Abstract
The aim of the Structural Biology Extensible Visualization Scripting Language (SBEVSL) project is to allow users who are experts in one scripting language to use that language in a second molecular visualization environment without requiring the user to learn a new scripting language. ConSCRIPT, the first SBEVSL release, is a plug-in for PyMOL that accepts RasMol scripting commands either as premade scripts or as line-by-line entries from PyMOL's own command line. The plug-in is available for download at http://sourceforge.net/projects/sbevsl/files in the ConSCRIPT folder.
Keywords: PyMOL, RasMol, ConSCRIPT, molecular visualization, structural bioinformatics
There are many excellent visualization programs used in structural biology [1]. Most come with their own scripting languages that permit both command-line control and the packaging of complex sets of display commands in a file. Coordination among multiple visualization programs is often necessary in the course of a single investigation and in combining results from multiple investigations. If two programs use different scripting languages, valuable time can be lost learning the new scripting language. The long-range goal of the Structural Biology Extensible Visualization Scripting Language (SBEVSL) [2] project is to make common molecular visualization tools available to the broadest possible audience, so that each user can use many programs, with knowledge of the scripting language for only one of those programs. Three of the most popular molecular visualization programs are Jmol [3], PyMOL [4], and RasMol [5, 6]. All are open source programs, making them ideal initial targets for establishing a common scripting language. Jmol, the most popular of the three, already uses a variant of the RasMol scripting language. Chime [7], a proprietary version of RasMol, also uses a variant of the RasMol scripting language. For many educators and scientists, PyMOL is the application of choice for preparing images and animations of their structures because of the beauty and quality of the images. Therefore, making the RasMol scripting language work with PyMOL will have a significant unifying impact. RasMol's scripting language is known for ease of use and a natural language interface. However, its interface was implemented in 1993 and does not take full advantage of today's graphics processing units. Nevertheless, it is still widely used on computers running Windows, Linux, or Macintosh operating systems. PyMOL is a much newer and more versatile 3D molecular visualization environment. Its graphics are much more advanced and have become the standard in today's captured images. However, its scripting language is based on object-oriented Python and can be difficult for someone unfamiliar with computer science to learn. It is also a complex language with many more commands than RasMol's simple language.
Design Principles
To create a RasMol command interpreter for PyMOL (the first goal of the SBEVSL project), we compared the command sets for RasMol and PyMOL, then created ConSCRIPT, a plug-in for PyMOL that accepts RasMol script files and commands, translates the commands, and executes them in PyMOL. The process involves searching for recognized, valid, RasMol script commands and performing their PyMOL equivalents, either as single commands or as a series of commands that produce the same result. Several commands that are most commonly used in RasMol are listed in Table I, shown along with their PyMOL equivalents.
Table I. A list of commonly used commands available in RasMol and their equivalents in PyMOL represented as either a single command or as a series of commands.
| RasMol | PyMOL |
|---|---|
| Backbone | Set Ribbon_sampling, 20 |
| Set Ribbon_smooth, 1 | |
| Set Ribbon_width, 0.02 | |
| Show ribbon, <selection> | |
| Background | Bg_color <color> |
| Cartoon | Cartoon rectangle |
| Show cartoon, <selection> | |
| Center | Center <selection> |
| Map generate | Map_new <name>, <type>, <grid>, <selection> |
| Isomesh/isodot <name>, <map>, <level>, <selection> | |
| Zoom | |
| Map zap | Delete <map> |
| Rotate | Rotate <axis> <angle> |
| Script | Run [script] |
| Select | Select <selection> |
| Spacefill | Show spheres, <selection> |
| Wireframe | Show sticks, <selection> |
As we considered which commands to translate, it became clear that a number of features in PyMOL should be added to RasMol to make the two programs more complementary. To overcome this difference and to facilitate movement between the programs, several new features have been implemented and are now supported in RasMol [8]:
SBEVSL movie commands [9].
Electron density maps [10].
Lee–Richards surface approximation by contouring pseudo-Gaussian electron densities [11].
We used a variety of coding methods to correlate RasMol commands to their PyMOL command equivalents. When a single command could be directly translated, we used simple code to recognize that command and perform its equivalent. For RasMol commands containing multiple parameters, the command was translated into a series of PyMOL commands, which were executed sequentially. Several commands required altering PyMOL's default settings to produce exactly the same results. Moreover, several commands remain untranslated for various reasons including lack of universal features to mimic a RasMol code in PyMOL, and intentionally untranslated commands that would only serve to confuse users, such as help, where returning the PyMOL help would be of no use to a ConSCRIPT user. The untranslated commands are Bulgarian, Chinese, English, French, Italian, Japanese, Russian, Spanish, Clipboard, Colourmode, Connect, Help, Load Inline, Molecule, Notoggle, Pause/Wait, Print, Renumber, Show, Star, Strands, and Structure.
Features
Script Loading Interface
Once ConSCRIPT is installed, it automatically loads whenever PyMOL opens. The user can load a RasMol script file by selecting ConSCRIPT from the Plug-in drop-down menu. This opens a dialog box that will enable the user to search for the script file. The converter sequentially executes the given RasMol commands on any protein already loaded in PyMOL. As an alternative, users can reinitialize PyMOL and include commands for loading the protein in the RasMol script.
Command Line Interface
With ConSCRIPT installed, users can enter RasMol commands directly on PyMOL's own command line. To execute RasMol commands from PyMOL's command line, begin the desired command with the token “VSL” followed by a space and then the desired command; users can also enter a Rasmol command prefaced by “r,” “R,” “rasmol,” or “RASMOL,” That command will then be converted using the same algorithm as the script converter and executed immediately. Requiring one of these tokens allows the user to switch between RasMol and PyMOL commands at will and prevents ambiguities between similar RasMol and PyMOL commands.
Implementation
The image of porcine pancreatic elastase in Fig. 1 was generated using a RasMol script in RasMol. The protein is shown in a cartoon representation with the catalytic triad and a bound inhibitor highlighted as wireframes. Table II contains the RasMol and PyMOL scripts used to produce Figs. 1, 2, and 3. The script used to produce Fig. 1 was then converted to a PyMOL script so as to create the same output from PyMOL, which is shown in Fig. 2. Then, the same RasMol script was used to reproduce the image from Fig. 1 using the ConSCRIPT plug-in for PyMOL, as seen in Fig. 3.
Fig. 1.
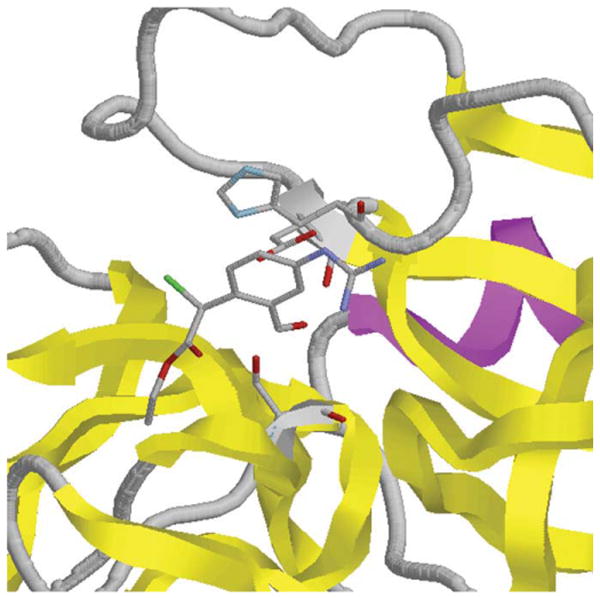
View of the active site of a serine protease with a bound inhibitor (porcine pancreatic elastase [12], PDB entry 8est) that was generated by RasMol using a RasMol script.
Table II. Translation of a set of RasMol commands into the scripting language of PyMOL using ConSCRIPT.
| Rasmol command | PyMOL equivalent |
|---|---|
| load “/path/to/file/8est.pdb” | load “/path/to/file/8est.pdb” rotate ×, 180 |
| background white | bg_color white |
| select all | select all |
| wireframe off | hide sticks, sele; hide lines, sele |
| restrict not water | hide everything, resn hoh |
| select helix | select ss h |
| cartoon 200 | show cartoon, sele |
| color magenta | color magenta, sele |
| select sheet | select ss s |
| cartoon 200 | set cartoon_rect_length, 0.8; cartoon rectangle; show cartoon, sele |
| color yellow | color yellow, sele |
| select protein and not helix and not sheet | select not (ss h or ss s) |
| trace | cartoon tube, sele; show cartoon, sele |
| select gis | select resn gis |
| wireframe 30 | set stick_radius, .24, sele; show sticks, sele |
| color cpk | no native pymol command for this |
| zoom 300 | zoom all −15 |
| select 57,102,195 | select resi 57+102+195 |
| wireframe 30 | set stick_radius, .24, sele; show sticks, sele |
| color cpknew | no native pymol command for this |
| translate y 10 | translate [0,−10,0] |
Fig. 2.
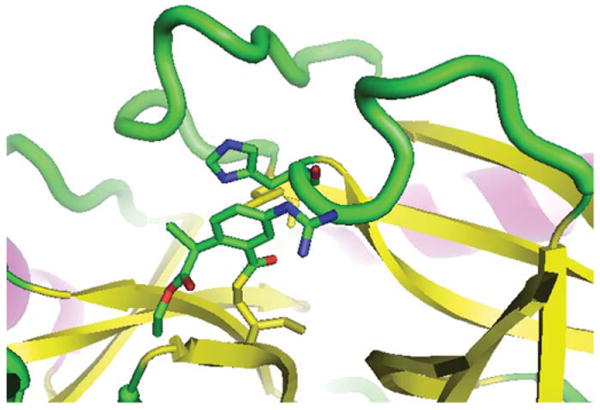
View of the active site of a serine protease with a bound inhibitor (porcine pancreatic elastase [12], PDB entry 8est) that was generated by PyMOL using a PyMOL script.
Fig. 3.
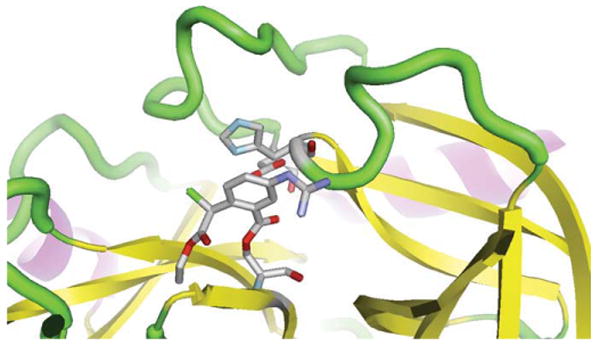
View of the active site of a serine protease with a bound inhibitor (porcine pancreatic elastase [12], PDB entry 8est) that was generated by PyMOL using a RasMol script translated by ConSCRIPT.
The conversion of RasMol scripts into PyMOL scripts presented many challenges. For example, the two programs load molecules in different orientations. ConScript counteracts this by rotating the molecule 180° about the x-axis immediately after it executes the load command. Also, the operation of the zoom command was different in scale and implementation (linear in RasMol vs. non-linear in PyMOL). Sticky selections are a feature present in RasMol, but not in PyMOL. Our solution to this is quite simple. PyMOL does not recall the last selection automatically, even though it may remember it. ConSCRIPT simply ensures that any command executed is executed on the previous selection by having a variable dedicated to the previous selection and forcing the PyMOL command to act on that selection. The largest difference is in the script format—RasMol uses natural language scripting, whereas PyMOL uses object-oriented code. Finally, there were some RasMol commands (including bond, connect, star, and strands) that did not have equivalents in PyMOL and could not be implemented at the level of a plug-in. The advantage of using a plug-in is that ConSCRIPT is able to work with a wide range of existing PyMOL installations. The plug-in will remain available, but the missing commands will be implemented within PyMOL proper in a future release of open source PyMOL by the SBEVSL project.
The plug-in is available for download as a compressed file in both .tar.gz or .zip formats at http://sourceforge.-net/projects/sbevsl/files in the ConSCRIPT folder. The compressed file contains the plug-in (ConSCRIPT.py), the licensing information, and a readme file in text, html, and pdf formats with instructions for installation in Mac OSX, Windows, and Linux environments. Information on creating publication quality figures using PyMOL commands is included in the readme.txt, readme.html, and readme.pdf files in the compressed file. It is important to note that clicking on the prominent green download arrow on this page (or the main SBEVSL page on SourceForge.net) may lead you to another file that is part of the SBEVSL project. You will need to open the ConSCRIPT folder on http://sourceforge.net/projects/sbevsl/files by clicking on the green triangle to the left of the ConSCRIPT folder. Similarly, you should open the latest ConSCRIPT folder (currently ConSCRIPT-2.1) and download the most recent .tar.gz or .zip file.
Acknowledgments
The authors thank the invaluable assistance of Frances C. Bernstein and current and former students who have worked at Dowling College and at RIT on the SBEVSL project. The people at Dowling College working with Herbert Bernstein have been Isaac Awuah Asiamah, Darina Boycheva, Georgi Darakev, Nikolay Darakev, Jonathan Ihm, John Jemilawon, Nan Jia, Petko Kamburov, Greg McQuillan, Daniel O'Brien, Matt Rousseau, Georgi Todorov, and Elena Zlateva. The people at RIT working with Paul Craig have been Brett Hanson, Charles Westin, and Corey Wischmeyer.
Footnotes
This work is supported in part by National Institute of General Medical Sciences grants 1R15GM078077-01 and 3R15GM078077-01S
References
- 1.Lesk AM, Bernstein HJ, Bernstein FC. In: Computational Structural Biology: Methods and Applications, Chapter 27. Schwede T, Peitsch M, editors. World Scientific Publishing Company; Hackensack, NJ: 2008. pp. 729–770. [Google Scholar]
- 2.Westin C, Hanson BR, Bernstein HJ, Awuah Asiamah I, Boycheva D, Darakev G, Darakev N, Jemilawon J, Jia N, Kamburov P, Todorov G, Craig PA, Mottarella S. SBEVSL: Communicating scripts between molecular visualization programs. American Crystallographic Association Annual Meeting; Salt Lake City, UT. July 21–26, 2007; 2007. p. E0003. [Google Scholar]
- 3.Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- 4.DeLano WL. The PyMOL molecular graphics system. [Accessed September 14, 2010];2009 available at: http://www.pymol.org.
- 5.Bernstein HJ. Recent changes to RasMol, recombining the variants. Trends Biochem Sci. 2002;25:453–455. doi: 10.1016/s0968-0004(00)01606-6. [DOI] [PubMed] [Google Scholar]
- 6.Sayle R, Milner-White EJ. RasMol: Biomolecular graphics for all. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 7.Symyx Solutions. [accessed April 30, 2010]; available at: http://www.symyx.com/downloads/
- 8.Bernstein HJ. README: RasMol 2.7.5. [accessed May 19, 2010];2009 available at: http://www.openrasmol.org/software/rasmol/
- 9.Darakev N, Bernstein HJ, Ihm J, McQuillan G, Zlteva E, Craig PA, Grell L. New molecular graphics movie scripting features under SBEVSL. American Crystallographic Association Annual Meeting; Toronto, Canada. July 25–30,2009; p. W0124. [Google Scholar]
- 10.Bernstein HJ, Awuah Asiamah I, Darakev G, Darakev N, Jemilawon J, Jia N, Kamburov P, McQuillan G, Todoro G. SAXS, Lee-Richards surfaces and a simplified command set for maps. American Crystallographic Association Annual Meeting; Knoxville, TN. May 31–June 5,2008; p. W0070. [Google Scholar]
- 11.Bernstein HJ, Craig PA. Efficient molecular surface rendering by linear-time pseudo-Gaussian approximation to Lee-Richards surfaces (PGALRS) J Appl Crystallogr. 2010;43:356–361. doi: 10.1107/S0021889809054326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powers JC, Oleksyszyn J, Narasimhan SL, Kam CM, Radhakrishnan R, Meyer EF. Reaction of porcine pancreatic elastase with 7-substituted 3-alkoxy-4-chloroisocoumarins: Design of potent inhibitors using the crystal structure of the complex formed with 4-chloro-3-ethoxy-7-guanidinoisocoumarin. Biochemistry. 1990;29:3108–3118. doi: 10.1021/bi00464a030. [DOI] [PubMed] [Google Scholar]


