoff-Schweizer RR, Schweizer HP. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am J Infect Control. 2003;31:124–127. doi: 10.1067/mic.2003.11. [DOI] [PubMed] [Google Scholar]
- Coleman FT, Mueschenborn S, Meluleni G, Ray C, Carey VJ, Vargas SO, Cannon CL, Ausubel FM, Pier GB. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc Natl Acad Sci U S A. 2003;100:1949–1954. doi: 10.1073/pnas.0437901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne MJ, Jr, Russell KS, Coyle CL, Goldberg JB. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J Bacteriol. 1994;176:3500–3507. doi: 10.1128/jb.176.12.3500-3507.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Napper J, Teter MA, Yu HD. Lipotoxin F of Pseudomonas aeruginosa is an AlgU-dependent and alginate-independent outer membrane protein involved in resistance to oxidative stress and adhesion to A549 human lung epithelia. Microbiology. 2009a;155:1028–1038. doi: 10.1099/mic.0.025833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Qiu D, Yu HD. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J Bacteriol. 2009b;191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Yu HD. Pseudomonas aeruginosa MucD regulates alginate pathway through activation of MucA degradation via MucP proteolytic activity. J Bacteriol. 2011;193:286–291. doi: 10.1128/JB.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Dikshit R, Konyecsni WM, Chakrabarty AM, Misra TK. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989;171:1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries CA, Ohman DE. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley J. The Elements. Clarendon Press; Oxford: 1988. [Google Scholar]
- Ernst RK, Adams KN, Moskowitz SM, Kraig GM, Kawasaki K, Stead CM, Trent MS, Miller SI. The Pseudomonas aeruginosa lipid A deacylase: selection for expression and loss within the cystic fibrosis airway. J Bacteriol. 2006;188:191–201. doi: 10.1128/JB.188.1.191-201.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Moskowitz SM, Emerson JC, Kraig GM, Adams KN, Harvey MD, Ramsey B, Speert DP, Burns JL, Miller SI. Unique lipid a modifications in Pseudomonas aeruginosa isolated from the airways of patients with cystic fibrosis. J Infect Dis. 2007;196:1088–1092. doi: 10.1086/521367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, Miller SI. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- Firoved AM, Boucher JC, Deretic V. Global genomic analysis of AlgU (sigma(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J Bacteriol. 2002;184:1057–1064. doi: 10.1128/jb.184.4.1057-1064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved AM, Deretic V. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J Bacteriol. 2003;185:1071–1081. doi: 10.1128/JB.185.3.1071-1081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved AM, Ornatowski W, Deretic V. Microarray analysis reveals induction of lipoprotein genes in mucoid Pseudomonas aeruginosa: implications for inflammation in cystic fibrosis. Infect Immun. 2004a;72:5012–5018. doi: 10.1128/IAI.72.9.5012-5018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved AM, Wood SR, Ornatowski W, Deretic V, Timmins GS. Microarray analysis and functional characterization of the nitrosative stress response in nonmucoid and mucoid Pseudomonas aeruginosa. J Bacteriol. 2004b;186:4046–4050. doi: 10.1128/JB.186.12.4046-4050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett ES, Perlegas D, Wozniak DJ. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU) J Bacteriol. 1999;181:7401–7404. doi: 10.1128/jb.181.23.7401-7404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JB, Gorman WL, Flynn JL, Ohman DE. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J Bacteriol. 1993;175:1303–1308. doi: 10.1128/jb.175.5.1303-1308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooderham WJ, Gellatly SL, Sanschagrin F, McPhee JB, Bains M, Cosseau C, Levesque RC, Hancock RE. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology. 2009;155:699–711. doi: 10.1099/mic.0.024554-0. [DOI] [PubMed] [Google Scholar]
- Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- Hancock RE, Mutharia LM, Chan L, Darveau RP, Speert DP, Pier GB. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head NE, Yu H. Cross-Sectional Analysis of Clinical and Environmental Isolates of Pseudomonas aeruginosa: Biofilm Formation, Virulence, and Genome Diversity. Infect Immun. 2004;72:133–144. doi: 10.1128/IAI.72.1.133-144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Kutchma AJ, Becher A, Schweizer HP. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- Knutson CA, Jeanes A. A new modification of the carbazole reaction: application to heteropolysaccharides. Anal Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Ohtsu I, Fujimura M, Fukumori F. A mutation in dnaK causes stabilization of the heat shock sigma factor sigma(32), accumulation of heat shock proteins and increase in toluene-resistance in Pseudomonas putida. Environ Microbiol. 2010 doi: 10.1111/j.1462-2920.2010.02344.x. [DOI] [PubMed] [Google Scholar]
- Leech AJ, Sprinkle A, Wood L, Wozniak DJ, Ohman DE. The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J Bacteriol. 2008;190:581–589. doi: 10.1128/JB.01307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol. 1999;34:305–316. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]
- Martin DW, Holloway BW, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993a;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Mudd MH, Deretic V. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol Microbiol. 1993b;9:497–506. doi: 10.1111/j.1365-2958.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A. 1993c;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, Hoiby N, Kharazmi A. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. beta-galactosidase assay. In: Miller JH, editor. Experiments in molecular genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- Mohr CD, Hibler NS, Deretic V. AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J Bacteriol. 1991;173:5136–5143. doi: 10.1128/jb.173.16.5136-5143.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz SM, Ernst RK. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Subcell Biochem. 2010;53:241–253. doi: 10.1007/978-90-481-9078-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman DE, Chakrabarty AM. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin E, Lehoux DE, Kukavica-Ibrulj I, Richard KL, Sanschagrin F, Lau GW, Levesque RC. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol. 2003;5:1294–1308. doi: 10.1046/j.1462-2920.2003.00542.x. [DOI] [PubMed] [Google Scholar]
- Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. PBAD-based shuttle vectors for functional analysis of toxic and highly-regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol. 2008a;74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology. 2008b;154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Eisinger VM, Rowen DW, Yu HD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology. 2005;151:2251–2261. doi: 10.1099/mic.0.27772-0. [DOI] [PubMed] [Google Scholar]
- Schurr MJ, Deretic V. Microbial pathogenesis in cystic fibrosis: co-ordinate regulation of heat-shock response and conversion to mucoidy in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:411–420. doi: 10.1046/j.1365-2958.1997.3411711.x. [DOI] [PubMed] [Google Scholar]
- Schurr MJ, Yu H, Boucher JC, Hibler NS, Deretic V. Multiple promoters and induction by heat shock of the gene encoding the alternative sigma factor AlgU (sigma E) which controls mucoidy in cystic fibrosis isolates of Pseudomonas aeruginosa. J Bacteriol. 1995;177:5670–5679. doi: 10.1128/jb.177.19.5670-5679.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh SJ, Silo-Suh L, Woods DE, Hassett DJ, West SE, Ohman DE. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam C, Missiakas D. Changes in lipopolysaccharide structure induce the sigma(E)-dependent response of Escherichia coli. Mol Microbiol. 2005;55:1403–1412. doi: 10.1111/j.1365-2958.2005.04497.x. [DOI] [PubMed] [Google Scholar]
- Tart AH, Wolfgang MC, Wozniak DJ. The Alternative Sigma Factor AlgT Represses Pseudomonas aeruginosa Flagellum Biosynthesis by Inhibiting Expression of fleQ. J Bacteriol. 2005;187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry JM, Pina SE, Mattingly SJ. Environmental conditions which influence mucoid conversion Pseudomonas aeruginosa PAO1. Infect Immun. 1991;59:471–477. doi: 10.1128/iai.59.2.471-477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T, Yamaguchi N, Michibata H. Chloride channel in vanadocytes of a vanadium-rich ascidian Ascidia sydneiensis samea. Comp Biochem Physiol B Biochem Mol Biol. 2003;136:91–98. doi: 10.1016/s1096-4959(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Ulanova M, Petersen TD, Ciofu O, Jensen P, Hahn-Zoric M, Hanson LA, Hoiby N. The clonal antibody response to Pseudomonas aeruginosa heat shock protein is highly diverse in cystic fibrosis patients. APMIS. 1997;105:449–456. [PubMed] [Google Scholar]
- Waters M. In: Toxicology of vanadium. Toxicology of trace elements. Goyer RA, Mehlman MA, editors. Wiley; New York: 1977. [Google Scholar]
- Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock RE, Brinkman FS. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 2009;37:D483–488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol. 2006;62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- Wood LF, Ohman DE. Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J Bacteriol. 2006;188:3134–3137. doi: 10.1128/JB.188.8.3134-3137.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LF, Ohman DE. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol. 2009a;72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- Wood LF, Ohman DE. Use of cell wall stress to characterize sigma(22) (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol. 2009b doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- Wozniak DJ, Ohman DE. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol. 1991;173:1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lu CD. Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa. J Bacteriol. 2007;189:3945–3953. doi: 10.1128/JB.00261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi EC, Hackett M. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst. 2000;125:651–656. doi: 10.1039/b000368i. [DOI] [PubMed] [Google Scholar]
- Yu H, Boucher JC, Hibler NS, Deretic V. Virulence properties of Pseudomonas aeruginosa lacking the extreme-stress sigma factor AlgU (sigmaE) Infect Immun. 1996;64:2774–2781. doi: 10.1128/iai.64.7.2774-2781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborina O, Holbrook C, Chen Y, Long J, Zaborin A, Morozova I, Fernandez H, Wang Y, Turner JR, Alverdy JC. Structure-function aspects of PstS in multi-drug-resistant Pseudomonas aeruginosa. PLoS Pathog. 2008;4:e43. doi: 10.1371/journal.ppat.0040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Lin S, Cotter RJ, Raetz CR. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- Zhu L, Lin J, Ma J, Cronan JE, Wang H. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob Agents Chemother. 2010;54:689–698. doi: 10.1128/AAC.01152-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski NA, Chakrabarty AM, Berry A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J Biol Chem. 1991;266:9754–9763. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.