Abstract
Calmodulin (CaM), a ubiquitous intracellular sensor protein, binds Ca2+ and interacts with various targets as part of signal transduction. Using hydrogen/deuterium exchange (H/DX) and a high resolution PLIMSTEX (Protein-Ligand Interactions by Mass Spectrometry, Titration, and H/D Exchange) protocol, we examined five different states of calmodulin: calcium-free, calcium-loaded, and three states of calcium-loaded in the presence of either melittin, mastoparan, or skeletal myosin light-chain kinase (MLCK). When CaM binds Ca2+, the extent of HDX decreased, consistent with the protein becoming stabilized upon binding. Furthermore, Ca2+-saturated calmodulin exhibits increased protection when bound to the peptides, forming high affinity complexes. The protocol reveals significant changes in EF hands 1, 3, and 4 with saturating levels of Ca2+. Titration of the protein using PLIMSTEX provides the binding affinity of Ca2+ to calmodulin within previously reported values. The affinities of calmodulin to Ca2+ increase by factors of 300 and 1000 in the presence of melittin and mastoparan, respectively. A modified PLIMSTEX protocol whereby the protein is digested to component peptides gives a region-specific titration. The titration data taken in this way show a decrease in the root mean square fit of the residuals, indicating a better fit of the data. The global H/D exchange results and those obtained in a region-specific way provide new insight into the Ca2+-binding properties of this well-studied protein.
1. INTRODUCTION
Calcium ions (Ca2+) play a vital role in biological functions of higher organisms, particularly in signal transduction [1–3]. Calcium channels regulate the [Ca2+] inside the cell to be in the range of 0.1–1.0 µM, from an extracellular concentration of 1.0 mM [4]. Of the over 500 Ca2+-binding proteins identified, many are responsible for monitoring the intracellular Ca2+ concentration and communicating this signal to a binding partner [5]. Calmodulin (CaM) is a highly conserved, small (148 amino acids in eukaryotes), acidic (pI 3.9 – 4.3), ubiquitous protein that functions as an intracellular sensor protein using Ca2+ [6–9]. CaM in its calcium-bound form can bind up to 300 target proteins and/or peptides in signal transduction pathways [4].
Calcium-binding proteins contain a helix-loop-helix motif [10] called an EF hand that consists of a nine-amino acid residue helix, a 12-residue loop, and then another eight-residue helix. The loop contains highly conserved Asp, Gly, and Glu in position 1, 6, and 12 [5]. CaM consists of two globular domains at the N- and C-termini, each containing two EF hands; the globular domains are connected by a central α-helix. The Ca2+-bound form of CaM is dumbbell-shaped with two Ca2+ ions bound in each domain. The structure of Ca2+ loaded CaM determined by X-ray crystallography reveals the overall dumbbell shape of the protein, including an α-helix connecting both domains [11, 12]. A more recent NMR structure reveals that the central linker is highly flexible [13]. The apo calmodulin NMR structure also revealed that the helical domains adopt a “closed conformation” by packing the hydrophobic residues. This packing is disrupted upon binding Ca2+, altering the structure of each domain and exposing the hydrophobic surfaces for binding to target proteins [14].
The nature of the binding of CaM to Ca2+ and to peptides and protein targets has been studied by mass spectrometry [15–20], calorimetry [21–23], and fluorescence [24–26]. They reveal that CaM binds Ca2+ in a sequential manner when a target protein or peptide is not present [27, 28]. Although the two EF hand pairs have ~50% sequence identity and are over 75% sequence similar, the C-terminal domain binds Ca2+ with 10-fold higher affinity than the N-terminal domain [4]. CaM undergoes a significant conformational change upon binding Ca2+, consistent with four EF hands that represent nearly 80% of the sequence (29 residues per EF hand and a total of four EF hands). In the presence of protein or peptide targets, the affinity of CaM for Ca2+ (Ki) increases significantly, possibly associated with a slower off-rate (koff) of Ca2+ when CaM binds a peptide or another protein (26–28).
We report here H/DX kinetics and PLIMSTEX at the peptide level to determine the site-specific (peptide level) affinity and conformational changes that take place in CaM upon Ca2+ and subsequent binding to three peptides. Given that the peptides, melittin [29–37], mastoparan [38–46] and MLCK [47–49] interact in an established way, our principal goal is to establish the outcome of H/DX and PLIMSTEX for studies of metal-containing proteins and their binding to peptides. This extended methodology builds on the global protein analysis reported earlier [20] and offers a higher resolution approach (peptide level) of H/DX kinetics and titrations. To carry out this research, we modified the original PLIMSTEX fitting algorithm [50] to incorporate the high resolution titration data.
2. EXPERIMENTAL
2.1 Materials
“Calcium-free” porcine calmodulin was from Ocean Biologics Co. (Edmonds, WA). Deuterium oxide, potassium chloride, formic acid, calcium chloride, melittin from honey bee venom (MW 2846), mastoparan from Vespula lewisii (MW 1478), acetonitrile, EGTA (ethylene glycol-bis(2-aminoethylether)-N,N,N',N'-tetra-acetic acid), HEPES [N-(2-hydoxyethyl)piperazine-N'-(2-ethanesulfonic acid)], and HEPES sodium salt were from Sigma-Aldrich (St. Louis, MO) at the highest purity available. MLCK (MW 2966) was purchased from AnaSpec (Fremont, CA). Immobilized pepsin on agarose was from Pierce (Rockford, IL). The MW of porcine calmodulin was determined by ESI mass spectrometry to be 16790 Da (see supplementary Figure S-1), indicating that the protein has two post-translational modifications, likely N-terminal acetylation (+42 Da) and trimethylation of Lys 115 (+42 Da).
2.2 H/D Exchange – Kinetics and Titration
Protein stock solutions (238 µM, 1 mg protein in 250 µL buffer) were prepared with 10 mM HEPES (pH = 7.4) and 150 mM KCl. H/DX kinetics experiments were conducted with apo CaM (containing 1 mM EGTA) and holo CaM (containing 2 mM Ca2+). Prior to titration, solutions were equilibrated for 1 h. To initiate exchange, 0.5 µL of the protein stock was diluted with 20 µL of D2O containing 10 mM HEPES and 150 mM KCl at 25 °C to give a solution that was > 97% D2O. After certain times, the exchange was quenched with ice-cold 1.0 M HCl to give a final pH of 2.0.
To examine D uptake for regions of the protein, 5 µL of immobilized pepsin on agarose was added to the quenched solution, digesting for 3 min at 0 °C and vortexing every 15 sec. The resulting mixture was briefly centrifuged (2–3 sec) so that the beads congregated at the bottom. The supernatant protein digest, including some undigested protein, was loaded on a C18 column (LC Packings, 1×15 mm, PepMap cartridge, Dionex Corp., Sunnyvale, CA) that was pre-equilibrated with 100 µL of 0.2% formic acid in water (0 °C). The column was washed with 300 µL of 0.2% formic acid in water (0 °C), back exchanging the labile sites of the peptides and protein, and the peptides were separated with a LC gradient (5% B to 40% B in 6 min, 40% B to 75% B in 2 min, 75% B to 5% B in 0.5 min, 5% B to 40% B in 0.5 min, then back to 5% B for equilibration) at a flow rate of 40 µL/min (Solvent A: 95% water, 5% acetonitrile containing 0.3% formic acid; Solvent B: 5% water, 95% acetonitrile containing 0.3% formic acid). To minimize back exchange, the incoming/outgoing LC solvent line, injection valve, and sample loop were submerged in ice/water slush (0 °C).
2.3 LC-ESI/MS Analysis with a Q-TOF Mass Spectrometer
All ESI mass spectra during the H/DX experiments were acquired in the positive-ion mode on a Waters (Micromass) Q-TOF Ultima (Manchester, U.K.) equipped with a Z-spray ESI source. The capillary voltage was 3.2 kV, cone voltage readback of 100 V, and the source and desolvation temperatures were 80 and 180 °C. The cone and desolvation gas flows were 40 and 400 L/h. The MS profile used for quadrupole transmission was from m/z 500, dwell for 5% of the scan time, ramp to m/z 1000 for 45% of the scan time, and then dwell at m/z 1000 for 50% of the scan time.
2.4 LC-ESI/MS-MS Analysis of Protein Digest
After 3 min of pepsin digestion, the solution containing the protein digest, including some undigested protein, was loaded onto a C18 custom-packed column (75 µm i.d., 10 cm length). The peptides were separated over 70 min using an Eksigent NanoLC-1D (Dublin, CA) with an LC gradient from 3–97% acetonitrile containing 0.1% formic acid at a flow rate of 260 nL/min with spray directly into the mass spectrometer using a PicoView PV-500 nanospray source (New Objective, Woburn, MA) attached to an LTQ-FTMS (Thermo, San Jose, CA), which afforded accurate mass and product-ion sequencing by MS/MS. The peptides were identified by searching against NCBI database on Mascot (Matrix Science, Oxford, UK), and each peptide was manually verified by de novo sequencing.
2.5 Data Analysis
For H/DX data collected on the Q-TOF, the protein mass spectrum at each exchange time point was deconvoluted with MaxEnt1 algorithm (MassLynx 4.0). The D level at each time point was determined by subtracting the centroided mass of the undeuterated protein from the that of the deuterated protein. The rate of back exchange was 1 D loss per min. Ion signals for the deuterated peptides were smoothed twice in MassLynx with a Savitsky-Golay algorithm and imported into Microsoft Excel as an x,y pair (mass, intensity). The centroid and width of the deuterium distribution for each peptide was analyzed using HX-Express software [51]. The back exchange occurs at the same rate as in the experiment in which exchange at the global level was examined. No corrections were made for back exchange because only relative D levels were compared. The experiments were in triplicate.
2.6 Kinetic and Titration Modeling
The H/DX kinetic data were fit with a fixed rate-constant binning model in which all exchangeable H’s were separated into four fixed rate-constant bins. The bins were chosen to span four orders of magnitude (10, 1, 0.1, and 0.01 min−1), representing rate constants that cover the experimental time scale and allowing comparison of the number of exchangeable H’s among different binding states of CaM [20]. The model was applied globally to full-length proteins and to component peptides in the digest. The number of exchangeable hydrogens was optimized by using the “Minimize” function in MathCAD to minimize the root mean square (RMS) of the residuals. Each trial (of a total of three) was fit separately, and the results were averaged and reported with ± one standard deviation.
The details of the global titration modeling were described previously [50]. To utilize the information from titration data at the peptide level, the protocol was modified. Traditionally, a set of parameters were chosen to fit the experimentally observed data. These included the deuterium uptake with no ligand present (D0), the deuterium uptake change upon each binding event (ΔDi), and the affinity constants expressed as βi’s where β1 = Ka1, β2 = Ka1Ka2, β3 = Ka1Ka2Ka3, etc. for each ligand binding event (where i = binding stoichiometry).
The details of the first two Ca2+ binding events cannot be resolved on the basis of our titration data, as was described previously [20]. Therefore, the values of Ka1, Ka2, ΔD1, and ΔD2 were held constant during the parameter search. In the modified protocol, the deuterium uptake as f([Ca2+]) was input for the full-length protein and the individual constituent peptides. The initial guesses for D0 and ΔD4 were set to the experimental values. The non-linear least squares fitting utilized the “Minimize” function in MathCAD to minimize the root mean square (RMS) of all inputs by optimizing the parameters of Ka3, Ka4, D0, ΔD3, and ΔD4.
3. Results and Discussion
3.1 Global H/D Exchange Kinetics of Calmodulin
We found with the pepsin digestion 30–50% protein remained in the digest, probably a result of the low T (0 °C). We took advantage of the incomplete digestion to obtain in the same experiment the D uptake data for the whole protein and for the peptides. The forward H/DX experiments were conducted over 60 min for (1) CaM with 1 mM EGTA, (2) CaM with 2 mM Ca2+, (3) CaM with 2 mM Ca2+ and 2.9:1 ratio of melittin:CaM, and (4) CaM with 2 mM Ca2+ and 2.9:1 ratio of mastoparan:CaM, and (5) CaM with 2 mM Ca2+ and 2.9:1 ratio of MLCK:CaM. The addition of EGTA ensures that the apo state was essentially 100%, whereas the latter samples contained sufficient Ca2+ to ensure that the protein is ~100% Ca2+-bound.
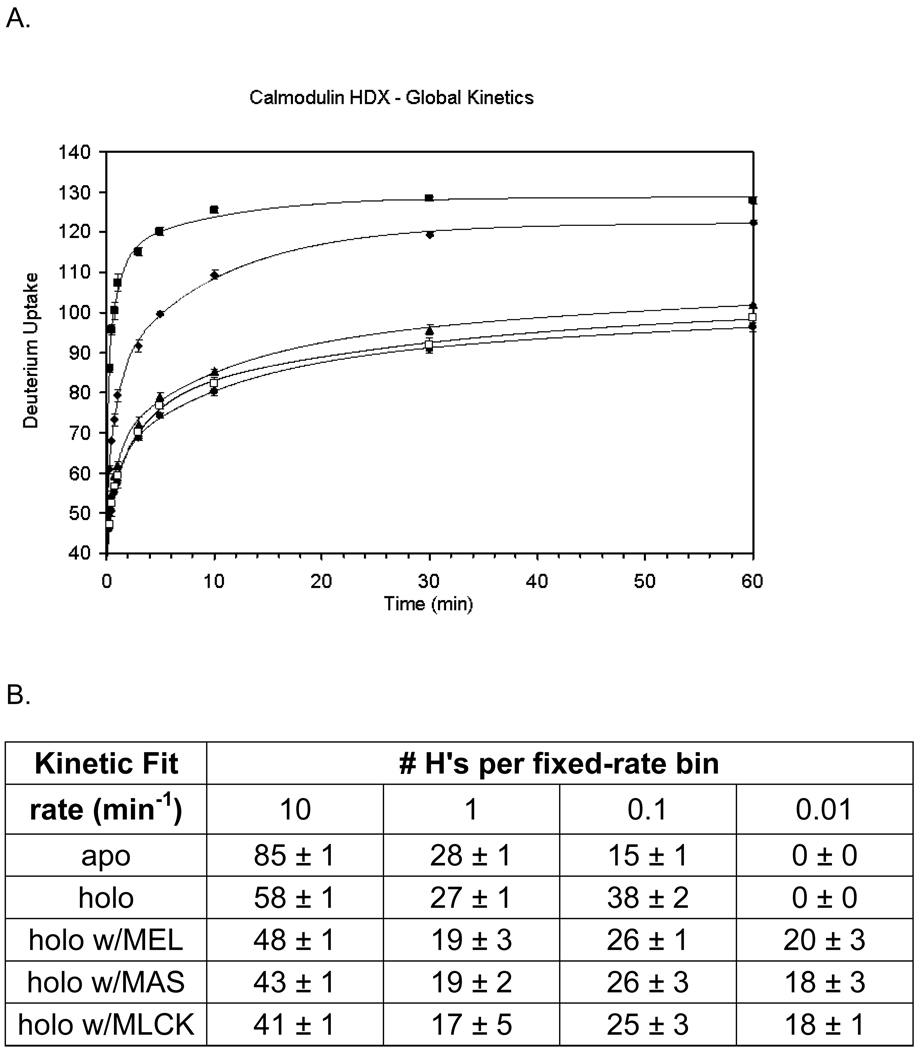
A plot of D uptake vs. time shows the protection change of apo CaM upon binding Ca2+ in the presence of melittin, mastoparan, and MLCK (Figure 1). The total number of exchangeable H’s for porcine CaM is 145, considering there are two Pro and that the terminal NH2 undergoes rapid back exchange. Given 97% D2O in all H/DX experiments, the maximum number of observable exchange events is 140. After 10 min of H/DX, apo CaM showed a mass shift of 125.3 ± 0.6 Da, indicating that ~ 90% of the amide sites were deuterated. This high level of exchange is consistent with the highly flexible, solvent-exposed, and dynamic nature of CaM in the absence of Ca2+. In the presence of Ca2+, the extent of HDX decreases by 10–15 amides, owing an increase in H bonding and/or a decrease in solvent accessibility (Figure 1). Upon addition of melittin at a 2.9:1 ratio of CaM in the presence of Ca2+, the extent of H/DX decreases from 109 ± 1 Da to 85 ± 1 Da. A decreased D uptake also occurs upon addition of mastoparan. The corresponding mass shift, from 109 ± 1 Da to 80 ± 1 Da shows that 29 amide sites are affected by mastoparan binding. Although melittin is larger than mastoparan (26 vs. 14 amino acids), the effect of mastoparan binding on HDX of CaM is more pronounced. MLCK (26 amino acids), a similar sized peptide as melittin, also introduces a significant decrease of the extent of H/DX from 109 ± 1 Da to 82 ± 1 Da. This difference is close to the one for mastoparan binding, perhaps indicates that the protein complexes share similar structures.
Figure 1.
(A) Global H/DX kinetics experiments: calmodulin with no Ca2+(1 mM EGTA) (squares), calmodulin with 2 mM Ca2+ (diamonds), 2:1 melittin:calmodulin with 2 mM Ca2+ (triangles), 2:1 mastoparan:calmodulin with 2 mM Ca2+ (circles) and 2:1 MLCK:calmodulin with 2 mM Ca2+ (open squares). H/DX was conducted over a 60 min time course with 97% D2O, 10 mM HEPES (pH 7.4), 150 mM KCl, and 4–5 µM calmodulin. (B) Results of the fixed-rate kinetic binning model for the four kinetics curves in A.
Kinetic modeling reveals (Figure 1B) that the apo state has 85 sites exchanging with a k = 10 min−1 whereas when loaded with Ca2+, the number decreases to 58, indicating that many fast-exchanging amide sites are affected by Ca2+ binding. Concomitantly, there is an increase from 15 to 38 that exchange with a rate constant of 0.1 min−1. There are further decreases in the number of exchanging amide hydrogens with a k = 10 min−1 upon binding melittin, mastoparan and MLCK. Overall, we see a net shift to lower exchange rate constants for some amides by as much 104, consistent with formation of a less flexible, more stabilized secondary structure upon Ca2+ and peptide binding.
3.2 H/DX Kinetics for Regions of Calmodulin
Pepsin digestion, which permits exploration of regions of the exchanging protein, affords ~76 unique peptides, which we identified from their product-ion spectra and accurate masses (ion trap/FT mass spectrometer LC/MS/MS with Mascot analysis). Although we were unable to observe all these peptides with the Q-TOF, most likely because it is less sensitive and was not equipped with nanospray, we found a sufficient number to afford good coverage.
3.3 Extents of HDX for the EF Hands
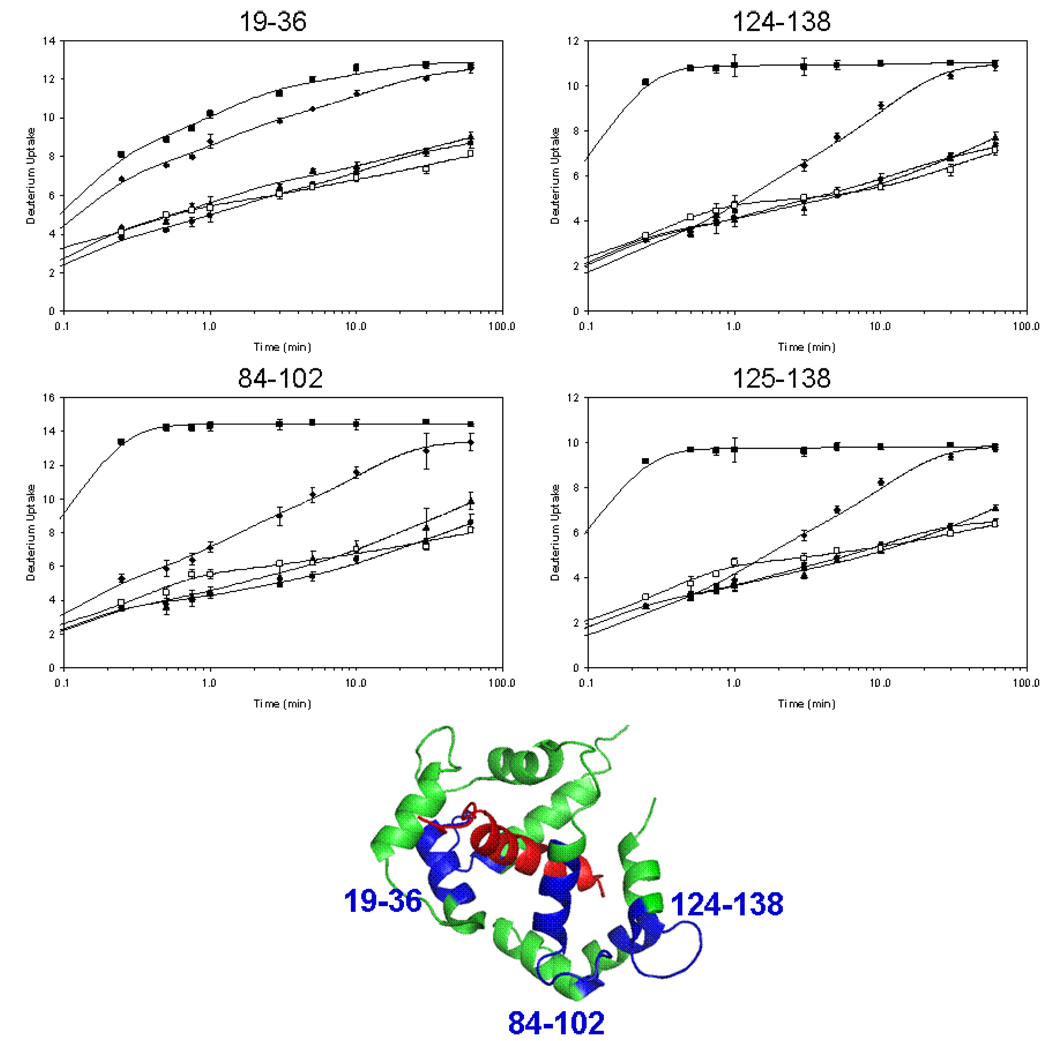
When CaM binds Ca2+ at saturating concentrations, the largest changes in H/DX occur for EF-hand 3 (residues 84–102) and 4 (residues 124–138, 125–138, and 125–140) (see Figure 2 kinetic data and structure of the protein/MLCK complex). By way of contrast, EF-hand 1 in the N-terminal domain shows only a small change in D uptake upon binding Ca2+. The S/N ratio representing the peptide containing EF-hand 2 was insufficient to analyze its D uptake. The region-specific exchange results show that a tighter, less exchangeable structure forms with Ca2+ binding in the C-terminal domain. This is consistent with the higher affinity for Ca2+ at the C-terminal than at the N-terminal domain [4].
Figure 2.
Local H/DX kinetics in the EF hand regions of CaM. Peptide 19–36 represents EF hand 1, 84–102 represents EF hand 3, and 124–138 and 125–138 represent EF hand 4. Four states are shown: CaM with no Ca2+ (squares), CaM with 2 mM Ca2+ (diamonds), 2:1 melittin:CaM with 2 mM Ca2+ (triangles), 2:1 mastoparan:CaM with 2 mM Ca2+ (circles) and 2:1 MLCK:CaM with 2 mM Ca2+ (open squares). H/DX was conducted over 60 min with 97% D2O, 10 mM HEPES (pH 7.4), 150 mM KCl, and 4–5 µM calmodulin. The curves were fit with a four fixed-rate binning model using exchange rate constants of 10, 1 0.1, and 0.01 min−1. The structure of CaM (green):4Ca2+ binding to MLCK (red) (PDB: 2BBM) is shown in center. Peptide regions that reported here are shown in blue.
There are also significant changes in D uptake upon binding Ca2+ in the presence of melittin, mastoparan and MLCK with respect to Ca2+ binding to CaM itself. At 60 min, the largest changes take place in EF hands 1 (peptide 19–36), 3 (peptide 84–102), and 4 (peptides 124–138, 125–138, and 125–140, data not shown for last peptide because it adds no additional insight). Interestingly, melittin, mastoparan and MLCK affect EF hands 1, 3, and 4 similarly, indicating that the Ca2+ binding regions do not distinguish these three peptides (i.e., each region becomes comparably stabilized upon binding). Moreover, by knowing the structure of 4Ca2+:CaM:MLCK [47], we can then predict that the structures of Ca2+-loaded CaM when bound to melittin, mastoparan and MLCK are nearly identical.
3.4 Extents of H/DX in the Linker Regions
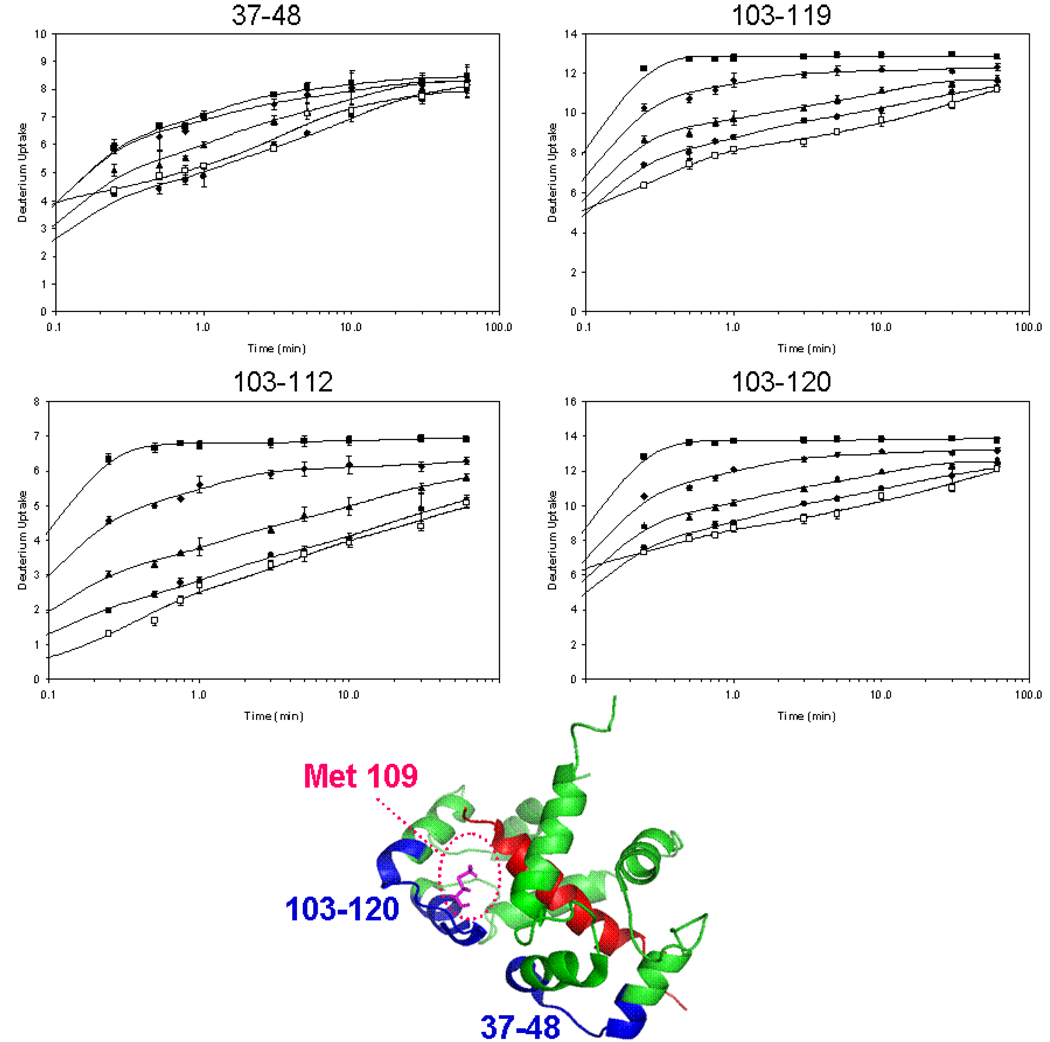
The linker regions between EF hands 1 and 2 (residues 37–48) and between EF hands 3 and 4 (residues 103–112, 103–119, 103–120, and 103–123) (see Figure 3 for the kinetic data and the structure of the protein/MLCK structure) show relatively little change upon Ca2+ binding. There are, however, differences in the D uptakes of peptide 37–48 between the Ca2+-loaded protein and in the presence of the peptides; binding to the peptides adds additional protection. Similar trends pertain for the linker between EF hands 3 and 4 (peptides 103–112, 103–119, 103–120, and 103–123) when melittin, mastoparan, or MLCK binds to Ca2+-loaded CaM. Mastoparan binding consistently adds protection to one additional amide site upon binding CaM. These linker regions show some specificity for mastoparan over melittin binding, and the approach may ultimately give site-specific peptide binding.
Figure 3.
Local H/D exchange kinetics experiments in the linker regions between EF hands of calmodulin. Peptide 37–48 represents the linker between EF hand 1 and 2 and peptides 103–112, 103–119, 103–120, and 103–123 (not shown) represent the linker between EF hand 3 and 4. Four states of CaM are shown: CaM with no Ca2+ (squares), CaM with 2 mM Ca2+ (diamonds), 2:1 melittin:CaM with 2 mM Ca2+ (triangles), 2:1 mastoparan:CaM with 2 mM Ca2+ (circles) and 2:1 MLCK:CaM with 2 mM Ca2+ (open squares). Met109 on the CaM structure, shown in pink, is in position to bind peptides.
There are no X-ray or NMR structures of the CaM:4Ca2+:MEL or MAS complexes, but there is an X-ray structure of CaM with the RS20 peptide (RGB ID 1QTX) and, more importantly, an NMR structure of CaM with MLCK (PDB:2BBM) [47] (Figures 2 and 3). Residues 37–48 and 103–120 of CaM nearly contact one another in the CaM:4Ca2+:MLCK structure even though they are on different domains of calmodulin. The methionines of these two regions of CaM may be involved in binding the peptides. In comparing the extents of D uptake in regions 103–112, 103–119, 103–120, and 103–123, a 1 Da or more difference in deuterium uptake upon binding melittin, mastoparan, and MLCK appears in all the peptide segments. Thus, the incremental protection occurring with the peptide binding must already occur in region 103–112, which contains Met 109 that is pointing directly at the MLCK peptide in the calmodulin:4Ca2+:MLCK complex structure (Figure 3).
3.5 Regions of CaM Not Affected by Binding
There are regions of CaM that are little affected by Ca2+ binding. Peptides 12–18, 69–72, and, to a lesser extent, peptides 117–128 and 141–148 show only small increases in protection upon binding Ca2+(see supplementary Figure S-2). Peptide 12–18 is part of the N-terminal tail and EF hand 1, and 69–72 is the beginning of the central α-helix connecting the two domains. Peptide 117–128 is part of EF hand 4, and 141–148 is the C-terminal tail.
A comparison of the overlap between a region that shows little change upon binding, peptide 117–128, and that showing a significant change, peptide 124–138, reveals that residues 129–138 are those affected by Ca2+ binding. These residues are part of EF-hand 4.
3.6 Calmodulin Binding to Ca2+ in the presence or absence of Melittin and Mastoparan
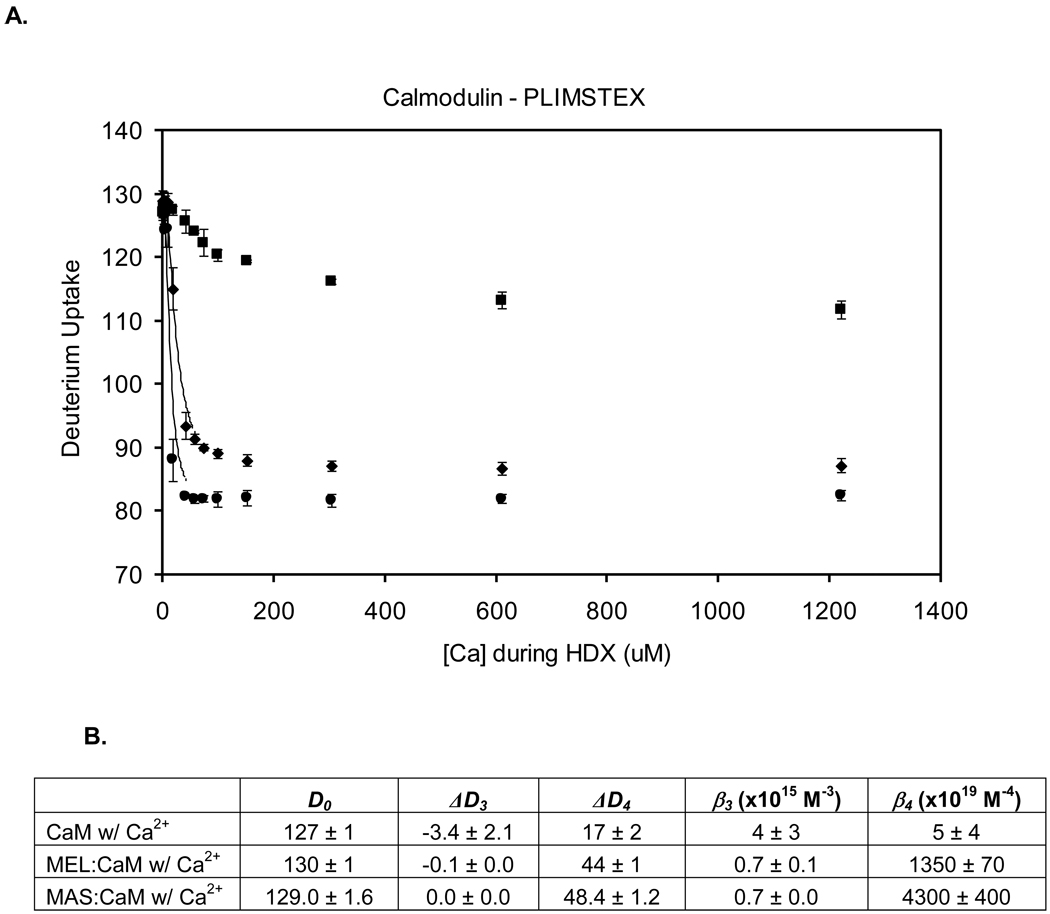
If there are differences in H/DX between a bound and unbound state of a protein, a PLIMSTEX determination will give the protein’s affinity for the ligand [50, 52]. We fit the experimental PLIMSTEX curves (Figure 4) by a non-linear least squares model to afford the affinity constants of CaM for Ca2+. The best fit determines the D uptake with no Ca2+ present (D0), the deuterium uptake change upon each binding event (ΔDi), and the affinity constants as βi’s where β1 = Ka1, β2 = Ka1Ka2, β3 = Ka1Ka2Ka3, and β4 = Ka1Ka2Ka3Ka4. Modeling the titration curve of CaM with Ca2+ using a five parameter search [20] gave D0, ΔD3, ΔD4, β3, and β4. The values of Ka1 and Ka2 were fixed at 5.0 × 104 M−1 and 1.6 × 106 M−1, respectively, for apo CaM titrated with Ca2+ [25], and the values of ΔD1 and ΔD2 were fixed at 0. Data from three trials were fit separately to determine the average and standard deviation. The experimental value of D0 is 127 ± 1 Da, which is in agreement with that from the modeling, (i.e., 127 ± 1 Da). The ΔDTOT for CaM titrated with Ca2+ is 15 ± 1 Da, in agreement with the modeling (i.e., 17 ± 2 Da). The Ca2+ binding affinity from PLIMSTEX, expressed as β4, is 5 ± 4 × 1019 M−4 (2.9 µM CaM, 10 mM HEPES, 150 mM KCl, pH 7.4, 25 °C). We determined again this value to test the long-term reproducibility of PLIMSTEX; the β4 from our previous study is 3.6 × 1019 M−4 (15 µM CaM, 50 mM HEPES, 100 mM KCl, pH 7.4, 21.5 °C) [20]. The value from NMR is 1.6 × 1020 M−4 (20–30 µM CaM, 2 mM Tris/HCl, pH 7.5, 25 °C, 150 mM KCl, and 25–30 µM Br2BAPTA) [25]. All the results are in good agreement.
Figure 4.
(A) Global H/DX titration experiments: CaM titrated with Ca2+ (squares), 2.9:1 melittin:CaM titrated with Ca2+ (diamonds), and 2.9:1 mastoparan:CaM titrated with Ca2+ (circles). H/DX was conducted at a constant 10 min with 97% D2O, 10 mM HEPES (pH 7.4), 150 mM KCl, and 4–5 M CaM. (B) Output of the PLIMSTEX titration modeling using a five parameter search including D0, ΔD3, ΔD4, β3, and β4.
The Ca2+ binding affinity of CaM and its extent of protection increase dramatically when melittin or mastoparan are present in solution. To obtain a fit, the same five-parameter search was used to model the data of CaM binding Ca2+ as was used in the absence of melittin and mastoparan with one exception: Ka1 and Ka2 were changed to 5.0 × 106 M−1 and 1.4 × 106 M−1, respectively [32], to reflect the values reported in the literature. These values were determined by tryptophan fluorescence at a lower binding temperature than our protocol (60 µM CaM, 1:1 ratio of CaM:MEL, 20 mM TES buffer, 100 mM NaCl, 1.0 mM EGTA at pH 7.0 and 7 °C). The experimental value of ΔDTOT is 42 ± 1 Da, which agrees with the fit parameter of 44 ± 1 Da, and the new affinity, expressed as β4, of CaM for Ca2+ in the presence of melittin has increased to 1.4 ± 0.1 × 1022 M−4 (2.9 µM CaM, 10 mM HEPES, and 150 mM KCl). This value agrees reasonably well with that determined previously by us under slightly different conditions; that is, β4, is 8.0 × 1022 M−4 (15 µM CaM, 50 mM HEPES, and 100 mM KCl) [53]. The binding affinity of CaM for Ca2+ in the presence of melittin is 300 times greater than in its absence.
Mastoparan also significantly affects β4, but this was not reported previously. Our result is 4.3 ± 0.4 × 1022 M−4 (2.9 µM CaM, 10 mM HEPES, and 150 mM KCl), showing also a significant increase in binding in the presence of mastoparan; β4 in the presence of mastoparan is nearly 1000 times greater than that without it and 3 times greater than the affinity in the presence of melittin.
MLCK also increases the β4; the value is 4.2 × 1022 M−4, as determined by using flow dialysis (5–10 µM CaM, 10 mM HEPES, 150 mM NaCl, and 1 mM MgCl2 at 25 °C) [54]. This value is nearly identical to the β4 in the presence of mastoparan, as described above. Although we didn’t measure β4 for Ca2+-loaded CaM in the presence of MLCK, the H/D kinetics at the global and peptide levels results also show that MLCK affects the binding of CaM with Ca2+ in approximately the same way as does mastoparan.
Once the affinity of Ca2+ is determined, we calculated the fractionally bound CaM species as a function of Ca2+, giving the concentration of CaM:xCa2+ (x = 1–4) species under various conditions. The fourth Ca2+ binding event triggers the largest change in D uptake and mimics the overall shape of the observed PLIMSTEX titration curve (see supplementary Figure S-3). The relative populations of CaM:1Ca2+ and CaM:2Ca2+ are relatively small. In the presence of melittin or mastoparan, the binding scenario changes significantly. The CaM:1Ca2+ and CaM:3Ca2+ are the lowest concentration species, whereas the CaM:2Ca2+ is intermediate. This suggests that once the second Ca2+ is bound, the third Ca2+ begins to bind and then yields to the high-affinity binding of the fourth Ca2+, as revealed by the low concentration of Ca2+ needed to form the CaM:4Ca2+:peptide complex. The fourth binding of Ca2+ produces the largest change in D uptake and contributes most to the shape of the observed PLIMSTEX curve for each peptide [20, 53].
3.5 Region-Specific H/DX Titration of Calmodulin
Our goal is to advance the PLIMSTEX protocol to a more region-specific model with the hope of obtaining site-specific affinity constants for any protein of interest. CaM affords an opportunity to test this prospect. CaM has four EF hands, three of which we can liberate by digestion for region-specific H/DX. We were able to follow a subset of eight peptides during the Ca2+ titration.
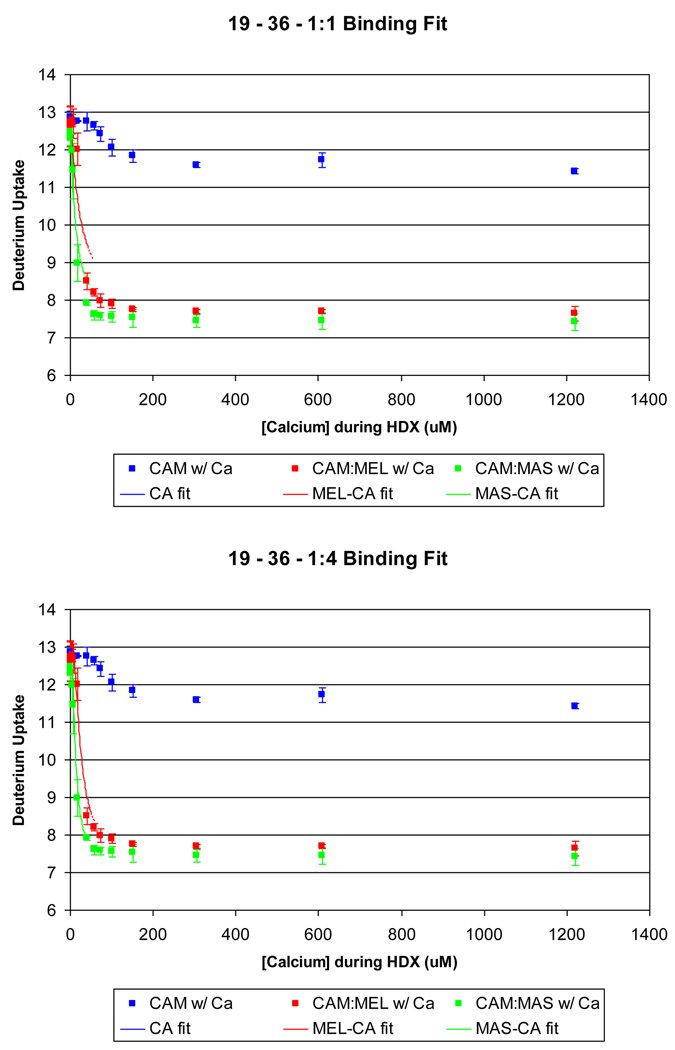
Perhaps the Ca2+ binding to each EF hand can be fit with a single affinity constant, treating each site with 1:1 binding stoichiometry. An attempt to do this with EF-hand 1 proved unsuccessful (Figure 5A). Although the fit is reasonable for Ca2+ binding apo calmodulin, it is poor for Ca2+ binding in the presence of melittin or mastoparan. The calculated curves from the fitting are shifted to higher concentrations of Ca2+. This shift and the poor agreement indicate that the change in D uptake for this EF hand is not independent of Ca2+ binding at other sites, a conclusion that is consistent with findings that Ca2+ binding to calmodulin is highly cooperative [5]. This same result was obtained for EF-hand 4 (data not shown).
Figure 5.
A comparison of a forced 1:1 fit (A) and the 1:4 fit (B) binding model of EF hand 1 (residues 19–36) for the following experiments: calmodulin titrated with Ca2+ (squares), 2.9:1 melittin:calmodulin titrated with Ca2+ (diamonds), and 2.9:1 mastoparan:calmodulin titrated with Ca2+ (circles). The fit for melittin and mastoparan was not successful with a 1:1 binding model. The 1:4 fit, however, models the observed experimental data correctly.
The lack of agreement prompted us to modify the existing PLIMSTEX model to incorporate the D uptake of component peptides into the fitting algorithm. Upon fitting the peptide deuterium shift using the four association constants (Ka1, Ka2, Ka3, Ka4) from the global data and then minimizing the RMS associated with the ΔD’s, we obtained a better fit of EF hand 1 (Figure 5 B), and of peptides representing EF hands 3 and 4 (see supplementary Figure S-4).
The linker regions (peptides 37–48, 103–112, 103–119, and 103–120) exhibit distinct differences for binding melittin vs. mastoparan (Figure S-5). A consistent deuterium difference of 1 Da is revealed after a minimal addition of Ca2+ to the solution. This is consistent with H/DX kinetics at the peptide level.
The modified PLIMSTEX model utilizes the deuterium shifts observed in the protein and peptide titration curves simultaneously, iterating through each to determine the affinity parameters. The expanded array of data improves the RMS of the global titration curve, most likely due to statistical power, when calmodulin is titrated with Ca2+. The RMS decreases from 0.40 to 0.16 for the apo CaM:Ca2+ titration. In this case the affinity output was different, whereas β4 from protein data is 4.0 × 1018 M−4, whereas with inclusion of the expanded array was 4.5 × 1019 M−4, an order of magnitude different. In the presence of melittin, the affinity (β4) was the same (1.4 × 1022 M−4), however the RMS decreased from 1.7 to 0.6. In the presence of mastoparan, the affinity (β4) was the same (4.3 × 1022 M−4). The RMS, however, decreased from 4.1 to 1.4, presumably because not only nine curves (one global and eight peptide-level), instead of one global protein curve, were monitored, but also because peptide MWs are more accurately measured than are protein MWs.
4. CONCLUSION
HD/X at the protein and peptide leves, utilizing kinetics and PLIMSTEX, affords a detailed binding picture of calmodulin binding to Ca2+. The presence of Ca2+ induces a conformational change observed in the EF hand regions that can be characterized by an overall stabilization of their secondary structures. In the presence of three model peptides, melittin, mastoparan, and MLCK, calmodulin changes even more its conformation when a Ca2+ signal is introduced. The D uptake in the EF hand regions is not specific to either peptide. The linker regions, on the other hand, show some specificity for mastoparan and MLCK, indicated by an increase in protection. These regions are known to contain hydrophobic residues that affect the extensive binding network of CaM. Incorporating the peptide titration data affords better fits for the affinity constants and changes in D uptake with ligand binding, suggesting a general approach to improve the accuracy and precision of PLIMSTEX. Although CaM has been studied extensively, the results presented here indicate that PLIMSTEX is a useful approach for these metal-binding peptides and that it can add to our knowledge about cooperativity in higher order binding systems. Moreover, the outcome shows that ligand/protein binding in the presence of potential binding partners, as it is in vivo, may be surprising different than ligand/protein alone in aqueous solution, the usual medium for biophysical measurements.
Supplementary Material
Acknowledgement
This research was supported by the National Center of Research Resources of the NIH, Grant No. 2P41RR000954.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Evenäs J, Malmendal A, Forsén S. Calcium. Current Opinion in Chemical Biology. 1998;2:293–302. doi: 10.1016/s1367-5931(98)80072-0. [DOI] [PubMed] [Google Scholar]
- 2.Brini M, Carafoli E. Calcium signalling: a historical account, recent developments and future perspectives. Cellular and Molecular Life Sciences. 2000;57:354–370. doi: 10.1007/PL00000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carafoli E. Calcium signaling: A tale for all seasons. Proceedings of the National Academy of Sciences. 2002;99:1115–1122. doi: 10.1073/pnas.032427999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vetter SW, Leclerc E. Novel aspects of calmodulin target recognition and activation. European Journal of Biochemistry. 2003;270:404–414. doi: 10.1046/j.1432-1033.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- 5.Yang JJ, Gawthrop A, Ye Y. Obtaining site-specific calcium-binding affinities of calmodulin. Protein and Peptide Letters. 2003;10:331–345. doi: 10.2174/0929866033478852. [DOI] [PubMed] [Google Scholar]
- 6.Kakiuchi S, Yamazaki R. Stimulation of the activity of cyclic 3',5'-nucleotide phosphodiesterase by calcium ion. Proceedings of the Japan Academy. 1970;46:387–392. [Google Scholar]
- 7.Cheung WY. Cyclic 3',5'-nucleotide phosphodiesterase. Demonstration of an activator. Biochemical and Biophysical Research Communications. 1970;38:533–538. doi: 10.1016/0006-291x(70)90747-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang JH, Waisman DM. Calmodulin and its role in the second-messenger system. Current Topics in Cellular Regulation. 1979;15:47–107. doi: 10.1016/b978-0-12-152815-7.50006-5. [DOI] [PubMed] [Google Scholar]
- 9.Wolff DJ, Brostrom CO. Properties and functions of the calcium-dependent regulator protein. Advances in Cyclic Nucleotide Research. 1979;11:27–88. [PubMed] [Google Scholar]
- 10.Kretsinger RH, Nockolds CE. Carp-muscle calcium-binding protein. Journal of Biological Chemistry. 1973;248:3313–3326. [PubMed] [Google Scholar]
- 11.Babu YS, Sack JS, Greenhough TJ, Bugg CE, Means AR, Cook WJ. Three-dimensional structure of calmodulin. Nature. 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 12.Babu YS, Bugg CE, Cook WJ. Structure of calmodulin refined at 2.2 A resolution. Journal of Molecular Biology. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- 13.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Solution structre of calcium-free calmodulin. Nature Structural and Molecular Biology. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Yuan T. Molecular mechanisms of calmodulin's functional versatility. Biochemistry and Cell Biology. 1998;76:313–323. doi: 10.1139/bcb-76-2-3-313. [DOI] [PubMed] [Google Scholar]
- 15.Hu P, Ye Q-Z, Loo J. Calcium stoichiometry determination for calcium binding proteins by electrospray ionization mass spectrometry. Analytical Chemistry. 1994;66:4190–4194. doi: 10.1021/ac00095a013. [DOI] [PubMed] [Google Scholar]
- 16.Lafitte D, Capony JP, Grassy G, Haiech J, Calas B. Analysis of the ion binding sites of calmodulin by electrospray ionization mass spectrometry. Biochemistry. 1995;34:13825–13832. doi: 10.1021/bi00042a014. [DOI] [PubMed] [Google Scholar]
- 17.Nemirovskiy O, Ramanathan R, Gross ML. Investigation of calcium-induced, noncovalent association of calmodulin with melittin by electrospray ionization mass spectrometry. Journal of the American Society for Mass Spectrometry. 1997;8:809–812. [Google Scholar]
- 18.Nousiainen M, Vainiotalo P, Feng X, Derrick PJ. Calmodulin-Rs20-Ca4 complex in the gas phase: electrospray ionization and Fourier transform ion cyclotron resonance. European Journal of Mass Spectrometry. 2001;7:393–398. [Google Scholar]
- 19.Steiner RF, Albaugh S, Fenselau C, Murphy C, Vestline M. A mass spectrometry method for mapping the interface topography of interacting proteins, illustrated by the melittin-calmodulin system. Analytical Biochemistry. 1991;196:120–125. doi: 10.1016/0003-2697(91)90127-f. [DOI] [PubMed] [Google Scholar]
- 20.Zhu MM, Rempel DL, Zhao J, Giblin DE, Gross ML. Probing Ca2+-induced conformational changes in porcine calmodulin by H/D exchange and ESI-MS: Effect of cations and ionic strength. Biochemistry. 2003;42:15388–15397. doi: 10.1021/bi035188o. [DOI] [PubMed] [Google Scholar]
- 21.Brokx RD, Lopez MM, Vogel HJ, Makhatadze GI. Energetics of target peptide binding by calmodulin reveals different modes of binding. Journal of Biological Chemistry. 2001;276:14083–14091. doi: 10.1074/jbc.M011026200. [DOI] [PubMed] [Google Scholar]
- 22.Gilli R, Lafitte D, Lopez C, Kilhoffer M-C, Makarov A, Briand C, Haiech J. Thermodynamic analysis of calcium and magnesium binding to calmodulin. Biochemistry. 1998;73:5450–5456. doi: 10.1021/bi972083a. [DOI] [PubMed] [Google Scholar]
- 23.Milos M, Schaer JJ, Comte M, Cox JA. Calcium-proton and calcium-magnesium antagonisms in calmodulin: microcalorimetric and potentiometric analyses. Biochemistry. 1986;25:6279–6287. doi: 10.1021/bi00368a067. [DOI] [PubMed] [Google Scholar]
- 24.VanScyoc WS, Shea M. Phenylalanine fluorescence studies of calcium binding to N-domain fragments of paramecium calmodulin mutants show increased calcium affinity correlates with increased disorder. Protein Science. 2001;10:1758–1768. doi: 10.1110/ps.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linse S, Helmersson A, Forsén S. Calcium binding to calmodulin and its globular domains. Journal of Biological Chemistry. 1991;266:8050–8054. [PubMed] [Google Scholar]
- 26.Kilhoffer M-C, Demaille JG, Gerard D. Tyrosine fluorescence of ram testis and octopus calmodulins. Effects of calcium, magnesium, and ionic strength. Biochemistry. 1981;20:4407–4414. doi: 10.1021/bi00518a027. [DOI] [PubMed] [Google Scholar]
- 27.Kilhoffer M-C, Kubina M, Travers F, Haiech J. Use of engineered proteins with internal tryptophan reporter groups and perturbation techniques to probe the mechanism of ligand-protein interactions: investigation of the mechanism of calcium binding to calmodulin. Biochemistry. 1992:8098–8106. doi: 10.1021/bi00149a046. [DOI] [PubMed] [Google Scholar]
- 28.Haiech J, Klee CB, Demaille JG, Haiech J. Effects of cations on affinity of calmodulin for calcium: ordered binding of calcium ions allows the specific activation of calmodulin-stimulated enzymes. Theoretical approach to the study of multiple ligand binding to a macromolecule. Biochemistry. 1981:3890–3897. doi: 10.1021/bi00516a035. [DOI] [PubMed] [Google Scholar]
- 29.Comte M, Maulet Y, Cox JA. Ca2+-dependent high-affinity complex formation between calmodulin and melittin. Biochemical Journal. 1983;209:269–272. doi: 10.1042/bj2090269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Y, Squier TC. Variable conformation and dynamics of calmodulin complexed with peptides derived from the autoinhibitory domains of target proteins. Biochemistry. 1996;35:6815–6827. doi: 10.1021/bi960229k. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger TC, Weissman L, Eisenberg D. The structure of melittin in the form I crystals and its implication for melittin's lytic and surface activities. Biophysical Journal. 1982;37:353–361. doi: 10.1016/S0006-3495(82)84683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maulet Y, Cox JA. Structural changes in melittin and calmodulin upon complex formation and their modulation by calcium. Biochemistry. 1983;22:5680–5686. doi: 10.1021/bi00293a035. [DOI] [PubMed] [Google Scholar]
- 33.Schulz DM, Ihling C, Clore GM, Sinz A. Mapping the topology and determination of a low-resolution three-dimensional structure of the calmodulin-melittin complex by chemical cross-linking and high-resolution FTICRMS: direct demonstration of multiple binding modes. Biochemistry. 2004;43:4703–4715. doi: 10.1021/bi036149f. [DOI] [PubMed] [Google Scholar]
- 34.Scaloni A, Miraglia N, Orru S, Amodeo P, Motta A, Marino G, Pucci P. Topology of the calmodulin-melittin complex. Journal of Molecular Biology. 1998;277:945–958. doi: 10.1006/jmbi.1998.1629. [DOI] [PubMed] [Google Scholar]
- 35.Caday CG, Steiner RF. The interaction of calmodulin with melittin. Biochemical and Biophysical Research Communications. 1986;135:419–425. doi: 10.1016/0006-291x(86)90011-2. [DOI] [PubMed] [Google Scholar]
- 36.Kataoka M, Head JF, Seaton BA, Engelman DM. Melittin binding causes a large calcium-dependent conformational change in calmodulin. Proceedings of the National Academy of Sciences. 1989;86:6944–6948. doi: 10.1073/pnas.86.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathur S, Badertscher M, Scott M, Zenobi R. Critical evaluation of mass spectrometric measurement of dissociation constants: accuracy and cross-validation against surface plasmon resonance and circular dichroism for the calmodulin-melittin system. Physcial Chemistry Chemical Physics. 2007;9:6187–6198. doi: 10.1039/b707946j. [DOI] [PubMed] [Google Scholar]
- 38.Malencik DA, Anderson SR. High affinity binding of the mastoparans by calmodulin. Biochemical and Biophysical Research Communications. 1983;114:50–56. doi: 10.1016/0006-291x(83)91592-9. [DOI] [PubMed] [Google Scholar]
- 39.Malencik DA, Anderson SR. Demonstration of a fluorometrically distinguishable intermediate in calcium binding by calmodulin-mastoparan complexes. Biochemical and Biophysical Research Communications. 1986;135:1050–1057. doi: 10.1016/0006-291x(86)91034-x. [DOI] [PubMed] [Google Scholar]
- 40.Yazawa M, Ikura M, Hikichi K, Ying L, Yagi K. Communication between two globular domains of calmodulin in the presence of mastoparan or caldesmon fragment. Journal of Biological Chemistry. 1987;262:10951–10954. [PubMed] [Google Scholar]
- 41.Matsushima N, Izumi Y, Matsuo T, Yoshino H, Ueki T, Miyake Y. Binding of both Ca2+ and mastoparan to calmodulin induces a large change in the tertiary structure. Journal of Biochemistry. 1989;105:883–887. doi: 10.1093/oxfordjournals.jbchem.a122773. [DOI] [PubMed] [Google Scholar]
- 42.Yoshino H, Minari O, Matsushima N, Ueki T, Miyake Y, Matsuo T, Izumi Y. Calcium-induced shape change of calmodulin with mastoparan studied by solution x-ray scattering. Journal of Biological Chemistry. 1989;264:19706–19709. [PubMed] [Google Scholar]
- 43.Ohki S-y, Yazawa M, Yagi K, Hikichi K. Mastoparan binding induces Ca2+-transfer between two globular domains of calmodulin: a proton NMR study. Journal of Biochemistry. 1991;110:737–742. doi: 10.1093/oxfordjournals.jbchem.a123650. [DOI] [PubMed] [Google Scholar]
- 44.Ohki S-y, Tsuda S, Joko S, Yazawa M, Yagi K, Hikichi K. 1H NMR study on amide proton exchange of calmodulin-mastoparan complex. Journal of Biochemistry. 1991;109:234–237. [PubMed] [Google Scholar]
- 45.Wolf T, Solomon B, Ivnitski D, Rishpon J, Fleminger G. Interactions of calmodulin with metal ions and with its target proteins revealed by conformation-sensitive monoclonal antibodies. Journal of Molecular Recognition. 1998;11:14–19. doi: 10.1002/(SICI)1099-1352(199812)11:1/6<14::AID-JMR382>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 46.Moorthy AK, Gopal B, Satish PR, Bhattacharya S, Bhattacharya A, Murthy MRN, Surolia A. Thermodynamics of target peptide recognition by calmodulin and a calmodulin analogue: implications for the role of the central linker. FEBS Letters. 1999;461:19–24. doi: 10.1016/s0014-5793(99)01380-0. [DOI] [PubMed] [Google Scholar]
- 47.Ikura GMC Mitsuhiko, Gronenborn Angela M, Zhu Guang, Klee Claude B, Bax Ad. Solution Structure of a Calmodulin-Target Peptide Complex by Multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 48.Hultschig C, Hecht H-Jg, Frank R. Systematic Delineation of a Calmodulin Peptide Interaction. Journal of Molecular Biology. 2004;343:559–568. doi: 10.1016/j.jmb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 49.Wendy AF, Michael JG, Peter MB. Role of the N-terminal region of the skeletal muscle myosin light chain kinase target sequence in its interaction with calmodulin. Protein Science. 1995;4:2375–2382. doi: 10.1002/pro.5560041116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu MM, Rempel DL, Gross ML. Modeling data from titration, amide H/D exchange, and mass spectrometry to obtain protein-ligand binding constants. Journal of the American Society for Mass Spectrometry. 2004;15:388–397. doi: 10.1016/j.jasms.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. Journal of the American Society for Mass Spectrometry. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 52.Zhu MM, Rempel DL, Du Z, Gross ML. Quantification of protein-ligand interactions by mass spectrometry, titration, and H/D exchange: PLIMSTEX. Journal of the American Chemical Society. 2003;125:5252–5253. doi: 10.1021/ja029460d. [DOI] [PubMed] [Google Scholar]
- 53.Zhu MM. Chemistry. St. Louis: Washington University in St. louis; 2004. Determination of protein-ligand interactions using H/D exchange and mass spectrometry; p. 358. [Google Scholar]
- 54.Persechini Anthony YK, Stemmer Paul M. Ca2+ Binding and Energy Coupling in the Calmodulin-Myosin Light Chain Kinase Complex. Journal of Biological Chemistry. 2000;275:4199–4204. doi: 10.1074/jbc.275.6.4199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.