Abstract
In the past decade, cell free DNA, or circulating cell free DNA, or cell free circulating DNA, isolated from body fluids such as plasma/serum/urine has emerged as an important tool for clinical diagnostics. The molecular biology of circulating cell free DNA is poorly understood but there is currently an increased effort to understand the origin, mechanism of its circulation, and sensitive characterization for the development of diagnostic applications. There has been considerable progress towards these goals using real time polymerase chain reaction technique (rt-PCR). More recently, new attempts to incorporate mass spectrometric techniques to develop accurate and highly sensitive high-throughput clinical diagnostic tests have been reported. This review focuses on the methods to isolate circulating cell free DNA from body fluids, their quantitative analysis and mass spectrometry based characterization in evolving applications as prenatal and cancer diagnostic tools.
Keywords: Circulating cell free DNA, electrospray ionization mass spectrometry (ESI-MS), matrix assisted laser desorption ionization mass spectrometry (MALDI-MS), short oligonucleotide mass analysis (SOMA), Hemolytic disease of the fetus and newborn (HDFN), non-invasive prenatal diagnostic (NIPD), hepatocellular carcinoma (HCC), polymerase chain reaction (RE-PCR)
1. Introduction
Recent discoveries in the field of genomics, in particular the comprehensive analyses of genomes for cancers such as lung and melanoma, provide promising new insights for developing a clinically relevant diagnostic method [1, 2]. A new era in which the identification of all possible mutations in the human genome would help to build diagnostics methods where one would be able to ask the specific questions “Do I have cancer?” or “Am I predisposed to cancer?’ and receive clear answers could be based strictly on the presence of critical circulating genetic biomarkers. This review outlines the potential utility of circulating cell free DNA as a “critical circulating genetic biomarker” and as an efficient diagnostic for prenatal genetic screening, oncology diagnosis and discovery, organ transplantation studies and other diseases. Apart from plasma, serum and urine, extracellular nucleic acids can be isolated from stool, cerebrospinal fluid, amniotic fluid, sputum, lymphatic and peritoneal fluids and bone-marrow but this review focuses on plasma/serum and urine sample due to the brevity of space.
Nucleic acids (DNA and RNA) in plasma were first reported in 1948 by Mandel and Metais [3] but remained largely forgotten until Leon and Shapiro reported in 1977 on the elevated circulating DNA concentrations in serum of cancer patients as compared to non malignant disease patients [4]. In important work in 1989, Stroum and Anker were the first to determine that the amount of DNA in plasma samples of various malignant diseases patients was higher than in normal healthy human beings and the origin of this DNA was traced to cancer cells using a test based on decreased strand stability [5]. Vasioukhin et al., in 1994 reported the detection of point mutations of the N-ras gene in the plasma DNA of 10 patients with myelodysplastic syndrome (MDS) or acute myelogenous leukemia (AML) using polymerase chain reaction (PCR) [6]. In that same year Sorenson et al., reported detection of mutated K-ras sequences in the serum of pancreatic cancer patients by polymerase chain reaction (PCR) [7]. It was in 2001 that Sozzi, et al. reported a detailed study involving plasma DNA from 84 patients with non-small cell lung cancer and 43 healthy controls and they were able to discriminate between lung cancer patients and healthy individuals on the basis of higher mean values of plasma DNA concentration in cancer patients using PCR [8].
Since these early reports, hundreds of papers have been published on circulating cell free nucleic acids in blood and have reported higher levels of CCFDNA in plasma/serum of patients with lung cancer, colorectal cancer, breast cancer, testicular germ cell cancer, non-Hodgkin’s lymphoma, clear cell renal cell carcinoma, gastric cancer, non small cell lung cancer, epithelial ovarian carcinoma, endometrial cancer and esophageal squamous cell carcinoma as compared to healthy individuals [9–19]. Elevated levels of CCFDNA have also been detected in other malignancies like systemic sclerosis, peritoneal dialysis, obstructive sleep apnea, acute pancreatitis, preeclampsia, severe aseptic inflammation and urinary tract infections [20–26]. A major breakthrough towards the development of prenatal diagnostic methods based on circulating cell free fetal DNA (CCFFDNA) was achieved by Lo, et al. in 1997 when they reported the discovery of fetal DNA in maternal plasma [27]. Later, Bianchi et al. in 2001 and in following years extensively investigated the isolation and detection of cell free fetal DNA from maternal blood using PCR [28–29]. To highlight the challenges associated with the analysis of fetal DNA in maternal plasma, in 2002 Invernizzi et al. published a report on the detection of CCFFDNA decades after pregnancy and delivery using real-time quantitative PCR assay by analyzing a specific Y chromosome sequence (the SRY gene) isolated from plasma DNA. These reported results were later contradicted in 2007 by Tomaiuolo et al. and attributed to a contamination during the extraction process as it is widely believed that all the CCFDNA is removed within a few hours after delivery [30–31].
With the discovery of CCFFDNA in maternal plasma by Lo et al., a number of research articles reported the isolation and detection of cell free circulating fetal DNA (CFCFDNA) from the plasma/serum of pregnant women and have used elevated levels of CFCFDNA for a wide variety of screening assays for the diagnosis of pregnancy complications and genetic disorders such as: sex-linked disorders, aneuploid pregnancies, preeclampsia, the RhD status of fetuses, spontaneous preterm delivery, fetal growth restriction, and single gene disorders such as beta-thalassaemia, congenital adrenal hyperplasia, achondroplasia, placental insufficiency and placental abruption [32–46].
The importance of the emergence of CCFDNA as a prominent prenatal diagnostic lies in its simple noninvasive sample collection. This is in contrast to traditional invasive procedures such as amniocentesis or chorionic villous sampling (CVS) [47]. Another body fluid in which circulating cell free DNA has started gaining prominence for clinical genetic diagnostic development is urine. Botezatu et al. in 2000 were the first to report transfer of some cell free DNA across the kidney barrier into urine and the goal of this study was to determine whether cell free DNA from the bloodstream crossed the kidney barrier. Nested PCR was performed and the male-specific DNA sequences were detected in 5 of 9 urine samples obtained from women who were transfused with male blood. Similarly, using PCR assay in 8 of 10 cases Y-specific sequence was detected in women pregnant with a male fetus at gestational age of 7–10 weeks and the K-ras mutations were detected in 5 of 8 urine samples of colon adenocarcinomas and pancreatic carcinomas patients. Thus in this report it was successfully demonstrated that urine could be a source of CCFDNA for future applications [48].
The unique specificity of targeting circulating genetic material in accessible biofluids was highlighted by Zhang and coworkers in which they reported a comparative study in which they were able to detect the Y chromosome specific sequences of the SRY gene (sex-determining region Y gene) in the cell free urine samples of female patients who had received renal transplants from male donors but not in cases in which the women had received female donor transplants. The reported detection of Y specific sequences by Zhang et al., in cases of renal transplants was followed up by Zhong et al. in 2001 who reported similar results. In this work they were able to detect the Y chromosome specific sequences from the urine samples of female kidney transplant patients who had received male kidneys by both nested and real-time PCR. However in the same research article Zhong et al. reported the failure to detect the Y chromosome-specific DNA sequences in urine samples from 8 women pregnant with male fetuses using a very sensitive nested PCR assay [49, 50].
Ying-Hsiu Su et al. in 2004 reported on the analysis of CCFDNA in which 150 to 250 base pair DNA was isolated from urine and used for the diagnosis of colorectal cancer and polyps that contain K-ras mutations. In this work, the nucleic acids were isolated from 15 ml of urine and found to have between 40 and 200 ng/ml CCFDNA and the samples were analyzed by the restriction-enriched polymerase chain reaction (RE-PCR) assay to detect codon 12 mutation of K-ras mutation. The authors report the identification of mutant K-ras sequences in the urine of 15 of the 18 urine samples (83.3%) from individuals with confirmed diagnosis of colorectal disease, and 19% of those with no diagnosis of colorectal disease [51].
In order to validate the use of CCFDNA as a noninvasive, robust marker for cancer detection in urine, CCFDNA isolated from 5 urine samples was compared by Bryzgunova et al. in 2006 with the CCFDNA isolated from the blood of breast cancer patients obtained 2 weeks after a course of combined anticancer therapy. Analysis by methylation-specific PCR of CCFDNA isolated from the blood of breast cancer patients and CCFDNA isolated from the corresponding urine confirmed the presence of methylated promoters of the same RASSF1A and RARβ2 genes in plasma and in the corresponding urine sample. In addition, in this report CCFDNA was isolated from 15 mL of urine from 19 healthy people independent of gender and showed concentration in the range of 6 ng/mL to 50 ng/mL and thus the concept of using CCFDNA from urine as a noninvasive test for cancer diagnostics was advanced[52].
With evolving improvements in isolation techniques for CCFDNA, several research groups have reported on the isolation of fetal DNA using CCFDNA. Botezatu et al. in 2000, Koide and coworkers in 2005 and Sandra et al. in 2007 reported isolating circulating cell free fetal DNA in maternal urine of pregnant women with the total amount (maternal and fetal) DNA estimated using a quantitative real-time PCR assay [48, 53, 54]. There were also contrasting reports published by Zhong and coworkers in 2001, Li and coworkers in 2003 and Illanes and coworkers in 2006 where they failed to isolate circulating cell free fetal DNA from the maternal urine of pregnant women [50, 55, 56]. The reason for this difference was later attributed to renal function, in particular glomerular permeability, the small size of DNA fragments, and the presence of urinary nucleases [57]. In addition, Ying-Hsiu Su et al. in 2008 reported an improved and enhanced detection method to isolate the trans-renal DNA from urine by using carboxylated magnetic beads which also separates high molecular weight DNA (>1kb) from low molecular DNA. With this DNA isolation and separation technique and using restriction-enriched polymerase chain reaction (RE-PCR) assay the codon 12 mutation of the K-ras gene was detected in urine samples of 36 patients with various colorectal diseases [58].
Recently Shekhtman et al. in 2009 isolated the CCFFDNA from maternal urine using a traditional silica-based method and a new technique based on adsorption of cell-free nucleic acids on Q-Sepharose resin. Using conventional and real-time PCR the presence of SRY gene sequences in urine of pregnant women was successfully detected in 78 of 82 women pregnant with male fetuses and false-positive results were also reported for 11 of 91 women pregnant with female fetuses which also detected SRY gene [59]. García Moreira and coworkers in 2009 have shown how isolated CCFDNA from urine acts as an acute rejection marker in renal transplantation and could also detect urinary tract infections [60]. The increased importance of urine as a sample for prenatal or disease diagnostic lies in its absolutely non-invasive collection which is uncontaminated with pathogens like human immunodeficiency virus (HIV) thus reducing the risk of infections from collection and lower protein content as compared to plasma. The quantities of CCFDNA isolated from urine samples do vary on a daily basis in terms of alcohol consumption, smoking, kidney and urethra disorder, menstruation cycle in women as well as when the sample is collected [61]. The reports on isolation of CCFDNA in urine have been few but significant and applications have been reported to diagnose genetic imperfections, fetus sex determination from maternal urine, urinary tract infection and cancer.
This article is an attempt to focus on the mechanism of origin of circulating cell free DNA, review various analytical protocols and characterization techniques available for CCFDNA analysis, to highlight mass spectrometry based methods for CCFDNA characterization and to present a wide range of applications for potential use of CCFDNA towards improving human health. Despite the seemingly limited application of mass spectrometry in this field to date, it is hoped that this thorough review will stimulate increased interested in the use of mass spectrometry to solve the challenging problem of low-level isolation, characterization and quantitation of CCFDNA. As such, there are many reviews which highlight the enormous advances made by various groups concerning the characterization of nucleic acids using electrospray ionization and MALDI techniques (62–64). We refer the reader to several high quality reviews for more details on the general use of mass spectrometry for oligonucleotide analysis.
1.1. Circulating cell-free DNA– origin and mechanisms of release into plasma and urine
The source and origin of CCFDNA into the plasma circulation has been a subject of speculation and the mechanism by which it is released into urine is not clear. Circulating cell free DNA has been reported in healthy, cancerous and nonmalignant disease populations as well as pregnant women. However, as compared to healthy controls, CCFDNA concentrations are higher in pregnant women and persons with a specific disease state. Various mechanisms have been proposed to explain this increased CCFDNA concentration including cell dying mechanisms -necrosis and apoptosis (programmed cell-death). Necrosis, however, failed to justify the observed initial decrease rather than increase in CCFDNA after radiation therapy [65, 66]. Apoptotic mechanisms have found support as one of the most important mechanism of origin for CCFDNA in plasma as a large number of cells are programmed to die on a daily basis. This explains the presence of CCFDNA in healthy individuals and in cancer patients and the disproportionate quantity of CCFDNA due to the high turnover of cancerous cells releasing CCFDNA into the plasma. Apoptosis as the mechanism of origin of CCFDNA has also found support from the reports that CCFDNA gives a ladder pattern when analyzed by gel electrophoresis similar to that shown by apoptotic cells. Along with the major hypothesis of CCFDNA derived from cellular apoptosis, active secretion of DNA from tumor cells, the presence of lymphocytes in the blood which contains DNA, nucleated cells, nucleosomes and other nucleoproteins add to the presence of CCFDNA in plasma [65–72].
With regard to the presence of fetal DNA in maternal plasma, Tjoa et al. in 2006 and Alberry et al. in 2007 and others have attributed the origin of circulating cell free fetal DNA in maternal plasma to the embryo’s apoptotic placenta cells (trophoblasts) and that circulating cell free fetal DNA comprises around 3–6% of the total cell-free DNA in maternal circulation. Cell free fetal DNA has been reported to consist of short DNA fragments of less than 193 base-pairs in length by Chan et al., which may also explain the distribution of fetal DNA in maternal plasma [27, 73–76].
The origin of circulating cell free DNA in urine is unclear but it is widely believed that small amounts of CCFDNA from the plasma cross the kidney barrier into the bladder and hence is named “transrenal DNA”. However urinary DNA could also originate from epithelial cells shed from the urinary organs, often lymphocytes and other white blood cells, while bacterial infection is also another source [48–57]. It is hypothesized that there is an approximately 70KD upper size limit of “transrenal DNA” which can be filtered from the blood through the kidney into urine for a normal functioning kidney, which corresponds to about 100 base pairs [77].
1.2. Isolation of cell free circulating DNA from plasma/serum/urine, protocols and commercial kits
The isolation of CCFDNA from complex fluids like plasma or urine in the presence of cellular matter represents a significant challenge. The inherently low concentration of CCFDNA (around 10–100 ng/mL) and the variable levels among individuals also adds to the problems associated with isolation of these molecules from body fluids. Along with the traditional method of phenol/chloroform liquid-liquid extraction, there are number of commercial kits which have been developed to address these problems and hence isolate and purify the CCFDNA from the body fluids. These are detailed in Table 1 [78–90].
Table 1.
various protocols/commercial kits available to isolate CCFDNA from plasma/urine
| DNA isolation kit | Sample source | Samples/volume used/(concentration range) DNA quantitation | References |
|---|---|---|---|
| ChargeSwitch® gDNA Kit | Serum | 74 samples/1 mL of sample/(8.09 vs. 0.82 ng/ml, respectively in healthy and cancer patients. | 12 |
| Minipreps DNA purification system, Promega | Urine | 27 samples/2–96 μg/L | 48 |
| Wizard DNA isolation kit | Urine | 36 samples/10–55 ng/mL | 58 |
| QIAamp DNA Blood Mini Kit | urine | 124 healthy samples & 55 patients with UTI/healthy samples yielded 0 to 147 GE/mL tr-DNA, from UTI patient group measured tr-DNA ranging from 8 to 87871 GE/mL. | 60 |
| QIAamp MiniElute Virus Spin Kit | Urine | 50 healthy samples yielded Tr.-DNA, median was 99 GE/mL | 60 |
| Phenol-chloroform method | Plasma | 77 | |
| Triton/Heat/Phenol protocol (THP) | Plasma/serum | 15 samples/500μL/The mean concentration of CFDNA was 4.73 ng/ml | 78 |
| QIAamp DNA Blood Mini Kit | Plasma | 45 samples and patients/I mL of sample used/concentration range 3.5–67.1 ng/mL | 79 |
| QIAamp DNA Blood Mini Kit | Plasma | 10 healthy donors/3.6 to 5.0 ng/ml | 80 |
| QIAamp UltraSens Virus Kit | Plasma | 142 samples/400–800 μL/In 41 healthy individuals (mean 13.9 ng/ml), In patients (mean 63.5 ng/ml). | 81 |
| Wizard Plus Mini-Prep DNA Purification System | Plasma/serum/urine | 20 samples/7.4 ng/mL (median)/31.7 ng/mL (median)/23.7 ng/mL (median) | 82 |
| Q resin based technique/Qiagen QIAquick column | Maternal Urine | 91 pregnant samples/mean concentration 196.5 genome equivalents/mL urine | 83 |
| Silica based technique/Qiagen QIAquick column | Maternal Urine | 91 pregnant samples/mean concentration 26.3 genome equivalents/mL urine | 83 |
| Norgen DNA isolation kit | Urine | 122 urine samples (16 from HBV infected patients, 74 from HCV infected patients, 32 from HCC post HCV patients, 10 urine samples from healthy Egyptian individuals were also used as a control) | 84 |
| NucleoSpin Plasma XS Kit | Maternal Plasma | 44 pregnant cases/median concentration 95.5 genome equivalents/mL | 85 |
| magnetic capture hybridization (MCH) method | Maternal Plasma | 1000 pregnant cases/95% detection limit was 286 pg/ml | 86 |
| NucliSens Magnetic Extraction system/QIAamp DSP Virus Kit (QDSP)/QIAamp DNA Blood Mini Kit | Maternal Plasma | 75 maternal samples/yield was 1.7 times more using the NMAG system, and 1.5 times more using the QDSP as compared to QIAMP. The total DNA yield was improved by a mean factor of 2.3 using the NMAG system, and by a mean factor of 1.3 using the QDSP. |
87 |
| QIAamp DSP Virus Kit | Maternal Plasma | 1000 pregnant cases/95% detection limit was 138 pg/ml | 87 |
| magnetic bead separation method | Maternal plasma | 15 samples/5 different DNA isolation protocols: two conventional, two column-based, and one magnetic-bead based/DNA isolation using the magnetic beads yielded the highest quantity of total DNA (2018.83 ± 4.09 GEq/mL), with 100% fetal DNA detection. | 88 |
| MagNA Pure LC Instrument | Plasma/serum | 87 blood donors and 50 healthy adults who had never donated blood, the concentration of cf-DNA in serum was about eightfold higher than that in plasma | 89 |
Circulating cell free DNA can be isolated from serum, plasma and urine using the classic technique of phenol/chloroform liquid-liquid extraction or by using various commercial kits available for specific body fluids. In general, for commercial kits, freshly collected samples in EDTA bottles or from samples stored at −80°C are centrifuged at 2500g for 15 minutes to remove cellular components. A further centrifugation at higher speed (>10000g) for an additional 10–15 minutes further assures sedimentation of cellular components until finally the supernatant containing CCFDNA is collected. To the collected supernatant which contains CCFDNA, a binding buffer is added which binds to the CCFDNA and is loaded onto a silica gel based membrane spin column. After a couple of washings, the bound CCFDNA is eluted by addition of elution buffer or 70% ethanol solution and the CCFDNA is concentrated for subsequent analysis.
While the procedure for the isolation of circulating cell free DNA from plasma and serum using commercial kits is routine, the various reagents and protocols reported for the different kits do vary in terms of the volume of the starting sample required, the final elution volume, the initial concentration of circulating cell free DNA present and the possible degradation of DNA during isolation. The complexities associated with the isolation of circulating cell free fetal DNA, which relates specifically to the limited quantity in circulation in maternal plasma for example, compounds the problems of efficient free fetal DNA extraction. Despite these challenges, the huge potential of CCFFDNA in prenatal diagnostic continues to drive research into CCFFDNA and its efficient isolation. To improve circulating cell free fetal DNA recovery, protocols and commercial kits available have been optimized to ensure maximum yield and purity of cell free fetal DNA. Efficient DNA extraction is crucial as the presence of fetal cell free DNA is limited to 3–6% in maternal circulation [27].
As compared to plasma and serum, there are few commercial kits which use urine as a sample to isolate CCFDNA. Previous work used either phenol-chloroform liquid-liquid extraction or commercial kits designed for plasma or serum. Because these kits are not optimized for the unique matrix of urine, reproducibility problems, isolated yields and matrix interferences were commonly reported. Recently, Norgen Biotek Corp. has introduced a urine DNA isolation maxi kit for the isolation of CCFDNA from urine. Also commercial kits for CCFDNA isolation from plasma and serum can be tailored to make them suitable for urine samples, however, the ambiguity about the presence of cell free fetal DNA in maternal urine may be a consequence of using these kits for an unintended sample matrix. This may be one explanation for the previous published reports about the inability to detect cell free fetal DNA from urine or its presence in low concentration [50, 55, 56]. The recent reports by Umansky et al. and Shekhtman et al. of the detection and analysis of circulating cell free fetal DNA from maternal urine using Q-resin–based methods with Qiagen QIAquick® columns holds promise for improved isolation methods of these important markers from urine [57, 59].
2. Quantification of circulating cell free DNA
A number of methods and techniques have been developed for the quantification of circulating cell free DNA. These include 32P-labeled radioimmunoassays [4], visual comparison with known standards (DNA DipStick Kit) [8], spectrophotometric determination [51] and the small sample size instrument platform NanoDrop® ND-1000 Spectrophotometer [89]. However, the signal contamination from single-stranded nucleic acids, proteins and RNA contributes to the poor specificity and decreased sensitivity for the quantitative analysis of low concentration CCFDNA. The development of DNA specific reagents such as Invitrogen’s PicoGreen® dsDNA quantitation reagent assays [90], real-time quantitative PCR assay (RT-qPCR) [9, 55, 91–97], digital PCR approach [98], and fluorometric PCR assay [99, 100] have decreased the sample size requirement and increased the reliability of CCFDNA identification and quantitation.
Recently, biological fluids like blood and urine have been directly assayed for CCFDNA without any prior DNA isolation using the commercial fluorescent SYBR® Gold stain [101] from Invitrogen. With the Invitrogen kit, real-time quantitative PCR, with a detection limit of picograms, has emerged as a powerful tool for CCFDNA detection and quantification. Some disadvantages of using RT-qPCR include its high cost per sample, the requirement of sufficient fluorescence signals for detection of the products, the absence of standard calibrators at known concentration and the limited instruments found mostly in clinical biochemistry laboratories. In our group we use Invitrogen’s Qubit® quantitation fluorometer for quantitation of cell free DNA which provides highly accurate and sensitive detection from as little as 1 μl of sample volume and measurement of DNA concentration as low as 0.2 ng/mL using Quant-iT dsDNA high Sensitivity assay[92, 102].
3. Characterization of circulating cell free DNA
The CCFDNA in body fluids like plasma and urine has been detected and characterized by techniques such as real-time quantitative polymerase chain reaction [55, 91–97], quantitative fluorescent PCR (QF-PCR) [98,99] and short oligonucleotide mass analysis (SOMA) based ESI-MS methods [103–111, Table 2]. In addition, methods based on matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry such as homogenous MassEXTEND (hME), single allele base extension reaction (SABER), and the allele-specific base extension reaction (ASBER) have been successfully used to characterize CCFDNA [112–129, Table 2]. The analysis of CCFDNA normally performed using qPCR suffers from sensitivity issues as well as the limited ability to detect gene mutations at preselected positions within a gene. Quantitative fluorescent PCR has been successfully used to overcome the low sensitivity limitations of RT-qPCR and has the added advantage of accuracy, speed and automation but still suffers from a requirement for a predefined selective sequence primer for unknown identification.
Table 2.
Mass Spectrometry analysis of CCFDNA from plasma/urine
| condition/source of CCFDNA | Gene involved/mutation | Technique used | Quantification | Remarks | Reference |
|---|---|---|---|---|---|
| Hepatocellular carcinoma (HCC)/Plasma | codon 249 in the p53 tumor suppressor gene | short oligonucleotide mass analysis (SOMA), an electrospray ionization- LCQ ion-trap mass spectrometer | DNA was extracted from plasma using a QIAamp Blood Kit (Qiagen). | 6 of 11 (55%) patients with mutation in the tumor tissue had a positive plasma sample. 10 plasma samples from healthy individuals were all negative. | 104 |
| Hepatocellular carcinoma (HCC)/plasma | codon 249 in the p53 tumor suppressor gene | HaeIII predigestion-SOMA- ESI- MS | 26 DNA samples from HCC and normal liver were analyzed by RFLP and SOMA. SOMA is about 2.5– 15-fold more sensitive than RFLP | 105 | |
| Hepatocellular carcinoma (HCC)/Plasma | TP53/codon 249 | HaeIII predigestion- SOMA- ESI- MS | Extracted from 100 μl (Chinese) and 300 μl (control sample) plasma, QIAamp Blood Kit | plasma samples of 16 liver cancer from china, 18 healthy US control plasma samples | 106 |
| Hepatocellular carcinoma (HCC)/plasma | TP53/codon 249 | PCR- SOMA- ESI-MS assay | Extracted from 100 μl of plasma, using QIAamp_ DNA Blood Mini Kit | 17 plasma samples, 29 biopsy specimens of HCC from The Gambia in West Africa. Results matched 88.2% between tumor and plasma samples. | 107 |
| Hepatocellular carcinoma (HCC)/plasma | hepatitis B virus (HBV) gene/1762T/1764A double mutation | PCR-ESI-MS- SOMA assay | A phenol- chloroform extraction followed by an ethanol precipitation. | Plasma samples were collected from 120 residents of Qidong, 18 plasma samples from healthy U.S. adults were used as controls. | 108 |
| Hepatocellular carcinoma (HCC)/plasma | TP53/codon 249 | PCR- ESI-MS- SOMA assay | Extracted from 200 μL of plasma, using the QIAamp DNA Blood Mini kit. | 89 HCC cases, 42 cirrhotic patients, 131 healthy controls, all from highly aflatoxin-exposed regions of The Gambia. | 109 |
| Liver Cancer/plasma | hepatitis B virus (HBV) gene/1762T/1764A mutation | PCR-ESI-MS- SOMA assay | A phenol- chloroform extraction followed by an ethanol precipitation | 515 plasma cases in Qidong, China Hepatitis B virus (HBV) DNA could be isolated in 355 (69%) of these samples | 110 |
| HCC diagnosis in plasma DNA | codon 249 of TP53 | PCR-ESI-MS- SOMA assay | 249 sample of aflatoxin-albumin adducts, 168 (67%) were positive. SOMA found 9/14 HCC positive from plasma. | 111 | |
| β-thalassaemia mutations/maternal plasma | Mutation- Deletion of CTTT at codons 41 and 42 of the β-globin gene, HBB | MassARRAY (PCR-MALDI - TOF MS) primer extended by hME protocol or SABER protocol | 10 mL of maternal and paternal blood, cell free DNA was extracted with the QIAamp Blood Kit | 12 at risk (thalassaemia major), 50 second-trimester control pregnancies. Maternal plasma is analyzed to confirm or excluding the presence of mutation in case the father is a carrier |
114 |
| Detection of paternally inherited SNP/maternal plasma. | fetal DNA detected in overwhelmi ng maternal DNA backgroun | MALDI-TOF MS- base extension reaction - PCR | 800 μl of plasma was used for DNA extraction, QIAamp DNA Blood Mini Kit | 16 plasma DNA samples were selected for the analysis | 115 |
| NIPD of the HbE mutation/maternal plasma | GAG3AAG missense mutation, codon 26 of the globin gene | MALDI-TOF 800 MS detect the primer extended by hME or ASBER protocol- PCR products. | 800 μL plasma DNA extracted by Nucleon Blood DNA extraction Kit and a QIAamp Blood Mini Kit. | 13 pregnant samples (HbE-negative), male partners (HbE carriers), mutation detected 100% (8 of 8) and a sensitivity of 80% (4 of 5). | 116 |
| Comparison of MALDI-TOF MS assay with Taman® rt- PCR assay/maternal plasma | fetal SRY gene sequence | MassARRAY (PCR-MALDI TOF MS) - hME protocol products vs. TaqMan rt- PCR assays | 500 μL of plasma for DNA isolation by use of the MagNa Pure DNA isolation instrument. | 97 maternal blood samples, MALDI-TOF MS assay sensitivity 95% (55/58), and the TaqMan real-time PCR assay Sensitivity 93% (54/58). | 118 |
| Genotyping fetal paternally inherited SNPs/Maternal plasma. | 41 paternally inherited fetal point mutations in fetus | Agarose gel electrophore (size fractionation)- MALDI-TOF MS detect the primer extended by hME protocol or SABER protocol - PCR products | 5–8 mL maternal plasma, cell free DNA isolated with Roche High Pure DNA Purification kit. | cf-DNA was extracted from 18 maternal plasma samples, 10 taken at term and 8 obtained early in the second trimester. | 119 |
| Genotyping paternally inherited SNP alleles/maternal plasma | Detection of Fetal-Globin Gene Mutations | Agarose gel electrophore (size fractionation) SYBR Green dye- peptide nucleic acid- mediated PCR- MALDI-TOF MS | Roche High Pure DNA Preparation kit, QIAEX II gel extraction kit | Cell-free DNA was extracted from 5 to 10 ml of maternal plasma. | 121 |
| NIPD of achondroplasia/maternal plasma | FGFR3 gene, fetal G1138A mutation | MALDI-TOF MS detect the primer extended by hME or SABER protocol - PCR products. | 18 mL maternal blood (first case) and 30 mL (second case). 9 mL of paternal blood, CFF DNA extracted by QIAamp Blood Kit | 2 pregnant women maternal plasma samples at risk for achondroplasia | 122 |
| Preimplantation genetic diagnosis (PGD)/maternal plasma | single-gene mutations | MALDI-TOF MS assay with SABER protocol or by the size- fractionation approach | 4 maternal plasma obtained from PGD-conducted pregnancies. The presence or absence of mutations was correctly detected in all cases | 124 | |
| paternally inherited fetal point mutation for β-thalassaemia/maternal plasma | β-globin gene-codon 39 mutation & IVSI-110 mutation | Agarose gel Electrophoresis (size fractionation)- PCR- MALDI- TOF MS | 1 early second trimester pregnancy maternal plasma at risk for β-thalassaemia | 125 | |
| Non-invasive prenatal diagnostic/maternal plasma | RASSF1 and SERPINB5 genes | EpiTYPER assay- MALDI- TOF MassCLEAVE reagent- PCR | CF-DNA was extracted from 500 μL of plasma using the Roche High Pure DNA Preparation Kit | 50 plasma samples (20 from pregnant and 30 from non-pregnant women), normal and pre-eclamptic pregnancies | 126 |
| foetal Rhesus D status/maternal plasma/serum samples | fetal RHD gene | MassARRAY system, MALDI- TOF MS detect the primer extended SABER -PCR products | automated Roche MagNA Pure™ system | CF-DNA was extracted from 1 mL of maternal plasma of 178 RHD-negative pregnant women. | 128 |
| KEL blood disorder/Maternal plasma | fetal KEL1 gene | MALDI-TOF MS-based SABER protocol | Then 800 μL of plasma, DNA isolation using MagNA Pure LC instrument | Sensitive detection of fetal KEL1 allele in 11/13 of the KEL1- positive samples. The paternal KEL1 allele determined in 94% of cases (30/32) | 129 |
| Donor SNPs, Renal Transplant Recipients/urine | Detection of Donor SNPs in the Urine of Renal Transplant Recipients/urine | PCR-MALDI- TOF MS | Extracted from 1 mL of recipient urine by Roche High Pure DNA Purification kit. | 4 cases, CF-DNA/mL isolated ~ 3000 genome equivalents in urine which is collected within 24h of transplantation | 130 |
| SNPs/Maternal plasma & Urine of Kidney Transplant Recipients | SNPs | Agarose gel electrophore (size fractionation)- PCR-MALDI- TOF MS | Plasma (5–7 mL) from pregnant women was used for DNA isolation. | 4 cases involving related living-donor kidney transplants | 131 |
3.1. ESI-Mass spectrometric Analysis of circulating cell free DNA
The analysis and characterization of CCFDNA isolated from plasma and serum samples of patients with hepatocellular carcinoma (HCC) has been reported using both a mass spectrometry based method (HPLC-ESI-MS-SOMA) and qPCR [103–111]. Hepatocellular carcinoma is a cancer of the liver and is one of the ten most frequent cancers worldwide, but it is a major public health problem in the East Asia area of People’s Republic of China and the Sub-Saharan Africa. The major causes of HCC are chronic infection with hepatitis B (HBV) and hepatitis C (HCV) viruses and exposure to aflatoxins from food. Ongoing genetic studies are evaluating critical genes which could be responsible for the mutations leading to liver cancer. The HPLC-ESI-MS-SOMA protocol used for CCFDNA screening represents a sensitive technique for identification of mutations in genes leading to HCC.
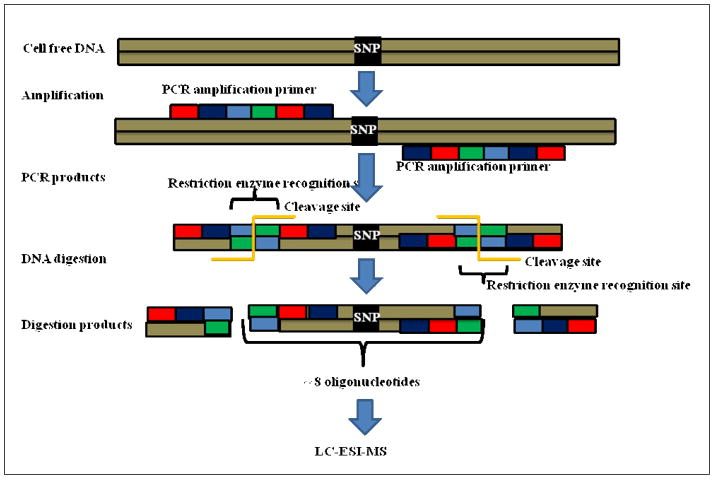
A vast clinical study which involved tissue and plasma samples from HCC patients, individuals at risk for HCC and healthy persons was conducted in the West African nation of Gambia and in Qidong county in the People’s Republic of China using HPLC ESI-MS-SOMA. The SOMA assay combines PCR amplification, restriction digestion, and electrospray ionization mass spectrometry. The SOMA method has been shown to be more sensitive than restriction fragment length polymorphism-PCR (RFLP-PCR) for detecting the specific mutations from circulating cell free DNA extracted from plasma or serum. In this assay, a PCR primer which has a restriction enzyme site is incorporated into the target sequence gene such that when the restriction digestion protocol is performed it attacks the restriction enzyme site thus removing the outer sides to PCR primer resulting in a ~8 base pairs oligonucleotide which is analyzed by HPLC ESI tandem mass spectrometry [Figure 1] [103]. The plasma samples showed elevated levels of the isolated circulating cell free DNA in cases of HCC patients or those at risk as compared to healthy controls. From the isolated CCFDNA, a specific codon 249 mutation in the p53 gene from the Gambian population and a double mutation at codons 130 and 131 of the HBV X gene from the People’s Republic of China population were identified.
Figure 1. SOMA [Short Oligonucleotides Mass Spectrometry].
SOMA is a technique in which the circulating cell free DNA having SNP and wild CCFDNA are amplified using PCR, and the PCR products formed have a restriction enzyme recognition sites. On application of restriction enzyme the digested products are formed which are characterized by LC-ESI-MS [Ref. 134, Used with Permission].
In 2001 Jackson and coworkers using the HPLC ESI-MS-SOMA protocol reported for the first time a relationship between mutations in codon 249 of the p53 gene in tumor tissue and plasma samples in HCC patients from Qidong and Shanghai. The free DNA isolated from plasma established SOMA as a sensitive method for detecting specific mutations in tumor patients [104]. Another study comparing the relative sensitivity for two techniques, PCR-RFLP and ESI based SOMA for the P53 gene codon 249 mutation in liver cancer patients samples from Qidong showed that the SOMA based method was 2.5 times more sensitive than RFLP, and becomes 15-fold more sensitive if the samples are predigested with HaeIII before application of the PCR SOMA protocol [105]. In 2003, Jackson et al. reported the early identification of a p53 tumor gene mutation at codon 249 from CCFDNA in samples from Qidong and demonstrated that these mutations were detected as early as 1 year prior to hepatocellular carcinoma (HCC) diagnosis by using SOMA method [106]. Similar results were reported from the Gambian population samples which also used the SOMA based method to show that cell free DNA in plasma can be used for early identification of HCC and cancer diagnosis and this was also correlated with the tissue sample diagnosis [107]. At the same time, the ESI-MS based SOMA method identified and compared double mutations in the HBV gene from both tissue and plasma samples collected from the Qidong population and proposed that cell free DNA can act as a biomarker for the diagnosis of HCC. The HBV double mutation was detected in 52.1% of all plasma samples after the diagnosis of liver cancer [108].
In an improvement to the HPLC ESI-MS-SOMA protocol, a novel internal standard plasmid step was incorporated into the SOMA method to quantify the P53 gene codon 249 mutation in the circulating cell free DNA isolated from plasma to differentiate individuals with hepatocellular carcinoma from cirrhotic patients and nondiseased control subjects on the basis of the amount of DNA isolated [109].
Despite the reported results for HCC in the Gambian and Chinese samples, progress in identifying mutations causing other cancers has been slow using HPLC ESI-MS SOMA. However, despite the lack of further follow up on their initial success, these early results point to a great potential for this protocol for studying the progression and regression of various cancers and there is a critical need to develop clinical diagnostics for such diseases. Mass spectrometry has an important role to play for rapid, sensitive methods which could be a critical factor for improving human health [110, 111].
3.2. MALDI-TOF-MS analysis of circulating cell free DNA
As compared to ESI-MS based SOMA where reports have been few and limited almost exclusively to HCC, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has emerged as a popular technique for analyzing point mutations in CCFDNA samples. MALDI-TOF MS analysis of circulating cell free DNA from maternal plasma or urine has emerged as an important technique for prenatal diagnosis, fetal genotyping and analyzing kidney transplant patients which previously were monitored by conventional invasive procedures like chorionic villus, amniocentesis and blood tests [112]. Similar to the ESI-MS-SOMA method, MALDI-TOF does not act as a stand-alone technique for the analysis of genetic alterations in circulating cell free DNA but incorporates protocols such as homogenous MassEXTEND (hME), single allele base extension reaction (SABER) or the allele-specific base extension reaction (ASBER) assays [Figure 2, Figure 3]. The hME, SABER or ASBER approaches involve a base extension of the DNA sequence of interests such that they have a different mass compared to the original DNA and hence are easily detected by mass spectrometry. The critical difference in the hME and SABER assays is in the DNA sequence being extended and terminated. In the case of hME, both fetal DNA as well as maternal DNA is extended whereas in SABER it is possible to amplify the fetal DNA sequence only in the presence of majority of maternal DNA. In the ASBER method, the 3′ end of the extension primer for ASBER is complementary to the fetal DNA sequence of interest and thus maternal DNA would be inhibited by this 3′ primer of ASBER due to template mismatch resulting in increased specificity and sensitivity over SABER [113–115].
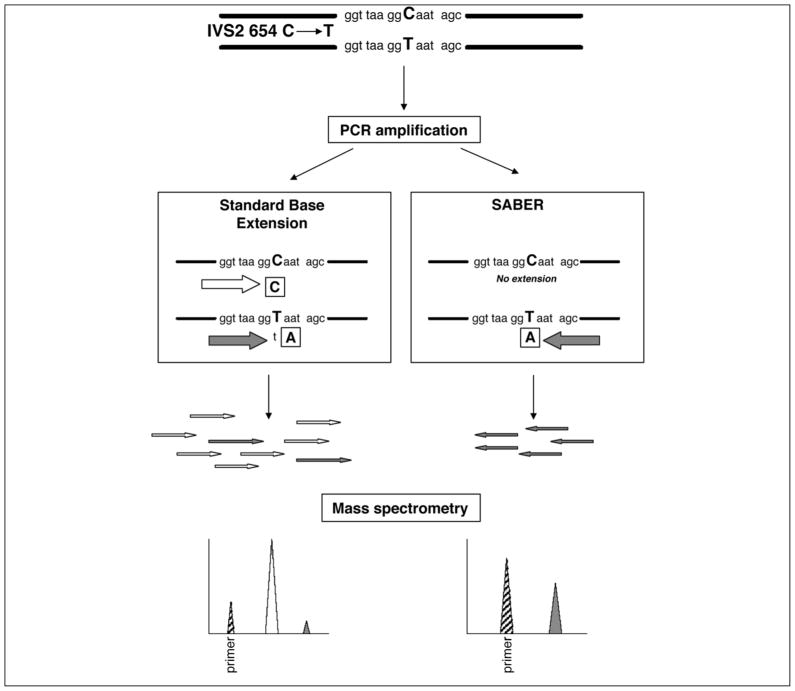
Figure 2. Schematic illustration of the SABER and standard MassARRAY assays.
Maternal plasma detection of the paternally inherited fetal-specific -thalassaemia mutation, IVS2654 C➝ T, is presented as an illustrative example. The standard protocol involves the base extension of both the mutant fetal allele (T allele) and the background allele (C allele), whereas the SABER method only extends the fetal-specific mutant allele. The base extension reactions are terminated by dideoxynucleotides, indicated in boxes. The extension products of the standard protocol include a predominance of the nonmutant allele (open arrows) with a small fraction of the fetal-specific mutant allele (filled arrows). The low abundance of the fetal allele (filled peak) is overshadowed by the nonmutant allele (open peak) on the mass spectrum. Because SABER only involves the extension of the mutant allele, the latter’s presence (filled peak) can be robustly identified from the mass spectrum. The striped peaks represent the unextended primer. (Ref. 114, Used with Permission)
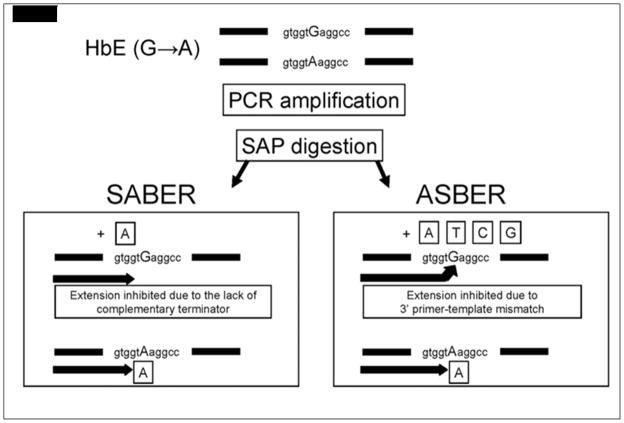
Figure 3.
Schematic diagram comparing the principles of the SABER and ASBER protocols. The site of the point mutation is indicated in capital letters. The boxed letter indicates the type of dideoxynucleotide terminator in the base extension reaction. (Ref. 116, Used with Permission)
Commercialized successfully by Sequenom Inc., many of the technologies and advancements relating to use of MALDI-TOF MS for single point mutation analysis coincided with the discovery of single nucleotide polymorphisms (SNPs) found during work involving the Human Genome Project. It is reported that SNPs occur as frequently as one in every 100–300 bases and thus a rapid instrumental technique with the capability to genotype a large number of these SNPs in one experiment is needed. Most importantly, the ability to identify a single base mutation which results in a small mass difference for the total oligonucleotide is difficult to resolve by qPCR. Thus, with the emergence of MALDI-TOF-MS and its high resolution capability, a rapid, sensitive method is available for SNP analysis [116].
In terms of MALDI application to the analysis of DNA, Sequenom Inc. has emerged as a leader in the field of genetic analysis and in particular for genotyping, methylation detection and quantitative gene expression analysis. Specifically, the MassARRAY method involves PCR amplification of the SNP region of interest, digestion with Shrimp Alkaline Phosphatase (SAP) restriction enzyme to remove unused nucleotides and a post PCR primer extension by deoxynucleotides and dideoxynucleotides, which forms a unique set of masses depending on the different alleles present. This mixture of extended primer alleles is then analyzed by a bench top MALDI-TOF MS. This process involves software to design primers like “SPECTRODESIGNER” or the recently introduced “iPLEX Gold”. The main difference in them lies in high throughput of “iPLEX Gold” which can design assays for 40 SNPs at a time.
To study DNA methylation, Sequenom introduced EpiTYPER a rapid, accurate and a high-throughput quantitative analysis method. In this method DNA undergoes sodium bisulphate treatment and PCR amplification followed by base specific cleavages using MassCLEAVE™ reagent. These cleavage products result in unique masses for methylated and unmethylated products which are analyzed by MALDI-TOS-MS MassARRAY system thus providing quantitative methylation estimates for these CpG sites. The single most important component of the MassARRAY method which makes this technique high throughput is the SpectroCHIP, a small microarray chip that can hold 384 SNP samples individually. Ten of these SpectroCHIP arrays can be loaded onto MassARRAY system thus providing analysis of 3840 SNPs samples in a single, automated, unattended run [116].
The emergence of Sequenom’s MALDI TOF MS system lies in its rapid and accurate analysis, high throughput nature, highly flexible system, appropriate bench size, sophisticated primer design software, beadless and label free reproducible primer extension, cost efficiencies and consistent reliable results. Despite the apparent secondary role of MALDI-TOF MS in these platforms, the ability to resolve small differences in mass as a result of a SNP much more efficiently then qPCR suggests a vital role for MS in this field. Several other single gene mutation disorders such as achondroplasia and β-thalassaemia, have been characterized using MALDI-TOF-MS based methods. In addition, the detection of hemolytic disease of the fetus and newborn (HDFN), detection of paternally inherited SNPs and complicated pregnancy-related disorders such as preimplantation genetic diagnosis (PGD) at an early stage of pregnancy using circulating cell free DNA from maternal plasma have also be developed [113–129].
3.2.1. MALDI-TOF-MS analysis of circulating cell free DNA as non-invasive prenatal diagnosis (NIPD)
Circulating cell free fetal DNA can be collected and detected starting from 5th week of gestation and is reported to completely clear from maternal plasma within hours of birth. It is increasingly becoming a reliable and accurate biomarker which holds promise as a non-invasive prenatal diagnostic (NIPD) tool for various genetic disorders [130]. The biggest challenge to an accurate clinical diagnosis is detection of SNPs or point mutations in fetal DNA in the presence of overwhelming quantities of maternal DNA. This challenge is made more complicated as fetal cell free DNA is reported to be smaller in size (numbers of base pairs) as compared to maternal cell free DNA. Paternally inherited SNPs are easy to detect from the maternal circulation due to absence of homologs in maternal plasma. In 2006 Li et al. reported that the detection of the sex determining region Y (SRY) in circulating cell free fetal DNA isolated from maternal plasma is more sensitive using MALDI-TOF MS as compared to real time PCR [118].
The smaller size of fetal DNA and its low relative quantity in maternal circulating cell free DNA led to studies which combined size fractionation and MALDI-TOF MS for improved detection of SNPs. By using agarose gel electrophoresis as an initial enrichment step for circulating cell free fetal DNA and combining hME or SABER and MALDI-TOF MS, 41 SNPs and fetal gene mutations were analyzed [119, 120]. It has also been reported that using MALDI-TOF-MS after size fractionation of circulating cell free DNA isolated from maternal plasma improves the detection of a paternally inherited codon 39 point mutation of the β-globin gene causing β-thalassaemia disorder [121]. In 2007 Li and coworkers reported a quantitative, specific and sensitive MALDI-TOF-MS analysis of size fractionated circulating cell free DNA isolated from maternal plasma of two pregnant women at risk for Achondroplasia, a genetic disorder of bone growth, most commonly leading to dwarfism. Achondroplasia results from two specific mutations which occur in the FGFR3 gene which limit the formation of bone from cartilage (a process called ossification) particularly in the long bones. Circulating cell free DNA was isolated and MALDI-TOF with SABER and hME protocol led to precise detection of the fetal G1138A mutation from both assays [122].
An important extension of the development of noninvasive prenatal diagnosis using cell free DNA isolated from maternal DNA was reported for preimplantation genetic diagnosis (PGD) pregnancies. Preimplantation genetic diagnosis is a screening assay specifically intended for cases in which a genetic risk to the embryo may result if one or both genetic parents have a known genetic abnormality [123]. The use of MALDI TOF MS for PGD screening is presently being used as a complementary technique to typical invasive methods but continues to show promise for this type of analysis. Li et al. have reported a MALDI-TOF MS assay using either SABER or the size-fractionation approach to detect single base mutations and paternally inherited gene mutations and other fetal gene mutations. Thus, in the future this technique could become the method of choice for PGD testing [12, 125].
With the emergence of epigenetics as an important field in genetic research, Bellido and coworkers recently reported the detection of epigenetic changes in circulating cell free fetal DNA as a potential biomarker tool for NIPD. In a case-controlled study of 20 pregnant women and 30 non-pregnant controls, the methylation patterns of MASPIN genes (RASSF1 and SERPINB5) were mapped using Sequenom’s MALDI-TOF system. While no significant differences were found in the methylation levels of RASSF1 and SERPINB5 genes from the plasma of non-pregnant and pregnant women, in this small study the promise of CCFFDNA as a biomarker for NIPD and other diseases predicated on epigenetic changes requires further research [126].
3.2.2. Non invasive genotyping of hemolytic disease of the fetus and newborn (HDFN) using CCFDNA and mass spectrometry
Hemolytic disease of the fetus and newborn (HDFN) is due to maternal alloantibodies directed against paternally inherited antigens on fetal red cells, such that these maternal alloantibodies cause immune-mediated destruction of fetal/newborn red blood cells thus presenting risk to the fetus/new born [127]. The most important mutation causing HDFN is a fetal RHD exon 7 mutation and, because of it, maternal Rhesus-D negative (RhD) antibodies are directed against paternally inherited antigens on fetal red blood cells. The detection of this fetal RHD exon 7 mutation is normally performed using rt-PCR but Grill et al. reported a method in 2009 using MALDI-TOF MS with SABER in which they were able to detect as little as 2.5% of RHD-positive cell free DNA in a background of RHD-negative genomic DNA in maternal plasma of 178 pregnant women. The MALDI assay was not only sensitive and accurate but also had a 96.1% agreement with PCR results. One of the major advantages of using these mass spectrometry platforms for detecting HDFN lies in the technique’s high-throughput nature. With multiple sampling points per MALDI experiment, there is an increased possibility of detecting numerous RhD alleles which could result in saving time and cost for blood group genotyping [128]. MALDI-TOF mass spectrometry using the SABER protocol has also been reported to be a useful technique to detect another single base substitution (C to T) in exon 6 of the fetal KEL1 gene causing HDFN. An accuracy of 94% and high sensitivity (20 pg mutant DNA detected in 580 pg wildtype DNA) from cell-free fetal DNA isolated from maternal plasma with no false positive results were reported. This fatal gene mutation has the potential to cause severe hemolytic transfusion reactions and it is vital to have a rapid, sensitive method to detect this genetic condition [129].
3.2.3. MALDI-TOF-MS analysis of circulating cell free DNA from urine
Bauer and Pertl highlighted the emerging importance of isolating circulating cell free DNA from urine in a recent Clinical Chemistry editorial and indicated that the origin of elevated levels of circulating cell free DNA could be traced to the presence of diseases like cancer, blood transfusions, fetal diseases in pregnant women and in kidney transplantation cases [57]. Although isolating CCFDNA from urine is technically challenging due to inherent problems from contamination from bladder tissue DNA and bacterial DNA as a result of infection, a fair amount of research is being conducted to develop urine based diagnostic methods based on CCFDNA. In 2005 it was reported that in renal transplant recipients, donor specific SNPs were detected and analyzed by MALDI-TOF-MS from the cell free DNA isolated from urine [130]. Similar results were reported in 2006 when Li et al. detected SNPs in the plasma of pregnant women and in the urine of kidney transplant recipients which were also analyzed by MALDI-TOF mass spectrometry. In this report, the authors noted improved results in analysis of paternally inherited SNPs by the size-fractionation approach [131]. Although mass spectrometry is inferior to methods such as PCR for sequencing, its potential when integrated with sequencing methods like PCR has been impressive. The incorporation of MS results in improved protocols and methods for gene analysis, increased accuracy and sensitivity, rapid analysis times, easy automation, no need for radioactive or fluorescent labels, and no constraints due to the secondary structure of isolated CCFDNA as compared to existing PCR methods.
4. Conclusions
The past few years have seen remarkable growth in the development of diagnostic methods for cancer and the monitoring of its progression and regression after chemotherapy using CCFDNA. This growth can be directly ascribed to advances made in the field of proteomics and genomics and the evolution of mass spectrometry within these fields. As a result of these improvements, a clearer understanding of the role of genetic mutations and cancer development for some diseases has been realized. More importantly, the realization that point mutations have a critical role in specific diseases has necessitated the development of sensitive methods of analysis for their detection. While a large number of reports involving cancer and the utility of rt-PCR for the analysis of CCFDNA from plasma have been reported, as highlighted in Table 3, one challenge for existing techniques such as rt-PCR or rt-qPCR remains the ability to detect CCFDNA in cases where the composition is unknown. In these instances, PCR type methods will not work because of the need for a specific primer to initiate the amplification process. However, given the utility and flexibility of HPLC-based MS methods, the analysis of short DNA sequences has become routine.
Table 3.
Recent applications of CCFDNA in early stage detection of cancer using PCR methods
| Cancer/source of CFF DNA/Gene involved | Extraction kit/Technique used | Quantification | Remarks | Reference |
|---|---|---|---|---|
| Lung Cancer/plasma | q- PCR | The healthy and cancer groups were 10.4 and 22.6 ng/ml, respectively. | 102 patients with lung cancer and 105 healthy individuals | 9 |
| Colorectal cancer/plasma | QIAamp DNA Mini Kit/q-PCR | The calculated values were between 22 and 3922 ng/ml for cancer patients and for healthy donors were significantly lower (5–16 ng/ml). | 55 patients with advanced colorectal cancer and 20 healthy individuals. | 10 |
| Breast cancer/plasma/Exon 7, P53 | Qiagen kit/q- PCR | CF DNA higher in advanced stage breast cancer patients than in controls (G.E. 18850 vs. 431) | 25 newly diagnosed untreated breast cancer patients and 25 healthy, 9 patients after chemotherapy. | 11 |
| Testicular cancer/serum/ACTB gene | ChargeSwitch ® gDNA Kit/rt- PCR | Testicular cancer compared to those in healthy individuals (8.09 vs. 0.82 ng/ml, respectively). | 74 patients with testicular cancer, (39 with seminoma and 35 with nonseminoma) and 35 healthy individuals | 12 |
| Gastric cancer/plasma | q- pcr | DNA concentrations in the short and long assays of the gastric cancer patients were significantly higher than those of the control group. | 53 patients with gastric cancer and 21 healthy individuals | 14 |
| Endometrial cancer/plasma/KRAS Codon 12 mutation | Sigma blood DNA isolation kit/enriched PCR-RFLP method | 200 μl blood plasma/CFDNA was detectable in 12 samples of Type I EC (13.8%) and in 8 samples of Type II (36.4%). | 109 patients with EC (87 patients with Type I and 22 Patients with Type II) | 18 |
| Colorectal carcinoma/urine/K-ras Codon 12 | Wizard DNA Isolation Kit/q- PCR | The calculated values were between 43 and 198 ng/ml for cancer patients. | 20 subjects with CRC or Adenomatous polyps and healthy individuals. | 51 |
| Colon cancer/urine/K-ras codon 12 | Wizard DNA Isolation Kit- CMB suspension/rt- PCR | The calculated values were between 10 and 95 ng/ml for cancer patients. | 5 volunteers with no known diseases and 36 patients colon cancer | 58 |
| Hodgkin’s and non-Hodgkin’s lymphomas/plasma | q- PCR | 142 patients with lymphomas, 41 healthy individuals | 81 | |
| Bladder cancer/urine | spin column- based method/rt- PCR | Median free DNA quantification did not differ statistically between bladder cancer patients and healthy subjects. | 45 bladder cancer patients and 87 healthy individuals | 92 |
| Primary breast cancer/blood serum | QIAamp Blood DNA Mini Kit/fluorescence- labeled PCR | The range of DNA concentrations was between 58 and 5317 ng/ml of serum with a mean value of 886 ng/ml and a median value of 519 ng/ml | 81 breast cancer patients | 100 |
| Ovarian carcinoma/plasma | Qiagen DNA Isolation Kits/rt-PCR | 19 patients with primary ovarian carcinoma and 12 healthy individuals | 132 | |
| Ovarian Cancer/plasma | Qiagen DNA extraction Mini kit/rt- PCR | EOC patients had a median preoperative CF DNA level of 10,113 GE/mL, compared with patients with benign ovarian neoplasms (~ 2365 GE/mL) and controls (~ 1912 GE/mL) | DNA was extracted from plasma of 164 women with invasive epithelial ovarian carcinoma (EOC), 49 with benign ovarian neoplasms, and 75 age-matched controls. | 133 |
With improved isolation methods for CCFDNA in urine, MS seems to be the perfect, flexible platform for the screening of these important circulating biomolecules. The incorporation of MS results in improved protocols and methods for gene analysis, increased accuracy and sensitivity, rapid analysis times, easy automation, no need for radioactive or fluorescent labels, and no constraints due to the secondary structure of isolated CCFDNA as compared to existing PCR methods. PCR-based methods also suffer from the inherent requirement that the sequence of interest for duplication be known. This requirement is mitigated completely with the ability to sequence an unknown oligonucleotide using tandem MS/MS techniques using either ESI or MALDI ionization. As was previously stated, the SOMA method has been shown to be more sensitive than restriction fragment length polymorphism-PCR (RFLP-PCR) for detecting the specific mutations from circulating cell free DNA extracted from plasma or serum. While there are distinct benefits to using MS for the analysis of CCFDNA, problems associated with the analysis of oligonucleotides do exist. For example, high salt content in the isolated samples can result in electrospray problems or the formation of multiply charged salt adducts which could complicate ESI MS interpretation. This salt effect also causes problems with the MALDI mechanism of ionization resulting in some cases of complete signal loss. Notwithstanding the challenges associated with the ionization of oligonucleotides, the expectation is that mass spectrometry can play an significant role in the analysis of CCFDNA as has been discussed. It has been the intent of this review to introduce the importance of cell free DNA to mass spectrometric researchers such that more effort should be invested to solve the challenges in making mass spectrometry the technique of choice for analyzing the cell free DNA.
Acknowledgments
This work was supported by National Institutes of Health grant numbers 1RO1 CA69390 and 1RO1 CA112231. This is Contribution No. 898 from the Barnett Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pleasance ED, et al. Nature. 2009;463:184. [Google Scholar]
- 2.Pleasance ED, et al. Nature. 2009;463:191. [Google Scholar]
- 3.Mandel P, Metais P, Les CR. Acad Sci Paris. 1948;142:241. [Google Scholar]
- 4.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Cancer Res. 1977;37:646. [PubMed] [Google Scholar]
- 5.Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic Oncology. 1989;46:318. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- 6.Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Br J Haematol. 1994;86:774. doi: 10.1111/j.1365-2141.1994.tb04828.x. [DOI] [PubMed] [Google Scholar]
- 7.Sorenson GD. Ann N Y Acad Sci. 2000;906:13. doi: 10.1111/j.1749-6632.2000.tb06582.x. [DOI] [PubMed] [Google Scholar]
- 8.Sozzi G, Conte D, Mariani L, Vullo SL, Roz L, Lombardo C. Cancer Res. 2001;61:4675. [PubMed] [Google Scholar]
- 9.Yoon K-A, Park S, Lee SH, Kim JH, Lee JS. J Mol Diagn. 2009;11:182. doi: 10.2353/jmoldx.2009.080098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarzenbach H, Stoehlmacher J, Pantel K, Goekkurt E. Annals of the New York Academy of Sciences. 2008;1137:190. doi: 10.1196/annals.1448.025. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharyya S, Raina V, Shukla NK, Shukla S, Kumar R, Hedau S, Kumar G, Bharti AC, Rath GK, Das BC. Journal of Clinical Oncology. 2009;27:e22213. [Google Scholar]
- 12.Ellinger J, Wittkamp V, Albers P, Perabo FGE, Mueller SC, von Ruecker A, Bastian PJ. The Journal of Urology. 2009;181:363. doi: 10.1016/j.juro.2008.08.118. [DOI] [PubMed] [Google Scholar]
- 13.Feng G, Li G, Zhao A, Gentil-Perret A, Genin C, Tostain J. Urology. 2010;75:262. [Google Scholar]
- 14.Sai S, Ichikawa D, Tomita H, Ikoma D, Tani N, Ikoma H, Kikuchi S, Fujiwara H, Ueda Y, Otsuji E. Anticancer Res. 2007;27:2747. [PubMed] [Google Scholar]
- 15.Zancan M, Galdi F, Di Tonno F, Mazzariol C, Orlando C, Malentacchi F, Agostini M, Maran M, Bianco PD, Fabricio ASC, Murer B, Pianon C, Gion M. Int J Biol Markers. 2009;24:147. doi: 10.1177/172460080902400304. [DOI] [PubMed] [Google Scholar]
- 16.Ellinger J, Ellinger N, Heukamp LC, Kahl P, Perabo FG, Büttner R, Müller SC, Von Ruecker A, Bastian PJ. European Urology Supplements. 2008;7:207. [Google Scholar]
- 17.Capizzi E, Gabusi E, Antonia DG, De Iaco P, Rosati M, Zamagni C, Fiorentino M. Diagnostic Molecular Pathology. 2008;17:34. doi: 10.1097/PDM.0b013e3181359e1f. [DOI] [PubMed] [Google Scholar]
- 18.Dobrzycka B, Terlikowski SJ, Mazurek A, Kowalczuk O, Niklinska W, Chyczewski L, Kulikowski M. Int J Cancer. 2010;XX:XXX. doi: 10.1002/ijc.25077. [DOI] [PubMed] [Google Scholar]
- 19.Tomochika S, Iizuka N, Watanabe Y, Tsutsui M, Takeda S, Yoshino S, Ichihara K, Oka M. Experimental and Therapeutic Medicine. 2010;1:89. doi: 10.3892/etm_00000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosca M, Giuliano T, Cuomo G, Doveri M, Tani C, Curcio M, Abignano G, De Feo F, Bazzichi L, Rossa AD, Valentini G, Bombardieri S. Clin Rheumatol. 2009;28:1437. doi: 10.1007/s10067-009-1245-5. [DOI] [PubMed] [Google Scholar]
- 21.Ozkaya O, Bek K, Bedir A, Açıkgöz Y, Özdemir T. Nephron Clin Pract. 2009;113:c258. doi: 10.1159/000235250. [DOI] [PubMed] [Google Scholar]
- 22.Shin C, Kim JK, Kim JH, Jung KH, Cho KJ, Lee CK, Lee SG. Psychiatry and Clinical Neurosciences. 2008;62:721. doi: 10.1111/j.1440-1819.2008.01876.x. [DOI] [PubMed] [Google Scholar]
- 23.Lazar L, Rigó J, Jr, Nagy B, Balogh K, Makó V, Cervenak L, Mézes M, Prohászka Z, Molvarec A. BMC Medical Genetics. 2009;10:120. doi: 10.1186/1471-2350-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gornik I, Wagner J, Gašparović V, Lauc G, Gornik O. Clinical Biochemistry. 2009;42:38. doi: 10.1016/j.clinbiochem.2008.09.121. [DOI] [PubMed] [Google Scholar]
- 25.Moreira VG, García BP, de la T, Martínez C, Menéndez FVA. Clinical Biochemistry. 2009;42:729. [Google Scholar]
- 26.Ioannis GF, Aspasia D, Konstantinos M, Athanasios ZJ, Christina V, Dimitrios K, Mastorakos G, Mitrakou A, Taxildaris K, Kanavakis E, Papassotiriou I. Clinical Chemistry. 2006;52:1820. doi: 10.1373/clinchem.2006.070417. [DOI] [PubMed] [Google Scholar]
- 27.Lo YMD, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW. Lancet. 1997;350:485. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 28.Bianchi DW, LeShane ES, Cowan JM. Clin Chem. 2001;47:1867. [PubMed] [Google Scholar]
- 29.Wataganara T, Bianchi DW. Ann N Y Acad Sci. 2004;1022:90. doi: 10.1196/annals.1318.015. [DOI] [PubMed] [Google Scholar]
- 30.Invernizzi P, Biondi ML, Battezzati PM, Perego F, Selmi C, Cecchini F, Podda M, Simoni G. Hum Genet. 2002;110:587. doi: 10.1007/s00439-002-0725-3. [DOI] [PubMed] [Google Scholar]
- 31.Tomaiuolo M, Greco P, Grandone E. Int J Gynecol Obstet. 2007;96:202. doi: 10.1016/j.ijgo.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 32.Zhong XY, Laivuori H, Livingston JC, Ylikorkala O, Sibai BM, Holzgreve W, Hahn S. Am J Obstet Gynecol. 2001;184:414. doi: 10.1067/mob.2001.109594. [DOI] [PubMed] [Google Scholar]
- 33.Spencer K, de Kok JB, Swinkels DW. Prenatal Diagnosis. 2003;23:580. doi: 10.1002/pd.647. [DOI] [PubMed] [Google Scholar]
- 34.Puszyk WM, Crea F, Old RW. Prenat Diagn. 2008;28:1. doi: 10.1002/pd.1902. [DOI] [PubMed] [Google Scholar]
- 35.Lo YM, Leung TN, Tein MS, Sargent IL, Zhang J, Lau TK. Clin Chem. 1999;45:184. [PubMed] [Google Scholar]
- 36.Lee T, Le Shane ES, Messerlian G, Canick JA, Farina A, Heber W. Am J Obstet Gynecol. 2002;187:1217. doi: 10.1067/mob.2002.127462. [DOI] [PubMed] [Google Scholar]
- 37.Zhong XY, Holzgreve W, Hahn S. BJOG. 2000;107:766. doi: 10.1111/j.1471-0528.2000.tb13338.x. [DOI] [PubMed] [Google Scholar]
- 38.Finning K, Martin P, Daniels G. Ann N Y Acad Sci. 2004;1022:119. doi: 10.1196/annals.1318.019. [DOI] [PubMed] [Google Scholar]
- 39.Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM. Am J Hum Genet. 1998;62:768. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Naro ED, Vitucci A, Zimmermann B, Holzgreve W, Hahn S. JAMA. 2005;293:843. doi: 10.1001/jama.293.7.843. [DOI] [PubMed] [Google Scholar]
- 41.Du J, Zou X, Pan Y, Lu G-X. Fertility and Sterility. 2009;92:S199. [Google Scholar]
- 42.Li Y, Holzgreve W, Page-Christiaens GCML, Gille JJP, Hahn S. Prenatal Diagnosis. 2004;24:896. doi: 10.1002/pd.1030. [DOI] [PubMed] [Google Scholar]
- 43.Sekizawa A, Sugito Y, Iwasaki M, Watanabe A, Jimbo M, Hoshi S, Saito H, Okai T. Clinical Chemistry. 2001;47:2164. [PubMed] [Google Scholar]
- 44.Farina A, LeShane ES, Romero R, Gomez R, Chaiworapongsa T, Rizzo N, Bianchi DW. American Journal of Obstetrics & Gynecology. 2005;193:421. doi: 10.1016/j.ajog.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 45.Nakib MA, Desbrière R, Bonello N, Bretelle F, Boubli L, Gabert J, Mozziconacci AL. Fetal Diagn Ther. 2009;26:24. doi: 10.1159/000236355. [DOI] [PubMed] [Google Scholar]
- 46.Zhong XY, Steinborn A, Sohn C, Holzgreve W, Hahn S. Prenat Diagn. 2006;26:1272. doi: 10.1002/pd.1613. [DOI] [PubMed] [Google Scholar]
- 47.Gurwitz D. The Lancet. 1997;350:1254. doi: 10.1016/S0140-6736(05)63490-3. [DOI] [PubMed] [Google Scholar]
- 48.Botezatu I, Serdyuk O, Potapova G, Alechina R, Arsenin S, Melkonyan H. Clin Chem. 2000;46:1078. [PubMed] [Google Scholar]
- 49.Zhang J, Tong KL, Li PK, Chan AY, Yeung CK, Pang CC, et al. Clin Chem. 1999;45:1741. [PubMed] [Google Scholar]
- 50.Zhong XY, Hahn D, Troeger C, Klemm A, Stein G, Thomson P, Holzgreve W, Hahn S. Annals of the New York Academy of Sciences. 2001;945:250. [PubMed] [Google Scholar]
- 51.Hsiu Su Y, Wang M, Brenner DE, Ng A, Melkonyan H, Umansky S, Syngal S, Block TM. Journal of Molecular Diagnostics. 2004;6:101. doi: 10.1016/S1525-1578(10)60497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryzgunova OE, Skvortsova TE, Kolesnikova EV, Starikov AV, Rykova EY, Vlassov VV, Laktionov PP. Annals of the New York Academy of Sciences. 2006;1075:334. doi: 10.1196/annals.1368.045. [DOI] [PubMed] [Google Scholar]
- 53.Koide K, Sekizawa A, Iwasaki M, Matsuoka R, Honma S, Farina A, et al. Prenat Diagn. 2005;25:604. doi: 10.1002/pd.1213. [DOI] [PubMed] [Google Scholar]
- 54.Majer S, Bauer M, Magnet E, Strele A, Giegerl E, Eder M. Prenat Diagn. 2007;27:1219. doi: 10.1002/pd.1875. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Zhong XY, Kang A, Troeger C, Holzgreve W, Hahn S. J Soc Gynecol Investig. 2003;10:503. doi: 10.1016/s1071-5576(03)00155-2. [DOI] [PubMed] [Google Scholar]
- 56.Illianes S, Denbow ML, Smith RP, Overton TG, Soothill PW, Finning K. Prenat Diagn. 2006;26:1216. doi: 10.1002/pd.1591. [DOI] [PubMed] [Google Scholar]
- 57.Bauer M, Pertl B. Clinical Chemistry. 2009;55:605. doi: 10.1373/clinchem.2008.121855. [DOI] [PubMed] [Google Scholar]
- 58.Su Y-H, Song J, Wang Z, Wang X-H, Wang M, Brenner DE, Block TM. Annals of the New York Academy of Sciences. 2008;1137:82. doi: 10.1196/annals.1448.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shekhtman EM, Anne K, Melkonyan HS, Robbins DJ, Warsof SL, Umansky SR. Clin Chem. 2009;55:723. doi: 10.1373/clinchem.2008.113050. [DOI] [PubMed] [Google Scholar]
- 60.Moreira VG, García BP, de la T, Martínez C, Menéndez FVA. Clinical Biochemistry. 2009;42:729. [Google Scholar]
- 61.Umansky SR, Tomei LD. Expert Rev Mol Diagn. 2006;6:153. doi: 10.1586/14737159.6.2.153. [DOI] [PubMed] [Google Scholar]
- 62.Banoub JH, Limbach PA. Mass Spectrometry of Nucleosides and Nucleic Acids. 2009. [Google Scholar]
- 63.Meng Z, Limbach PA. RNA. 2007;13(2):295. doi: 10.1261/rna.272507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bakhtiar R, Nelson RW. Biochemical Pharmacology. 2000;59(8):891. doi: 10.1016/s0006-2952(99)00317-2. [DOI] [PubMed] [Google Scholar]
- 65.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD. Cancer Res. 2001;61:1659. [PubMed] [Google Scholar]
- 66.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. Clin Chim Acta. 2001;313:139. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 67.Vaart MVD, Pretorius PJ. Clinical Chemistry. 2007;53:2215. doi: 10.1373/clinchem.2007.092734. [DOI] [PubMed] [Google Scholar]
- 68.Ivancic-Jelecki J, Brgles M, Šantak M, Forcic D. Journal of Chromatography A. 2009;1216:2717. doi: 10.1016/j.chroma.2008.10.087. [DOI] [PubMed] [Google Scholar]
- 69.Chang CP-Y, Chia R-H, Wu T-L, Tsao K-C, Sun C-F, Wu JT. Clinica Chimica Acta. 2003;327:95. doi: 10.1016/s0009-8981(02)00337-6. [DOI] [PubMed] [Google Scholar]
- 70.Saukkonen K, Lakkisto P, Pettila V, Varpula M, Karlsson S, Ruokonen E, Pulkki K. Clinical Chemistry. 2008;54:1000. doi: 10.1373/clinchem.2007.101030. [DOI] [PubMed] [Google Scholar]
- 71.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Pancreas. 1998;17:89. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 72.Pathak AK, Bhutani M, Kumar S, Mohan A, Guleria R. Clinical Chemistry. 2006;52:1833. doi: 10.1373/clinchem.2005.062893. [DOI] [PubMed] [Google Scholar]
- 73.Tjoa ML, Davies TC, Boskovic OS, Bianchi DW, Burton GJ. Am J Path. 2006;169:400. doi: 10.2353/ajpath.2006.060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alberry M, Maddocks D, Jones M. Prenat Diagn. 2007;27:415. doi: 10.1002/pd.1700. [DOI] [PubMed] [Google Scholar]
- 75.Chan KCA, Zhang J, Hui ABY, Wong N, Lau TK, Leung TN, Lo KW, Huang DWS, Lo YMD. Clin Chem. 2004;50:88. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 76.Bischoff FZ, Lewis DE, Simpson JL. Hum Reprod Update. 2005;11:59. doi: 10.1093/humupd/dmh053. [DOI] [PubMed] [Google Scholar]
- 77.Jen J, Wu L, Sidransky D. Ann NY Acad Sci. 2000;906:8. doi: 10.1111/j.1749-6632.2000.tb06581.x. [DOI] [PubMed] [Google Scholar]
- 78.Xue X, Teare MD, Holen I, Zhu YM, Woll PJ. Clinica Chimica Acta. 2009;404:100. doi: 10.1016/j.cca.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 79.Altimari A, D’Errico Grigioni A, Benedettini E, Gabusi E, Schiavina R, Martinell A, Morselli-Labate AM, Martorana G, Grigioni WF, Fiorentino M. Am J Clin Pathol. 2008;129:756. doi: 10.1309/DBPX1MFNDDJBW1FL. [DOI] [PubMed] [Google Scholar]
- 80.Suzuki N, Kamataki A, Yamaki J, Homma Y. Clinica Chimica Acta. 2008;387:55. doi: 10.1016/j.cca.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 81.Hohaus S, Giachelia M, Massini G, Mansueto G, Vannata B, Bozzoli V, Criscuolo M, D’Alò F, Martini M, Larocca LM, Voso MT, Leone G. Annals of Oncology. 2009;20:1408. doi: 10.1093/annonc/mdp006. [DOI] [PubMed] [Google Scholar]
- 82.Su Y-H, Wang M, Brenner DE, Norton PA, Block TM. Ann N Y Acad Sci. 2008;1137:197. doi: 10.1196/annals.1448.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shekhtman EM, Anne K, Melkonyan HS, Robbins DJ, Warsof SL, Umansky SR. Clinical Chemistry. 2009;55:723. doi: 10.1373/clinchem.2008.113050. [DOI] [PubMed] [Google Scholar]
- 84.http://www.norgenbiotek.com/display-resource.php?ID=143
- 85.Sifakis S, Zaravinos A, Maiz N, Spandidos DA, Nicolaides KH. American Journal of Obstetrics & Gynecology. 2009;201:472.e1. doi: 10.1016/j.ajog.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 86.Legler TJ, Liu Z, Heermann K-H, Hempel M, Gutensohn K, Kiesewetter H, Pruss A. Transfusion and Apheresis Science. 2009;40:153. doi: 10.1016/j.transci.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 87.Clausen FB, Krog GR, Rieneck K, Dziegiel MH. Prenat Diagn. 2007;27:6. doi: 10.1002/pd.1605. [DOI] [PubMed] [Google Scholar]
- 88.Jorgez CJ, Dang DD, Simpson JL, Lewis DE, Bischoff FZ. Genetics in Medicine. 2006;8:615. doi: 10.1097/01.gim.0000241904.32039.6f. [DOI] [PubMed] [Google Scholar]
- 89.Zhong XY, Hahn S, Kiefer V, Holzgreve W. Ann Hematol. 2007;86:139. doi: 10.1007/s00277-006-0182-5. [DOI] [PubMed] [Google Scholar]
- 90.Beau-Faller M, Gaub MP, Schneider A, Ducrocq X, Massard G, Gasser B, et al. Int J Cancer. 2003;105:361. doi: 10.1002/ijc.11079. [DOI] [PubMed] [Google Scholar]
- 91.Ozkaya O, Bek K, Bedir A, Açıkgöz Y, Özdemir T. Nephron Clin Pract. 2009;113:c258. doi: 10.1159/000235250. [DOI] [PubMed] [Google Scholar]
- 92.Zancan M, Galdi F, Di FT, Mazzariol C, Orlando C, Malentacchi F, Agostini M, Maran M, Del PB, Fabricio AS, Murer B, Pianon C, Gion M. The International journal of biological markers. 2009;24:147. doi: 10.1177/172460080902400304. [DOI] [PubMed] [Google Scholar]
- 93.Schwarzenbach H, Stoehlmacher J, Pantel K, Goekkurt E. Annals of the New York Academy of Sciences. 2008;1137:190. doi: 10.1196/annals.1448.025. [DOI] [PubMed] [Google Scholar]
- 94.Schwarzenbach H, Alix-Panabières C, Müller I, Letang N, Vendrell J-P, Rebillard X, Pantel K. Clin Cancer Res. 2009;15:1032. doi: 10.1158/1078-0432.CCR-08-1910. [DOI] [PubMed] [Google Scholar]
- 95.Hohaus S, Giachelia M, Massini G, Mansueto G, Vannata B, Bozzoli V, Criscuolo M, D’Alò F, Martini M, Larocca LM, Voso MT, Leone G. Ann Oncol. 2009;20:1408. doi: 10.1093/annonc/mdp006. [DOI] [PubMed] [Google Scholar]
- 96.Chang HW, Tsui KH, Shen LC, Huang HW, Wang SN, Chang PL. Int J Biol Markers. 2007;22:287. doi: 10.1177/172460080702200408. [DOI] [PubMed] [Google Scholar]
- 97.Zimmermann B, Zhong XY, Holzgreve W, Hahn S. Methods in Molecular Medicine. 2007;132:43. doi: 10.1007/978-1-59745-298-4_5. [DOI] [PubMed] [Google Scholar]
- 98.Sikora A, Zimmermann BG, Rusterholz C, Birri D, Kolla V, Lapaire O, Hoesli I, Kiefer V, Jackson L, Hahn S. Clin Chem. 2010;56:136. doi: 10.1373/clinchem.2009.132951. [DOI] [PubMed] [Google Scholar]
- 99.Cirigliano V, Voglino G, Cañadas MP, Marongiu A, Ejarque M, Ordofiez E, Plaja A, Massobrio M, Todros T, Fuster C, Campogrande M, Egozeue J, Adinolfi M. Mol Hum Reprod. 2004;10:839. doi: 10.1093/molehr/gah108. [DOI] [PubMed] [Google Scholar]
- 100.Schwarzenbach H, Pantel K, Kemper B, Beeger C, Otterbach F, Kimmig R, Kasimir-Bauer S. Breast Cancer Research. 2009;11:R71. doi: 10.1186/bcr2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goldshtein H, Hausmann MJ, Douvdevani A. Ann Clin Biochem. 2009;46:488. doi: 10.1258/acb.2009.009002. [DOI] [PubMed] [Google Scholar]
- 102.http://www.invitrogen.com/site/us/en/home/brands/Product-Brand/Qubit/qubit-fluorometer.html 04/21/2010
- 103.Housby JN, editor. Mass Spectrometry and Genomic Analysis. 2000. p. 76. [Google Scholar]
- 104.Jackson PE, Qian GS, Friesen MD, Zhu YR, Lu P, Wang JB, Wu Y, Kensler TW, Vogelstein B, Groopman JD. Cancer Res. 2001;61:33. [PubMed] [Google Scholar]
- 105.Qian GS, Kuang SY, He X, Groopman JD, Jackson PE. Cancer Epidemiol Biomarkers Prev. 2002;11:1126. [PubMed] [Google Scholar]
- 106.Jackson PE, Kuang S-Y, Wang J-B, Strickland PT, Muñoz A, Kensler TW, Qian G-S, Groopman JD. Carcinogenesis. 2003;24:1657. doi: 10.1093/carcin/bgg101. [DOI] [PubMed] [Google Scholar]
- 107.Szymánska1 K, Lesi OA, Kirk GD, Sam O, Taniere P, Yves Scoazec J, Mendy M, Friesen MD, Whittle H, Montesano R, Hainaut P. Int J Cancer. 2004;110:374. doi: 10.1002/ijc.20103. [DOI] [PubMed] [Google Scholar]
- 108.Yuan Kuang S, Jackson PE, Bing Wang J, Xing Lu P, Muñoz A, Sun Qian G, Kensler TW, Groopman JD. PNAS. 2004;9:3575. doi: 10.1073/pnas.0308232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lleonart ME, Kirk GD, Villar S, Lesi OA, Dasgupta A, Goedert JJ, Mendy M, Hollstein MC, Montesano R, Groopman JD, Hainaut P, Friesen MD. Cancer Epidemiol Biomarkers Prev. 2005;14:2956. doi: 10.1158/1055-9965.EPI-05-0612. [DOI] [PubMed] [Google Scholar]
- 110.Chen JG, Kuang SY, Egner PA, Lu JH, Zhu YR, Wang JB, Zhang BC, Chen TY, Muñoz A, Kensler TW, Groopman JD. Cancer Epidemiology, Biomarkers & Prevention. 2007;16:1213. doi: 10.1158/1055-9965.EPI-06-0905. [DOI] [PubMed] [Google Scholar]
- 111.Szymañska K, Guo Chen J, Cui Y, Gong YY, Turner PC, Villar S, Wild CP, Parkin DM, Hainaut P. Cancer Epidemiol Biomarkers Prev. 2009;18:1638. doi: 10.1158/1055-9965.EPI-08-1102. [DOI] [PubMed] [Google Scholar]
- 112.Zhong XY, Holzgreve W. Transfus Med Hemother. 2009;36:263. doi: 10.1159/000223098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ding C, Chiu RWK, Lau TK, Leung TN, Chan LC, Chan AYY, Charoenkwan P, Ng ISL, yang Law H, Ma ESK, Xu X, Wanapirak C, Sanguansermsri T, Liao C, Ai MATJ, Chui DHK, Cantor CR, Lo YMD. PNAS. 2004;101:10762. doi: 10.1073/pnas.0403962101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ding C. Methods Mol Biol. 2008;444:253. doi: 10.1007/978-1-59745-066-9_20. [DOI] [PubMed] [Google Scholar]
- 115.Tsang JCH, Charoenkwan P, Chow KCK, Jin Y, Wanapirak C, Sanguansermsri T, Lo YMD, Chiu RWK. Clinical Chemistry. 2007;53:2205. doi: 10.1373/clinchem.2007.095133. [DOI] [PubMed] [Google Scholar]
- 116.http://www.sequenom.com 04/21/2010
- 117.Vogel N, Schiebel K, Humeny A. Transfus Med Hemother. 2009;36:253. doi: 10.1159/000225089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li Y, Holzgreve W, Kiefer V, Hahn S. Clinical Chemistry. 2006;52:2311. doi: 10.1373/clinchem.2006.076257. [DOI] [PubMed] [Google Scholar]
- 119.Li Y, Wenzel F, Holzgreve W, Hahn S. Electrophoresis. 2006;27:3889. doi: 10.1002/elps.200600084. [DOI] [PubMed] [Google Scholar]
- 120.Li Y, Hahn S, Holzgreve W. Annals of the New York Academy of Sciences. 2006;1092:285. doi: 10.1196/annals.1365.024. [DOI] [PubMed] [Google Scholar]
- 121.Li Y, Holzgreve W, Hahn S. Methods Mol Biol. 2008;444:239. doi: 10.1007/978-1-59745-066-9_19. [DOI] [PubMed] [Google Scholar]
- 122.Li Y, Page-Christiaens GCML, Gille JJP, Holzgreve W, Hahn S. Prenatal Diagnosis. 2007;27:11. doi: 10.1002/pd.1608. [DOI] [PubMed] [Google Scholar]
- 123.Flinter FA. BMJ. 2001;322:1008. doi: 10.1136/bmj.322.7293.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Y, Altarescu G, Renbaum P, Geva TE, Lahad EL, Margalioth EJ, Zhong X, Hahn S, Holzgreve W. Reproductive biomedicine Online. 2009;19:714. doi: 10.1016/j.rbmo.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 125.Li Y, Naro ED, Vitucci A, Grill S, Zhong XY, Holzgreve W, Hahn S. Fetal Diagn Ther. 2009;25:246. doi: 10.1159/000223442. [DOI] [PubMed] [Google Scholar]
- 126.Bellido ML, Radpour R, Lapaire O, Bie ID, Hösli I, Bitzer J. Biolreprod. 2010;109:082271. doi: 10.1095/biolreprod.109.082271. [DOI] [PubMed] [Google Scholar]
- 127.Illanes S, Soothill P. Expert Review of Hematology. 2009;2:577. doi: 10.1586/ehm.09.45. [DOI] [PubMed] [Google Scholar]
- 128.Grill S, Banzola I, Li Y, Rekhviashvili T, Legler TJ, Müller SP, Zhong XY, Hahn S, Holzgreve W. Arch Gynecol Obstet. 2009;279:533. doi: 10.1007/s00404-008-0774-5. [DOI] [PubMed] [Google Scholar]
- 129.Li Y, Finning K, Daniels G, Hahn S, Zhong X, Holzgreve W. Prenat Diagn. 2008;28:203. doi: 10.1002/pd.1936. [DOI] [PubMed] [Google Scholar]
- 130.Li Y, Hahn D, Holzgreve W, Hahn S. Clin Chem. 2005;51:1903. doi: 10.1373/clinchem.2005.055038. [DOI] [PubMed] [Google Scholar]
- 131.Li Y, Hahn D, Wenzel F, Holzgreve W, Hahn S. Ann NY Acad Sci. 2006;1075:144. doi: 10.1196/annals.1368.019. [DOI] [PubMed] [Google Scholar]
- 132.Kamat AA, Sood AK, Dang D, Gershenson DM, Simpson JL, Bischoff FZ. Annals of the New York Academy of Sciences. 2006;1075:230. doi: 10.1196/annals.1368.031. [DOI] [PubMed] [Google Scholar]
- 133.Kamat AA, Baldwin M, Urbauer D, Dang D, Han LY, Godwin A, Karlan BY, Simpson JL, Gershenson DM, Coleman RL, Bischoff FZ, Sood AK. Cancer. 2010;00:XXX. doi: 10.1002/cncr.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Laken SJ, Jackson PE, Kinzler KW, Vogelstein B, Strickland PT, Groopman JD, Friesen MD. Nature Biotechnology. 1998;16:1352. doi: 10.1038/4333. [DOI] [PubMed] [Google Scholar]