Abstract
Hematopoietic stem cells recipients remain susceptible to opportunistic viral infections including herpes simplex virus type-1 (HSV-1). The purpose of this investigation was to analyze susceptibility of human mesenchymal stem cells (hMSCs) to HSV-1 infection and identify the major entry receptor. Productive virus infection in hMSCs was confirmed by replication and plaque formation assays using a syncytial HSV-1 KOS (804) virus. To examine the significance of entry receptors, RT-PCR and antibody-blocking assays were performed. RT-PCR data showed the expression of gD receptors: nectin-1, 3-O sulfotransferase-3 (3-OST-3), and HVEM. Antibody-blocking assay together with heparinase treatment suggested an important role for HS and 3-O-sulfated heparan sulfate (3-OS HS), but not nectin-1 or HVEM, in mediating HSV-1 entry and spread in hMSCs. Taken together, our results provide strong evidence demonstrating that HSV-1 is capable of infecting hMSCs and HS and 3-OS HS serve as its entry receptors during this process.
1. Introduction
Herpes simplex virus type-1 (HSV-1) is a ubiquitous pathogen associated with facial cold sores [1, 2], but severe complications especially in neonates and immunocompromised patients may result in corneal blindness, retinitis, and encephalitis [3, 4]. The virus can also cause severe opportunistic infections in the patients following allogeneic stem cell transplant [5–8]. Such infections could be donor derived or due to the reactivation of an endogenous latent virus [9]. In addition, the drug-resistant HSV-1 is also an emerging concern to promote viral infections after allogeneic stem cell transplantation [10]. It has been proposed that human mesenchymal stem cells (hMSCs) may be susceptible to infection if infused in patients with HSV-1 viremia and may compromise the host's defense against infectious agents [7, 11].
HSV-1 infects host cells through initial attachment to cell surface heparan sulfate (HS) followed by fusion of the virion envelope with the plasma membrane of the host cells [12, 13]. The current model suggests that entry of virus requires four HSV glycoproteins (gB, gD, gH, and gL) [14–19] and at least one cellular receptor for glycoprotein D (gD) [20–22]. The receptors for HSV-1 gD include a member of the TNF-receptor family named HVEM [23, 24], a member of the immunoglobulin superfamily commonly known as nectin-1 [25, 26] and 3-O sulfated heparan sulfate (3-OS HS) [27]. The later form of HS is enzymatically modified by multiple D-glucosaminyl 3-O-sulfotransferase (3OST) isoforms. Among the known 3-OSTs isoforms all mediate HSV-1 entry [27–31] and cell-to-cell fusion [22, 32] except 3-OST-1 isoform.
Although HSV-1 infections lead to significant morbidity and mortality in transplant recipient [6, 10, 11], the susceptibility of human mesenchymal stems cells (hMSCs) to HSV-1 entry and key entry mediator that allows HSV-1 infection is poorly understood. In this study, we investigated the HSV-1 infection of hMSCs and the role of HSV-1 entry receptors in this process. The finding that HSV-1 can induce cytopathic effect and productively replicate in hMSCs in high titer provides new information on the susceptible nature of target cell population present within the host. We also demonstrated that HS and modified form of HS (3-O-sulfated heparan sulfate; 3-OS HS) is a major determinant of HSV-1 entry and spread into hMSCs.
2. Materials and Methods
2.1. Cell Culture and Viruses
Wild-type Chinese hamster ovarian-K1 (CHOK1) cells were grown in Ham's F-12 medium (Gibco/BRL, Carlsbad, Calif, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen), and penicillin and streptomycin (Gibco/BRL). HeLa cells were grown in DMEM media supplemented with 10% fetal bovine serum (Invitrogen), and penicillin and streptomycin (Gibco/BRL). African green monkey kidney (Vero cells) were grown as previously described [33]. Human mesenchymal stem cells (hMSCs) were purchased from Fisher Scientific (SV3010201) grown in Hyclone Advance STEM expansion media (Fisher Scientific, SH30875KT). Passage 4 hMSCs were used in all experiments described.
2.2. Plasmids
The HSV-1 (KOS) glycoprotein expressing plasmids used were pPEP98 (gB), pPEP99 (gD), pPEP100 (gH), and pPEP101 (gL) [18]. Other plasmids used in this study include pCalifGT7 (T7 RNA polymerase) and pT7EMCLuc (luciferase gene).
2.3. HSV-1 Entry Assay
Viral entry assays were based on quantitation of β-galactosidase expressed from the viral genome [27]. Cells (hMSCs, HeLa) were plated at 2 × 104 per well in 96-well plates at least 16 h prior to infection. After 6-h postinfection, β-galactosidase assays were performed using a soluble substrate o-nitrophenyl-β-D-galactopyranoside (ONPG at 3.0 mg/mL) assay. The enzymatic activity was measured at 410 nm using a microplate reader (BioTek Instruments Inc. EL×808 absorbance microplate reader, Vt, USA). Viral entry was further confirmed by 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) staining. The hMSCs were grown in Lab-Tek chamber slides. After 6 hr of infection with the reporter virus, the cells were washed with PBS and fixed with 2% formaldehyde and 0.2% glutaradehyde at room temperature for 15 min. The cells were then washed with PBS and permeabilized with 2 mM MgCl2, 0.01% deoxycholate and 0.02% nonidet NP-40 for 15 min. After rinsing with PBS, 1.5 mL of 1.0 mg/mL X-gal in ferricyanide buffer was added to each well, and the blue color developed in the cells was examined. Microscopy was performed using 40x objective of the inverted microscope (Olympus IX50).
2.4. Fluorescent Microscopy of Viral Entry
Cultured monolayers of hMSCs were grown overnight in media containing 10% FBS on chamber slides (Lab-Tek). The cells were either mock infected or infected with green fluorescent protein (GFP)-tagged HSV-1 (VP26-GFP) at 50 pfu/cell in serum-free media OptiMEM (Invitrogen). The GFP was fused with the HSV-1 UL35 open reading frame (ORF) which encodes a 12-kDa capsid protein designated VP26 [34]. VP26 is located on the outer surface of the capsid, specifically on the tips of the hexons that constitute the capsid shell. At 90 min post-infection, cells were fixed and stained with 10 nm rhodamine-conjugated phalloidin for F-actin. Images of immunofluorescent labeled cells were acquired using a Olympus IX50 inverted fluorescence microscope.
2.5. Effect of Actin Depolymerizing Agents on HSV-1 Entry in hMSCs
In order to demonstrate the significance of actin cytoskeleton network during HSV-1 entry into hMSCs, the effects of actin depolymerizing agents on entry of HSV into hMSCs were examined. Monolayer cultures of hMSCs (approximately 106 cells) cultured in a 96-well plate were pretreated with the indicated concentrations of agents for 1 hr at room temperature with cytochalasin D (Cyto D) and latrunculin B (Lat B) (Sigma) or mock-treated as a control. Cells were infected with the lacZ+ HSV-1 KOS (gL86) at 50 PFU/cell for 6 h at 37°C. ONPG entry assay was performed to estimate the enzymatic activity at 410 nm by spectrophotometer.
2.6. Kinetics of Viral Plaque Assay
Confluent layers of hMSCs (approximately 106) in six-well dishes were infected with HSV-1 (804) strain at 0.01 PFU/cell or mock infected for 2 hrs at 37°C. After removal of inoculum, monolayers were overlaid with DMEM containing 2.5% heat inactivated calf serum and incubated at 37°C. At different time points: 0, 12, 24, and 36 hr, the cells were fixed by using fixative buffer at room temperature for 20 min, followed by Giemsa staining for 45 min. The cells were again washed five times in PBS, and the numbers of plaques were counted. The images were taken by using an Olympus IX50 inverted fluorescence microscope. Virus replication was further examined by quantitative plaque assay. A monolayer of cultured hMSCs (approximately 4 × 106 cells per 25-mL flask) was infected (MOI, 0.01) with HSV-1(KOS) or mock infected with PBS alone for 2 h at 37°C. Vero cells mock infected of infected with the HSV-1 KOS under similar conditions were used as negative and positive controls, respectively. After removal of the inoculum, monolayers were overlaid with DMEM containing 2.5% heat-inactivated calf serum and incubated at 37°C until the time of harvest (12 to 48 h). Infectious virus titers were determined on Vero cells cultured in triplicate by using an overlay of medium containing methylcellulose. In order to block secondary plaque formation, human immunoglobulin G (IgG; Sigma) was added to the inoculum. The cells were washed with PBS buffer, fixed in alcohol, and stained with Giemsa stain. Infectivity was recorded as the number of PFU. Virus attachment was determined by using Olympus IX50 inverted fluorescence microscope
2.7. HSV-1 gD Interference Assay
Cultured hMSCs were transfected with Lipofectamine 2000 (Invitrogen) with an HSV-1 gD expression plasmid (pPEP99) [18] or a control plasmid (pCDNA3) in six-well dishes (1.0 μg of plasmid DNA total per well). The interference assay was performed as described previously [35].
2.8. Detection of gD Receptors by Reverse Transcription (RT)-PCR Analysis
Total RNA was isolated from cultured hMSCs using a QIAGEN RNeasy kit with DNAase in column treatment (QIAGEN Corp., Valencia, Calif, USA). SUPERSCRIPT II reverse transcriptase (Invitrogen Corp.) was used for RT to generate cDNAs. PCR amplification of cDNAs was done with primers 5′-GGAGACAATACCCTCATTCA-3′ and 5′-TATTTACATCCTCCCACAGC-3′ for HVEM, and primers 5′-TCCTTCACCGATGGCACTATCC-3′ and 5′-TCAACACCAGCAGGATGCTC-3′ for nectin-1. The 3-OST-3 sequences were amplified with primers 5′-CAGGCCATCATCATCGG-3′ and 5′-CCGGTCATCTGGTAGAA-3′. PCR was performed using ThermoStart DNA polymerase (Fisher cat no. AB0908) under the following conditions: 95°C 30′′, 52°C 30′′, 72°C 30′′, and 32 cycles for HVEM; 95°C 1′, 62°C 1′, 72°C 2′, and 35 cycles for Nectin-1; 95°C 1′, 55°C 1′, 72°C 2′, and 32 cycles for 3-OST. The expected sizes of the PCR products were 582 bps for HVEM, 738 bps for nectin-1, and 736 bps for 3-OST-3. The products were separated by electrophoresis on an agarose gel soaked in gel green solution and imaged using Bio-Rad Molecular Imager Gel Doc XR+ system.
2.9. Antibody Blocking Assay
Antibody blocking assay was performed as previously described [36]. hMSCs plated in 96-well plate were preincubated at room temperature with twofold dilutions of Anti-HVEM, anti-nectin-1 antibody (PRR1), and a control antibody (α-TGFbR II) for 90 min. Cells were then challenged with identical doses of HSV-1 (gL86) at 5 × 105 pfu per well at 37°C. After 6 hours, the cells were washed twice with PBS and treated for 1 min with 0.1 M citrate buffer (pH 3.0). The substrate, ImmunoPure ONPG was prepared in PBS buffer with nonionic detergent, and β-galactosidase activity was read at 410 nm. The experiment was repeated three times with similar results.
2.10. Heparinase Treatment
Cultured hMSCs plated on 96-well plates were washed with Mg2+- and Ca2+-free PBS and incubated at room temperature for 90 min with heparinase I (1.5 U mL−1; Sigma) or PBS alone and incubated at room temperature. The cells were then washed with PBS and used for HSV-1 entry [33].
2.10.1. Surface Immunofluorescence Assay
To detect the binding of HSV-1 to the surface of hMSCs, cultured monolayers of cells (approximately 106) were grown overnight on glass bottom culture dishes (Mat Tek Corp.). The cells were then treated with heparinases I (1.5 U mL−1) or with PBS buffer for 45 min, exposed to GFP-tagged HSV-1 K26GFP for 30 min at 4°C, and fixed with acetone. The cells were then stained with phalloidin (F-actin) and mounted on glass slides before examination under an Olympus IX50 inverted fluorescence microscope.
2.10.2. Glycoprotein D (gD)-Fc Binding Assay
Cultured hMSCs were plated overnight into 96-well dishes (approximately 4 × 104 cells/well). They were treated with heparinases I or with PBS buffer as described above for 2 h. This was followed by incubation with gD-Fc (1 μg/mL) for 1 h and fixation at room temperature with 2% formaldehyde and 0.02% glutaraldehyde for 15 min. The cells were washed with PBS containing 3% BSA (Sigma) and then incubated at room temperature sequentially with a biotinylated secondary antibody against rabbit IgG (Sigma) at a 1 : 500 dilution in PBS-3% BSA for 45 min and AMDEX streptavidin-conjugated horseradish peroxidase (Amersham) at a 1 : 20,000 dilution for 30 min. Following washing in PBS, the cells were incubated with 50 μL 3,3′,5,5′-tetramethylbenzidine (Sigma) in 50 mM phosphate-citrate buffer. Products were quantitated by use of a microplate reader (BioTek Instruments Inc. EL×808 absorbance microplate reader, Vt, USA) to measure optical density at 650 nm (OD650).
2.10.3. HSV-1 Glycoprotein-Induced Cell Fusion Assay
In this experiment, the CHO-K1 cells designated effector cells were cotransfected with plasmids expressing four HSV-1(KOS) glycoproteins: pPEP98 (gB), pPEP99 (gD), pPEP100 (gH), and pPEP101 (gL), along with plasmid pT7EMCLuc, which expresses the firefly luciferase gene under the control of the T7 promoter [22]. Cultured hMSCs considered target cells were cotransfected with pCalifGT7, which expresses T7 RNA polymerase with the chicken actin promoter and the CMV enhancer [18]. Effector cells expressing pT7EMCLuc and pCDNA3 (devoid of any glycoproteins) and target hMSCs transfected with T7 RNA polymerase alone were used as negative controls. In either case, the total amount of DNA used for transfection was kept constant. After 16 h, target and effector cells were mixed in a 1 : 1 ratio and then replated in 24-well dishes. Activation of the reporter luciferase gene, as a measurement of cell fusion, was examined by reporter lysis assay (Promega) at 24 h postmixing by using a Sirius luminometer (Berthold detection systems). Cell fusion was also visualized for multinucleated giant cells or polykaryocytes by microscopy following Giemsa (Fluka) staining at 24 h postmixing. Photographs of representative cells were imaged using an Olympus IX50 inverted microscope.
3. Results
3.1. Human Mesenchymal Stem Cells (hMSCs) Are Susceptible to HSV-1 Entry
To determine HSV-1 entry, confluent monolayers of cultured hMSCs were infected with serial dilutions of recombinant HSV-1(KOS) gL86 [27], which expresses β-galactosidase upon entry into cells. HeLa cells that are naturally susceptible to HSV-1 entry were used as a positive control. Entry of HSV-1 was measured after 6 h of viral infection. As shown in Figure 1(a), similar to HeLa cells, HSV-1 entered hMSCs in a dose-dependent manner. By contrast, CHO-K1 cells without any receptor remained resistant to HSV-1 entry. Further confirmation of HSV-1 entry was established using an X-gal assay. No β-gal activity was observed in mock infected hMSCs (Figure 1(b), panel A), while a dosage of virus sufficient to infect 100% of hMSCs at 10 PFU/cell (Figure 1(b), panel (B)) also led to a nearly complete infection of hMSCs. In order to obtain more direct and visual evidence of HSV-1 entry, GFP-tagged HSV-1(K26GFP) [34] was used to infect cultured hMSCs, and fluorescence microscopy was used to visualize the virions inside the hMSCs stained with rhodamine phalloidin, which specifically labels F-actin (Figure 1(c), panel (B)). The GFP colocalization with F-actin was seen all over the cell (Figure 1(c), panel (B)). This result correlated with the observed susceptibility of hMSCs to HSV-1 entry during X-gal assay.
Figure 1.
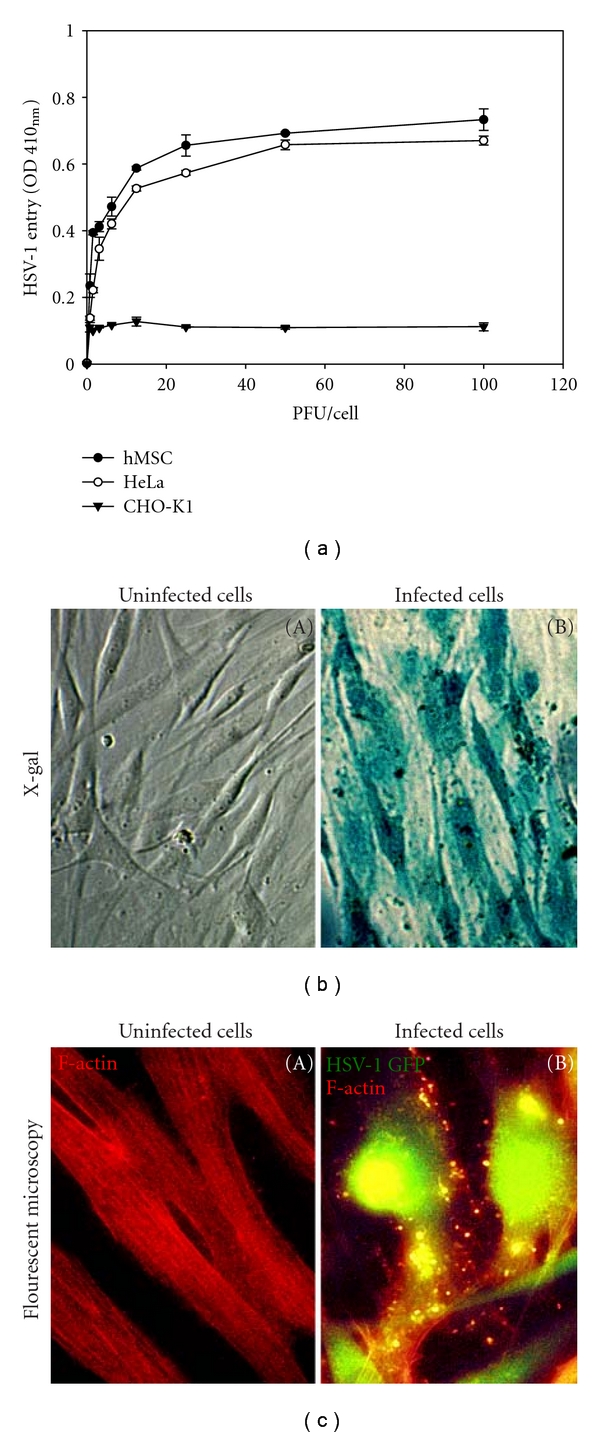
Herpes simplex virus type-1 (HSV-1) entry into cultured human mesenchymal stem cells (hMSCs). (a) Dosage response analysis of HSV-1 entry into hMSCs. Recombinant HSV-1(KOS) gL86 enters hMSCs and naturally susceptible HeLa cells but not wild-type CHO-K1 cells in a dosage-dependent manner. Data represent the mean ± the standard deviation of results from triplicate wells in a representative experiment. (b) β-galactosidase staining of HSV-1 entry. Cultured hMSCs that were exposed to HSV-1(KOS) gL86 (panel (B)) were 100% stained blue, indicating 100% viral infection, whereas uninfected cells did not show any staining. (c) Fluorescence imaging of HSV-1 entry. Confluent monolayer culture of hMSCs were mock infected (panel (A)) or infected with GFP-tagged HSV-1 (K26-GFP) at 50 PFU/cell (panel (B)) and stained with TRITC-conjugated phalloidin (red color), which binds to F-actin. HSV-1 infected cells showed both green fluorescence from HSV-1 capsid and red fluorescence from F-actin expression (green and red overlapping areas appear yellow), whereas mock-treated cells only showed red fluorescence. The images were taken on fluorescent microscope at 20x magnification.
3.2. Effect of Cytoskeleton Rearrangements in hMSCs during HSV-1 Infection
Our previous findings have shown that HSV-1 entry into natural target cells leads to change in actin cytoskeleton [36–38]. We therefore decided to examine whether cytoskeleton changes played any significant role in HSV-1 entry into hMSCs. To address this issue, fluorescent microscopy of hMSCs exposed to GFP-tagged HSV-1 virions at 30 min postinfection was performed. Quite noticeably, some GFP-tagged virus particles were found to attach at filopodia-like structures in rhodamine-phalloidin stained hMSCs (Figures 2(a) and 2(b)). In order to demonstrate the significance of actin network and filopodia during HSV-1 entry into hMSCs, we used the actin inhibitors latrunculin B (Lat B) or cytochalasin D (Cyto D). Both agents prevent cytoskeleton changes by blocking actin polymerization [39, 40]. As shown in Figures 2(c) and 2(d), pretreatment of hMSCs with Lat B and Cyto D resulted in significant inhibition of HSV-1 entry, suggesting that cytoskeleton modification may be needed during the initial phase of HSV-1 infection.
Figure 2.
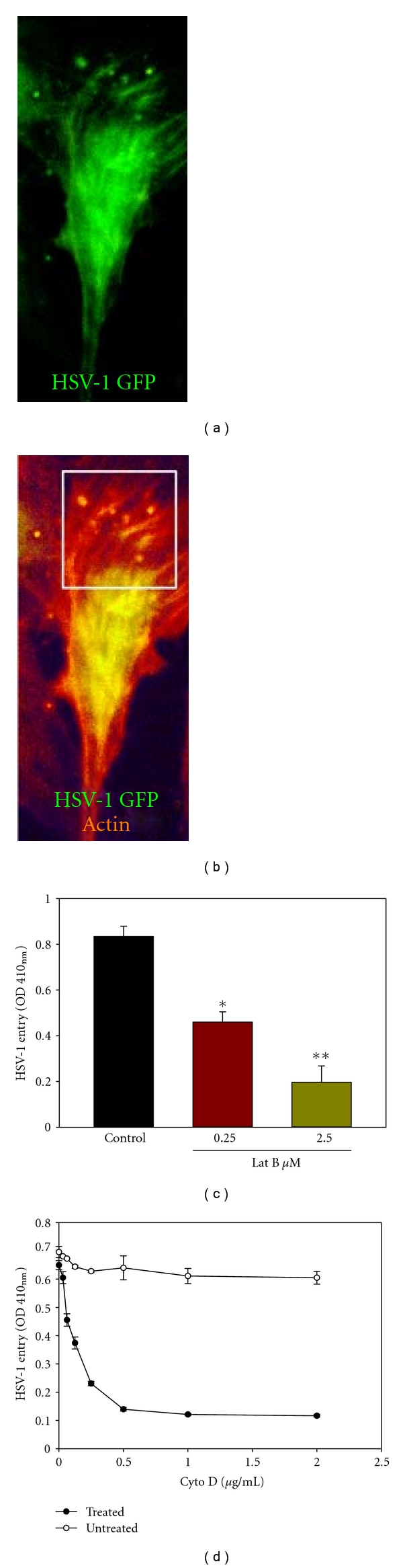
HSV-1 entry into hMSCs is cytoskeleton dependent. (a) An infected cell showing the virus particles (green). Cells were infected with HSV-1 (K26GFP) at 50 PFU/cell and imaged 45 min postinfection. (b) Virus particles colocalize with F-actin in filopodia-like structures (boxed area). F-actin was stained by rhodamine phalloidin (red). Areas where virus particle (green) and F-actin overlap appear yellow. (c) HSV-1(KOS) gL86 entry into hMSCs (50 PFU/cell) was inhibited by actin depolymerizing agent Latrunculin B (Lat B) in a dose-dependent manner. (d) HSV-1(KOS) gL86 entry into hMSCs (50 PFU/cell) was inhibited by actin depolymerizing agent Cytochalisin D (Cyto D) in a dose-dependent manner. The mock-treated hMSCs were used as a control. Viral entry was quantitated 6 h after infection at 410 nm using a spectrophotometer *P < .05, **P < .01.
3.3. HSV-1 Replicates in Cultured hMSCs
Because HSV-1 was able to enter cultured hMSCs, we next evaluated whether this entry led to a productive virus replication. Initially, microscopy was used to obtain visual evidence of HSV-1 replication. Syncytial plaque forming HSV-1 (KOS) 804 virus [41] was used for infecting cultured hMSCs, and the virus was allowed to replicate. Cells were fixed at different time points and Giemsa stained. The cytopathic effect in the form of plaque formation increased significantly overtime in virus-infected hMSCs as seen in Figures 3(a) and 3(b). Furthermore, to assess viral replication, the infectious yields of virus were determined by plaque assays with Vero cells. As shown in Figure 3(c), inoculum harvested from infected hMSCs produced a larger number of plaques overtime. In contrast, CHO-K1 cells infected with identical doses of the same virus failed to produce infectious virions. These results, together with those of the entry assay, show that entry of HSV-1 into cultured hMSCs leads to productive infection.
Figure 3.
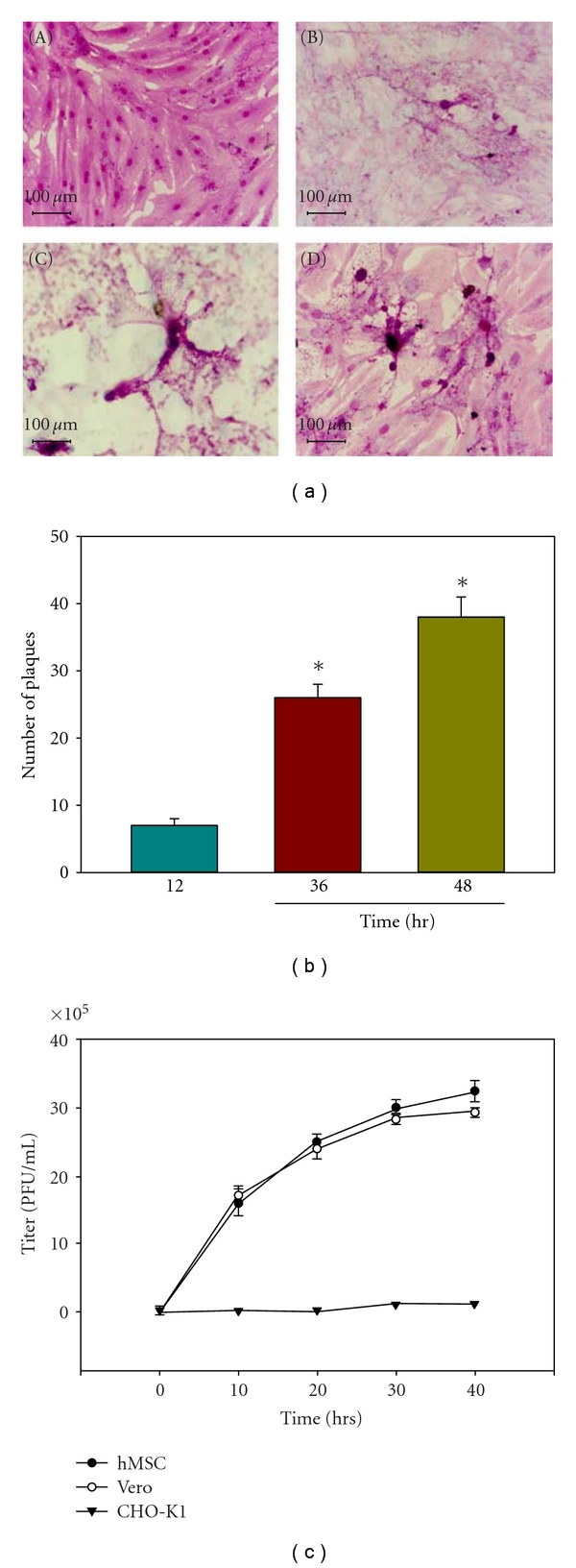
HSV-1 replicates in infected hMSCs. (a) Visualization and quantification of HSV-1 replication in cultured hMSCs. Confluent monolayers of hMSCs (5 × 106) were infected with HSV-1 KOS (804) virus at 0.01 PFU/cell were fixed and Giemsa stained at 0 hr (panel (A)) 12 hr (panel (B)) 24 hr (panel (C)) and 48 hr (panel (D)) postinfection. The numbers of plaques were visualized. (b) The number of plaques formed postinfection increased in a time-dependent manner. Error bars represent standard deviations. (*P < .05), one way ANOVA. (c) Infectious yields of HSV-1 during viral infection were quantified. Confluent monolayers of Vero and hMSCs were infected with HSV-1 at 0.01 PFU per cell for 90 min at 37°C. Inoculums were harvested from both the cells at 10–40 h postinfection. The infectious virus-titer (PFU per milliliter) determined in triplicates in Vero cell by plaque assay indicates that the viral titer in cultured hMSCs increased overtime. Data represent the mean ± the standard deviation of results in triplicate wells in a representative experiment.
3.4. Expression of HSV-1 gD in Cultured hMSCs Renders Resistance to HSV-1 Entry
In order to determine if the gD receptors play a role in human hMSCs infection by HSV-1, a gD-mediated interference assay was used [35]. This assay is based on the principle that cells that are normally susceptible to viral entry become resistant upon expression of viral gD because of sequestration of gD receptors by cell-expressed gD [35]. To carry out the assay, hMSCs were transiently transfected with an HSV-1 gD-expressing plasmid (or an equal amount of the empty vector, pCDNA3, as a control), followed by infection with serial dilutions of β-galactosidase-expressing HSV-1(KOS) gL86. As shown in Figure 4, HSV-1 entry into gD-expressing hMSCs was approximately 2.0-fold lower compared to the pCDNA3 control. Thus, entry into hMSCs is likely mediated by gD receptors expressed on the cell surface in a gD-dependent manner.
Figure 4.
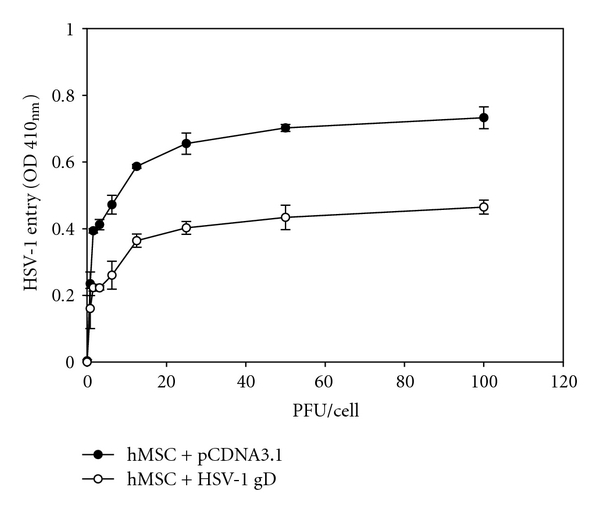
HSV-1 glycoprotein D (gD) receptors are important for infection of hMSCs. Cultured hMSCs were transfected with an HSV-1 gD expression plasmid (white dot) or with the empty vector pCDNA3.1 (black dot) and infected with HSV-1 at two fold dilution. Reporter enzyme activity, as a measurement of viral entry, was measured with spectrophotometer at 410 nm.
3.5. Identification of gD Receptors Expressed in Cultured hMSCs
Reverse transcriptase (RT)-PCR analysis was performed to determine the identity of gD receptors potentially expressed in hMSCs. Gene-specific primers for nectin-1, 3-OST-3 and HVEM were used. As shown in Figure 5(a), products of the expected sizes for cDNAs encoding nectin-1, 3-OST-3 and HVEM were all detected, indicating that all three genes are normally expressed in hMSCs.
Figure 5.
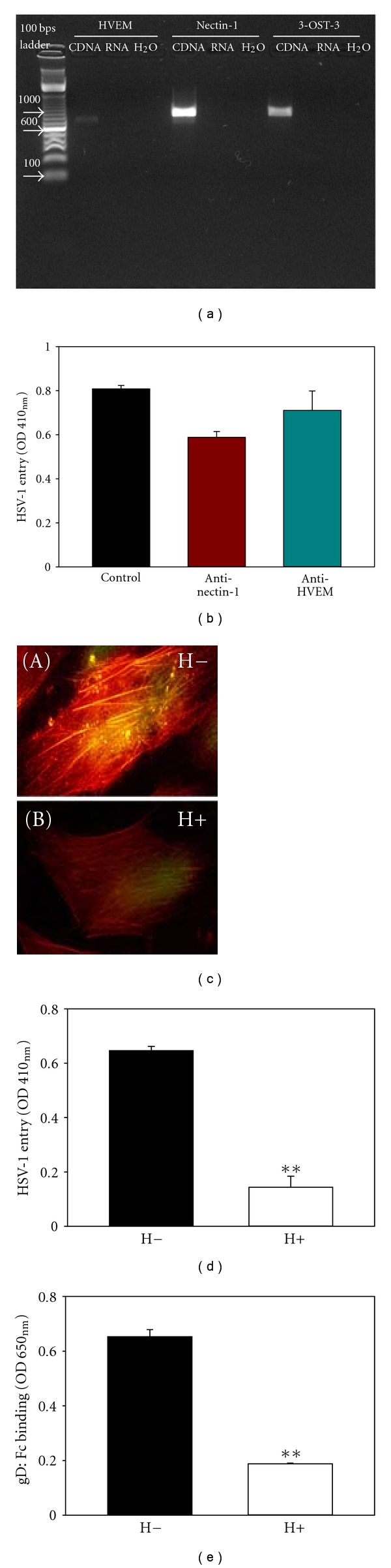
3-OS HS but not Nectin-1 or HVEM serves as the critical gD receptor in mediating HSV-1 entry into hMSCs. (a) Expression of HSV-1 glycoprotein D (gD) receptors in hMSCs. RT-PCR analysis of nectin-1, 3-OST-3, and HVEM demonstrated that all 3 genes are normally expressed by hMSCs. No products were detected in the H2O or RNA lanes in which H2O or the same RNA that was used in generating cDNA templates was used as PCR template to control for potential genomic DNA contamination. The products were separated by electrophoresis on an agarose gel. (b) HSV-1 entry into hMSCs was not significantly affected by anti-nectin-1 or anti-HVEM antibody treatment. Cells were preincubated with anti-nectin-1 and anti-HVEM antibodies (1 : 10 dilution) followed by infection with reporter HSV-1 (KOS) gL86 virus. β-galactosidase activity was recorded 6 h later to determine HSV-1 entry. The experiment was repeated three times with similar results. (c) Visualization of heparinase treatment on HSV-1 binding into hMSCs. Cultured hMSCs were mock-treated with PBS (H−) or heparinase I (1.5 U mL−1) (H+), followed by exposure to GFP-tagged HSV-1 K26GFP virus at 50 pfu/cell for 30 min at 4°C. Cells were then stained with rhodamine phalloidin that recognizes F-actin (red). Only mock-treated but not heparinase-treated cells showed HSV-1 GFP binding, which appeared yellow when its green fluorescence overlaps with red fluorescence. Virus attachment was determined by using Olympus IX50 inverted fluorescence microscope. (d) Effect of heparinase treatment on HSV-1 entry into hMSCs. Cells were treated with heparinase I (1.5 U mL−1) (white bar) or mock-treated with PBS alone (black bar), followed by exposure to HSV-1 KOS gL86 at 50 pfu/cell. Viral entry was quantitated 6 hr later by ONPG assay. Viral entry was reduced more than 3 folds in cells pretreated with heparinase compared to mock-treated cells (**P < .01, t test). Data shown are the means of triplicate measures. (e) Effect of heparinase treatment on HSV-1 gD binding to hMSCs. The cells were either pretreated with a mixture of heparinase-I (treated at 1.5 U mL−1; white bar) or mock-treated (untreated; black bar), followed by exposure to a soluble recombinant form of HSV-1 gD (gD:Fc). Binding of gD:Fc was detected by using HRP-tagged antibody against IgG:Fc and subsequent HRP enzyme detection. Asterisks indicate significant difference from the control treatment (**P < .01, t test). Error bars represent standard deviation (SD).
3.6. Anti-Nectin-1 and Anti-HVEM Antibody Do Not Significantly Affect HSV-1 Entry in hMSCs
To determine which receptor were important for HSV-1 entry into hMSCs, previously established receptor specific antibodies were used [33, 36]. As shown in Figure 5(b), antibody against nectin-1 affected HSV-1 entry by 20%, while HVEM had no significant effect on HSV-1 entry suggesting that neither of these receptors plays a critical role in mediating HSV-1 infection of hMSCs.
3.7. Significance of HS and 3-OST-3 as the Mediator of HSV-1 Entry and gD Binding to Cultured hMSCs
Expression of 3-OST-3 raised the possibility that 3-OS HS could be a mediator of HSV-1 entry into hMSCs. Since cells that use 3-OS HS as the major entry receptor become significantly more resistant to HSV-1 upon removal of HS from the surface [37, 38], we examined the effect of HS-degrading enzyme heparinase-I (1.5 U mL−1) on HSV-1K26GFP binding and entry into hMSCs. This enzyme is known to cleave off both heparin and HS chains containing 1–4 linkages between glucosamine and O-sulfated iduronic acid residues [42]. As shown in Figure 5(c), heparinase-treated hMSCs (H+) were resistant to HSV-1 binding (panel B) compared to the mock-treated cells (H-, panel A). Similar resistance to HSV-1 KOS gL86 entry was also observed as a result of heparinase treatment (Figure 5(d)). Next, we tested whether a soluble recombinant form of HSV-1 gD or gD-Fc [29, 35] would bind cultured hMSCs and whether this binding could be adversely affected by removal of HS from the surface using cell enzyme-linked immunosorbent assay [26, 28, 33]. Cells either treated with heparinases or mock-treated were allowed to bind equal amount of gD, and binding was detected with a spectrophotometer. Figure 5(e) shows that gD binding to mock-treated hMSCs was not affected; however, the binding of gD to heparinase-treated hMSCs was reduced by about 75%. This finding strongly supports that gD-binding receptors were specifically removed upon heparinase treatment in hMSCs, indicating that HS and 3-OST-3 play critical roles in mediating HSV-1 entry through gD binding to cultured hMSCs.
3.8. Requirement of 3-OS HS for HSV-1 Glycoprotein-Induced Cell-to-Cell Fusion of Cultured hMSCs
To further verify that 3-OS HS is indeed the mediator of the membrane fusion that occurs during HSV-1 entry into hMSCs, we used a quantitative and efficient cell-to-cell fusion assay [14, 22]. It has been shown that, similar to the membrane fusion that occurs during entry, cells expressing HSV-1 glycoproteins gB, gD, gH, and gL fuse with cells expressing gD receptors [18], and thus cell-to-cell fusion mimics the minimum requirement for entry. First, we studied the significance of HS in the fusion of hMSCs with HSV-1 glycoproteins expressing effector CHO-K1 cells. HS dependence is a way to determine whether 3-OS HS is the principal mediator of membrane fusion since, unlike nectin-1 and HVEM, which do not require HS for membrane fusion, 3-OS HS-mediated cell fusion is highly dependent on the expression of HS on the cell surface [22]. As expected, fusion of hMSCs with effector cells expressing the HSV-1 glycoproteins was significantly reduced upon heparinase treatment (Figure 6(a)), and a similar result was seen with CHO-K1 expressing 3-OST-3 (data not shown), suggesting that expression of HS and 3-OS HS in hMSCs are required for effective fusion with glycoprotein expressing-CHO-K1 cells. In a separate set of experiments, heparinase treatment also reduced multinucleated giant cell formation or polykaryons in hMSCs cocultured with effector CHO-K1 cells expressing HSV-1 glycoproteins (Figures 6(b) and 6(c)). Taken together, our results demonstrate that HS and 3-OS HS are required for HSV-1 glycoprotein-induced cell to cell fusion in cultured hMSCs, further implying their significant role in mediating HSV-1 entry and spread into hMSCs.
Figure 6.
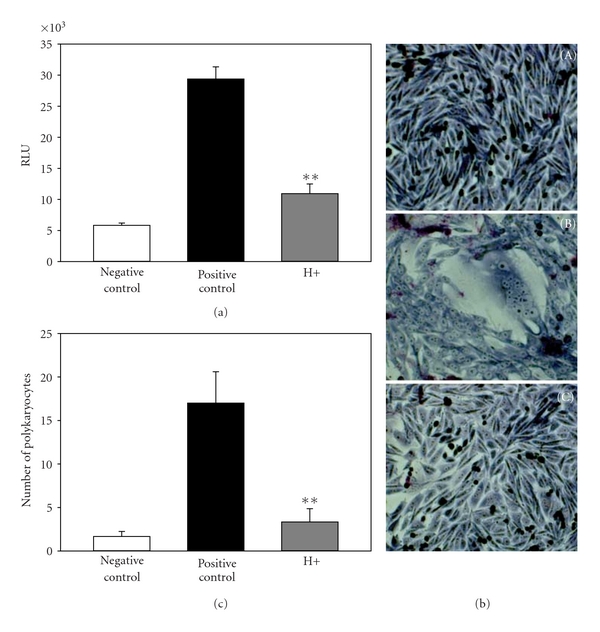
Effect of heparinase on quantitative HSV-1 glycoprotein-induced fusion with hMSCs. (a) In this experiment, the effector CHO-K1 cells were transfected with HSV-1 glycoproteins gB, gD, gH, and gL, and T7 RNA polymerase. The target hMSCs were transfected with plasmids expressing luciferase reporter gene. A luciferase reporter assay was performed 24 h after the two cell populations were mixed together. Cell fusion was measured in relative luciferase units (RLUs). Expression of gD receptors in hMSCs results in the fusion of CHO-K1 cells with HSV-1 glycoprotein expressing cells (black bar), while in a negative control coculture of target cells with effector cells devoid of all four glycoproteins-encoding genes, no luciferase activity was recorded (white bar). Coculture of heparinase (H+) pretreated-target cells with effector cells resulted in significant decrease in cell fusion (grey bar). (b). The effects of heparinase on multinucleated polykaryocytes formation were visualized. Multinucleated cells or polykaryocytes were microscopically observed with the hMSC target cells cocultured with effector CHO-K1 cells expressing HSV-1 glycoproteins (panel (B)) but not with CHO-K1 cells expressing control vector pDream2.1 (panel (A)) or with the pretreatment of target hMSCs with heparinase-I (1.5 U mL−1) before mixing with effector cells (panel (C)). Both type of cells were cocultured and stained with Giemsa at 24 h postmixing. Shown are photographs of representative cells (Olympus IX50 inverted microscope) after 24 h postmixing. (c) Quantitative determination of the number of fused polykaryocytes formed after coculture of target hMSCs cocultured with the effector CHO-K1 cells expressing HSV-1 glycoproteins in presence (grey bar) or absence (black bar) of heparinase. Effector CHO-K1 cells expressing empty vector (pCDNA3.1) cultured with target hMSCs was used as a negative control (white bar). The data shown are the means of triplicate measures with three independent experiments. Asterisks indicate significant difference from other treatments (**P < .01, t test), and error bars represent standard deviation (SD).
4. Discussion
HSV-1 infections remain a significant problem for allogeneic stem cell recipients. Understanding viral entry into hMSCs in a healthy donor or in a recipient host is important for two reasons. First, productive viral infection into hMSCs may trigger immune response during or after stem cell transplant and, second, the infected population of hMSCs may become large reservoir of virions which may constantly expand both immune response and the viral transmission to the localized cells or tissues or may find the new host cells and tissues in the recipient. Therefore, viral receptor on these cells could determine the type of the infection, nature of immune response, and viral transmission. We cultured hMSCs derived from the adipose tissue of healthy donors to develop an in vitro model for studies of HSV-1 entry and replication and for identification of cellular mediators of entry. Although all the three HSV-1 gD-receptors (nectin-1, 3-OST-3 and HVEM) were detected, however, antibodies against nectin-1 and HVEM failed to block significantly HSV-1 entry (Figures 5(a) and 5(b)). Therefore, our focus shifted to 3-OS HS as a potential entry receptor. Adding further strength to this possibility, heparinase enzymes completely abolished both HSV-1 attachment (Figure 5(c)) and entry into cultured hMSCs (Figure 5(d)). In addition, the gD binding exhibited sensitivity to enzyme treatment (Figure 5(e)) and the effect of heparinase to viral spread during cell to cell fusion assay was also significantly inhibited in cultured hMSCs (Figure 6). Similar effect of heparinase treatment on HSV-1 entry and fusion has been shown in primary cultures of corneal fibroblasts obtained from human eye donors or in the cells exclusively expressing 3-OST isoforms [28, 29, 33]. Taken together, we speculate one interesting possibility that although all the three known gD-receptors are expressed in hMSCs but HSV-1 preferentially uses HS/3-OS-HS to gain entry could be related for two main reasons. First HS and 3-OS HS being larger in size are widely distributed and expressed all over the surface of the cells. Second they are also easily accessible to HSV-1 unlike nectin-1 which is localized in cell junctions. This is also possible that nectin-1 and HVEM receptors are involved in other complex interactions that do not support HSV-1 entry into hMSCs.
In summary, we conclude HS and 3-OS HS play a key role during HSV-1 infection into hMSCs. The finding provides new information on susceptible nature of hMSCs population within the host cell and contributes the broader significance of HS and 3-OS HS receptor in HSV-1 infection. Because HS also play a key role in signaling and immune activation [43], it is possible that they might play a dual role allowing both HSV-1 infection and facilitating the inflammation to further damage hMSCs. This unique feature of HS/3-OS HS in hMSCs can be exploited to develop the agent that blocks HSV-1 gD interaction with 3-OS HS receptor to suppress both viral entry/spread and immune response. Finally, hMSCs have potential to differentiate into many other cell-types including neurons. Interestingly, HS is known to mediate the proliferation and differentiation in mesenchymal stem cells [44]. Therefore it will be interesting to evaluate the expression pattern of gD-receptors in differentiated stem cells including neuronal stem cells where HSV-1 can easily infect and establish latency. Similarly the strategic development to isolate population of stem cell lines that does not support HSV-1 infection may further suppress the HSV-1-induced morbidity and mortality and bring enhanced success of stem cell transplantation.
Acknowledgments
The authors acknowledge kind support of Doctors Spear (Northwestern University, Chicago), Shukla (University of Illinois at Chicago), and Prashant Desai (Johns Hopkins University) for the reagents. This work was supported by research grants from the Western University of Health Sciences (WUHS) N12587 to V. Tiwari, and IDEA Award to F. Spors. Y. X. Zhao was supported by NIH Grant no. 1SC3GM094078-01. M. Marquez and F. Alencastro are equally contributing authors.
References
- 1.Whitley RJ, Kimberlin DW, Roizman B. Herpes simplex viruses. Clinical Infectious Diseases. 1997;26:79–109. doi: 10.1086/514600. [DOI] [PubMed] [Google Scholar]
- 2.Whitley RJ, Roizman B. Herpes simplex viruses. In: Richman DD, Whitely RJ, Hayden FG, editors. Clinical Virology. New York, NY, USA: Churchill Livingstone; 1997. pp. 380–382. [Google Scholar]
- 3.Corey L, Spear PG. Infections with herpes simplex viruses. The New England Journal of Medicine. 1998;314:p. 686–689, p. 749–757. doi: 10.1056/NEJM198603133141105. [DOI] [PubMed] [Google Scholar]
- 4.Whitley RJ, Kimberlin DW, Roizman B. Herpes simplex viruses. Clinical Infectious Diseases. 1998;26(3):541–553. doi: 10.1086/514600. [DOI] [PubMed] [Google Scholar]
- 5.Hirsh MS. Herpes group virus infections in the compromised host. In: Robin RH, Young LS, editors. Clinical Approach to Infection in the Compromised Host. 3rd edition. New York, NY, USA: Plenum Medical Book; 1998. pp. 379–392. [Google Scholar]
- 6.Nichols WG, Boeckh M, Carter RA, Wald A, Corey L. Transferred herpes simplex virus immunity after stem-cell transplantation: clinical implications. Journal of Infectious Diseases. 2003;187(5):801–808. doi: 10.1086/367894. [DOI] [PubMed] [Google Scholar]
- 7.Shiley K, Blumberg E. Herpes viruses in transplant recipients: HSV, VZV, human herpes viruses, and EBV. Infectious Disease Clinics of North America. 2010;24(2):373–393. doi: 10.1016/j.idc.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Yoshikawa T. Significance of human herpesviruses to transplant recipients. Current Opinion in Infectious Diseases. 2003;16(6):601–606. doi: 10.1097/00001432-200312000-00014. [DOI] [PubMed] [Google Scholar]
- 9.Razonable RR, Eid AJ. Viral infections in transplant recipients. Minerva Medica. 2009;100(6):479–501. [PubMed] [Google Scholar]
- 10.Chen Y, Scieux C, Garrait V, et al. Resistant herpes simplex virus type 1 infection: an emerging concern after allogeneic stem cell transplantation. Clinical Infectious Diseases. 2000;31(4):927–935. doi: 10.1086/314052. [DOI] [PubMed] [Google Scholar]
- 11.Guimaraes ALS, Gomes CC, da Silva LM, et al. Association between oral HSV-1 and survival in allogeneic hematopoietic stem cell transplanted patients. Medicina Oral, Patologia Oral y Cirugia Bucal. 2009;14(2):E62–E68. [PubMed] [Google Scholar]
- 12.Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. The Journal of Clinical Investigation. 2001;108(4):503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spear PG, Eisenberg RJ, Cohen GH. Three classes of cell surface receptors for alphaherpesvirus entry. Virology. 2000;275(1):1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 14.Browne H, Bruun B, Minson T. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. Journal of General Virology. 2001;82(6):1419–1422. doi: 10.1099/0022-1317-82-6-1419. [DOI] [PubMed] [Google Scholar]
- 15.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. Journal of Virology. 1992;66(1):341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis-Poynter N, Bell S, Minson T, Browne H. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. Journal of Virology. 1994;68(11):7586–7590. doi: 10.1128/jvi.68.11.7586-7590.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muggeridge MI. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. Journal of General Virology. 2000;81(8):2017–2027. doi: 10.1099/0022-1317-81-8-2017. [DOI] [PubMed] [Google Scholar]
- 18.Pertel PE, Fridberg A, Parish ML, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology. 2001;279(1):313–324. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 19.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. Journal of Virology. 1998;72(1):873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allison TT, Montgomery RI, Warner MS, Geraghty RJ, Spear PG. Contributions of gD receptors and glycosaminoglycan sulfation to cell fusion mediated by herpes simplex virus 1. Virus Research. 2001;74(1-2):39–45. doi: 10.1016/s0168-1702(00)00244-6. [DOI] [PubMed] [Google Scholar]
- 21.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. Journal of Virology. 1998;72(12):9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. A role for 3-O-sulfated heparan sulfate in cell fusion induced by herpes simplex virus type 1. Journal of General Virology. 2004;85(4):805–809. doi: 10.1099/vir.0.19641-0. [DOI] [PubMed] [Google Scholar]
- 23.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87(3):427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 24.Tiwari V, Clement C, Scanlan PM, Kowlessur D, Yue BYJ, Shukla D. A role for herpesvirus entry mediator as the receptor for herpes simplex virus 1 entry into primary human trabecular meshwork cells. Journal of Virology. 2005;79(20):13173–13179. doi: 10.1128/JVI.79.20.13173-13179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of Alphaherpesviruses Mediated by Poliovirus Receptor-Related Protein 1 and Poliovirus Receptor. The FEBS Journal. 2008;280(5369):1618–1618. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 26.Shukla D, Dal Canto MC, Rowe CL, Spear PG. Striking similarity of murine nectin-1alpha to human nectin-1alpha (HveC) in sequence and activity as a glycoprotein D receptor for alphaherpesvirus entry. The Journal of Virology. 2000;74(24):11773–117781. doi: 10.1128/jvi.74.24.11773-11781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shukla D, Liu J, Blaiklock P, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99(1):13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 28.O’Donnell CD, Tiwari V, Oh MJ, Shukla D. A role for heparan sulfate 3-O-sulfotransferase isoform 2 in herpes simplex virus type 1 entry and spread. Virology. 2006;346(2):452–459. doi: 10.1016/j.virol.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Tiwari V, O’Donnell CD, Oh MJ, Valyi-Nagy T, Shukla D. A role for 3-O-sulfotransferase isoform-4 in assisting HSV-1 entry and spread. Biochemical and Biophysical Research Communications. 2005;338(2):930–937. doi: 10.1016/j.bbrc.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 30.Xia G, Chen J, Tiwari V, et al. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. Journal of Biological Chemistry. 2002;277(40):37912–37919. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 31.Xu D, Tiwari V, Xia G, Clement C, Shukla D, Liu J. Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1. Biochemical Journal. 2005;385(2):451–459. doi: 10.1042/BJ20040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Donnell CD, Shukla D. A novel function of heparan sulfate in the regulation of cell-cell fusion. Journal of Biological Chemistry. 2009;284(43):29654–29665. doi: 10.1074/jbc.M109.037960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Little SP, Schaffer PAA. Expression of the syncytial (syn) phenotype in HSV-1, strain KOS: genetic and phenotypic studies of mutants in two syn loci. Virology. 1981;112(2):686–702. doi: 10.1016/0042-6822(81)90314-7. [DOI] [PubMed] [Google Scholar]
- 34.Clement C, Tiwari V, Scanlan PM, Valyi-Nagy T, Yue BYJT, Shukla D. A novel role for phagocytosis-like uptake in herpes simplex virus entry. Journal of Cell Biology. 2006;174(7):1009–1021. doi: 10.1083/jcb.200509155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spector I, Shochet NR, Blasberger D, Kashman Y. Latrunculins—novel marine macrolides that disrupt microfilament organization and affect cell growth: I. Comparison with cytochalasin D. Cell Motility and the Cytoskeleton. 1989;13(3):127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- 36.Tiwari V, Oh MJ, Kovacs M, Shukla SY, Valyi-Nagy T, Shukla D. Role for nectin-1 in herpes simplex virus 1 entry and spread in human retinal pigment epithelial cells. The FEBS Journal. 2008;275(21):5272–5285. doi: 10.1111/j.1742-4658.2008.06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. Journal of Virology. 1998;72(9):7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geraghty RJ, Jogger CR, Spear PG. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology. 2000;268(1):147–158. doi: 10.1006/viro.1999.0157. [DOI] [PubMed] [Google Scholar]
- 39.Tiwari V, ten Dam GB, Yue BYJT, van Kuppevelt TH, Shukla D. Requirements of 3-O-sulfated heapran sulfate during cell fusion in primary cultures of human corneal fibroblasts. The FEBS Letters. 2007;581(23):4468–4472. doi: 10.1016/j.febslet.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiwari V, Clement C, Xu D, et al. Role for 3-O-sulfated heparan sulfate as the receptor for herpes simplex virus type 1 entry into primary human corneal fibroblasts. Journal of Virology. 2006;80(18):8970–8980. doi: 10.1128/JVI.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schafer DA. Regulating actin dynamics at membranes: a focus on dynamin. Traffic. 2004;5(7):463–469. doi: 10.1111/j.1600-0854.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 42.Ernst S, Langer R, Cooney CL, Sasisekharan R. Enzymatic degradation of glycosaminoglycans. Critical Reviews in Biochemistry and Molecular Biology. 1995;30(5):387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 43.Teixe T, Nieto-Blanco P, Vilella R, Engel P, Reina M, Espel E. Syndecan-2 and -4 expressed on activated primary human CD4+ lymphocytes can regulate T cell activation. Molecular Immunology. 2008;45(10):2905–2919. doi: 10.1016/j.molimm.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 44.Dombrowski C, Song SJ, Chuan P, et al. Heparan sulfate mediates the proliferation and differentiation of rat mesenchymal stem cells. Stem Cells and Development. 2009;18(4):661–670. doi: 10.1089/scd.2008.0157. [DOI] [PubMed] [Google Scholar]


