The ABH8 protein (also called ALKBH8) is a member of the AlkB (alkylated DNA repair protein) family of nonheme iron/ α-ketoglutarate (αKG)-dependent dioxygenases.[1] Other members of this protein family are known to catalyze oxidative demethylation to repair damaged DNA/RNA.[2] AlkB-like proteins exist in viruses, bacteria, and eukaryotic species.[3] Nine mammalian homologues of AlkB (ABH1-ABH8, and FTO, a protein associated with fat mass and obesity) have been identified through bioinformatics studies.[1b,4] Four of the homologues (ABH1, ABH2, ABH3, and FTO) show demethylation activity for N-methylated bases, in either DNA or RNA, through the hydroxylation of the N-methyl group and subsequent release of an aldehyde from the oxidized intermediate.[5] ABH2, which repairs m1A, m3C, and εA (1,N 6-ethenoadenine) in dsDNA, guards mammalian genomic DNA against methylation damage.[6] ABH3 appears to be involved in RNA repair.[7] FTO was first identified in a fused-toes malformation phenotype resulting from a mouse genome deletion,[8] and was later shown to significantly affect energy homeostasis and lead to obesity.[9] FTO has the highest activity in demethylating m3T in ssDNA and m3U in RNA;[4,10] however, the link between its biochemical activity and physiological phenotype is yet to be discovered. Aside from ABH1, ABH2, ABH3, and FTO, the biochemical activities of the other AlkB homologues are still unknown. Very recently Tet1, another iron(II)/αKG-dependent dioxygenase, has been shown to hydroxylate the 5-methyl group of m5C in dsDNA to form 5-hydroxymethylcytosine (hm5C) in certain neural and stem cells. This discovery has fueled speculation that this modification is important in epigenetics.[11]
ABH8 has been identified in high eukaryotes. This protein contains an N-terminal RNA recognition motif (RRM) and a predicted C-terminal methyltransferase domain fused to the central AlkB homologous domain (Figure 1a). This configuration is conserved from insects (e.g., B. mori), to worms (e.g., C. elegans), to humans. Together with the observation that ABH8 is exclusively located in the cytoplasm, the potential role of ABH8 in the regulation of RNA function through controlled methylation/demethylation has been suggested.[1a] However, the demethylation of methylated bases by ABH8 has never been reported. We have cloned, expressed, and purified several constructs of truncated ABH8, having both RRM and AlkB domains, from both humans (hABH8) and mice (mABH8), and tested potential demethylation activity against m1A, m3C, m1G, m3T, m6A, and εA. No activity was observed under standard conditions with iron and αKG cofactors (see the Experimental Section; data not shown).
Figure 1.
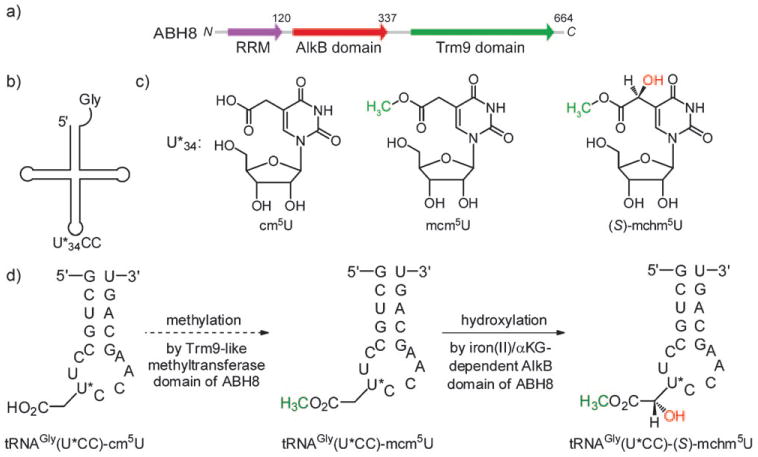
Distinct domains and proposed function of ABH8. a) ABH8 contains three domains: the N-terminal RNA recognition motif (RRM; blue), the middle iron/αKG-dependent AlkB domain (red), and the C-terminal methyltransferase domain (green). The C-terminal domain is homologous to a tRNA modification methyltransferase Trm9 in yeast. b) The anticodon wobble-base U*34 in higher eukaryotes tRNAGly(U*CC) can be hypermodified. c) Examples of hypermodification on U*34: 5-carboxymethyluridine (cm5U), 5-methoxycarbonylmethyluridine (mcm5U), and 5-methoxycarbonylhydroxymethyluridine (mchm5U). d) The proposed sequential modification of the wobble U*34 in a 17-mer RNA that contains the anticodon stem–loop of tRNAGly(U*CC) by the two catalytic domains of ABH8.
Subsequently, we noticed through sequence alignment that the methyltransferase domain of ABH8 is homologous to the yeast Trm9 protein (Figure 1a and Figure 1 in the Supporting Information), which is a methyltransferase for several yeast tRNAs. Trm9 catalyzes the methylation of 5-carboxymethyluridine (cm5U) to form 5-methoxycarbonyl-methyluridine (mcm5U), and methylation of of 5-carboxylmethyl-2-thiouridine (cm5s2U) to give 5-methoxycarbonyl-methyl-2-thiouridine (mcm5s2U) at the anticodon wobble position.[12] This tRNA modification modulates the decoding selectivity of the tRNA for a subset of codons, thereby affecting translation in a codon-specific manner;[13] this data prompted us to propose that the Trm9-like domain in ABH8 may carry out a similar methylation in tRNA, and that the AlkB domain in ABH8 may add another modification at the same anticodon wobble nucleotide. Therefore, the AlkB domain in ABH8 might modify either cm5U or mcm5U in tRNAs. After searching the known tRNA modifications, we noticed that 5-(S)-carboxyhydroxymethyluridine ((S)-chm5U) and 5-(S)-methoxycarbonylhydroxymethyluridine ((S)-mchm5U), which are the α-hydroxylation products of cm5U and mcm5U, respectively, had already been identified at the anticodon wobble position of tRNAGly(U*CC) (U* is the modified uridine) in the silkworm, Bombyx mori (Figure 1b,c).[14] The ABH8 gene is conserved among silkworms and mammals (see Figure 2 in the Supporting Information), and the sequence of tRNAGly(U*CC) is also conserved (see Figure 3 in Supporting Information). Therefore, we set out to test the idea that the AlkB domain of ABH8 in high eukaryotes catalyzes the hydroxylation of cm5U or mcm5U into chm5U or mchm5U, respectively, in tRNAGly(U*CC) (Figure 1 d).
A truncated form of mABH8 (1-340) containing the RRM and AlkB domain with an N-terminal His6 tag was expressed and purified from E. coli (see Figure 4 in the Supporting Information). This construct (mABH81−340) gives the most stable ABH8 protein, therefore it was then employed in our activity studies. The conserved iron-binding ligands in ABH8 are His237, Asp239, and His292 as determined from sequence alignment with the AlkB family proteins (see Figure 2 in the Supporting Information). Two mutant forms of mABH81−340, in which either His237 or Asp239 was mutated to Ala, were also constructed. The mutant proteins were expressed and purified in the same way as the wild-type mABH81−340. To evaluate the potential modification of cm5U or mcm5U at the wobble base of tRNAGly(U*CC), we chemically synthesized the phosphoramidite of mcm5U (see Figure 5 in the Supporting Information). By using solid-phase synthesis, the phosphoramidite was incorporated into a 17-mer stem–loop RNA, corresponding to the sequence of the anticodon stem-loop of tRNAGly(U*CC) conserved in high eukaryotes, with mcm5U at the U* position. After using different deprotection strategies, the 17-mer RNA oligonucleotides containing either the mcm5U or cm5U modification were obtained (Scheme 1 and Figure 5 in the Supporting Information). These 17-mer RNA oligonucleotides were purified by HPLC methods and characterized as having the correct mass by using MALDI-MS methods (see Figure 6 in the Supporting Information).
The two 17-mer RNAs were treated with purified mABH81−340 in the presence of iron and αKG. The RNAs before and after the reaction with mABH8 were digested with nuclease P1 and alkaline phosphatase, and the resulting single nucleosides were analyzed by HPLC methods. Whereas mABH81−340 did not show activity against cm5U in the 17-mer RNA, it catalyzed the hydroxylation of mcm5U in the 17-mer RNA (Figure 2a). The peak representing the product having the added hydroxy group was observed in the high-resolution mass spectrum (HRMS calcd., 333.09286; found, 333.09311; Figure 2b). MS/MS analysis of the enzymatic product shows the exact fragmentation pattern as seen for the synthesized mchm5U standard, thereby confirming the location of the hydroxy group (see Figures 7 and 8 in the Supporting Information). The chemically synthesized diastereomers of the nucleoside mchm5U can be separated, and the absolute configuration at the asymmetric C51 atom was determined by comparing the CD spectra of the diastereomers (see Figure 9 in the Supporting Information) with the previously reported CD spectra of the same compounds.[15] The S diastereomer, which was discovered in B. mori., shows the same HPLC retention time when compared to the product obtained from the ABH8-catalyzed reaction, whereas the R diastereomer does not (see Figure 10 in the Supporting Information), suggesting that the enzymatic hydroxylation of mcm5U by ABH8 is also stereoselective. Three different control experiments—1) in the absence of either iron or αKG cofactors, 2) with the active site mutation or either mABH81–340 H237A or mABH81–340 D239A, and 3) with other AlkB family proteins such as ABH2, ABH3 or FTO, all of which were run under the same conditions—failed to produce (S)-mchm5U (data not shown). Thus, this reaction is specific to ABH8 and requires iron(II) and αKG cofactors.
Figure 2.

Hydroxylation of mcm5U in a 17-mer tRNAGly(U*CC) anticodon stem-loop. a) The chemically synthesized 17-mer RNA oligonucleotide resembling the tRNAGly(U*CC) anticodon stem-loop was treated with mABH81−340 in the presence of iron(II) and α-KG. The HPLC traces show the appearance of a peak for the product (S)-mchm5U when mcm5U is converted in the presence of the enzyme. b) HRMS data of the product (S)-mchm5U confirms the addition of a hydroxy group to mcm5U. MES = 2-(N-morpholino)ethanesulfonic acid.
The AlkB family of proteins are thought to be engaged in oxidative de-modification (e.g., demethylation) in biological systems. We show herein that ABH8 exhibits hydroxylase activity and modifies mammalian tRNA. Thus, members of this family of proteins could also perform modification functions on nucleic acids. ABH8 has been shown to affect cancer progression.[16] This protein is highly expressed in bladder cancer, and over-expression of ABH8 was proposed to effect an increased reactive oxygen species (ROS) production inside these cancer cells. In yeast, the tRNA-modification function of Trm9 changes the codon preference and alters the translational efficiency of mRNAs with increased abundance of cognate codons read by the modified tRNAs.[13b] In B. mori, it has been shown that the additional hydroxylation modification of mcm5U into mchm5U has a significant impact upon the affinity of tRNAGly(U*CC) to GGA and GGG codons.[17] This oxidation modification by ABH8 may therefore affect the translation of certain mRNAs.
In summary, we have presented herein the identification of a unique biochemical activity of ABH8 distinct from those of other proteins from the AlkB family. The discovery of the tRNA modification chemistry catalyzed by the AlkB domain of ABH8 may help elucidate the exact cellular function of ABH8 and its potential roles in cancer. Furthermore, studies on the structure and function of ABH8 are ongoing in our laboratory.[18]
Experimental Section
Assays for the hydroxylation reaction of RNA oligonucleosides: In a typical reaction, the RNA substrate (1 nmol) was added to 50 μL of 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 7.5). The solution containing RNA oligonucleotides was heated at 55°C for 3 min, and then slowly cooled to room temperature, after it had been was cooled in an ice bath. The following components were then added to reach the target concentration: KCl (100 mm), MgCl2 (2 mm), RNasin (0.2 U μL−1, Promega), l-ascorbic acid (2 mm), α-KG (300 μm), (NH4)2Fe(SO4)2·6H2O (150 μm), and mABH8 (1 nmol). The reaction mixture was incubated for 12 h at 16°C. The reaction was quenched by the addition of ethylene diamine tetraacetic acid (EDTA) to a final concentration of 1 mM.
RNA digestion and HPLC assay: After the hydroxylation reaction, 10% of 3M NaOAc pH 5.3 was added to the reaction mixture, after which isopropanol (2.5 times the volume of the crude mixture) was added to precipitate the 17-mer RNA. The pallet was washed with 75 % ethanol once and then dried by air. The resulting pallet was dissolved in 100 μL Milli-Q H2O and then digested into nucleosides using nuclease P1 and alkaline phosphatase. The digestion solution was analyzed by using a high-performance liquid chromatography (HPLC) system having a C18 column (150 × 4.6 mm). The eluent comprised buffer A1 (water containing 5 mM ammonium acetate) and buffer B1 (5 mM ammonium acetate, 60% of acetonitrile, 40% water and 0.01% trifluoroacetic acid (TFA)); A1/ B1 ratios ranged from 100:0 to 90:10 (v/v) with a flow rate of 1 mL min−1 at room temperature. The detection wavelengths were set at 260 nm and 280 nm. The fraction containing the digested product was collected and lyophilized for HRMS and MS–MS analysis. For LC–MS analysis, the digestion solution was run on the same C18 column using buffer A2 (water containing 0.1 % TFA) and buffer B2 (acetonitrile containing 0.08 % TFA).
Supplementary Material
Scheme 1.

Synthesis of mcm5U phosphoramidite (5), and incorporation of mcm5U and cm5U into a tRNAGly(U*CC) anticodon stem–loop oligomer sequence (RNA-I and RNA-II). Reaction conditions: a) Ac2O, Py, RT, overnight; b) BzCl, DMAP, NEt3, 0.5 h; c) CH2(COMe)2, DBU, THF, RT, overnight; d) NaOMe, MeOH, 50°C, 1 h; e) DMTr, Py, RT, overnight; f) TBDMSCl, Im, Py, RT, 10 h; g) (iPr2N)P(Cl)OCH2CH2CN, iPr2NEt, THF, RT, 4 h; h) standard solid-phase RNA synthesis and subsequent deprotection under ultramild conditions (see the Supporting Information). Bz=benzoyl, DBU=1,8-diazabicyclo[5.4.0]undec-7-ene, DMAP=4-dimethylaminopyridine, Im=imidazole, Py=pyridine, TBDMS=tert-butyldimethylsilyl, THF=tetrahydrofuran.
Footnotes
This work was supported by the U.S. National Institute of Health (GM071440 to C.H.) and by an NIH EUREKA award (GM088599 to C.H. and T.P.). Q.D. was supported by the Chicago Biomedical Consortium (CBC). W.Z. was partially supported by the American Recovery and Reinvestment Act NIGMS Administrative Supplementary grant (3R01M071440-05S1). We thank Dr. Leslie M. Hicks for performing mass spectrometry analysis and Dr. Stephen B. H. Kent for the use of the LC mass spectrometer.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201001242.
Contributor Information
Ye Fu, Department of Chemistry, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA), Fax: (+1) 773-702-0805.
Qing Dai, Dr., Department of Biochemistry and Molecular Biology, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA)
Wen Zhang, Department of Chemistry, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA), Fax: (+1) 773-702-0805.
Jin Ren, Dr., Department of Chemistry, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA), Fax: (+1) 773-702-0805
Tao Pan, Prof., Email: taopan@uchicago.edu, Department of Biochemistry and Molecular Biology, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA); Institute for Biophysical Dynamics, The University of Chicago (USA).
Chuan He, Prof., Email: chuanhe@uchicago.edu, Department of Chemistry, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA), Fax: (+1) 773-702-0805; Institute for Biophysical Dynamics, The University of Chicago (USA).
References
- 1.a) Aravind L, Koonin EV. Genome Biol. 2001;2:RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Loenarz C, Schofield CJ. Nat Chem Biol. 2008;4:152. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 2.a) Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Proc Natl Acad Sci USA. 2002;99:16660. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Falnes PO, Johansen RF, Seeberg E. Nature. 2002;419:178. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]; c) Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Nature. 2002;419:174. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 3.Bratlie MS, Drablos F. BMC Genomics. 2005;6:1. doi: 10.1186/1471-2164-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerken T, et al. Science. 2007;318:1469. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishina Y, Duguid EM, He C. Chem Rev. 2006;106:215. doi: 10.1021/cr0404702. and reference therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Ringvoll J, et al. EMBO J. 2006;25:2189. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Aas PA, et al. Nature. 2003;421:859. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 7.Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO. Mol Cell. 2004;16:107. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Peters T, Ausmeier K, Ruther U. Mamm Genome. 1999;10:983. doi: 10.1007/s003359901144. [DOI] [PubMed] [Google Scholar]
- 9.a) Dina C, et al. Nat Genet. 2007;39:724. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]; b) Scott LJ, et al. Science. 2007;316:1341. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Frayling TM, et al. Science. 2007;316:889. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Bruning JC, Ruther U. Nature. 2009;458:894. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 10.Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C. FEBS Lett. 2008;582:3313. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahiliani M, et al. Science. 2009;324:930. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalhor HR, Clarke S. Mol Cell Biol Res Commun. 2003;23:9283. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Maraia RJ, Blewett NH, Bayfield MA. Nat Chem Biol. 2008;4:162. doi: 10.1038/nchembio0308-162. [DOI] [PubMed] [Google Scholar]; b) Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Mol Cell. 2007;28:860. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawakami M, Takemura S, Kondo T, Fukami T, Goto T. J Biochem. 1988;104:108. doi: 10.1093/oxfordjournals.jbchem.a122403. [DOI] [PubMed] [Google Scholar]
- 15.Barbara N, Malkiewicz A. Nucleosides Nucleotides. 1989;8:1499. [Google Scholar]
- 16.Shimada K, Nakamura M, Anai S, De Velasco M, Tanaka M, Tsujikawa K, Ouji Y, Konishi N. Cancer Res. 2009;69:3157. doi: 10.1158/0008-5472.CAN-08-3530. [DOI] [PubMed] [Google Scholar]
- 17.Kawakami M, Tsonis P, Nishio K, Takemura S. J Biochem. 1980;88:1151. doi: 10.1093/oxfordjournals.jbchem.a133069. [DOI] [PubMed] [Google Scholar]
- 18.During the preparation of this manuscript, it was been shown that the methyltransferase domain of ABH8 is indeed responsible for converting cm5U into mcm5U in the wobble position of several tRNA species, which additionally suggests that the hydroxylation activity of AlkB domain of ABH8 showed herein could be the physiological activity of ABH8 in vivo: Songe-Møller L, van den Born E, Leihne V, Vågbø CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes Pø, Klungland A. Mol Cell Biol Res Commun. 2010;30:1814. doi: 10.1128/MCB.01602-09.Fu D, Brophy JAN, Chan CTY, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. Mol Cell Biol Res Commun. 2010;30:2449. doi: 10.1128/MCB.01604-09.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


