Abstract
Biofilms grow on various surfaces and in many different environments, a phenomenon that constitutes major problems in industry and medicine. Despite their importance little is known about the viscoelastic properties of biofilms and how these depend on the chemical microenvironment. Here, we find that the mechanical properties of Pseudomonas aeruginosa (P.a.) biofilms are highly robust towards chemical perturbations. Specifically, we observe that P.a. biofilms are able to fully regain their initial stiffness after yielding is enforced, even in the presence of chemicals. Moreover, only trivalent ions and citric acid significantly affect the biofilm elasticity, the first of which also alter the texture of the material. Finally, our results indicate that biofilm mechanics and bacteria viability inside the biofilm are not necessarily linked which suggests that targeting bacteria alone might not be sufficient for biofilm removal strategies.
Introduction
Biofilms have been defined as a community of microbes that adhere to each other and/or to a surface1, 2. Biofilm architecture is provided by extracellular polymeric substances (EPS), a mix of polysaccharides, proteins, lipids, and nucleic acids3–5. The EPS embeds cells (Fig. 1A), makes up to 50–90% of the total organic material in biofilms6 and provides increased resistance to antibiotics and environmental stress7. Biofilms are materials with intriguing properties: they are stable towards external mechanical forces as imposed by, for example, flowing water or peristaltic, which presumably causes constant and major reorganization of the biofilm structure. At the same time, they provide a sufficiently soft and loose environment that accommodates bacteria movement or proliferation within8. Accordingly, the viscoelastic properties of biofilms must be carefully balanced and – once established – need to be quite robust against external perturbations to sustain efficient biofilm growth. Although often exposed to mechanical shear forces, biofilms are able to grow on a wide variety of surfaces, causing problems in industry9–11. Furthermore, biofilms are involved in numerous human infections and diseases, and they can contaminate foreign body materials12. Biofilms survive in many different environments, and their biophysical properties can vary significantly between different biofilm types, and also during biofilm maturation13.
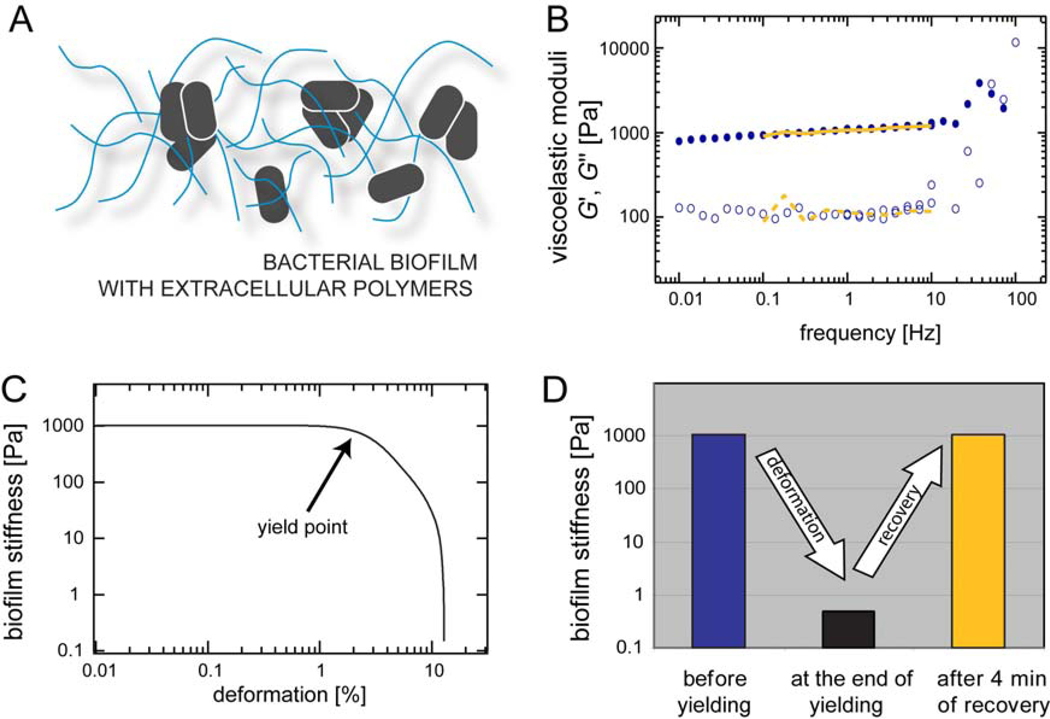
Figure 1.
(A) Schematic representation of a bacterial biofilm. Bacteria grow in distinct localized colonies and secrete extracellular polymers that create a 3-dimensional scaffold around them. (B) The linear viscoelastic response of a P.a. biofilm is determined over a broad range of frequencies. We compare the viscoelastic response before and 4 min after the biofilm was forced to yield. The elastic modulus G’(f) is depicted by closed symbols and full lines, the viscous modulus G”(f) is depicted by open symbols and dashed lines. (C) Mechanical response of a P.a. biofilm at large deformations. Beyond a critical deformation the biofilm yields and its apparent stiffness is decreased by more than three orders of magnitude. (D) Schematic representation of the rupture/recovery process as shown in detail in (B) and (C). The depicted stiffness values are taken from graphs (B) and (C).
Despite their importance, little is known about the biophysical properties that render biofilms so versatile and sturdy. A suitable model organism that forms biofilms on various surfaces is Pseudomonas aeruginosa (P.a.), a human opportunistic pathogen, which can colonize the mucus of cystic fibrosis patients, catheters, and contact lenses14–16. Here, we probe the viscoelastic properties of Pseudomonas biofilms and ask to what extent they are influenced by external chemical stimuli and large shear forces. We demonstrate that these biofilms show a remarkable viscoelastic behavior, which allows them to fully regain their initial stiffness after yielding is enforced. Surprisingly, this recovery process is highly robust towards chemical perturbations such as ions, polyelectrolytes, organic molecules, or pH changes. Furthermore, we observe that among these chemicals trivalent ions such as Fe(III) and Al3+, but not divalent ions, significantly enhance the biofilm elasticity and alter the texture of the material as well. Conversely, we identify citric acid as a potent fluidization agent for pre-formed biofilms. Last, these effects appear to be a more generic biofilm property as they are not limited to the Pseudomonas aeruginosa wild type but also apply to five other Pseudomonas variants.
Results
1. A Pseudomonas aeruginosa biofilm can be treated as a viscoelastic material with defined mechanical properties
We first ask if P.a. biofilms can be treated like any other polymer-based substance with well-defined material properties. These material properties typically combine elastic and viscous aspects which are parameterized by the viscoelastic moduli, the storage modulus G’(f) and the loss modulus G”(f). The storage modulus describes the elastic properties of the material while the loss modulus represents its viscous properties. The viscoelastic response of a polymer based material depends on the time scale of an applied deformation. Therefore, viscoelastic moduli are typically determined over a broad range of frequencies.
We observe that in the linear response regime, i.e. at small deformations (corresponding to torques ~1 µNm), the viscoelastic frequency spectrum of a P.a. biofilm exhibits two distinct regimes (Fig. 1B). At high frequencies, i.e. above ~30 Hz, the loss modulus seems to dominate over the storage modulus. In analogy to many other polymer networks17, this could indicate that in this regime, the viscoelastic response of P.a. biofilms is dictated by the properties of individual biofilm polymers. At lower frequencies, i.e. below 10 Hz, the storage modulus G’(f) dominates (Fig. 1B). At these time scales, the material displays a collective response to external mechanical stimuli and thus behaves predominantly as an elastic material. Furthermore, in this regime both moduli are relatively independent of frequency. The observed plateau allows us to subsume the linear biofilm properties into a single material constant, the plateau modulus G0. For simplicity, G0 = G’(1 Hz) will be used as a typical biofilm elasticity value for the remainder of this article. We note that the plateau moduli of P.a. biofilms from distinct growth batches show relatively low variation in their absolute values, i.e. 〈G0〉 = (2.0 ± 0.9) kPa as obtained from N = 20 different growth batches. In combination, these findings suggest that despite their biological and structural complexity, P.a. biofilms can indeed be treated as a viscoelastic material with well-defined properties.
2. Pseudomonas aeruginosa biofilms yield at high deformations but are able to fully regain their initial stiffness
In their natural environment, biofilms are often exposed to large forces as, for example, created by shear flow in tubes or pipes. At high forces, Hooke’s law does not apply anymore and the viscoelastic response of polymer materials becomes non-linear until the material finally yields18–20. Thus, we next probe the mechanical response of P.a. biofilms in the non-linear regime, i.e. at high deformations. We apply a constant shear rate of dγ/dt = 12.5%/s to the P.a. biofilm and measure its resulting stress state σ. From the corresponding smoothed stress-strain relation the tangent modulus K = dσ/dγ is calculated, which is a suitable parameter for describing the stiffness of biopolymer networks in the non-liner regime21. At small deformations, we observe a highly similar elasticity value as obtained from the linear frequency spectrum. However, above a critical deformation, i.e. around a few percent, the biofilm elasticity starts to drop by more than three orders of magnitude (Fig. 1C), which we refer to as yielding. After such a drastic yielding event one might expect that the material properties of the P.a. biofilm would be significantly and permanently altered. Yet, the opposite behavior is observed here: if biofilms are left undisturbed for a few minutes, they fully recover and their viscoelastic frequency spectra almost perfectly match the initial conditions, i.e. before the yielding step was conducted (Fig. 1D, Fig. 1B).
3. Pseudomonas aeruginosa biofilms are robust towards many chemical perturbations
The biofilms investigated so far have been grown on standard LB-agar. However, in their natural environment, be it either in the human body or in pipes, tubes, and catheters, P.a. biofilms encounter various chemical conditions. We now aim to quantify the influence of different chemicals on the viscoelastic properties of P.a. biofilms. We choose a spectrum of chemical agents that was suggested to affect biofilm properties such as growth or development. The tested molecules and sample conditions range from mono- and divalent ions, polyelectrolytes and distinct pH levels to potential nutrients and are listed in table 1. We add 5% (v/w) of a certain chemical agent to the P.a. biofilm, which is then gently stirred with a pipette tip to homogenize the sample. As a control, we first test the impact of adding 5% (v/w) water to the biofilm. This procedure entails a softening of the material by ~40% which is significant but still a relatively weak effect as its magnitude is comparable to the variation in biofilm elasticities between different growth batches. We use this value as a base line for all further experiments (dashed line in Fig. 2A).
Table 1.
List of chemicals used in this study and summary of their effects on P.a. biofilms
| Chemical | Mode of action | Effect on elasticity * |
Effect on recovery |
Effect on viability ** |
|---|---|---|---|---|
| NaCl | osmotic stress inducer, inhibitor of biofilm formation34 | NO | NO | NO |
| KH2PO4 | osmotic stress inducer | NO | NO | NO |
| CaCl2 | regulator of alginate gel elasticity35 | NO | NO | NO |
| CuSO4 | bactericidal36, inhibitor of normal biofilm growth36 | NO | NO | NO |
| FeCl2 | non-metabolizable analog of FeCl3 | − | NO | NO |
| FeCl3 | necessary metabolite of bacterial growth37 | +++ | NO | NO |
| Al2(SO4)3 | trivalent ion, control for Fe(III) | +++ | NO | NO |
| pH | control for pH changes induced by other chemicals | NO | NO | NO |
| Salicylic acid | quorum sensing inhibitor38 | NO | NO | NO |
| Citric acid | ion chelating agent, putative biofilm removal agent39, antimicrobial agent40, 41 | − − | NO | YES |
| EDTA | ion chelating agent, putative biofilm removal agent39, inhibitor of biofilm formation42 | − | NO | NO |
| EGTA | calcium chelating agent | − | NO | NO |
| Lactoferrin | iron chelating protein, antimicrobial43 | NO | NO | NO |
| Spermidine | polyelectrolyte | NO | NO | NO |
| Gentamycin | bactericidal: disrupts protein synthesis | NO | NO | NO |
| Colistin | bactericidal: disrupts the integrity of bacterial outer membrane | NO | NO | NO |
| Ofloxacin | bactericidal: disrupts separation of replicated DNA | NO | NO | YES |
| Amylase | enzyme present in the oral cavity | NO | NO | NO |
| Ethanol | antimicrobial agent | NO | NO | NO |
| Triton | detergent | NO | NO | NO |
| Bleach | antimicrobial agent | NO | NO | YES |
| Glucose | nutrient | NO | NO | NO |
| Urea | potential biofilm removal agent39 | NO | NO | NO |
| Xylitol | disrupts biofilm function43 | NO | NO | NO |
| Hexandiol | interferes with hydrophobic interactions | NO | NO | NO |
chemicals with a weakening effect (reduction of biofilms stiffness by ~90%) are marked with (−); chemicals with a strong weakening effect (reduction of biofilm stiffness by ~99 %) are marked with (− −); chemicals inducing a fortification effect are marked with (+); we only detect very strong fortification effects and those are marked as (+++) accordingly
chemicals are defined effective if they reduce bacterial viability by more than 95%
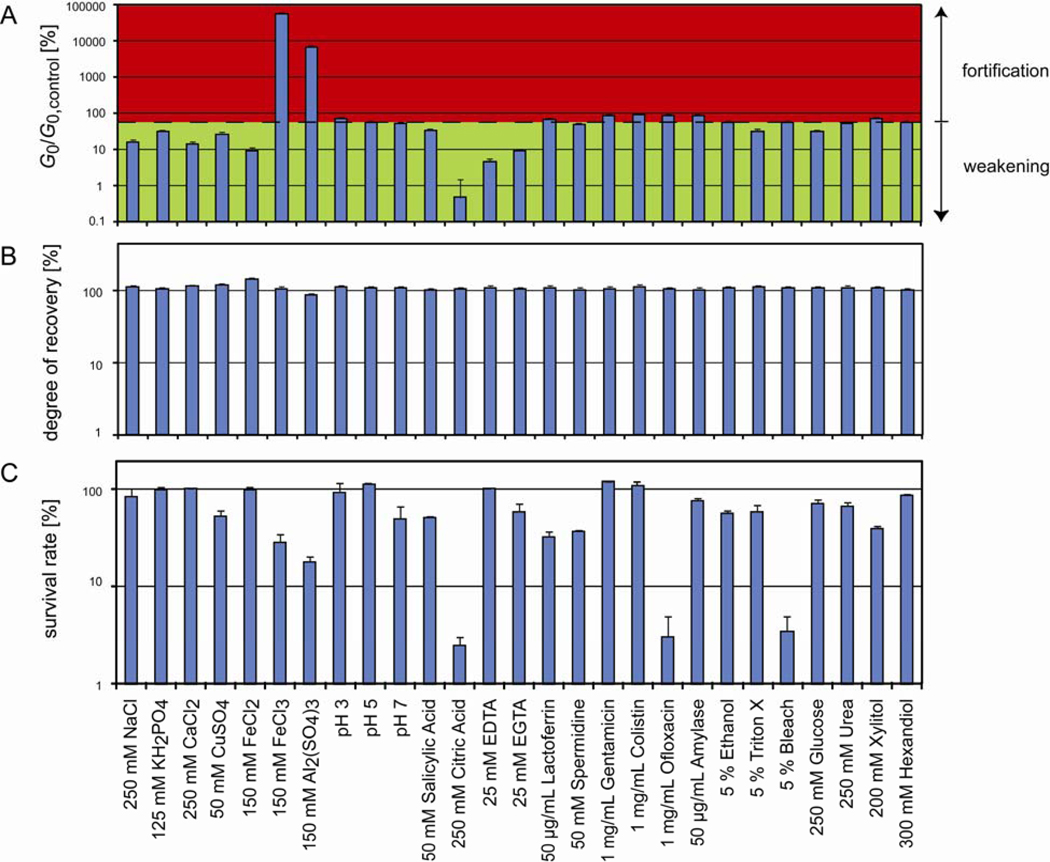
Figure 2.
Influence of different chemical environments on the properties of P.a. biofilms. (A) Among all listed conditions, mainly the addition of Fe(III), Al3+ or citric acid significantly alters the elasticity of the biofilm. The dashed base line indicates the elasticity of a P.a. biofilm modified with 5% (v/w) water only. (B) For all conditions tested in this study, the biofilm shows an almost complete recovery after mechanical yielding. (C) Survival rate of P.a. bacteria as determined from serial dilutions of biofilms that have been challenged with different chemicals. With our experimental conditions, mainly citric acid, ofloxacin and bleach significantly reduce bacteria viability. All measurements are performed in duplicates, the error bars denote the error of the mean.
Next, we analyze P.a. biofilms that have been challenged with different chemical agents (Fig. 2A). Surprisingly, our screen does not return major changes in the biofilm elasticity for most of the conditions tested. However, we observe that citric acid is highly potent in further fluidizing the biofilm compared to treatment with water alone, as 250 mM citric acid reduces the biofilm elasticity by almost 99.5%. In contrast, the addition of 150 mM Fe(III) balances the softening invoked by the dilution with water and induces a strong fortification effect, i.e. a ~500fold increase in the biofilm elasticity G0. This fortification effect of Fe(III) can be countered by adding citric acid together with the Fe(III) ions (Fig. 3C). This indicates that Fe(III) and citric acid either act antagonistically or form an inactive complex that does not affect the biofilm elasticity anymore. A similar enhancement of the biofilm elasticity as observed for Fe(III), albeit less pronounced, can be achieved by the addition of aluminum sulfate, which also forms trivalent ions (Al3+) in aqueous solutions. Strikingly, iron ions in their reduced form, Fe(II), as well as other divalent ions such as Ca2+ and Cu2+ have the opposite effect and cause a slight but significant weakening of the biofilm elasticity compared to the addition of water alone (Fig. 2A).
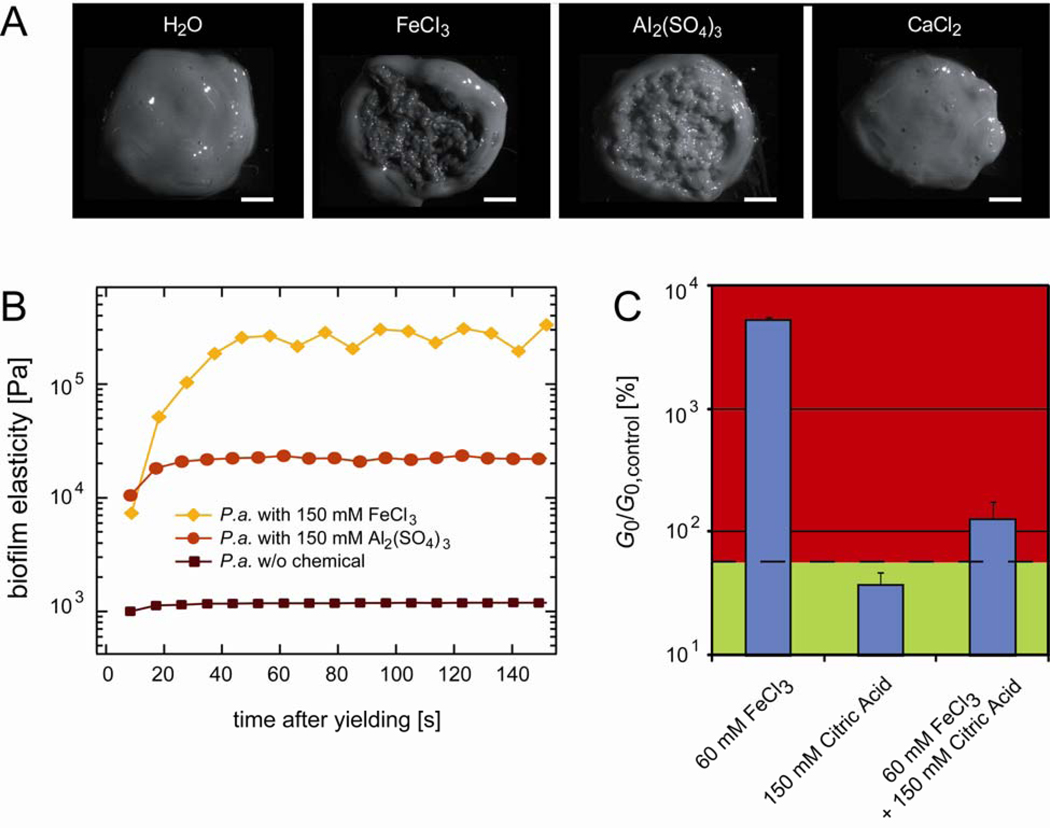
Figure 3.
Response of Pseudomonas aeruginosa biofilms to trivalent ions. (A) Texture of collected P.a. biofilms after a drop of a chemical solution is added to the center of the biofilm and distributed by gentle stirring. 250 mM Ca2+ ions do not alter the biofilm texture compared to the control, whereas 150 mM Fe(III) and 150 mM Al3+ evoke a granular, sand-like texture. The scale bars denote 2 mm. (B) The speed of the mechanical recovery process of P.a. biofilms depends on the biofilm elasticity. PAO1 biofilms without any chemical additions fully recover within ~20 s after yielding. This recovery time increases up to ~1min if the biofilm is treated with 150 mM Al3+ or Fe(III), respectively. (C) Fe(III) and citric acid act antagonistically and inhibit each other. For all data shown, 5% (v/w) of an aqueous solution containing different amounts of chemicals were added to the biofilm material. The dashed base line indicates the elasticity of a P.a. biofilm modified with 5% (v/w) water only.
Together with increasing the elasticity of the biofilm, we observe that both Fe(III) and Al3+ also change the texture of the P.a. biofilm. With both substances, the material acquires a more granular, sand-like quality (Fig. 3A). In contrast, divalent ions such as Ca2+ do not induce such an alteration in the biofilm texture, which is consistent with our rheological results. It is important to note that the change in biofilm texture forms immediately when the ions are mixed into the biofilm. This indicates that this change in biofilm morphology probably does not require a metabolic response from the bacteria within the biofilm. Instead, the external chemical stimulus seems to directly act on the biofilm polymers or on their interaction with the bacteria.
We next test if the alterations in biofilm elasticity induced by the addition of certain chemicals are due to an antimicrobial effect of these chemicals. For this purpose, we determine the survival rate of bacteria embedded in the biofilm by challenging them with different chemicals (see Methods). By normalizing the number of bacterial colonies to that from chemically unchallenged biofilm material we determine the bacterial survival rate for each chemical modification. We observe that 250 mM citric acid, 1 mg/mL ofloxacin and 5% (v/w) bleach reduce the bacterial viability by >95% whereas all other chemicals show only weak or no effects (Fig. 2C). It is important to recall that neither ofloxacin nor bleach had a significant influence on the elasticity of P.a. biofilms. This demonstrates that bacteria viability and biofilm mechanics are not necessarily linked. For comparison, citric acid not only substantially fluidizes the biofilm material but has also a strong anti-microbial effect on the bacteria inside the biofilm matrix.
In a biofilm, bacteria interact with the biofilm polymers, which in turn might also bind to each other. The remarkable self-healing properties of the biofilm as observed in Fig. 1D suggest that most of these interconnections as well as the interactions between bacteria and biofilm polymers are transient and can thus be easily reestablished once they are ruptured. These transient polymer-polymer or polymer-bacteria interactions, in turn, might offer various possibilities to interfere with the biofilm integrity by chemical perturbations. Thus, we next aim to screen the influence of the same chemical perturbations on the self-mending properties of P.a. biofilms. To quantify the degree of mechanical recovery we determine the ratio of the P.a. biofilm elasticity G0 before and after yielding. As depicted in Fig. 2B, none of the conditions tested can suppress or even significantly weaken the self-mending abilities of P.a. biofilms. This robustness of the biofilm is startling and underlines its resistance towards mechanical or chemical removal. However, as depicted in Fig. 3B, in the presence of 150 mM Al3+ or Fe(III), i.e. with increasing biofilm stiffness, the time required for complete recovery increases from ~20 s (w/o added chemical) up to ~1 min.
4. Other Pseudomonas biofilms show similar properties as Pseudomonas aeruginosa biofilms
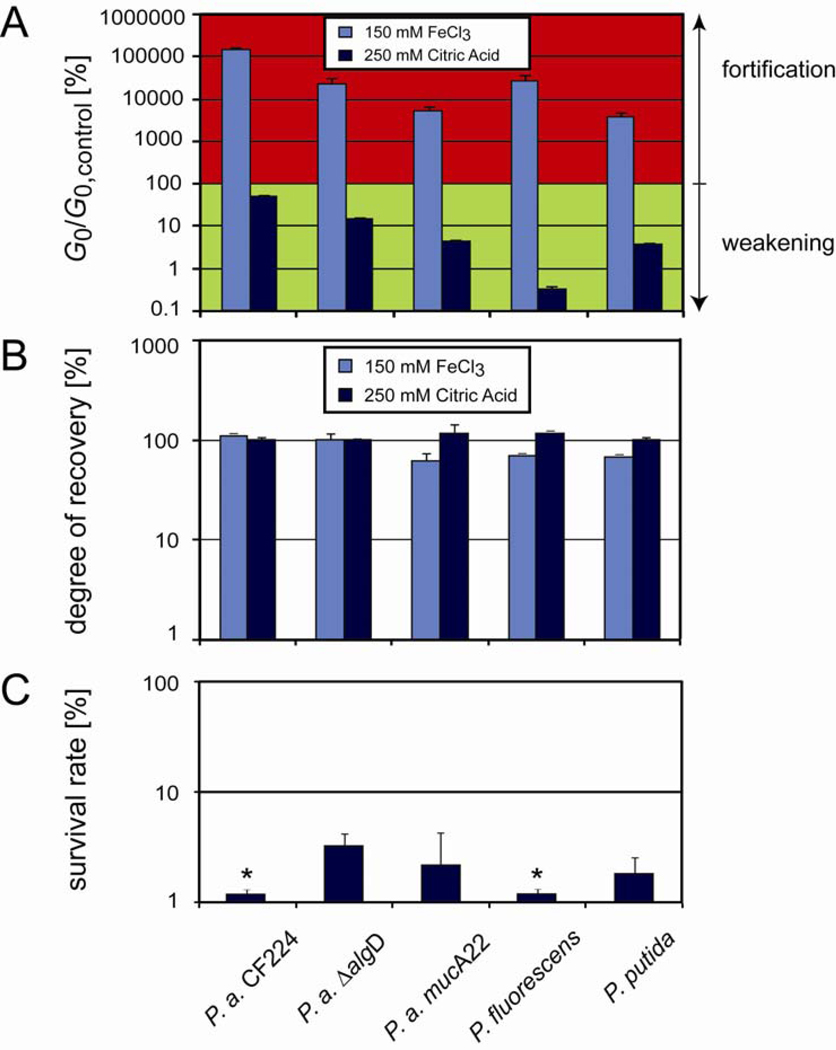
Last, we test if the mechanical robustness observed for Pseudomonas aeruginosa PAO1 biofilms also applies to other Pseudomonas strains. We first examine a Pseudomonas aeruginosa strain (P.a. CF224) that has been isolated from the mucosal surface of a cystic fibrosis patient, and two P.a. mutants for which the expression level of alginate, a component of the EPS, is altered (P.a. ΔalgD and P.a. mucA22). Furthermore, we also study biofilms formed by two different Pseudomonas species, P. fluorescens and P. putida. All these strains are listed in table 2. The immediate formation of a granular texture after exposure to Fe(III) is observable for all Pseudomonas strains (data not shown), similar to what we show for the PAO1 strain in Fig. 3A. In addition, all Pseudomonas strains also show a pronounced fortification in their elastic moduli in the presence of 150 mM Fe(III) and a significant fluidization in presence of 250 mM citric acid (Fig. 4A). Yet, all Pseudomonas variants tested here are able to fully recover their initial stiffness after yielding is enforced, both in the absence (data not shown) and presence of 150 mM Fe(III) and 250 mM citric acid, respectively (Fig. 4B). Moreover, we find that citric acid reduces the viability of biofilm bacteria by >95% for all Pseudomonas variants (Fig. 4C) similar to what we find for PAO1 in Fig. 2C. In conclusion, all major effects we describe for the PAO1 strain can also be identified for five other Pseudomonas strains.
Table 2.
List of bacterial strains used for this study
| Bacterial strain | Characteristics | Described in |
|---|---|---|
| P. aeruginosa PAO1 | wound isolate | reference 44 |
| P. aeruginosa CF224 | cystic fibrosis isolate | – |
| P. aeruginosa ΔalgD | mutant unable to produce alginate | reference 45 |
| P. aeruginosa mucA22 | mucoid mutant | reference 45 |
| P. fluorescens PFO1 | soil isolate | reference 46 |
| P. putida KT2440 | soil isolate | reference 47 |
Figure 4.
Response of other Pseudomonas biofilms to chemical and mechanical treatment. (A) The fortification and weakening effects of Fe(III) and citric acid observed for Pseudomonas aeruginosa can also be identified for other Pseudomonas strains, yet at different efficiencies. (B) Almost full mechanical recovery after yielding is also observed for those other Pseudomonas strains, even in the presence of Fe(III) and citric acid. Measurements are performed in duplicates, the error bars denote the error of the mean. (C) Survival rate of different Pseudomonas bacteria as determined from serial dilutions of biofilms that have been challenged with 250 mM citric acid. In all cases, the bacteria viability is efficiently reduced. All measurements are performed in duplicates, the error bars denote the error of the mean. Bars labelled with a ‘*’ represent survival rates that are smaller than 1%.
Summary and discussion
In this study, we aimed at characterizing the viscoelastic properites of Pseudomonas aeruginosa biofilms and their response to chemical treatment and large shear forces. We find that biofilms from various Pseudomonas strains are able to efficiently recover from mechanical damage. One implication of this observation is that, high shear forces, as they for example occur in water pipes, might transiently fragment biofilms22, 23, however, these biofilm fragments could fuse with neighboring biofilm parts and will thereby be dispersed rather than removed.
Moreover, we find that the viscoelastic properties of PAO1 biofilms remain unchanged for most of the treatments tested here. This information is relevant for biofilm removal strategies, which employ mechanical force, the use of chemicals, or a combination of both. As we show here, even if the viscoelastic properties of biofilms might sometimes be sensitive towards certain chemicals24–26, their recovery abilities are slowed down at best. This demonstrates the urgent need to identify and/or develop new chemical agents for biofilm removal.
For such an endeavor, it is a priori not clear, which biofilm component is the most promising target. One strategy might be to reduce bacterial viability inside the biofilm matrix. However, our results indicate that, whereas some chemicals kill most bacteria inside a biofilm matrix, the material properties of the biofilm may nevertheless remain unharmed. Thus, the remaining biofilm layer could serve as a “hibernation” site for a few surviving bacteria which would lead to a continuation of biofilm growth once the chemical challenge subsides.
Another possibility could be to target certain biopolymers in the EPS. Some studies suggest that alginate is a key component of the EPS and a good model system for understanding the viscoelastic properties of bacterial biofilms24, 27. Indeed, the viscosity of alginate gels increases with increasing Fe(III) concentrations28, which is in qualitative agreement with our results. However, the viscoelasticity of alginate gels can also be enhanced by Ca2+ ions27, which is qualitatively different from what we observe here for PAO1 biofilms. Furthermore, our results on the ΔalgD mutant, which lacks the expression of alginate, demonstrate that alginate is not sufficient to explain the mechanical properties of PAO1 biofilms. This conclusion would also be consistent with the recent finding that the structural profiles and antibiotic-resistance of wild-type PAO1 and ΔalgD mutant biofilms are identical29.
Upon addition of trivalent ions such as Fe(III) or Al3+ we have observed the immediate formation of a granular structure in Pseudomonas biofilms. A similar aggregation event has been described before as “bioflocculation” and was suggested to arise from interactions between trivalent ions such as Fe(III) and negatively charged groups in the EPS of sludge30, 31. Fe(III) can form complexes with citric acid, and indeed we find here that a simultaneous application of Fe(III) and citric acid does not change the biofilm mechanics. One might speculate that, in the absence of citric acid or other iron chelating agents, Fe(III) ions form similar complexes with certain biofilm biopolymers, leading to a local aggregation of biofilm material.
Here, we have identified citric acid to be both a potent fluidization agent and antimicrobial chemical for various pre-formed Pseudomonas biofilms. It remains to be shown if this anti-biofilm activity of citric acid also extends to other biofilm forming bacteria. We speculate that the effectiveness of other antimicrobial agents could be enhanced when the biofilm stability is weakened by, for example, citric acid.
Material and methods
Strains and growth conditions
In this study, biofilms formed by different bacterial strains are characterized. The biological characteristics of these strains are listed in table 1. All strains were first grown in liquid Luria Bertani (LB) medium for 16 hrs at 37 °C for P. aeruginosa and at 30 °C for both P. fluorescens and P. putida. The bacteria solutions were then diluted and plated on LB media containing 1.5% agar. At high cellular density a structured slimy community (a “biofilm”) was formed after 2 days of incubation at 37 °C – similarly to what has been described previously32. P. fluorescens and P. putida were grown in the same conditions as described above but at 30 °C.
Rheometry
Rheological measurements were performed using a commercial rheometer (AR-G2, TA instruments, New Castle, USA) with a 20 mm plate-plate geometry and 300 µm plate separation in strain-controlled mode. Biofilms were collected from agar plates by gentle scraping similar to what has been described in ref. 33. After collecting, the biofilm material was pooled, transferred into tubes, and then loaded into the rheometer. Rheological measurements were conducted at 22 °C. Frequency spectra are obtained using small torques (~ 1 µNm) to guarantee linear response. For non-linear measurements (yielding tests) a constant shear rate of 12.5%/s was applied for 8 s and the resulting stress-strain relation was analyzed. After rupture, samples were left undisturbed for 4 min to allow for recovery before their elasticity was probed again by another frequency sweep. The experimental errors for these rheological measurement are small (< 10%) as confirmed by multiple frequency sweep measurements on the same sample. To analyze the effect of chemicals on the viscoelastic properties of the biofilms, the samples were transferred to a sterile tube and weighed. Chemical modifications were performed by adding 5% (v/w) of a certain chemical to the sample and gentle mixing with a pipette-tip. We prepared our chemical stock solutions at as high concentrations as possible to maximize putative effects. All chemicals were incubated with the biofilm for one hour before their effect on the viscoelastic properties of the biofilm were analyzed by rheometry.
Biofilm imaging and viability assay
For biofilm imaging, a SteREO Lumar V12 microscope (Zeiss) equipped with a NeoLumar S 0.8× objective was used. Pseudomonas aeruginosa was grown as described previously and the biofilm was then collected onto a plastic plate. Before imaging, a drop of either water or of a solution of CaCl2, Al2(SO4)3 or FeCl3 was spotted in the middle of the specimen and gently mixed into the biofilm using a pipette tip.
For testing bacterial viability, scraped biofilm material was challenged with different chemicals for ~1 h, then serially diluted in combination with bead bashing and vortexing, and plated onto LB agar. After incubation over night, the number of colony forming units were counted and compared to those from unchallenged biofilms that were diluted and plated the same way.
Acknowledgements
We thank the McKinley lab at MIT for sharing their rheology equipment. K. Foster provided us with the PAO1 and KT2440 strains, K. Mathee the ΔalgD and mucA22 strain, R. Kolter CF224 and S.B. Levy PFO1, which is gratefully acknowledged. We thank Samuel Francis for conducting pilot experiments for this study. This project was funded by NIH (P50-GM068763 and P30-ES002109). OL was supported by a postdoc fellowship, and RB by a short time research fellowship, from the German Academic Exchange Service (DAAD).
References
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappinscott HM. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Monds RD, O'Toole GA. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Branda SS, Vik A, Friedman L, Kolter R. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Mayer C, Moritz R, Kirschner C, Borchard W, Maibaum R, Wingender J, Flemming HC. Int. J. Biol. Macromol. 1999;26:3–16. doi: 10.1016/s0141-8130(99)00057-4. [DOI] [PubMed] [Google Scholar]
- 5.Flemming HC, Wingender J. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 6.Donlan RM. Emerg. Infect. Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall-Stoodley L, Stoodley P. Cellular microbiology. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 8.Tolker-Nielsen T, Brinch UC, Ragas PC, Andersen JB, Jacobsen CS, Molin S. J. Bacteriol. 2002;182:6482–6489. doi: 10.1128/jb.182.22.6482-6489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiirola M, Lahtinen T, Vuento M, Oker-Blom C. J. Ind. Microbiol. Biotechnol. 2009;36:929–937. doi: 10.1007/s10295-009-0571-6. [DOI] [PubMed] [Google Scholar]
- 10.Raulio M, Wilhelmson A, Salkinoja-Salonen M, Laitila A. Food Microbiol. 2009;26:437–443. doi: 10.1016/j.fm.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi H, Miya S, Igarashi K, Suda T, Kuramoto S, Kimura B. J. Food Prot. 2009;72:1476–1480. doi: 10.4315/0362-028x-72.7.1476. [DOI] [PubMed] [Google Scholar]
- 12.Fux CA, Costerton JW, Stewart PS, Stoodley P. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Lau PCY, Dutcher JR, Beveridge TJ, Lam JS. Biophys. J. 2009;96:2935–2948. doi: 10.1016/j.bpj.2008.12.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson DM, Cavanagh HD. Clin. Ophthalmol. 2008;2:907–917. doi: 10.2147/opth.s3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campodonico VL, Gadjeva M, Paradis-Bleau C, Uluer A, Pier GB. Trends Mol Med. 2008;14:120–133. doi: 10.1016/j.molmed.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickel JC, Ruseska I, Wright JB, Costerton JW. Antimicrob. Agents Chemother. 1985;27:619–624. doi: 10.1128/aac.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubinstein M, Colby RH. Polymer Physics. Oxford University Press; 2003. [Google Scholar]
- 18.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nature. 2005;435:191–194. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 19.Lieleg O, Claessens MMAE, Bausch AR. Soft Matter. 2010;6:218–225. [Google Scholar]
- 20.Korstgens V, Flemming HC, Wingender J, Borchard W. J. Microbiol. Methods. 2001;46:9–17. doi: 10.1016/s0167-7012(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 21.Semmrich C, Larsen RJ, Bausch AR. Soft Matter. 2008;4:1675–1680. doi: 10.1039/b800989a. [DOI] [PubMed] [Google Scholar]
- 22.Stoodley P, Cargo R, Rupp CJ, Wilson S, Klapper I. J. Ind. Microbiol. Biotechnol. 2002;29:361–367. doi: 10.1038/sj.jim.7000282. [DOI] [PubMed] [Google Scholar]
- 23.Dunsmore BC, Jacobsen A, Hall-Stoodley L, Bass CJ, Lappin-Scott HM, Stoodley P. J. Ind. Microbiol. Biotechnol. 2002;29:347–353. doi: 10.1038/sj.jim.7000302. [DOI] [PubMed] [Google Scholar]
- 24.Korstgens V, Flemming HC, Wingender J, Borchard W. Water Sci. Technol. 2001;43:49–57. [PubMed] [Google Scholar]
- 25.Stoodley P, Jacobsen A, Dunsmore BC, Purevdorj B, Wilson S, Lappin-Scott HM, Costerton JW. Water Sci. Technol. 2001;43:113–120. [PubMed] [Google Scholar]
- 26.Chen X, Stewart PS. Appl. Microbiol. Biotechnol. 2002;59:718–720. doi: 10.1007/s00253-002-1044-2. [DOI] [PubMed] [Google Scholar]
- 27.Wloka M, Rehage H, Flemming HC, Wingender J. Colloid Polym. Sci. 2004;282:1067–1076. [Google Scholar]
- 28.Sreeram KJ, Shrivastava HY, Nair BU. Biochim. Biophys. Acta-Gen. Subj. 2004;1670:121–125. doi: 10.1016/j.bbagen.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O'Toole GA, Parsek MR. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang HF, Sun BS, Zhao XH, Gao ZH. Sep. Purif. Technol. 2008;63:341–347. [Google Scholar]
- 31.Yu G-H, He P-J, Shao L-M. Bioresource Technology. 2009;100:3193–31998. doi: 10.1016/j.biortech.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Mayer C, Moritz R, Kirschner C, Borchard W, Maibaum R, Wingender J, Flemming HC. International journal of biological macromolecules. 1999;26:3–16. doi: 10.1016/s0141-8130(99)00057-4. [DOI] [PubMed] [Google Scholar]
- 33.Shaw T, Winston M, Rupp CJ, Klapper I, Stoodley P. Physical Review Letters. 2004;93 doi: 10.1103/PhysRevLett.93.098102. 098102. [DOI] [PubMed] [Google Scholar]
- 34.Bazire A, Diab F, Jebbar M, Haras D. J Ind Microbiol Biotechnol. 2007;34:5–8. doi: 10.1007/s10295-006-0087-2. [DOI] [PubMed] [Google Scholar]
- 35.Korstgens V, Flemming HC, Wingender J, Borchard W. Water Sci Technol. 2001;43:49–57. [PubMed] [Google Scholar]
- 36.Harrison JJ, Turner RJ, Joo DA, Stan MA, Chan CS, Allan ND, Vrionis HA, Olson ME, Ceri H. Antimicrob Agents Chemother. 2008;52:2870–2881. doi: 10.1128/AAC.00203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cornelis P. Appl Microbiol Biotechnol. 2010;86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 38.Yang L, Rybtke MT, Jakobsen TH, Hentzer M, Bjarnsholt T, Givskov M, Tolker-Nielsen T. Antimicrob Agents Chemother. 2009;53:2432–2443. doi: 10.1128/AAC.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittaker C, Ridgway H, Olson BH. Appl Environ Microbiol. 1984;48:395–403. doi: 10.1128/aem.48.2.395-403.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laury AM, Alvarado MV, Nace G, Alvarado CZ, Brooks JC, Echeverry A, Brashears MM. J Food Prot. 2009;72:2208–2211. doi: 10.4315/0362-028x-72.10.2208. [DOI] [PubMed] [Google Scholar]
- 41.Nagoba BS, Gandhi RC, Hartalkar AR, Wadher BJ, Selkar SP. Burns. 2010;36:1242–1247. doi: 10.1016/j.burns.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 42.O'May CY, Sanderson K, Roddam LF, Kirov SM, Reid DW. J Med Microbiol. 2009;58:765–773. doi: 10.1099/jmm.0.004416-0. [DOI] [PubMed] [Google Scholar]
- 43.Ammons MC, Ward LS, Fisher ST, Wolcott RD, James GA. Int J Antimicrob Agents. 2009;33:230–236. doi: 10.1016/j.ijantimicag.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holloway BW. J. Gen. Microbiol. 1995;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 45.Stapper AP, Narasimhan G, Ohman DE, Barakat J, Hentzer M, Molin S, Kharazmi A, Hoiby N, Mathee K. J. Med. Microbiol. 2004;53:679–690. doi: 10.1099/jmm.0.45539-0. [DOI] [PubMed] [Google Scholar]
- 46.Compeau G, Alachi BJ, Platsouka E, Levy SB. Appl. Environ. Microbiol. 1998;54:2432–2438. doi: 10.1128/aem.54.10.2432-2438.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Regenhardt D, Heuer H, Heim S, Fernandez DU, Strompl C, Moore ERB, Timmis KN. Environ. Microbiol. 2002;4:912–915. doi: 10.1046/j.1462-2920.2002.00368.x. [DOI] [PubMed] [Google Scholar]