SUMMARY
GATA-1-dependent transcription is essential for erythroid differentiation and maturation. Suppression of programmed cell death is also thought to be critical for this process; however, the link between these two features of erythropoiesis has remained elusive. Here, we show that the POZ-Krüppel family transcription factor, LRF (also known as Zbtb7a/Pokemon), is a direct target of GATA1 and plays an essential anti-apoptotic role during terminal erythroid differentiation. We find that loss of Lrf leads to lethal anemia in embryos, due to increased apoptosis of late stage erythroblasts. This programmed cell death is Arf- and p53-independent and is instead mediated by up-regulation of the pro-apoptotic factor Bim. We identify Lrf as a direct repressor of Bim transcription. In strong support of this mechanism, genetic Bim-loss delays the lethality of Lrf-deficient embryos and rescues their anemia-phenotype. Thus, our data defines a key transcriptional cascade for effective erythropoiesis, whereby GATA-1 suppresses BIM-mediated apoptosis via LRF.
INTRODUCTION
Production of red blood cells (RBCs) is normally maintained at a constant level by a finely tuned regulation of erythropoiesis. During terminal maturation, mammalian erythroblasts extrude their nuclei and give rise to RBCs. Over the past twenty years a wealth of experimental evidence has unveiled the role of critical genes during erythropoiesis (Godin and Cumano, 2002). In particular, lineage specific transcription factors and cytokines regulate erythroid cell fate by activating or inactivating key factors for cellular differentiation and survival. The nuclear protein GATA1 is highly expressed in the erythroid lineages and activates the transcription factor Eklf (Crossley et al., 1994; Welch et al., 2004) along with many other erythroid specific genes. Disruption of the Gata1 gene in mice causes embryonic lethality due to defects in yolk sac primitive erythropoiesis (Fujiwara et al., 1996). Furthermore, hematopoietic differentiation of Gata1 null ES cells in vitro revealed a block of differentiation at the proerythroblast stage due to developmental arrest and apoptosis (Weiss et al., 1994), indicating that Gata1 also plays a key role in definitive erythropoiesis. How GATA1 regulates differentiation and survival of immature erythroblasts is not fully understood. The GATA1 target EKLF acts primarily as a transcriptional activator and induces multiple erythroid specific genes (Hodge et al., 2006). Eklf knockout mice die of severe anemia by 16.5 d.p.c. due to defective definitive erythropoiesis (Perkins et al., 1995). Erythropoietin (EPO) and its receptor (EPO-R) are also essential for RBC production and deliver key signals for erythroid cell survival (Richmond et al., 2005; Spivak, 2005). EPO activates multiple signaling pathways and induces the anti-apoptosis factor Bcl-XL (Richmond et al., 2005; Socolovsky et al., 1999). Both Epo and Epo-R knockout mice are embryonic lethal due to a lack of definitive erythropoiesis (Wu et al., 1995).
LRF (encoded by the ZBTB7a gene, formerly described as POKEMON (Davies et al., 1999; Maeda et al., 2005), FBI-1 (Pessler et al., 1997) and OCZF (Kukita et al., 1999) is a transcription factor, which belongs to the POK (POZ/BTB and Krüppel) protein family. POK proteins bind DNA through C-terminal Krüppel-type zinc fingers and recruit co-repressor complexes through the N-terminal POZ/BTB domain (Stogios et al., 2005). POK proteins act as transcriptional repressors and play important roles in cellular differentiation and oncogenesis (Costoya et al., 2004; He et al., 2005). We previously reported that Lrf can act as a proto-oncogene through repression of Arf: specifically, it can trigger lymphomagenesis in the mouse and it is markedly over-expressed in non-Hodgkin’s lymphoma cells (Maeda et al., 2005). We have also shown that LRF critically regulates B versus T lymphocyte fate decisions by opposing the Notch signaling pathway. Conditional loss of Lrf in adult HSCs results in the aberrant development in the BM of CD4/CD8 double positive T cells, at the expense of B cell development (Maeda et al., 2007). Here we show that Lrf exerts an essential role for the biology of erythroid precursors. We demonstrate that Lrf acts downstream of Gata1 by suppressing Bim-mediated apoptosis, thus ensuring continued production of RBCs.
RESULTS
Lrf is required for definitive fetal erythropoiesis
Zbtb7a+/− mutants (Maeda et al., 2005) were seemingly healthy and fertile, but inter-crossing did not produce any live Zbtb7a−/− progeny. When we analyzed embryos from timed pregnancies of Zbtb7a+/− intercrosses, we found that until 12.5 days post coitum (d.p.c.) the null embryos were present at normal Mendelian ratio and most of them had no gross morphological abnormalities (not shown). However, by 14.5 d.p.c the peripheral blood (PB) from Zbtb7a−/− embryos revealed large numbers of erythroblasts at the polychromatophilic to orthochromic stage, as though their further maturation was blocked: indeed, there were very few mature red cells (Fig. 1C, top). All Zbtb7a−/− embryos died between 15.5 and 16.5 d.p.c. This was likely due to anemia as indicated by severe pallor and a marked reduction in the hematocrit (Fig. 1A and 1B). Moreover, touch preparations of 14.5 d.p.c fetal liver (FL) demonstrated a lack of enucleated erythrocytes and a preponderance of immature precursors (Fig. 1C, bottom). Since the 14.5 d.p.c. FL cell compartment consists mainly of erythroblasts, we obtained from each genotype total FL cell counts; these were significantly reduced in Zbtb7a−/− embryos (Fig. 1D). Furthermore, differential counts demonstrated in Zbtb7a−/− a severe decrease in polychromatophilic and orthochromic erythroblasts, as well as in erythrocytes (Fig. 1E). Stages of erythroid development can be characterized by Flow cytometric analysis (FACS) on the basis of expression of Ter119 and CD71. As shown in Supplementary Fig. 1, the CD71+Ter119− population (R1) consists mainly of proerythroblasts and early basophilic erythroblasts, while the CD71+Ter119+ population (R2) is comprised of basophilic erythroblasts and early polychromatophilic erythroblasts. The CD71−Ter119+ population (R3) is a more heterogeneous population and consists of poly/orthochromatophilic erythroblasts and enucleated erythrocytes. FACS analysis revealed a severe defect in terminal erythroid differentiation in Zbtb7a−/− FLs (Fig. 1F). Proportions of R2 and R3 were significantly decreased in Zbtb7a−/− FLs, while R1 was barely affected (Fig. 1F). This observation was more evident in later embryonic days (14.5 d.p.c), in which exponential expansion of mature erythroblasts occurs (Fig. 1F). In addition, purified Zbtb7a−/− erythroblasts from each stage, particularly the later ones, exhibited a less condensed chromatin pattern compared to wild type embryos (Supplementary Fig. 1). Therefore, loss of Lrf causes lethal embryonic anemia with impaired erythroid maturation. This severe anemia is not due to lack of erythroid progenitors, as shown by colony-assays from 13.5 d.p.c. FL (Fig. 1G). However, whereas Zbtb7a+/+ colonies were largely comprised of mature erythrocytes, CFU-E colonies from Zbtb7a−/− mutants did not contain mature enucleated cells (Fig. 1G right). In order to verify the cell intrinsic nature of this phenotype, we introduced an Lrf retroviral vector in Zbtb7a−/− early erythroblasts, followed by differentiation in vitro. As shown in Fig. 1H, Lrf almost completely rescued the differentiation defect in Zbtb7a−/− cells. Only Lrf infected erythroblasts (GFP positive), but not GFP negative cells, successfully differentiated into Ter119 positive small mature erythroblasts in vitro.
Figure 1.
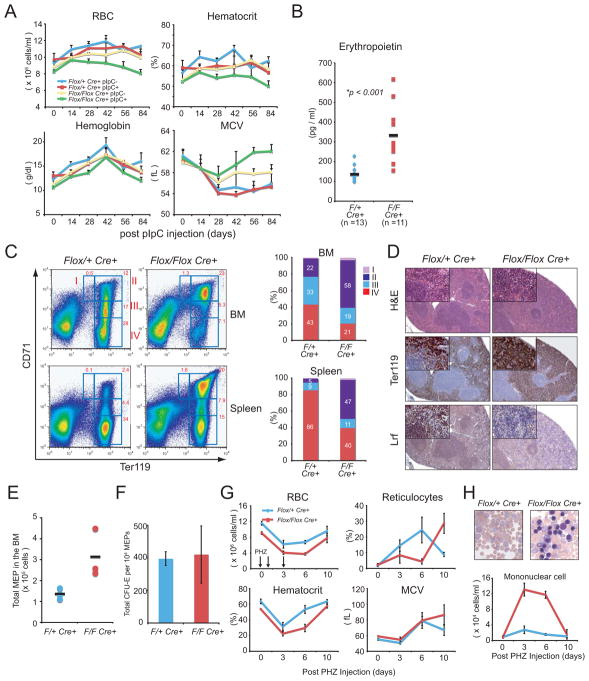
Loss of Lrf results in embryonic lethality due to severe anemia. (A) Picture of Zbtb7a+/+ and Zbtb7a−/− 15.5 d.p.c embryos. (B) Hematocrit of Zbtb7a+/+, Zbtb7a+/− and Zbtb7a−/− littermate embryos at 15.5 d.p.c. (C) PB and FL touch preparation from Zbtb7a+/+ and Zbtb7a−/− littermate embryos at 14.5 d.p.c. (D) Total cell numbers per FL were counted for each genotype at different embryonic days. (E) Differential counts on FL cells from Zbtb7a+/+ and Zbtb7−/− littermate embryos. (F) Representative FACS profiles of 12.5 d.p.c and 14.5 d.p.c. FLs from Zbtb7a+/+ and Zbtb7a−/− embryos. Bar graph represents proportions of each population in 12.5 d.p.c. FLs. (G) Colony counts of in vitro differentiated FL precursors derived from 13.5 d.p.c littermate embryos (left). Pictures demonstrate cytospin preparations of CFU-E colonies from Zbtb7a+/+ (top) and Zbtb7a−/− FLs (bottom). (H) Lrf add-back into Zbtb7a−/− erythroblasts rescued impaired differentiation phenotype in vitro. Both Zbtb7a+/+ and Zbtb7a−/− immature erythroblasts (Ter119−Gr1−B220−) were isolated from 12.5 d.p.c FLs and infected with either MSCV-Puro-IRES-GFP (PIG) empty vector or PIG-Lrf vector (Maeda et al., 2005). We then induced erythroid differentiation in vitro and subsequently analyzed for Ter119 expression and cell size (Forward scatter: FSC) by FACS. All error bars indicate s.d.
Conditional ablation of Lrf in the adult causes an EPO-unresponsive macrocytic anemia
To further elucidate the role of LRF in erythropoiesis, we next produced Lrf conditional knockout mice (Maeda et al., 2007) and made use Mx1-Cre transgenic mice, in which Cre recombinase can be induced specifically in HSCs by the administration of polyinosinic-polycytidylic acid (pIpC) (Kuhn et al., 1995). In Zbtb7aflox/floxMx1Cre+ mice, RBC and hematocrit gradually decreased after pIpC administration, whereas their MCV increased, suggesting that erythropoiesis was abnormal not only quantitatively but also qualitatively (Fig. 2A). From measurement of serum EPO levels one month after pIpC injection we found that EPO levels in pIpC-treated Zbtb7aflox/floxMx1Cre+ mice were in fact high (Fig. 2B), reflecting an appropriate physiological response to the anemia caused by loss of Lrf. FACS analysis in BM and spleen demonstrated that mature erythroblasts and erythrocytes (R IV) (Socolovsky et al., 2001) were dramatically decreased, while the early erythroblast compartment (R II) was expanded in Lrf conditional knockout mice (Fig. 2C); thus these mice displayed an erythroid defect specifically at the transition from R II to III/IV, which corresponds to basophilic erythroblasts and poly/orthochromatic erythroblasts, respectively, consistent with our findings in Lrf-knockout embryos. Splenomegaly in pIpC-treated Zbtb7aflox/floxMx1Cre+ mice is most likely due to a compensatory expansion of the early erythroid compartment which is negative for Lrf (Fig. 3D and Supplementary Fig. 2A). In the BM of these mice absolute numbers of MEPs (megakaryo-erythrocytic progenitors) were increased (Fig. 2E) and they gave rise to comparable numbers of CFU-E colonies as the controls (Fig. 2F), suggesting that loss of Lrf does not affect erythroid lineage commitment.
Figure 2.
Ablation of Lrf in the hematopoietic compartment of adult mice results in macrocytic anemia and ineffective erythropoiesis. (A) We examined 4 different groups of mice according to genotype and treatment; Zbtb7aFlox/+Mx1cre+ PBS-treated (blue), Zbtb7aFlox/+Mx1cre+ pIpC-treated (red), Zbtb7aFlox/FloxMx1cre+ PBS-treated (yellow) and Zbtb7aFlox/FloxMx1cre+ pIpC-treated (green). RBC counts, Hematocrit, Hemoglobin and MCV in PB were measured by a hematology analyzer. The average of 5 animals was plotted at each time point with error bars. (B) Serum EPO level was measured one month after pIpC injection. (C) Representative FACS profiles for Ter119 and CD71 expression in BM and Spleen one month after pIpC administration (left). Bar graphs represent percent proportion of each population in BM and Spleen (right). (D) Representative H&E staining of spleen sections (top), demonstrating expansion of red pulp formation in the spleen of pIpC-treated Zbtb7aFlox/FloxMx1cre+ mice (middle panels for Ter119 and bottom panels for Lrf staining). (E) Absolute numbers of megakaryocyte/erythroid progenitors (MEPs) in the BM one month after pIpC administration. Black bars resent the average cell counts of three mice. (F) Erythroid colony forming capacity per 10,000 flow-sorted MEPs was assessed in three independent mice for each genotype. (G) RBC counts, Hematocrit, and MCV were measured in the PB on indicated days after PHZ treatment. Reticulocytes were counted on PB smear slides upon new methylene blue staining. (H) Robust increase of PBMNCs in PHZ-treated, pIpC pre-treated, Zbtb7aFlox/FloxMx1cre+ mice (bottom). Pictures demonstrate Wright-Giemsa staining of PB on day 3. All error bars indicate s.d.
Figure 3.
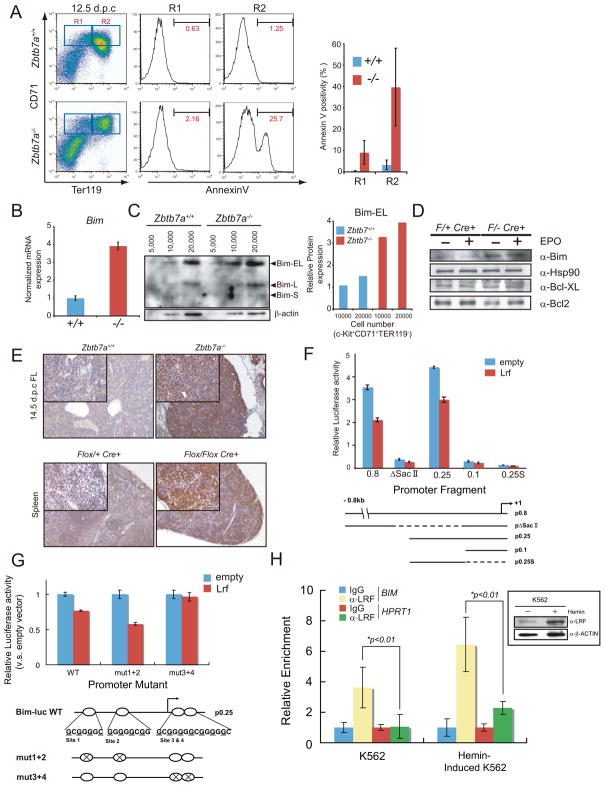
Loss of Lrf leads to increased apoptosis accompanied by high Bim expression. (A) Representative FACS profiles of the triple staining for Ter119, CD71 and Annexin V (left). Bar graph demonstrates the average of % Annexin V positivity in R1 and R2 cell population. (B) Bim mRNA was increased in Zbtb7a−/− FL erythroblasts. c-Kit+CD71+Ter119− erythroblasts were flow sorted and RNA was extracted. cDNA was subsequently synthesized after DNAse treatment and levels of Bim and Hprt1 transcripts were measured by q-RT-PCR. Bar graph represents normalized expression level of Bim mRNA with error bars. (C) Bim protein was up-regulated in Zbtb7a−/− FL erythroblasts. Different numbers of c-Kit+CD71+Ter119− erythroblasts (5,000, 10,000 and 20,000) were directly flow-sorted into protein sample buffer and Western blot was subsequently performed using anti-Bim antibody (left). Bar graph represents normalized Bim protein level to the corresponding β-actin protein levels in c-Kit+CD71+Ter119− erythroblasts from 12.5 d.p.c FLs (right). (D) High Bim protein expression in pIpC-treated Zbtb7aFlox/−Mx1cre+ CD71+ splenic erythroblasts. CD71+splenic erythroblasts were harvested from PHZ treated, pIpC-pretreated, Zbtb7aFlox/+Mx1cre and Zbtb7aFlox/−Mx1cre+ mice. Erythroblasts were then serum starved and subsequently stimulated with EPO. Cells were harvested 10 min after EPO administration and subsequently utilized for experiments. (E) IHC analysis for Bim in FL and Spleen. FLs were isolated from littermate embryos at 14.5 d.p.c. Spleens were collected one month after pIpC treatment. Both low power (x100 magnification) and high power (insets, x400 magnification) are shown. (F) Transrepression assays in 293 cells transfected with various luciferase reporter constructs of the murine proximal Bim promoter. (G) Identification of an essential LRF-binding site in the murine Bim promoter. The putative LRF-binding sites in the Bim promoter were mutagenized and subsequently used for reporter assays. Schematic representations of mutated promoter and expression control for the reporter assay are shown (bottom). Underlined bases of the LRF-binding sites were mutated to Adenine. Luciferase reporter plasmids were transfected into 293 cells. Luciferase activity was measured 24 hours after transfection using the Dual-Luciferase Reporter Assay system. Lrf did not repress the promoter activity of mut3+4 reporter. (H) Quantitive ChIP assay of the BIM promoter both in K562 cells and in induced K562 cells by Hemin. Primer sets were designated to amplify the proximal human BIM promoter and primers designated to the 5′ promoter of HPRT1 were used to detect non specific interactions. The chromatin was sonicated to 200–600bp. A Western blot demonstrates that LRF is upregulated in induced K562 cells by Hemin. All error bars indicate s.d.
Anemia triggered by conditional inactivation of Lrf was even more evident in a transplant model (Supplementary Fig. 2B). As expected in the Zbtb7a−/− FL, donor-derived HSCs were intact and donor-derived MEPs were expanded upon Lrf inactivation (not shown). However, recipients with BM reconstituted from Zbtb7aFlox/−Mx1cre+ cells, developed severe macrocytic anemia upon pIpC administration (Supplementary Fig. 2C). Taken together, these data support the notion that the anemia in Lrf conditional knockout mice is caused by a cell intrinsic mechanism.
We next examined how Lrf conditional knockout mice would respond to a severe hemolytic stress. Upon treatment with the potent hemolytic agent phenylhydrazine (PHZ: see Socolovsky et al., 2001), Lrf-ablated mice (Supplementary Fig. 2D and E) developed more severe anemia than control mice; also, they had a delayed reticulocyte response (Fig, 2G) and a striking increase of PB mononuclear cells (MNCs), which consisted almost exclusively of nucleated erythroblasts (Fig. 2H). We conclude that upon hemolytic stress Lrf conditional knockout mice were able to mobilize immature nucleated erythroblasts from BM and spleen into the peripheral blood, but they suffered even more anemia than control mice.
Loss of Lrf triggers Arf/p53 independent apoptosis during late stages of erythroid development
To further elucidate the mechanism by which loss of Lrf causes ineffective erythropoiesis, we first analyzed the cell cycle in Zbtb7a−/− FLs. As shown in Supplementary Fig. 2F, we found in FLs of these mice a relative increase of cells in G1 and a concomitant decrease of cells in S and G2/M. Next we looked for evidence of apoptosis and found a dramatic increase of Annexin V positive apoptotic cells (Fig. 3A); this was so pronounced, particularly in the later stage of erythroid differentiation (R2), that we suggest it could account for the lethal anemia of Zbtb7a−/− embryos.
Since loss of the tumor suppressor Arf reverted the senescence phenotype in Zbtb7a−/− MEFs (mouse embryonic fibroblasts) (Maeda et al., 2005) and p19Arf mRNA was only detectable in Zbtb7a−/− CD71+Ter119− erythroblasts by RT-PCR (not shown), we next set out to examine whether loss of Arf could bypass apoptosis and rescue the anemia phenotype in Zbtb7a−/− embryos. We intercrossed Zbtb7a+/−p19Arf+/− double heterozygous mice and analyzed FL erythropoiesis in 15.5 d.p.c. embryos. As shown in Supplementary Fig. 3A, Zbtb7a−/−p19Arf−/− embryos still demonstrated a block of differentiation at the R2 stage and total numbers of Zbtb7a−/−p19Arf−/− FL cells were comparable to Zbtb7a−/−p19Arf+/+ embryo (Supplementary Fig. 3B).
The tumor suppressor p53 inhibits cellular proliferation by inducing cell cycle arrest and apoptosis in response to cellular stresses such as DNA damage and hypoxia (Fridman and Lowe, 2003). Furthermore, we found that p53 itself is up-regulated in Zbtb7a−/− fetal liver erythroblasts (see below). We therefore tested whether genetic inactivation of p53 could rescue the anemia phenotype of Zbtb7a−/− mice. As shown in Supplementary Fig. 3C, Zbtb7a−/−p53−/− FLs still displayed a block of differentiation. These data suggest that the massive apoptosis at late stages of erythroid differentiation caused by loss of Lrf occurs in a p19Arf/p53 independent fashion.
Intact EPO signaling cascade in Lrf deficient erythroblasts
EPO plays a key role in terminal erythroid differentiation by preventing apoptosis in mature erythroblasts (Spivak, 2005). To investigate whether the loss of Lrf impairs EPO signaling, we examined activation status of signaling molecules upon EPO stimulation, such as STAT5, ERK/MAPK, PI3K/AKT and Bcl-XL (Richmond et al., 2005, Socolovsky et al., 1999). Mice were treated with PHZ and CD71+ splenic erythroblasts were harvested using magnetic beads. Cells were serum-starved for 2 hours and then stimulated with EPO. We next assessed the phosphorylation status of Stat5 by western blot and FACS. As shown in Supplementary Fig. 4A and 4B, activation of Stat5, ERK/MAPK and PI3K/AKT was induced normally upon EPO stimulation in pIpC-treated Zbtb7aflox/−Mx1Cre+ erythroblasts, indicating that upstream EPO signaling is intact in the absence of Lrf.
Furthermore, Bcl-XL was abundantly expressed in pIpC-treated Zbtb7aflox/−Mx1Cre+ erythroblasts (Supplementary Fig. 4C). Of note, the anti-apoptotic protein Mcl1 was comparably expressed as well, while Bcl-2 protein was slightly reduced in Zbtb7aflox/−Mx1Cre+ erythroblasts. Thus, erythroid apoptosis observed in Lrf conditional knockout mice is not due to an impaired EPO/EPO-R signaling pathway.
Up-regulation of the pro-apoptotic factor Bim in the absence of Lrf
In order to understand how loss of Lrf results in erythroblast apoptosis we studied mRNA expression profiles in the R1 population of flow-sorted CD71+Ter119− FL erythroblasts (Supplementary Fig. 5A): this population was selected for analysis because it consists of cells at a differentiation stage that did not exhibit overt cellular degeneration. A total of 189 probe sets were up or down-regulated more than 1.5 fold in Zbtb7a−/− R1 cells as compared to Zbtb7a+/+ and Zbtb7a+/− cells. From the list of genes thus compiled (see Supplementary Fig. 6), we selected genes that have been implicated in cellular apoptosis, differentiation, proliferation and the cell cycle (Supplementary Fig. 5A). Many genes known to be involved in definitive erythropoiesis (Godin and Cumano, 2002) were expressed normally in Zbtb7a−/− R1 cells (Supplementary Fig. 5B). Interestingly, the BH3 only protein Bim (O’Connor et al., 1998) (encoded by Bcl2l11 gene), a major apoptosis inducer in hematopoietic cells (Strasser et al., 1995), was found to be markedly up-regulated with two independent probe sets in Zbtb7a−/− R1 cells (Supplementary Fig. 5A). Since apoptosis of Zbtb7a−/− FL erythroblasts was not reverted by Arf/p53 loss and Bim is known to function in an Arf/p53-independent apoptosis pathway (Egle et al., 2004; Hemann et al., 2005), we investigated further the role of Bim in the defective erythropoiesis observed in the absence of Lrf. We found high Bim mRNA expression and an approximately three-fold up-regulation of Bim protein in flow-sorted c-Kit+CD71+Ter119− Zbtb7a−/− erythroblasts (Fig. 3B and Fig. 3C). Furthermore, Bim-EL protein levels were significantly elevated in CD71+ pIpC-treated Zbtb7aflox/−Mx1Cre+ splenic erythroblasts irrespective of EPO signals, while Bcl-XL protein level was similar to that of control cells (Fig. 3D). Markedly elevated Bim expression was also observed by Immunohistochemistry in both Zbtb7a−/− 14.5 FL and pIpC-treated Zbtb7aflox/floxMx1Cre+ spleens (Fig. 3E), indicating that Bim up-regulation is a characteristic feature of Lrf loss both in the embryo and in the adult.
Bim is a direct transcriptional target of Lrf
Up-regulation of Bim mRNA in Lrf deficient erythroblasts was due to an increase in transcription of the Bim gene rather than enhanced Bim mRNA stability, as reduction of Lrf expression via siRNA-mediated knockdown in erythroblasts did not affect Bim mRNA stability during erythroid differentiation (Supplementary Fig. 7A). In multiple cell types, Bim is regulated by the transcription factor Foxo3a downstream the PI3K/Akt pathway (Dijkers et al., 2000; Stahl et al., 2002). We therefore examined whether Bim upregulation in Zbtb7a−/− erythroblasts might be accompanied by an accumulation of Foxa3a protein in the nucleus. As shown in Supplementary Fig 7B and C, no significant increase of nuclear Foxo3a protein was observed by IHC in either Zbtb7a−/− 15.5 FL or pIpC-treated Zbtb7aflox/−Mx1Cre+ spleens, indicating that Bim upregulation in Zbtb7a−/− erythroblasts does not depend on Foxo3a.
Since we knew that LRF can act as a transcriptional repressor (Maeda et al., 2005), we next tested whether Lrf directly represses Bim transcription. To this end, we generated a set of luciferase reporter constructs containing regions of the proximal Bim promoter, and we assessed the ability of Lrf to repress reporter activity. As shown in Fig. 3F, Lrf efficiently repressed activity of an 800 bp region of the Bim proximal promoter. Furthermore, by generating a series of deletions and truncations of this 800bp fragment, we were able to identify a 250 bp region upstream the Bim transcription start site that had promoter activity and was still sensitive to Lrf-mediated repression (Fig. 3F). Within this 250 bp proximal promoter region we identified 4 potential LRF binding sites (Maeda et al., 2005) (Fig. 3G). Although mutation of any single LRF binding site alone did not impair Lrf-mediated repression (data not shown), mutation of two adjacent sites close to the transcription start site abrogated Lrf-mediated repression (Fig. 3G). Further, Lrf was found to directly bind to oligonucleotides that contain these putative LRF binding sites in the Bim gene as demonstrated by electrophoretic mobility shift assays. In addition, Lrf antibodies were found to super-shift the Lrf-oligonucleotide complex (Supplementary Fig. 8D and E). These data support a model whereby Lrf directly represses Bim expression through a tandem-binding site in the Bim proximal promoter region.
Chromatin immunoprecipitation experiments (ChIP) further demonstrated a direct binding of LRF on the Bim promoter. We designed two primer sets; one to amplify the proximal human BIM promoter and another to amplify the 5′ promoter of HPRT1 as a negative control. LRF antibody specifically precipitated the proximal BIM promoter sequences from chromatin preparations of K562 cells by hemin treatment (Rutherford et al, 1979). Under these conditions LRF is up-regulated, and an enhanced enrichment of LRF at the BIM promoter was detected (Fig. 3H). We conclude that LRF directly binds to the BIM promoter and inhibits its expression during erythroid differentiation.
Bim deficiency rescues the anemia observed in Lrf knockout mice
In order to define more clearly the functional role of the Lrf-Bim interaction in suppressing apoptosis in erythroblasts, mice heterozygous for both Lrf and Bim genes (Zbtb7a+/−Bim+/−) were intercrossed to obtain Lrf/Bim double knockout embryos (Zbtb7a−/−Bim−/−). As shown in Fig. 4A, while Zbtb7a−/−Bim+/+ 15.5 d.p.c embryos were overtly pale, Zbtb7a−/−Bim−/− embryos appeared grossly normal. Bim deficiency significantly increased total numbers of FL (Fig. 4B) and mature erythroblast (R3) cells in 12.5 d.p.c and 14.5 d.p.c. embryos (Fig. 4C and D, and Supplementary Fig. 8A). In the double knockout mice Annexin V positive apoptotic cells were nearly absent (Fig. 4E and F, and Supplementary Fig. 8B and C), providing proof that removal of Bim is sufficient to abrogate erythroblast apoptosis. Of note, Zbtb7a−/− embryos heterozygous for Bim (Zbtb7a−/−Bim+/−) displayed an intermediate phenotype (Figs. 4B-F). Thus, the rescue from fetal anemia of Lrf knockout mice through loss of Bim is gene-dosage dependent.
Figure 4.
Loss of Bim restores erythropoiesis in Zbtb7a−/− FL. (A) Picture of Zbtb7a+/+Bim+/+, Zbtb7a+/+Bim+/− and Zbtb7a+/+Bim+/− 15.5 d.p.c. embryos. (B) Total cell numbers per fetal liver (FL) were counted for each genotype at different embryonic days. Three litters were isolated for each time point and average numbers are presented with standard deviation. (C) Representative FACS profiles of 14.5 d.p.c. FLs from Zbtb7a+/+Bim+/+, Zbtb7a+/+Bim+/− and Zbtb7a+/+Bim+/− 14.5 d.p.c. embryos (D) Bar graph represents proportions of each population in 14.5 d.p.c. FLs. Three embryos were obtained and analyzed for each genotype. (E) Representative FACS profiles of the staining for Annexin V in R2 cell population. (F) Bar graph demonstrates the average of % Annexin V positivity in R1 and R2 cell population. Three FLs were obtained for each genotype. All error bars indicate s.d.
These data prompted us to analyze the effect of Bim loss on lethality seen in Zbtb7a−/− embryos. To this end, Zbtb7a+/−Bim+/− mice were intercrossed, and their progenies were analyzed at different embryonic days. Zbtb7a−/− embryos were alive at 14.5 d.p.c. irrespective of Bim genotype, but none of the Zbtb7a−/−Bim+/+ nor Zbtb7a−/−Bim+/− progenies were delivered (Supplementary Table 1). However, three Zbtb7a−/−Bim−/− pups out of 35 offspring analyzed were identified on the day of birth (P0), indicating that complete loss of the Bim gene prolonged the survival of Zbtb7a−/− embryos (Supplementary Table 1). These three Zbtb7a−/−Bim−/− mice were nevertheless paler than controls at birth and subsequently died postnatally.
Overall, Bim loss was sufficient to restore nearly normal erythropoiesis in Zbtb7a−/− FL up to 14.5 d.p.c. and partially rescued lethality in Zbtb7a−/− embryos. Our genetic analysis thus indicates that excessive Bim expression is a primary cause of defective erythropoiesis in Zbtb7a−/− FLs.
Lrf is a direct transcriptional target of Gata1
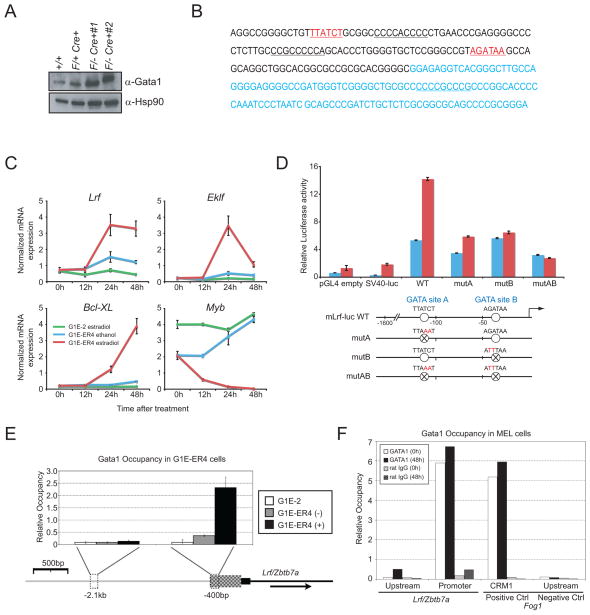
The Gata1 transcription factor plays a central role in erythroid gene expression (Cantor and Orkin, 2002; Fujiwara et al., 1996) and protects erythroblasts from apoptosis (Weiss and Orkin, 1995, Gregory et al., 1999). We next examined Gata1 expression in pIpC-treated Zbtb7aflox/−Mx1Cre+ CD71+ erythroblasts. As shown in Fig. 5A, Gata1 protein is abundantly expressed in pIpC-treated Zbtb7aflox/−Mx1Cre+ CD71+ erythroblasts.
Figure 5.
Lrf is a Gata1 downstream target. (A) Western blot for Gata1 in Zbtb7a+/+, Zbtb7aFlox/+Mx1cre and Zbtb7aFlox/−Mx1cre+ CD71+ splenic erythroblasts. Gata1 was abundantly expressed in the absence of Lrf. (B) Sequence of 450 bases of the predicted promoter region of Zbtb7a gene. Consensus EKLF CACCC boxes are underlined; consensus GATA1 binding motifs are depicted in red and underlined; exon 1 sequence is shown in blue. (C) G1E-ER4 is a Gata1-ablated erythroblast line expressing an estradiol-inducible form of GATA1. G1E-ER4 cells and its parental cell line G1E-2 were stimulated with 10−7 M estradiol or ethanol vehicle and RNA was extracted at the indicated time after stimulation. mRNA levels of Lrf, Eklf, Bcl-XL and Myb were measured by q-RT-PCR and normalized to the corresponding Hprt1 mRNA levels. (D) Two GATA elements in the Zbtb7a gene act cooperatively to enhance promoter activity in erythroid cells. (E) Quantitative ChIP assay at the Lrf promoter in Gata1-null parental (G1E-2), uninduced (−), and induced (+) G1E-ER4 cells using anti-Gata1 antibody. The Y-axis shows Gata1 occupancy, relative to a standard curve of the relevant input sample. Below, diagram of the 5′ end of the Zbtb7a gene. Thin black line indicates intron 1; black box represents exon 1; checkered box represents the predicted promoter region, which contains 2 consensus GATA1 binding sites and 3 consensus EKLF CACCC boxes; thin grey line represents the upstream intergenic region. Dashed boxes show regions amplified by quantitative real-time PCR for ChIP. Arrow indicates direction of transcription. (F) Quantitative ChIP assay using anti-GATA1 antibody in MEL cells chemically induced to differentiate for either 0 hours or 48 hours. Data from immunoprecipitations performed with normal rat IgG are shown as controls. All error bars indicate s.d.
LRF was recently identified as a EKLF-induced gene both in fetal liver cells and erythroid cell lines (Hodge et al., 2006). Since GATA1 is a transcriptional activator of EKLF (Crossley et al., 1994), and EKLF is indispensable for fetal erythropoiesis (Hodge et al., 2006; Perkins et al., 1995), we speculated that LRF is a key downstream target of the GATA1/EKLF transcriptional cascade enabling survival of differentiating erythroblasts. Indeed, while examining the promoter regions of the mouse Zbtb7a gene, we found two putative GATA binding sites and three EKLF consensus sequences (Fig. 5B). Of note, the two putative GATA binding sites are fully conserved in the promoter region of the human ZBTB7A gene (not shown). To test whether Gata1 can activate Lrf expression, we used the Gata1 null erythroid cell line G1E (Gregory et al., 1999). These cells proliferate as immature erythroblasts and undergo terminal maturation when Gata1 activity is restored. Upon activation of a drug-inducible form of Gata1, Lrf mRNA was up-regulated approximately 4 fold (Fig. 5C). We also observed up-regulation of Eklf and Bcl-XL and down-regulation of Myb, as previously reported (Fig. 5C) (Gregory et al., 1999; Welch et al., 2004).
We next examined whether Gata1 up-regulates Lrf transcription via two putative GATA binding sites within the Lrf promoter. Dual-Luciferase Reporter Assay experiments demonstrated that two Gata elements in the Zbtb7a gene act cooperatively to enhance promoter activity in murine erythroleukemia (MEL) cells (Fig. 5D). To further elucidate the mechanism by which Gata1 induces Lrf, we performed ChIP experiments. While we could not detect Eklf binding to the Zbtb7a promoter (Supplementary Fig. 9A and B), anti-Gata1 antibody specifically precipitated the ~400bp proximal Zbtb7a promoter sequences, which contains the putative Gata1 binding sites, from extracts prepared from the induced G1E2-ER4 cells, but not from the parental cell line G1E2 (Rylski et al., 2003) (Fig. 5E). As a further control, we could also efficiently enrich the Fog1 promoter sequence using anti-Gata1 antibody (Welch et al., 2004) (Supplementary Fig. 9C). We obtained similar results in ChIP assays using MEL cells, an independent experimental system for in vitro erythroid differentiation (Fig. 5F). These data indicate that Gata1 directly binds to the Zbtb7a proximal promoter in vivo and activates Lrf expression.
We next asked whether Lrf-overexpression restores terminal erythroid differentiation in the absence of Gata1 using G1E-ER4 cells. We performed all experiments in the presence of EPO, since G1E-ER4 cells underwent apoptosis in the absence of EPO irrespective of Lrf levels (not shown). Retroviral overexpression of Lrf in G1E-ER4 cells led to an increase of Ter119 positive erythrocytes (Fig. 6A) accompanied by a reduction in Bim mRNA levels (Supplementary Fig. 10A). Lrf-overexpressing G1E-ER4 cells formed hemoglobinized erythroid colonies without Gata1 induction when seeded in methylcelluose plates (Supplementary Fig. 10B). Conversely, siRNA-mediated Lrf knockdown resulted in increase of apoptosis revealed by morphological examination (Supplementary Fig. 10C) and Annexin V staining (Supplementary Fig. 10D). Successful Lrf knockdown in siRNA-treated cells was confirmed by western blot (Supplementary Fig. 10E). Furthermore, as expected, we observed a significant increase of apoptotic erythroblasts when Bim was ectopically expressed in differentiating G1E-ER4 cells (Supplementary Fig. 10F and G). Of note, the pro-apoptotic effects of Lrf knock down as well as Bim overexpression became evident once differentiation was initiated by Gata1 induction. This suggests that Bim repression by Lrf is chiefly required in late stages of erythroid differentiation, which is in complete agreement with our findings in Lrf knockout mice.
Figure 6.
Lrf overexpression restores erythroid differentiation in Gata1 deficient erythroblasts. (A) Proportions of Ter119 positive G1E-ER4 cells upon Lrf overexpression without estradiol treatment (left). Representative FACS profiles for Ter119 expression is demonstrated with isotype control (middle). Pictures demonstrate cytospin preparations of empty vector-or Lrf-expressing G1E-ER4 cells (right). (B) Gata1 WT and null ES cells expressing either empty vector or Lrf were induced erythroid differentiation on OP9 stromal layers. Floating hematopoietic cells were harvested and analyzed. Representative FACS profiles of the staining for Annexin V in Ter119 positive cell population (left). Bar graph demonstrates the average of % Annexin V positivity in Ter119 positive cell fraction (right). (C) Sequential analysis for the numbers of floating blood cells derived from Gata1 null ES cells expressing either empty vector (blue) or Lrf (red). (D) May-Giemsa staining of floating erythrocytes (x400 magnification). (E) Proposed model of the role for LRF in terminal erythroid differentiation. We propose that inhibition of apoptosis during erythroid terminal differentiation is dependent on both the EPO/BCL-XL and the LRF/BIM pathways which are activated in response to the transcription factor GATA1. Loss of the tumor suppressor Arf reverted the senescence phenotype in Zbtb7a−/− MEFs (Maeda et al., 2005). All error bars indicate s.d.
To confirm our findings in primary cells, we next took advantage of an OP9/mouse embryonic stem cell (ESC) co-culture system. We overexpressed Lrf in Gata1 null ES cells and examined erythroid differentiation (Weiss et al., 1994). In this system, ES cells give rise to a nonadherent hematopoietic cell population that includes erythrocytes (Umeda et al., 2004, Umeda et al., 2006). Nonadherent cells were harvested serially and Annexin V positivity within the Ter119 positive fraction was analyzed. Lrf overexpression significantly augmented their survival (Fig. 6B) and increased the production of nonadherent cells, which consisted mainly of Ter119 erythroblasts (Fig. 6C and D, and Supplementary Fig. 10H). Thus, overexpression of Lrf partially overrides erythroblast apoptosis and maturation arrest caused by lack of Gata1.
Discussion
The transcription factor LRF has previously been identified as a regulator of the important tumor suppressor ARF, and cells lacking Lrf have proved refractory to malignant transformation (Maeda et al., 2005). LRF also plays an essential role in the B versus T lymphoid cell-fate decision (Maeda et al., 2007): thus, the gene encoding Lrf, Zbtb7a, belongs to the select group of genes that have a crucial role in both oncogenesis and development. In this work we have shown that the latter role includes a function that is essential for erythropoiesis; and we have been able to define the mechanism whereby this function is fulfilled.
First, we have established that LRF plays a key role in fetal and adult erythropoiesis. Zbtb7a−/− embryos die of severe anemia by 16.5 d.p.c; and in adult mice conditional inactivation of Lrf in HSCs results in an EPO-unresponsive macrocytic anemia due to a cell intrinsic defect. Through targeted inactivation a number of other genes have been previously identified as key factors in definitive erythropoieisis (Godin and Cumano, 2002). Upon EPO-R activation, STAT5 is phosphorylated, translocates to the nucleus and induces transcriptional activation of target genes. Inactivation of both isoforms of Stat5 in mice (Stat5a−/−Stat5b−/−) results in inefficient erythropoiesis in both the fetus and the adult organism (Socolovsky et al., 2001). We find similar phenotypic features in Lrf conditional knockout mice, despite the fact that Lrf null erythroblasts have an intact EPO-STAT5 signaling pathway. Indeed, Bcl-XL, a downstream target of Stat5, is normally induced upon EPO stimulation in the absence of Lrf (Supplementary Fig. 4C); and the MAPK and PI3K pathways, which are also downstream of the EPO receptor, are activated normally in Lrf null erythroblasts (Supplementary Fig. 4B). Thus, the role of LRF in terminal erythroid differentiation does not appear to be related to the EPO signaling cascade.
We have shown previously that LRF blocks the Notch pathway at the level of HSC/progenitor cells to regulate B versus T lineage fate decision (Maeda et al., 2007). However, the defect in erythroid differentiation in Lrf knockout mice was not rescued by inhibition of Notch activity by treatment in vivo with a Gamma Secretase Inhibitor (GSI) (Supplementary Fig. 11). Furthermore, a Notch activation signature is not present in Lrf knockout MEP as compared to the HSC/CLPs (Common Lymphoid Progenitors) (Maeda et al., 2007). Thus, failure of erythropoiesis upon loss of Lrf cannot be accounted for by aberrant Notch activation.
Our next key finding was that Lrf loss causes apoptosis of erythroblasts, mostly at the transition from the basophilic to the polychromatophilic stage. The ARF/p53 pathway is critical in the induction of apoptosis, cell cycle arrest and cellular senescence in response to oncogenic stress (Lowe and Sherr, 2003). High p53 expression induces apoptosis in fetal liver erythroblasts and erythroid-specific inactivation of the mouse Mdm2 gene, a key negative regulator of p53, leads to increased apoptosis in erythroblasts and embryonic lethality due to a severe anemia (Mdm2lox/loxEpoRcre+) (Maetens et al., 2007). However, neither Arf loss nor p53 loss could rescue the defects in Lrf null erythroblasts (Supplementary Fig. 3 A-C): in other words, apoptosis of Lrf null erythroblasts was Arf/p53 independent.
The mechanism whereby Lrf loss produces apoptosis was clarified by analyzing the gene expression profile of Lrf-null cells by microarray. We focused on the fact that in Lrf-null cells the pro-apoptotic factor Bim was markedly up-regulated at the level of both mRNA and protein (Fig. 3B-E). Reporter assays and ChIP experiments demonstrated that LRF physically binds to the BIM promoter and directly represses transcription of this gene (Fig. 3F-H). Probably the most convincing evidence that Lrf function is linked to Bim comes from our genetic analysis: if Bim is knocked out, the severe erythroid cell phenotype of Zbtb7a−/− mutants, including lethal anemia, is almost completely reversed (Fig. 4 and Supplementary Fig. 8). The rescue is gene-dosage dependent, since loss of even one Bim allele provides a significant survival advantage to Zbtb7a−/− erythroblasts (Fig. 4 and Supplementary Fig. 8). Our data indicate that “fine-tuning” of Bim levels via Lrf is key for erythroblast survival. BIM was already known to function in an ARF/p53 independent apoptosis pathway (Egle et al., 2004; Hemann et al., 2005). An important implication of our work is that both p53- and Bim-dependent apoptotic pathways must be suppressed for effective erythroid differentiation.
Finally, we have discovered, to our surprise that LRF is a novel direct transcriptional target of GATA1. Lrf mRNA expression was induced upon Gata1 re-expression in a Gata1 null erythroblast cell line (Fig. 5C), and by ChIP analysis Gata1 is found to directly bind to the Lrf promoter in vivo (Fig. 5E and F). Gata1 expression is barely detectable at HSC/progenitor stages and is up-regulated during erythroid lineage commitment, being expressed at the highest levels in CFU-E and in proerythroblasts (Suzuki et al., 2003). Gata1 null embryonic stem cells fail to produce mature erythroid cells in vivo in chimeric mice (Pevny et al., 1991) due to massive apoptosis of erythroblasts (Weiss et al., 1994), demonstrating Gata1’s essential role as a survival factor during this differentiation process. In this context, our study identifies LRF as a key downstream target of GATA-1. In agreement with this notion, forced Lrf over-expression in Gata1 null cells led to Bim downregulation and prolonged the survival of Gata1 null erythroblasts (Fig. 6 and Supplementary Fig. 10H), enabling erythroid cells to undergo terminal maturation at least to some extent.
The differentiation and maturation of erythroid cells is a highly regulated process: in its terminal stages, it exhibits features of apoptosis (Weiss and Orkin, 1995; Gregory et al., 1999; Socolovsky et al., 1999; Yoshida et al., 2005). If apoptosis is abnormally triggered at earlier stages of differentiation, the output of mature erythrocytes will be compromised: this underlies the phenomenon of intramedullary destruction of red cell precursors known as ineffective erythropoiesis, which causes anemia in inherited disorders such as the congenital dyserythropoietic anemias (Rivella et al., 2009; Wu et al., 2005). Here we have shown that control of the pro-apoptotic protein BIM is critical for normal erythropoiesis: and this control is exerted by LRF. If we consider hematopoiesis as a whole, we can speculate that while BIM is essential in lymphoid cells for preventing auto-immunity (Bouillet et al., 1999), it is essential that BIM activity be curbed in erythroid cells: this notion fits well with the important finding that production of LRF itself is activated by GATA1, a master gene of erythropoiesis. Finally, our findings also have important implications for the role of LRF in tumorigenesis as they identify yet another critical mechanism (the ability to repress BIM with the consequent survival advantage) whereby LRF can lend to tumor cells oncogenic potential.
EXPERIMENTAL PROCEDURES
Mice
Generation of conventional (Zbtb7a+/−) and conditional (Zbtb7aFlox/+) Lrf knockout mice was described elsewhere (Maeda et al., 2007). Mx1-Cre transgenic mice (Gu et al., 1994), Bim (Bcl2l11) knockout mice (Bouillet et al., 1999) and the congenic strain that carries the CD45.1 antigen (B6.SJL-PtprcaPepcb/BoyJ) were purchased from Jackson laboratory.
Analysis of embryonic PB and colony forming assay
PB was collected with a micropipette from the carotid arteries of embryos and smears were prepared for Wright-Giemsa staining. Hematocrit was determined using microhematocrit tubes (Microcaps, Drummond Scientific). FL colony forming assays in methylcellulose were performed as previously described (Wang et al., 1998). For the CFU-E colony assay in Lrf conditional knockout mice, 10,000 flow-sorted the MEPs were cultured in EPO-containing methylcellulose plates (MethoCult M3334, StemCell Technologies) for 2 days and colony numbers were subsequently counted.
FACS and cell purification
After blocking non-specific antibody binding by incubating with FcBlock (BD), FL and BM cells were incubated with fluorochrome-conjugated (or with biotin-conjugated) antibodies. FACS analysis was performed in FACScan, FACSCalibur or CyAn (DAKO), followed by analysis with FlowJo software (Tree Star). Cell sorting was performed on a MoFlo at MSKCC and BIDMC Flow Cytometry Core Facility using DAPI for live/dead discrimination. Antibodies for FACS and cell sorting were purchased from eBioscience unless otherwise indicated. FACS analysis for Phospho-Stat5 was performed according to the phospho-FACS protocol from lab of Dr. Kevin Shannon (http://www.ucsf.edu/kmslab/resources/resources.html) using AlexaFluor647 conjugated anti-PhosphoStat5 antibody (pY694, clone47, BD). For cell cycle analysis, DRAQ5 (Axxora) was added to a final concentration of 10 μM and cells were subsequently incubated for 10 min at room temperature. For Annexin V staining, FL cells were suspended with 1 x Annexin Biding Buffer (BD) containing 2% FBS, stained with FITC-anti-CD71, PE-anti-Ter119 antibodies and APC-Annexin V (BD) following the manufacturer’s protocol.
Conditional inactivation of Lrf and PB analysis in adult mice
Cre recombinase was induced at the HSC level as previously described (Gu et al., 1994). PB samples were collected from the retro-orbital sinus with heparinized capillary tubes (Fisher Scientific) under isoflurane anesthesia. RBC counts, Hematocrit, Hemoglobin and MCV were measured using an Adivia 120 hematology analyzer (Bayer). Serum erythropoietin level was measured using Mouse/Rat Erythropoietin Quantikine ELISA Kit (R&D systems).
PHZ treatment and collection of splenic erythroblasts
Phenylhydrazine (PHZ) was purchased from Sigma and injected subcutaneously three times at the dose of 40 mg/kg as previously described (Socolovsky et al., 2001). For CD71+ splenic erythroblast collection, the spleen was isolated at day 4 after PHZ stimulation and cells were then incubated with anti-CD71 biotin conjugated antibody (C2, BD), followed by incubation with anti-Biotin MicroBeads (Miltenyi Biotec). The cell suspension was subsequently applied onto MACS separation LS columns (Miltenyi Biotec) for positive selection.
Microarray analysis
One Zbtb7a+/+, one Zbtb7a+/− and two Zbtb7a−/− 12.5 d.p.c FLs were isolated from littermate embryos. RNA of CD71+Ter119− erythroblasts was extracted according to the manufacturer’s specifications. cDNA synthesis, cRNA labeling and hybridization onto GeneChip Mouse Genome 430A2.0 chips (Affymetrix) was performed as previously described (Costoya et al., 2004). Since Zbtb7a+/− mice do not demonstrate any gross defect, we divided samples into two groups. Data sets from one Zbtb7a+/+ and one Zbtb7a+/− samples were defined as a control group and the data sets from two Zbtb7a−/− samples were defined as a knockout group. Raw expression data on each chip was generated utilizing the Affymetrix Microarray Suite 5.0. Genes, whose normalized data value were greater or less by more than 1.5 fold between two groups, were selected and further evaluated statistically (Parametric test using Welch’s approximate t-test, with p-value cutoff of 0.05) using GeneSpring software (SiliconGenetics).
In vitro hematopoietic differentiation of mouse embryonic stem (ES) cells
Wild-type and Gata1 null ES cells (Weiss et al., 1994; Penvy L et al., 1991) were retrovirally infected with mock or Lrf-expressing vector followed by selection with puromycin on gelatinized dish. In vitro differentiation of ESCs, cell sorting and colony-forming assays were performed as reported previously (Umeda et al., 2004; Umeda et al., 2006). Floating hematopoietic cells that emerged after cell sorting were processed every 3 days for May-Giemsa staining as described previously (Umeda et al., 2004, Umeda et al., 2006).
Identification of Potential cis-Regulatory Elements
To identify regions to be analyzed by ChIP, we examined the Lrf genomic locus in the UCSC Genome Browser along with a custom track showing erythroid predicted cis-regulatory modules (preCRMs) (Wang et al., 2006). Erythroid preCRMs are regions of high regulatory potential (Elnitski et al., 2003) containing at least one conserved consensus GATA1 binding motif (WGATAR). A 435-bp preCRM was identified immediately upstream of the start of transcription. Examination of the preCRM in the murine Lrf promoter revealed 2 consensus GATA1 binding motifs (WGATAR) and 3 consensus EKLF binding motifs (CCNCNCCCN).
Lrf promoter Reporter Assay
2 mg of Lrf-PGL-4 reporter plasmid + 0.2 mg renilla luciferase reporter plasmid (pRL) were transfected into murine erythroleukemia (MEL) cells using Lipofectamine LTX+ Plus reagents (Invitrogen). Where indicated, MEL cells were induced toward erythroid maturation by addition of 5 mM hexamethylene bisacetamide (HMBA) for 72 hours prior to transfection. Luciferase activity was measured 24 hours after transfection using the Dual-Luciferase Reporter Assay system (Promega). Reported luciferase activities are normalized for transfection efficiency according to renilla activities.
Bim promoter Reporter Assay
pGL3-mBim0.8 reporter plasmid, containing 800 bp region immediately upstream of exon 1 of mouse Bim gene, was generated from the p0.8 vector (kind gift from Dr. Jerry M. Adams) (Bouillet et al., 2001). Truncated mutants (pΔSacII, p0.25, p0.1 and p0.25S) were generated by enzyme digestion. Mutagenesis was then by standard methods using a Change-IT mutagenesis kit (USB Corporation, Cleveland, OH).
Supplementary Material
Acknowledgments
We thank Agnes Viale and Julia Zhao for the help of microarray experiments; Irena Linkov and Katia Manova for IHC analysis; Bernessa Vassall for May-Giemsa staining; Jia-Hui Dong for help on mice work; Jan Hendrikx and other MSKCC Flow Cytometry core facility members for assistance with FACS analysis and cell sorting; Yu Yao for help on Lrf promoter Reporter assay; Ainara Egia for histology and immunohistrochemistry assistance; Nicola Hawe, Carmela Gurrieri, Jose Costoya, Francesco Piazza, Luipa Khandker, Ilhem Guernah, Linda DiSantis, Stuart Megan, Thomas Naughton and other P.P.P. lab members for assistance, advice and helpful discussion. We also thank Margaret VanMeter for providing us with the Phospho Stat5 FACS protocol. This work is supported in part by the NCI grant CA-102142 to P.P.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P, Eyre HJ, Sutherland GR, Adams JM. Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm Genome. 2001;12:163–168. doi: 10.1007/s003350010242. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Crossley M, Tsang AP, Bieker JJ, Orkin SH. Regulation of the erythroid Kruppel-like factor (EKLF) gene promoter by the erythroid transcription factor GATA-1. J Biol Chem. 1994;269:15440–15444. [PubMed] [Google Scholar]
- Davies JM, Hawe N, Kabarowski J, Huang QH, Zhu J, Brand NJ, Leprince D, Dhordain P, Cook M, Morriss-Kay G, Zelent A. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 1999;18:365–375. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L, Hardison RC, Li J, Yang S, Kolbe D, Eswara P, O’Connor MJ, Schwartz S, Miller W, Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22:9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin I, Cumano A. The hare and the tortoise: an embryonic haematopoietic race. Nat Rev Immunol. 2002;2:593–604. doi: 10.1038/nri857. [DOI] [PubMed] [Google Scholar]
- Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood. 1999;94:87–96. [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, Weiss M, Grimmond S, Perkins A. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–3370. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Kukita A, Kukita T, Ouchida M, Maeda H, Yatsuki H, Kohashi O. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood. 1999;94:1987–1997. [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetens M, Doumont G, Clercq SD, Francoz S, Froment P, Bellefroid E, Klingmuller U, Lozano G, Marine JC. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood. 2007;109:2630–2633. doi: 10.1182/blood-2006-03-013656. [DOI] [PubMed] [Google Scholar]
- O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, Huang DC. Bim: a novel member of the Bcl-2 family that promotes apoptosis. Embo J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D’Agati V, Orkin SH, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- Pessler F, Pendergrast PS, Hernandez N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol Cell Biol. 1997;17:3786–3798. doi: 10.1128/mcb.17.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Pitto L, Simili M, Mariani L, Riccardi L, Ciucci A, Rizzo M, Evangelista M, Mercatanti A, Pandolfi PP, Rainaldi G. The proto-oncogene LRF is under post-transcriptional control of MiR-20a: implications for senescence. PLoS ONE. 2008;3:e2542. doi: 10.1371/journal.pone.0002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivella S. Ineffective erythropoiesis and thalassemias. Curr Opin Hematol. 2009;16:187–194. doi: 10.1097/MOH.0b013e32832990a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TD, Chohan M, Barber DL. Turning cells red: signal transduction mediated by erythropoietin. Trends Cell Biol. 2005;15:146–155. doi: 10.1016/j.tcb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Rutherford TR, Clegg JB, Weatherall DJ. K562 human leukaemic cells synthesise embryonic haemoglobin in response to haemin. Nature. 1979;280:164–165. doi: 10.1038/280164a0. [DOI] [PubMed] [Google Scholar]
- Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, Blobel GA, Weiss MJ. GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol. 2003;23:5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Huang DC, Krammer PH, Cory S. Bcl-2 and Fas/APO-1 regulate distinct pathways to lymphocyte apoptosis. Embo J. 1995;14:6136–6147. doi: 10.1002/j.1460-2075.1995.tb00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Suwabe N, Ohneda O, Obara N, Imagawa S, Pan X, Motohashi H, Yamamoto M. Identification and characterization of 2 types of erythroid progenitors that express GATA-1 at distinct levels. Blood. 2003;102:3575–3583. doi: 10.1182/blood-2003-04-1154. [DOI] [PubMed] [Google Scholar]
- Umeda K, Heike T, Yoshimoto M, Shiota M, Suemori H, Luo HY, Chui DH, Torii R, Shibuya M, Nakatsuji N, Nakahata T. Development of primitive and definitive hematopoiesis from nonhuman primate embryonic stem cells in vitro. Development. 2004;131:1869–1879. doi: 10.1242/dev.01065. [DOI] [PubMed] [Google Scholar]
- Umeda K, Heike T, Yoshimoto M, Shinoda G, Shiota M, Suemori H, Luo HY, Chui DH, Torii R, Shibuya M, et al. Identification and characterization of hemoangiogenic progenitors during cynomolgus monkey embryonic stem cell differentiation. Stem Cells. 2006;24:1348–1358. doi: 10.1634/stemcells.2005-0165. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang Y, Cheng Y, Zhou Y, King DC, Taylor J, Chiaromonte F, Kasturi J, Petrykowska H, Gibb B, et al. Experimental validation of predicted mammalian erythroid cis-regulatory modules. Genome Res. 2006;16:1480–1492. doi: 10.1101/gr.5353806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc Natl Acad Sci U S A. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Krishnamurti L, Kutok JL, Biernacki M, Rogers S, Zhang W, Antin JH, Ritz J. Evidence for ineffective erythropoiesis in severe sickle cell disease. Blood. 2005;106:3639–3645. doi: 10.1182/blood-2005-04-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Liu X, Jaenisch R, Lodish HF. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 1995;83:59–67. doi: 10.1016/0092-8674(95)90234-1. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kawane K, Koike M, Mori Y, Uchiyama Y, Nagata S. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature. 2005;437:754–758. doi: 10.1038/nature03964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.