Abstract
The intestinal microbiota plays an important role in modulation of mucosal immune responses. To seek interactions between intestinal epithelial cells (IEC) and commensal bacteria, we screened 49 commensal strains for their capacity to modulate NF-κB. We used HT-29/kb-seap-25 and Caco-2/kb-seap-7 intestinal epithelial cells and monocyte-like THP-1 blue reporter cells to measure effects of commensal bacteria on cellular expression of a reporter system for NF-κB. Bacteria conditioned media (CM) were tested alone or together with an activator of NF-κB to explore its inhibitory potentials. CM from 8 or 10 different commensal species activated NF-κB expression on HT-29 and Caco-2 cells, respectively. On THP-1, CM from all but 5 commensal strains stimulated NF-κB. Upon challenge with TNF-α or IL-1β, some CM prevented induced NF-κB activation, whereas others enhanced it. Interestingly, the enhancing effect of some CM was correlated with the presence of butyrate and propionate. Characterization of the effects of the identified bacteria and their implications in human health awaits further investigations.
1. Introduction
The adult human gut is populated with a large number of commensal bacteria known to influence many aspects of the host gut physiology, including immunity, development, and homeostasis. There is considerable clinical and experimental evidence showing that commensal gut bacteria contribute to immune homeostasis by altering microbial balance or by interacting with the gut immune system [1]. However, cellular and molecular mechanisms by which individual members of the commensal microbiota contribute to immune homeostasis have not been completely elucidated.
Intestinal epithelial cells (IEC) are the first point of contact for bacteria within the gut lumen, and they interact with the gut immune system. Consequently, IEC have a pivotal function in bacteria-host communication. Bacterial signatures generally activate signaling cascades that can trigger proinflammatory gene transcription through specific receptors (e.g., Toll-like receptors) expressed on apical and/or basolateral surface of epithelial cells. This mechanism is largely controlled by the transcriptional factor NF-κB [2]. NF-κB is a dimeric DNA binding protein whose major form is represented by the association of p65 and p50 proteins. In steady state, NF-κB is locked in the cytoplasm by an inhibitory protein of the IκB family. Upon receptor activation, IκB is phosphorylated by the IκB kinase complex (IKK) before undergoing degradation by the proteasome. Then, free NF-κB translocate to the nucleus to turn on a large number of genes involved in proinflammatory processes at the site of infection or tissue damage.
Obviously, the intestinal epithelium does not trigger inflammatory responses against commensal bacteria. Interestingly, the mechanisms allowing commensal micro-organisms to be tolerated by the intestinal mucosa are far from being completely understood. Numerous studies have suggested an involvement of active processes causing a functional downregulation of the inflammatory response that is generally obtained by interference with the NF-κB pathway. Indeed, previous reports showed the suppression of inflammatory responses in epithelial cells by commensal bacteria either through direct interaction leading to alteration in Toll-like receptors (TLRs) signaling to NF-κB [3, 4] or through direct inhibition of NF-κB transcriptional activity [5]. In addition, some secreted bacterial factors resulting from commensals or probiotics activity have been found to exert anti-inflammatory effects on IEC [6–10].
Furthermore, recent work has ascribed a critical role for NF-κB signaling in maintenance of homeostatic immuno-inflammatory function in the gut [11]. Indeed epithelial NF-κB preserves integrity of the gut epithelial barrier and coordinates antimicrobial actions of the innate and adaptive immune systems. Accordingly, both deficiency in, and hyperactivation of, this transcription factor are underlying factors in chronic inflammatory bowel diseases [12–16].
Finally, determining the microbial factors that positively or negatively regulate this key pathway is of great clinical and scientific interest. Thus, in the present study, we examined the effect of 49 commensal bacteria on the modulation of NF-κB pathway in human IEC models bearing a stable NF-κB-SEAP reporter system. Conditioned media (CM) from these bacteria were tested either alone or in combination with an activator of NF-κB signaling (TNF-α or IL-1β) to identify its inhibitory and enhancing potentials. To compare response profiles between IEC and immune cells, all CM were also tested on human monocyte cell line THP-1 bearing the same NF-κB reporter system.
2. Material and Methods
2.1. Cell Culture and Reagents
HT-29 cells were grown in DMEM (Sigma) with 2 mM L-glutamine, 50 IU/mL penicillin, 50 μg/mL streptomycin and 10% heat-inactivated fetal calf serum (FCS-Lonza) in a humidified 5% CO2 atmosphere at 37°C. Caco-2 were cultured in DMEM (Sigma) supplemented with the same amounts of glutamine and antibiotics, 20% heat-inactivated FCS and 1x nonessential amino acids (Invitrogen). The THP-1 blue CD14+ NF-κB reporter clone was obtained from Invivogen and used according to the manufacturer's instruction.
2.2. Commensal Strains and Preparation of Conditioned Media
The 49 commensal strains were grown in anaerobic condition at 37°C using the Hungate culture method [17]. Screened strains and corresponding growth media are listed in Table 1.
Table 1.
Bacteria strains, references, and growth media.
| Designation | Collection reference | Medium | Phylum |
|---|---|---|---|
| Atopobium parvulum | DSM 20649T | M104 | A |
| Bifidobacterium adolescentis | ATCC 15703 | M58 | A |
| Bifidobacterium angulatum | ATCC 27535-CIP 104167 | M58 | A |
| Bifidobacterium animalis ssp. animalis | DSM 20104 | M58 | A |
| Bifidobacterium bifidum | DSM20082/JCM1254 | M58 | A |
| Bifidobacterium breve 1 | DSMZ 20091 | M104 | A |
| Bifidobacterium breve 2 | ATCC 15701-89/23 | M104 | A |
| Bifidobacterium catenulatum | ATCC27539-CIP104175 | M58 | A |
| Bifidobacterium choerinum | DSM 20434 | M58 | A |
| Bifidobacterium dentium 1 | ATCC 15423 | M104 | A |
| Bifidobacterium dentium 2 | ATCC 27534-89/20 | M104 | A |
| Bifidobacterium gallicum | ATCC 49850-CIP 103417 | M58 | A |
| Bifidobacterium infantis | DSM20088/ATCC15697 | M58 | A |
| Bifidobacterium longum | ATCC 15707-NCTC 11818 | M58 | A |
| Bifidobacterium pseudocatenulatum | DSM 20438-ATCC 27919 | M58 | A |
| Bifidobacterium ruminantium | ATCC 49390 | M58 | A |
| Collinsella aerofaciens | ATCC 25986 | BHI** | A |
| Propionibacterium acnes | B15 | M104 | A |
| Bacteroides caecae | ATCC 43185-CIP 104201T | BHI | B |
| Bacteroides dorei | DSM 17855 | M104 | B |
| Bacteroides fragilis | B6-AL 2553 | BHI | B |
| Bacteroides ovatus | ATCC 8483-CIP103756 | M104 | B |
| Bacteroides thetaiotaomicron | ATCC 29148-VPI5482 | M104 | B |
| Bacteroides uniformis | ATCC 8492 | M104 | B |
| Bacteroides vulgatus | ATCC 8482-CIP 103714 | BHI | B |
| Parabacteroides distasonis | CIP104284-ATCC8503 | BHI | B |
| Parabacteroides johnsonii | DSM 18315 | M104 | B |
| Prevotella copri | DSM 18205 | M104 | B |
| Blautia/Clostridium coccoides | ATCC 2936 | BHI | F |
| Blautia producta/Ruminococcus productus | DSMZ 2950-92/46 | M104 | F |
| Blautia/Ruminococcus hansenii | DSM 20583 | M104 | F |
| Butyrivibrio fibrisolvens | DSM3071 | BHI | F |
| Clostridium leptum | ATCC29065-DSM753 | BHI | F |
| Clostridium nexile 96/2 | ATCC 27757-DSM 1787 | BHI | F |
| Clostridium paraputrificum | G12PR-X73445 | BHI | F |
| Clostridium sardiniensis | I11PR | M78 | F |
| Clostridium sordellii | ATCC 9714 | BHI | F |
| Clostridium sporosphaeroides | ATCC 25781 | M78 | F |
| Dorea/Eubacterium formicigenerans | ATCC 27755 | L-DON* | F |
| Eubacterium rectale | ATCC 33656-CIP105953 | BHI | F |
| Faecalibacterium prausnitzii | L2-6 (Duncan) | BHI | F |
| Roseburia faecis | DSM 16840 | BHI | F |
| Roseburia hominis | DSM 16839 | M58 | F |
| Roseburia intestinalis | DSM 14610 | M58 | F |
| Ruminococcus gnavus | FRE1 | L-DON | F |
| Ruminococcus lactaris | ATCC 29176 | L-DON | F |
| Ruminococcus obeum | ATCC 29174 | L-DON | F |
| Ruminococcus torques | ATCC 27756 | M104 | F |
| Selenomonas ruminantium | ATCC19205-DSM2872 | L-DON | F |
*Yeast extract-CaCl2—sodium thioglycholate—pyruvic acid.
**Brain heart infusion + yeast extract and hemine.
A: Actinobacteria; B: Bacteroidetes; F: Firmicutes.
At the end of the incubation period, bacterial cultures were centrifuged at 5,000 xg for 10 minutes. Bacteria conditioned media (CM) were then collected and filtered on 0.2 μm PES filters. Noninoculated bacteria culture medium served as control.
2.3. Analyses of NF-κB Activation—SEAP Reporter Assay
Construction and validation of the NF-κB reporter clones HT-29/kb-seap-25 and Caco-2/kb-seap-7 have been described previously [18]. For each experiment, Caco-2/kb-seap-7 and HT-29/kb-seap-25 reporter clones were seeded at 50,000 cells per well, into 96-wells plates and incubated 24 hours. Then, cells were stimulated for 24 hours with 10 μL of each tested bacteria CM, for a final volume of 100 μL per well (i.e., 10% vol/vol), in the presence or absence of TNF-α or IL-1β (10 ng/mL final, for HT-29 and Caco-2, resp.).
THP-1 reporter cells were seeded at 100,000 cells per well, into 96-wells plates and stimulated with 10% (vol/vol) of each tested bacteria CM. Cells were then incubated 24 hours prior to quantification of alkaline phosphatase (SEAP).
SEAP in the supernatant was revealed using the Quanti-Blue reagent (Invivogen) according to the manufacturer's protocol and quantified at 655 nm OD. All measurements were performed using a microplate reader (Infinite 200, Tecan).
2.4. Statistical Analysis
Results are expressed as mean ± SD. Data were analyzed using Student's t test.
3. Results
3.1. Effect of Bacteria CM on NF-κB Activation in IEC and Monocyte Models
Out of 49 bacteria CM, 8 and 10 CM significantly activated the NF-κB reporter system on HT-29 and Caco-2 reporter cells, respectively (Figures 1(a) and 1(b)). In fact, the 2 cell lines were largely unresponsive to the vast majority of the tested bacteria CM. Active bacteria CM identified on both epithelial cells belonged to Clostridium sardiniensis, Selenomonas ruminantium, Roseburia hominis, Roseburia intestinalis, Butyrivibrio fibrisolvens, Roseburia faecis, and Faecalibacterium prausnitzii. Bacteroides uniformis activated NF-κB only on HT-29/kb-seap-25, while a small, nonsignificant stimulation was observed on Caco-2 (P > .05). Clostridium paraputrificum and Parabacteroides distasonis induced NF-κB activation specifically in Caco-2/kb-seap-7. Although statistically significant, these observed stimulations were lower than the one observed with IL-1β on Caco-2/kb-seap-7 cells (mean 1.03 ± 0.17) or with TNF-α on HT-29/kb-seap-25 (mean 0.92 ± 0.18).
Figure 1.
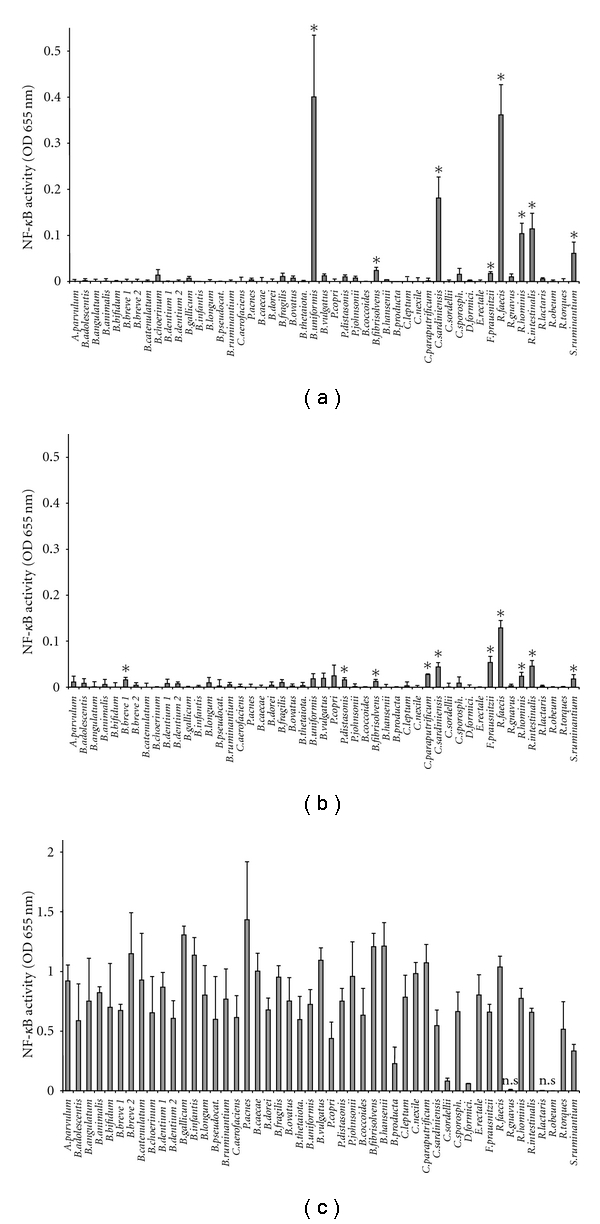
Comparison of the stimulatory effect of 49 commensal bacteria conditioned media on HT-29/kb-seap-25, Caco-2/kb-seap-7, and THP-1-blue. NF-κB activity is expressed as OD 655 nm. Data are presented as mean +/− SD of 3 independent experiments performed in HT-29 (a) and Caco-2 (b) and of 2 independent experiments performed in THP-1 (c). Control is noninoculated bacteria medium and its activity was normalized to 0 (represented by the X-axis). *P < .05 compared to control. For THP-1 only: ns: not significant. All other values are significantly different from control (P < .05).
Conversely, almost all bacteria CM activated NF-κB on THP-1 blue reporter cells except those from Ruminococcus gnavus, Ruminococcus obeum, and Ruminococcus lactaris (Figure 1(c)). Stimulation levels measured in HT-29 and Caco-2 were weak in comparison to those obtained in THP-1. Lipopolysaccharide (LPS) used as control in THP-1 cells showed a lower stimulatory effect than most activating CM (mean 0.38 ± 0.13).
3.2. Effect of Bacteria CM on Activated NF-κB in IEC
In order to explore the inhibitory and/or enhancing potentials of commensal bacteria CM on NF-κB activation, cotreatment with the proinflammatory cytokines TNF-α (10 ng/mL) or IL-1β (10 ng/mL) was performed on HT-29 and Caco-2, respectively (Figure 2). The positive control of NF-κB activation was noninoculated bacteria culture medium combined with the stimulatory cytokine. The value of NF-κB activity measured for the positive control was normalized to 0. The values obtained for each strain were expressed as percentage of activation relative to that of the positive control. For example, Selenomonas ruminantium CM combined with TNF-α induced NF-κB activity 21.6% higher than the one obtained with the positive control (i.e., noninoculated bacteria medium combined with TNF-α).
Figure 2.
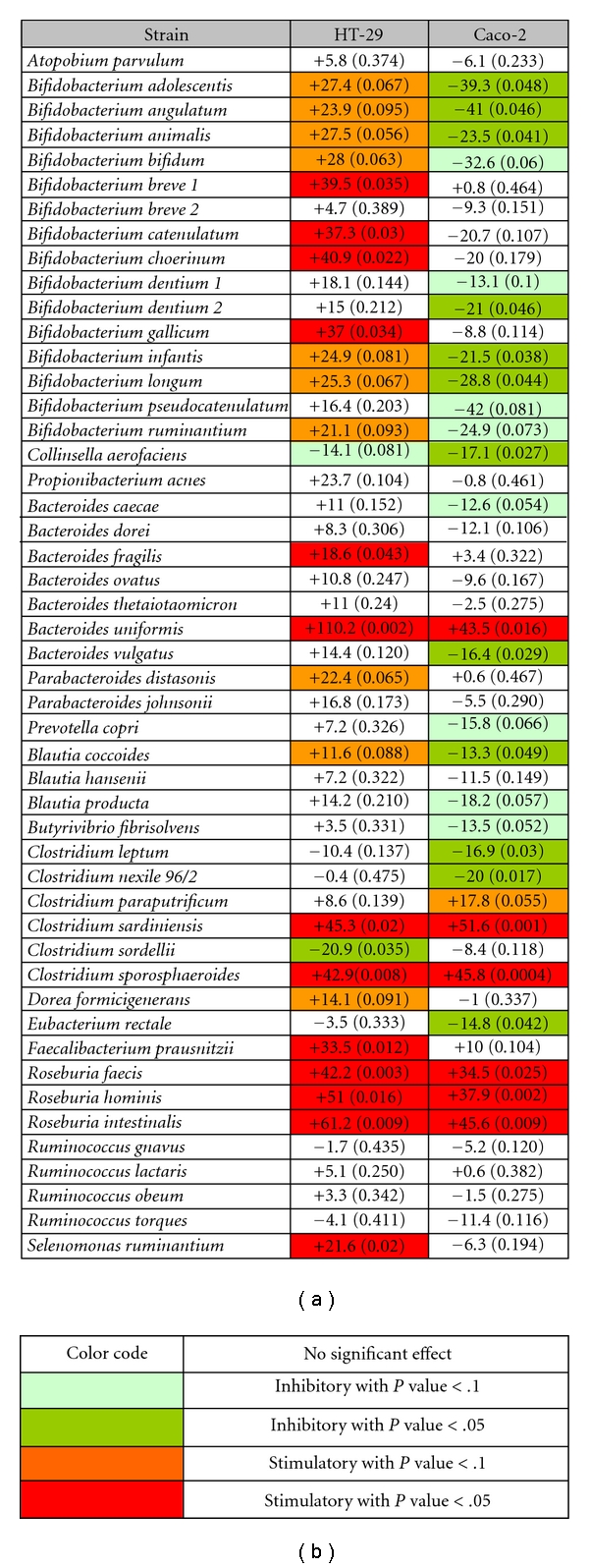
(a) Effect of bacteria CM on activated NF-κB in HT-29 and Caco-2 reporter cells. NF-κB activity is expressed as a relative percentage compared to the positive control (normalized to 0). Positive controls are cells treated with noninoculated bacteria culture medium and the stimulatory cytokine. The corresponding P value is indicated in parenthesis and inhibitory and stimulatory strains are identified following the color code shown in (b).
A different behavior of the 2 epithelial models in response to the bacteria CM could also be observed (Figure 2). Indeed on HT-29, CM overall enhanced NF-κB activity more than in Caco-2. Interestingly, a large majority of Bifidobacteria CM were stimulatory on HT-29 while inhibitory on Caco-2.
Some bacteria CM had a similar effect on the 2 reporter cells (Figure 2). Induced NF-κB activation was restrained in the 2 cell lines only by Colinsella aerofasciens, with inhibition rates of 14.1% (P = .081) and 17.1% (P = .027) for HT-29 and Caco-2, respectively. Furthermore, the 3 species of Roseburia as well as Clostridium sardiniensis, Clostridium sporosphareoides, and Bacteroides uniformis enhanced NF-κB activation significantly in both cell lines.
We also observed that some CM had effects only on one reporter cell line such as CM from Selenomonas ruminantium, Faecalibacterium prauznitzii, Bacteroides fragilis, Parabacteroides distasonis, and Bifidobacterium breve 1. These CM increased NF-κB activity in HT-29/kb-seap-25 but exhibited nonsignificant effects on Caco-2/kb-seap-7. Similarly, Eubacterium rectale, Clostridium nexile 96/2, Clostridium leptum, Blautia coccoides, and Bacteroides vulgatus exerted significant inhibitory effects only on Caco-2.
3.3. Effect of Short Chain Fatty and Organic Acids on NF-κB Activation in IEC
Commensal bacteria are known to produce a panel of acids during their metabolic activity, especially short chain fatty acid (SCFA), which could interfere with NF-κB response.
Therefore, we evaluated the effects of acetic, butyric, propionic, lactic, succinic, and formic acids on NF-κB in IEC, either alone or on cytokine-activated cells (Figure 3). The results presented were obtained on HT-29 although similar observations were performed on Caco-2 (data not shown).
Figure 3.
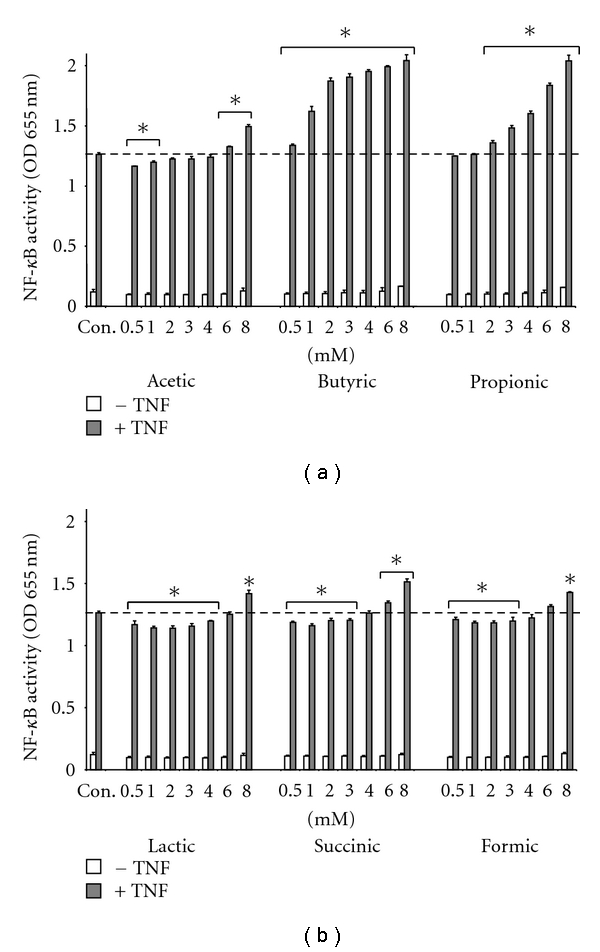
Effect of SCFA and organic acids on NF-κB activation. HT-29 cells were treated with acetic, butyric, propionic, lactic, succinic, and formic acids either alone or in combination with TNF-α (10 ng/mL) for 24 hours. Results are expressed as OD 655 nm. Data is representative of 1 experiment out of 3 performed. *P < .05 compared to TNF control.
None of the acids had an effect on HT-29 or Caco-2 when used alone. However, butyrate and propionate produced a dose-dependent hyperactivation of NF-κB on TNF-α activated cell. The other acids induced a small but significant stimulatory effect only at the highest concentration (6–8 mM final) and a very small but yet significant inhibitory effect of NF-κB at the lowest concentrations. Since butyrate and propionate are likely to act on activated NF-κB signaling, we quantified SCFA in the CM by HPLC and examined their associations with NF-κB activity. We found that out of 49 bacteria CM, 19 contained butyrate, propionate, or both (Figure 4).
Figure 4.
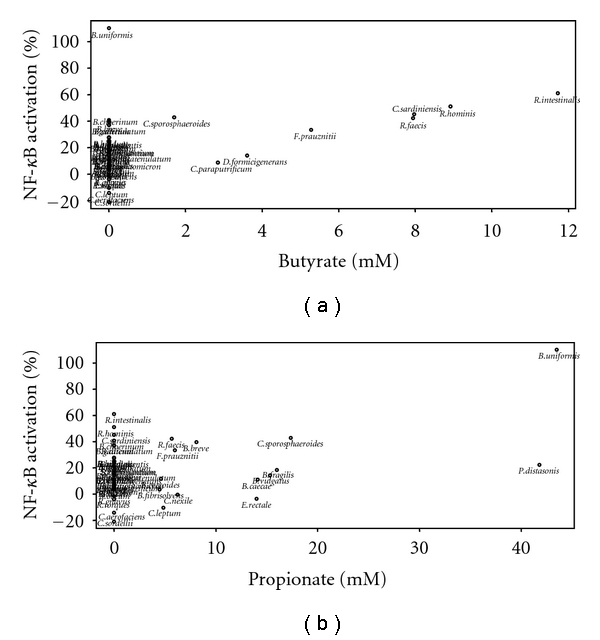
Correlation between butyrate or propionate concentration of the bacteria CM and NF-κB activity. NF-κB activity measured in HT-29 in response to cotreatment with bacteria CM and TNF-α (10 ng/mL) based on the amount of butyrate (a) and propionate (b) present in each bacteria CM. NF-κB activity is expressed as a relative percentage (values from Figure 2). Butyrate and propionate amounts were determined by HPLC and expressed in mM. A Spearman correlation test was performed: Butyrate versus NF-κB activity: r = 0.76 (P = .036). Propionate versus NF-κB activity: r = 0.49 (P = .075). Propionate (Bacteroidetes only) versus NF-κB activity: r = 0.81 (P = .005).
Figure 4 represents a plot of the amount of butyrate (Figure 4(a)) or propionate (Figure 4(b)) present in each bacteria CM (X-axis) with the NF-κB activity measured in HT-29 in response to cotreatment with bacteria CM and TNF-α (see Figure 2). A Spearman correlation test was performed by taking into account butyrate and propionate concentrations greater than 1 mM. A significant positive correlation (r = 0.76, P = .036) was observed between butyrate concentration and NF-κB activity, suggesting that the butyrate-producing strains might have exerted their stimulatory effect through the butyrate released during growth. Similarly, most of the propionate-producing bacteria were also stimulatory on HT-29, but the correlation between propionate concentration and NF-κB activity was not significant (r = 0.49, P = .075). However, the correlation became highly significant when only strains from the Bacteroidetes phylum (r = 0.81, P = .005) were included in the analyses. This suggests that the effect of the other propionate-producing strains might be due to other active metabolites that are different from propionate.
4. Discussion
In this study, we aimed at identifying commensal strains deemed capable of regulating eukaryotic cell signaling focusing on the NF-κB signaling pathway, which is largely involved in immune and inflammatory responses.
In the IEC models HT-29 and Caco-2, the majority of bacteria CM had no effect on NF-κB, contrasting with the results obtained with the monocyte cell line THP-1. This observation may be explained by the differences in the expression of receptors specialized in recognition of microbial structures, especially TLRs. Indeed, THP-1 reporter clone as well as the parental cell line express a complete range of TLRs and all five surface TLRs (TLR1/2, TLR2, TLR6/2, TLR4, and TLR5) are present and functional [18–20], resulting in increased sensitivity to a wide range of microbe-associated molecular patterns. Conversely, HT-29 and Caco-2 do not express all TLRs. Parental HT-29 cell line as well as the reporter clone HT-29/kb-seap-25 mainly express a functional TLR5, which is responsible for detection of flagellin [21]. Caco-2 parental cell line was also described to be responsive to TLR5 stimulation [22, 23], but the reporter clone Caco-2/kb-seap-7 that was obtained and used throughout our study is poorly sensitive [18]. In addition, unresponsiveness toward stimulation of other TLRs such as TLR2 and TLR4 was reported for parental intestinal epithelial cell lines [24–26] and was also observed with our IEC reporter clones [18].
Furthermore, the fact that HT-29/kb-seap-25 is sensitive to flagellin explains the basal stimulatory effect displayed by the CM from flagellated commensal strains such as Roseburia spp., Selenomonas ruminantium and Butyrivibrio fibrisolvens [27–29]. These flagellated commensals also stimulated NF-κB in Caco-2/kb-seap-7, but these cells are less sensitive to flagellin than HT-29 which explains the weak response obtained. Stimulation by Bacteroides uniformis and Faecalibacterium prausnitzii CM on both cell lines was not likely due to a putative flagellin expression since these bacteria are not motile [30, 31], which is not the case with Clostridium sardiniensis [32].
Clostridium paraputrificum, Parabacteroides distasonis, and Bifidobacterium breve 1 CM stimulated Caco-2 but not HT-29, supporting the assertion that the effect is independent of TLR5 recognition. Since Caco-2 reporter cells are not sensitive to TLRs stimulation [18], the specific stimulatory effect of these 3 bacteria CM need further evaluation. In addition, the lack of response of THP-1 toward CM from 3 different strains of Ruminococcus is an intriguing observation that also deserves attention.
The 2 reporter cells used in this study were stimulated by the commensal bacteria CM combined with a known NF-κB activator. This challenge was performed in order to identify both the stimulatory as well as the inhibitory contributors of NF-κB signaling activity. The 2 proinflammatory cytokines TNF-α and IL-1β were used to activate NF-κB on HT-29 and Caco-2, respectively, [18]. Under these conditions, some CM remarkably dampened NF-κB activation in these cells with the inhibition occurring mainly in IL-1β-activated Caco-2.
Several Bifidobacterium strains were described for their capacity to dampen NF-κB activation in vitro in different cell models, including Caco-2 and HT-29 [33, 34]. Consistent with this, few Bifidobacterium CM reduced NF-κB activation on Caco-2, but were ineffective on HT-29. In contrast, in the present study, CM from Bifidobacterium strains enhanced NF-κB activation by TNF on HT-29. However, in our study, we measured the transcriptional activity of NF-κB instead of the product of one gene controlled by NF-κB. It is well known that NF-κB controls the activation of several genes not only involved in inflammatory processes, but also in tissue protection and homeostasis. For example, production of human β-defensin in IEC, which is NF-κB-dependent [35], is stimulated by probiotics, including the Bifidobacterium-containing mixture VSL#3 [36]. Obviously, this dual effect of Bifidobacterium strains on NF-κB signaling requires further examination.
Butyrate (or butyric acid) and propionate, 2 products of bacterial fermentation, enhanced NF-κB activation induced by TNF-α or IL-1β in our reporter cells. As such, butyrate-producing bacteria stimulated NF-κB activity and a strong correlation has been found between bacteria CM butyrate concentration and NF-κB activity.
However some propionate-producing bacteria, such as Eubacterium rectale and Clostridium leptum did not enhance NF-κB, suggesting the existence of other metabolites that may counter the stimulatory effect of propionate on activated epithelial cells.
It is noteworthy that although butyrate is classically known for preventing NF-κB activation in IEC [37–39], some recent studies suggest that butyrate also promotes NF-κB transcriptional activity in IEC [40, 41]. In addition, butyrate has been shown to promote human β-defensin expression, whose gene transcription is controlled by NF-κB [42].
5. Conclusion
The mechanisms underlying the inhibitory and stimulatory effects of NF-κB signaling in monocyte and IEC models by nonbutyrate-producing and nonflagellated bacteria strains remain to be explored. The cell-based screening method employed in the present study provides a rapid identification of potentially interesting commensal species; however, their effects require further confirmation and characterization using other techniques of NF-κB detection. Moreover, the potential implication of these commensal bacteria and their host cells regulating properties in human health and disease may need to be evaluated.
Acknowledgments
The authors thank Chantal Bridonneau for providing the bacteria strains and Pascal Courtin for HPLC analysis. They are very grateful to Mihai Covasa for revising the paper for English.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nature Reviews Immunology. 2008;8(6):411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 3.Neish AS, Gewirtz AT, Zeng H, et al. Prokaryotic regulation of epithelial responses by inhibition of IκB-α ubiquitination. Science. 2000;289(5484):1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 4.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shutting of PPAR-γ and ReIA. Nature Immunology. 2004;5(1):104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 6.Menard S, Candalh C, Bambou JC, et al. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut. 2004;53:821–828. doi: 10.1136/gut.2003.026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heuvelin E, Lebreton C, Grangette C, Pot B, Cerf-Bensussan N, Heyman M. Mechanisms involved in alleviation of intestinal inflammation by bifidobacterium breve soluble factors. Plos One. 2009;4(4) doi: 10.1371/journal.pone.0005184. Article ID e5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaci G, Lakhdari O, Doré J, et al. The commensal Streptococcus salivarius inhibits NF-κB pathway in human intestinal epithelial cells. doi: 10.1128/AEM.03021-10. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrof EO, Kojima K, Ropeleski MJ, et al. Probiotics inhibit nuclear factor-κB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology. 2004;127(5):1474–1487. doi: 10.1053/j.gastro.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasparakis M. Regulation of tissue homeostasis by NF-κB signalling: implications for inflammatory diseases. Nature Reviews Immunology. 2009;9(11):778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- 12.Nenci A, Becker C, Wullaert A, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446(7135):557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 13.Zaph C, Troy AE, Taylor BC, et al. Epithelial-cell-intrinsic IKK-β expression regulates intestinal immune homeostasis. Nature. 2007;446(7135):552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 14.Eckmann L, Nebelsiek T, Fingerle AA, et al. Opposing functions of IKKβ during acute and chronic intestinal inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(39):15058–15063. doi: 10.1073/pnas.0808216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinbrecher KA, Harmel-Laws E, Sitcheran R, Baldwin AS. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. Journal of Immunology. 2008;180(4):2588–2599. doi: 10.4049/jimmunol.180.4.2588. [DOI] [PubMed] [Google Scholar]
- 16.Schreiber S, Nikolaus S, Hampe J. Activation of nuclear factor κB inflammatory bowel disease. Gut. 1998;42(4):477–484. doi: 10.1136/gut.42.4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hungate RE. The anaerobic mesophilic cellulolytic bacteria. Bacteriological reviews. 1950;14(1):1–49. doi: 10.1128/br.14.1.1-49.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakhdari O, Cultrone A, Tap J, et al. Functional metagenomics: a high throughput screening method to decipher microbiota-driven NF-κB modulation in the human gut. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0013092. Article ID e13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarember KA, Godowski PJ. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. Journal of Immunology. 2002;168(2):554–561. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- 20.Remer KA, Brcic M, Sauter KS, Jungi TW. Human monocytoid cells as a model to study Toll-like receptor-mediated activation. Journal of Immunological Methods. 2006;313(1-2):1–10. doi: 10.1016/j.jim.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi F, Smith KD, Ozinsky A, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410(6832):1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 22.Steiner TS, Nataro JP, Poteet-Smith CE, Smith JA, Guerrant RL. Enteroaggregative Escherichia coli expresses a novel flagellin that causes IL-8 release from intestinal epithelial cells. Journal of Clinical Investigation. 2000;105(12):1769–1777. doi: 10.1172/JCI8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogushi KI, Wada A, Niidome T, et al. Salmonella enteritidis FliC (flagella filament protein) induces human β-defensin-2 mRNA production by Caco-2 cells. Journal of Biological Chemistry. 2001;276(32):30521–30526. doi: 10.1074/jbc.M011618200. [DOI] [PubMed] [Google Scholar]
- 24.Melmed G, Thomas LS, Lee N, et al. Human intestinal epithelial cells are broadly unresponsive to Toll-like receptor 2-dependent bacterial ligands: implications for host-microbial interactions in the gut. Journal of Immunology. 2003;170(3):1406–1415. doi: 10.4049/jimmunol.170.3.1406. [DOI] [PubMed] [Google Scholar]
- 25.Otte JM, Cario E, Podolsky DK. Mechanisms of cross Hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology. 2004;126(4):1054–1070. doi: 10.1053/j.gastro.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Furrie E, Macfarlane S, Thomson G, Macfarlane GT. Toll-like receptors-2,–3 and–4 expression patterns on human colon and their regulation by mucosal-associated bacteria. Immunology. 2005;115(4):565–574. doi: 10.1111/j.1365-2567.2005.02200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duck LW, Walter MR, Novak J, et al. Isolation of flagellated bacteria implicated in Crohn’s disease. Inflammatory Bowel Diseases. 2007;13(10):1191–1201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- 28.Haya S, Tokumaru Y, Abe N, Kaneko J, Aizawa SI. Characterization of lateral flagella in Selenomonas ruminantium. Applied and Environmental Microbiology. 2011;77(8):2799–2802. doi: 10.1128/AEM.00286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. International Journal of Systematic and Evolutionary Microbiology. 2002;52(5):1615–1620. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- 30.Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. International Journal of Systematic and Evolutionary Microbiology. 2002;52(6):2141–2146. doi: 10.1099/00207713-52-6-2141. [DOI] [PubMed] [Google Scholar]
- 31.Eggerth AH, Gagnon BH. The bacteroides of human feces. Journal of Bacteriology . 1933;25(4):389–413. doi: 10.1128/jb.25.4.389-413.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Maegawa T, Karasawa T, Ozaki E, Nakamura S. Clostridium sardiniense Prévot 1938 and Clostridium absonum Nakamura et al. 1973 are heterotypic synonyms: evidence from phylogenetic analyses of phospholipase C and 16S rRNA sequences, and DNA relatedness. International Journal of Systematic and Evolutionary Microbiology. 2005;55(3):1193–1197. doi: 10.1099/ijs.0.63271-0. [DOI] [PubMed] [Google Scholar]
- 33.Bai AIP, Ouyang Q, Zhang W, Wang CH, Li SF. Probiotics inhibit TNF-α-induced interleukin-8 secretion of HT29 cells. World Journal of Gastroenterology. 2004;10(3):455–457. doi: 10.3748/wjg.v10.i3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Candela M, Perna F, Carnevali P, et al. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. International Journal of Food Microbiology. 2008;125(3):286–292. doi: 10.1016/j.ijfoodmicro.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 35.Uehara A, Fujimoto Y, Fukase K, Takada H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Molecular Immunology. 2007;44(12):3100–3111. doi: 10.1016/j.molimm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Schlee M, Harder J, Koten B, Stange EF, Wehkamp J, Fellermann K. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clinical and Experimental Immunology. 2008;151(3):528–535. doi: 10.1111/j.1365-2249.2007.03587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inan MS, Rasoulpour RJ, Yin L, Hubbard AK, Rosenberg DW, Giardina C. The luminal short-chain fatty acid butyrate modulates NF-κB activity in a human colonic epithelial cell line. Gastroenterology. 2000;118(4):724–734. doi: 10.1016/s0016-5085(00)70142-9. [DOI] [PubMed] [Google Scholar]
- 38.Yin L, Laevsky G, Giardina C. Butyrate suppression of colonocyte NF-κB activation and cellular proteasome activity. Journal of Biological Chemistry. 2001;276(48):44641–44646. doi: 10.1074/jbc.M105170200. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A, Wu H, Collier-Hyams LS, Kwon YM, Hanson JM, Neish AS. The bacterial fermentation product butyrate influences epithelial signaling via reactive oxygen species-mediated changes in cullin-1 neddylation. Journal of Immunology. 2009;182(1):538–546. doi: 10.4049/jimmunol.182.1.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adam E, Quivy V, Bex F, et al. Potentiation of tumor necrosis factor-induced NF-κB activation by deacetylase inhibitors is associated with a delayed cytoplasmic reappearance of IκBα. Molecular and Cellular Biology. 2003;23(17):6200–6209. doi: 10.1128/MCB.23.17.6200-6209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blais M, Seidman EG, Asselin C. Dual effect of butyrate on IL-1β-mediated intestinal epithelial cell inflammatory response. DNA and Cell Biology. 2007;26(3):133–147. doi: 10.1089/dna.2006.0532. [DOI] [PubMed] [Google Scholar]
- 42.Schwab M, Reynders V, Loitsch S, Steinhilber D, Schröder O, Stein J. The dietary histone deacetylase inhibitor sulforaphane induces human β-defensin-2 in intestinal epithelial cells. Immunology. 2008;125(2):241–251. doi: 10.1111/j.1365-2567.2008.02834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


