Abstract
Methionine sulfoxide reductases (Msr) belong to a gene family that contains one MsrA and three MsrBs (MsrB1, MsrB2, and MsrB3). We have identified all four of the genes that are expressed in mouse embryonic stem cell cultures. The vital cellular functions of the Msr family of genes are to protect cells from oxidative damage by enzymatically reducing the oxidized sulfide groups of methionine residues in proteins from the sulfoxide form (–SO) back to sulfide thus restoring normal protein functions as well as reducing intracellular reactive oxygen species (ROS). We have performed studies on the Msr family genes to examine the regulation of gene expression. Our studies using real-time RT-PCR and Western blotting have shown that expression levels of the four Msr family genes are under differential regulation by anoxia/reoxygenation treatment, acidic culture conditions and interactions between MsrA and MsrB. Results from these in vitro experiments suggest that although these genes function as a whole in oxidative stress protection, each one of the Msr genes could be responsive to environmental stimulants differently at the tissue level. J. Cell. Biochem.
Keywords: MSR GENE REGULATION, ANOXIA, ACIDOSIS, MOUSE STEM CELLS
Embryonic stem cells, with their remarkable capability of being totipotent, have the potential of providing adult tissue repair. Tissue damage, including cell death, can result from ischemia and reperfusion due to the accumulation of high levels of free radicals in the cells. This in turn can cause oxidization and functional impairment directly or through signal transduction pathways such as c-Jun N-terminal kinase (JNK), and p38 mitogen-activated protein kinase (MAPK) [Ueda et al., 2002]. In infarcted tissues such as in patients suffering from stroke or myocardial infarct, the local environment in the organ is stressed due to hypoxia and acidosis from lack of a blood supply resulting in hypoxia. It has been proposed by Webster and coworkers [Kubasiak et al., 2002] that hypoxia and acidosis are the two conditions, when present simultaneously, that can dramatically induce cell death in cardiomyocytes. With the development of autologous/allogeneic stem cell transplantation, how to improve the survival of stem cells in the harsh hypoxic and acidic environment is a critical issue for the success of tissue repair. The responses of stem cells to the local oxidative stress and the local acidic environment are not well understood.
One of the important self-defense mechanisms for stem cells to protect their vital cell constituents from free radical damage is the methionine sulfoxide reductase (Msr) system [Weissbach et al., 2002]. Free radicals can oxidize cellular components including proteins, lipids, and nucleic acids and thus impair certain cellular functions. The amino acid, methionine (met), either as a free molecule or as a residue inside a peptide, can be oxidized to methionine sulfoxide (Met-(O)). Numerous proteins have been shown to have impaired function due to this structural change [Abrams et al., 1981; Taggart et al., 2000; Jones et al., 2008; Shao et al., 2008]. An enzyme that can reduce methionine sulfoxide in proteins was first discovered in Escherichia coli more than 20 years ago. It is now apparent that there is a family of enzymes, referred to as methionine sulfoxide reductases (Msr) that can carry out the reduction of Met-(O) to Met. Two types of Msr genes, MsrA and MsrB, have been identified that are specific for reducing epimers Met(O)-S and Met(O)-R, respectively [Weissbach et al., 2002]. Both MsrA and MsrBs have been shown to protect cells against oxidative damage, which suggests a possible role of these genes in a large number of age-related diseases [Gabbita et al., 1999; Pal et al., 2007; Brennan and Kantorow, 2008].
As one of the cell protective mechanisms against oxidative damage, it is extremely important to understand whether the Msr system is modulated by local environmental conditions which stem cells encounter during their migration towards or staying within the damaged tissues. It is also unknown whether altered expression levels of one enzyme in the Msr family would affect the other(s), and/or whether there exists a co-regulation mechanism for different Msr genes. Currently, there are very few reports concerning Msr regulation in mammalian models, including studies on mice fed with selenium-deficient diets [Moskovitz, 2007; Uthus and Moskovitz, 2007; Cabreiro et al., 2008]; however, it remains unclear what factors control Msr gene expression at the molecular level. In the present studies, we have shown that the individual Msr genes in this family are under different regulation mechanisms. Depletion of oxygen and an acidic culture environment affect Msr gene expression, at least at the mRNA level, with the most significant response observed in MsrB3, indicating a non-housekeeping activity for this particular gene. Downregulation of MsrB3 at the transcriptional level was also noted when MsrA mRNA was knocked down by MsrA specific siRNA.
MATERIALS AND METHODS
MOUSE EMBRYONIC STEM CELL CULTURE
The mouse embryonic stem (MES) cells (CCE-24) [Narayanan et al., 1993] were routinely grown on 0.1% gelatin-coated dishes in Dubecco's modified Eagle's medium (DMEM) containing 15% heat-inactivated fetal bovine serum (catalog #10100, Invitrogen, Carlsbad, CA), 10 ng/ml human leukemia inhibitory factor (LIF) (LIF2010, Millipore, Billerica, MA), and monothioglycerol (Sigma, St. Louis, MO) at 4.5 × 10−4 M. The cells were grown on tissue culture plates coated with 0.1% gelatin (Sigma, St. Louis, WA) and routinely split every 2 days at 1:4–1:10 and immunostained for stem cell specific markers SSEA-1 (Mab4301) and SSEA-4 (Mab4304, Millipore) to ensure no differentiation has taken place. Only cells within the 20th passage were used for our studies.
ANOXIA/REOXYGENATION TREATMENT
The anoxic treatment of mouse embryonic stem cells was performed by incubating the cells in an anaerobic chamber (Sheldon Manufacturing, Inc., Cornelius, OR) supplied with 90% nitrogen gas, 5% hydrogen gas, and 5% carbon dioxide at 37°C. Cells were removed after selected time periods and incubated again in a regular cell culture incubator at 37°C for specific times.
SIRNA TRANSFECTION
MsrA specific siRNAs were commercially synthesized by Qiagen (Valencia, CA). After doing transfection pilot experiments, one of the three MsrA specific siRNAs showed a very significant reduction of MsrA mRNA levels in the mouse embryonic stem cells. This siRNA is numbered as 168.
The sequences are:
sense: CCAUGAAUCAUUUGCCAAAUCGCUU;
antisense: AAGCGAUUUGGCAAAUGAUUCAUGG.
The negative control siRNA (numbered as 882c) sequences are:
sense: CCAGCACUAACACCCAUCCCACAAA;
antisense: UUUGUGGGAUGGGUGUUAGUGCUGG.
For each transfection sample, siRNA-Lipofectamine™ 2000 (Invitrogen) complexes were prepared as follows: (a) dilute 2 μg siRNA in 250 μl of DMEM (no serum). Mix gently. (b) Dilute 5 μl Lipofectamine™ 2000 in 250 μl of DMEM (no serum). Mix gently and incubate for 15 min at room temperature. (c) After the 15-min incubation, combine the diluted siRNA and the diluted Lipofectamine™ 2000 (total volume ~505 μl). Mix gently and incubate for 15 min at room temperature to form complexes. (d) During the 15-min incubation, dissociate mouse embryonic stem cells by trypsinization, prepare a single cell suspension, and determine cell counts. Plate 5 × 105 cells per well in 6-well plates in a DMEM complete medium (with serum) in suspension. (e) Add the ~505 μl of siRNA-Lipofectamine™ 2000 complexes to each well containing cells and medium. Mix gently by rocking the plate back and forth. Cells were incubated at 37°C in a humidified CO2 incubator for 24 h before the samples were collected for RNA and protein extraction as well as H2O2 treatment. Medium was changed at 24 h and the cultures were continued for another 2 days. At each 24-h time point, samples were collected for RNA and protein extractions.
REAL-TIME RT PCR
Total RNA was extracted using TRI Reagent (catalog #9424, Sigma, MO) from mouse embryonic stem cells after various combinations of anoxia/reoxygenation treatments or MsrA siRNA transfection, following the manufacturer's protocol. Particularly for samples with only anoxia treatments, to avoid reoxygenation of cultured cells, TRI Reagent solution was pretreated in a hypoxia chamber for 2 h before cells were lysed with the solution inside the chamber. After pretreatment with RNase-free DNase I, 2 μg of total RNA was used for cDNA syntheses with a ThermoScript RT-PCR System (Invitrogen). Real-time RT-PCR experiments were performed on a Capillary Lightcycler (Roche) machine using a Roche Fast Start SYBR Green I Kit following the manufacturer's instructions. Specific amplification of desired genes was confirmed by calculating melting temperatures (Tm) for the products from the melting peak curve (–dF/dT vs. temperature). All the amplicons were collected and confirmed again by agarose gel electrophoresis and sequencing. A standard curve of cross-point versus log concentration (copies) was created using one of the cDNA samples with serial dilutions or with known concentrations of plasmid DNA with a MsrA gene insert. Negative controls were included using cDNAs synthesized the same way as above but with no reverse transcriptase added. Each cDNA sample was run in triplicate. The data were averaged and standard deviations were calculated. The beta-actin gene was used as a standard control. Primers for real-time PCR experiments were designed from two consecutive exons flanking a piece of intron to avoid amplification from contaminated genomic DNA. Primer sequences used in these studies are as follows:
MsrA-for: 5′-TCTGGGTCTTGAAAGGAGTGTA;
MsrA-rev: 5′-AGGTATTGCTGGTGGTAGTCTTC;
amplicon size: 395 bp.
MsrB1-for: 5′-CTGGCTGGGAGCTTTTGCCTGTC
MsrB1-rev: 5′-CGGGGATGAGTGTGCGTACTTCG
amplicon size: 160 bp.
MsrB2-for: 5′-CGGCTGGCCTTCATTTTCTGAGG
MsrB2-rev: 5′-CAGGCCTCCTGCAAACAGTCAGG
amplicon size: 244 bp.
MsrB3-for: 5′-GCTGGCCTGCCTTCCATGATGTG
MsrB3-rev: 5′-CCGCTCTCTTTGATGCCGCTTCC
amplicon size: 234 bp.
MMP2-for: 5′-ACCTGGATGCCGTCGTGGACCTG
MMP2-rev: 5′-CGCCAGGCTGCTTCACATCCTTC
amplicon size: 210 bp.
MMP9-for: 5′-TGGTGTGCCCTGGAACTCACACG
MMP9-rev: 5′-GACTTGCACTGCACGGTTGAAGCA
amplicon size: 215 bp.
β-actin-for: 5′-CCACTGCCGCATCCTCTTCCTC
β-actin-for: 5′-CAGCAATGCCTGGGTACATGGTG
amplicon size: 249 bp.
TREATMENT OF THE STEM CELLS WITH H2O2
After transfection with MsrA specific siRNA and negative control siRNA for 24 h, the culture medium for the mouse embryonic stem cells was replaced with fresh 100 μl DMEM complete medium without phenol red and cultured in a 37°C incubator for an additional hour. Differing concentrations of H2O2 in DMEM complete medium without phenol red were prepared to final concentrations of 0, 25, 50, 100, 150, 200, 250, 300 μM H2O2. The cells were placed in new medium prepared at different concentrations of H2O2 and continued in culture for an additional 24 h.
The cultures were then subjected to a MTT assay to test cell survival using the Vybrant Cell Proliferation Assay kit (Invitrogen) following the manufacturer's instructions.
WESTERN BLOTTING
Protein samples were heat-denatured at 95°C for 5 min before loading onto an SDS–polyacrylamide reducing gel (4–12%) (Invitrogen) and run using standard conditions. Proteins were transferred to nitrocellulose membranes which were subsequently blocked with a 5% (w/v) BSA in TBST (0.05% Tween-20) for 1 h at room temperature. The blot was incubated for 1.5 h at room temperature in the anti-MsrA polyclonal antibody (catalog #ab16803, Abcam, Cambridge, MA) diluted 1:4,000 in blocking buffer. The blot was incubated again for 40 min in horseradish peroxidase (HRP) labeled secondary antibody diluted 1/5,000 in blocking buffer. The blot was again rinsed and washed in TBST solution with 0.05% Tween-20 before ECL detection with an ECL kit (RPN 2108, Amersham, Buckinghamshire, UK). After exposure on X-ray film for MsrA protein, the blot was stripped in 1M NaOH solution for 10 min at room temperature and hybridized again using antibody to beta-actin (catalog #ab6276, Abcam) for normalization. The protein band densitometry was analyzed using a gel imaging system (Alpha Innotech Corp., San Leandro, CA).
RESULTS
CONFIRMATION OF THE MAINTAINED POTENCY TO DIFFERENTIATE INTO MULTIPLE ADULT CELL TYPES FROM MOUSE EMBRYONIC STEM CELLS IN FEEDER-LAYER-FREE CULTURE CONDITIONS
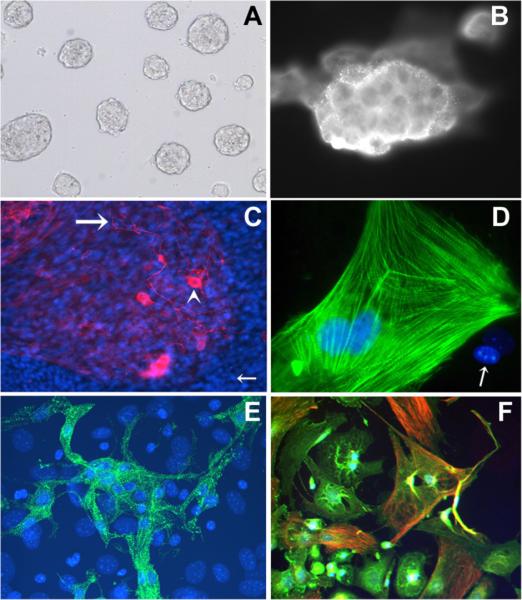
To avoid the interference of feeder-layer embryonic fibroblast cells in the stem cell studies, we have employed a feeder-layer-free culture system for mouse embryonic stem cells [Narayanan et al., 1993]. Cells were maintained at low passage number and cultured on gelatin-coated culture dishes with LIFs added to the medium. Cells in these conditions maintain their typical stem cell morphology, growing into round colonies with smooth edges (Fig. 1A). To confirm that the cells used for subsequent studies carry the genuine stem cell characteristics, we routinely stained the cells for embryonic stem cell specific markers and tested the capability of the cells to differentiate into various adult cell types at the same cell passages we used for the studies of Msr gene expression regulation. The cells under these culture conditions remain undifferentiated and stain positively with the stage-specific embryonic antigen-1 (SSEA-1) antibody (Abcam) (Fig. 1B). From these immunostaining results and cell counts, we have estimated only 5–10% SSEA-1 non-stainable cells in the population. After the induction of embryoid body formation from stem cell aggregation and subsequent autonomic cell differentiation for 2–4 weeks [Muller et al., 2000], we have successfully stained the differentiated cells by various cell linage markers and confirmed multi-potency of the cells using our feeder-layer-free culture conditions. Capability of these cells to differentiate into neurons and cardiac muscle cells were of particular interest to us since strokes and heart attacks are probably the two most prominent health problems that result from ischemia and both could potentially benefit from future stem cell therapy. Differentiated cells can be stained with neurofilament antibody for neuronal cells (Fig. 1C), cardiac troponin T for cardiomyocytes (Fig. 1D), fast skeletal troponin T monoclonal antibodies for skeletal muscle troponin T (Fig. 1E), or desmin and α-actinin for general muscle cell types (Fig. 1F).
Fig. 1.
Confirmation of stem cell identity and multipotent differentiation capability of the cultured mouse embryonic stem cells (MESCs) in feeder-layer-free culture system. A: Phase-contrast image of cultured MESCs without a feeder layer maintaining their typical round colonies with smooth edges indicating lack of differentiation. B: Positive staining with stem cell specific marker SSEA-1. C: Positive staining for the neuronal cell marker: neurofilament (NF). Antibodies to NF stain both cell bodies (arrowhead) and axons (large arrow). Nuclei are stained blue with DAPI (small arrow). D: Positive staining for the cardiac cell marker: cardiac Troponin T (cTnT) (green stain). Organized myofibrils are seen. E: Positive staining for the skeletal muscle cell marker: fast skeletal Troponin T (fsTnT) (green stain). DAPI, a blue nuclear fluorescent dye shows nuclear staining. F: Positive staining for both a-actinin (green) and desmin (red), markers for mesoderm-derived cell types. Magnifications for Figure 1 taken are: (A) 100×; (B) 250×; (C) 100×; (D) 600×;(E) 250×;(F) 250×.
RESPONSES OF MSR GENE EXPRESSION TO ANOXIA/REOXYGENATION TREATMENTS
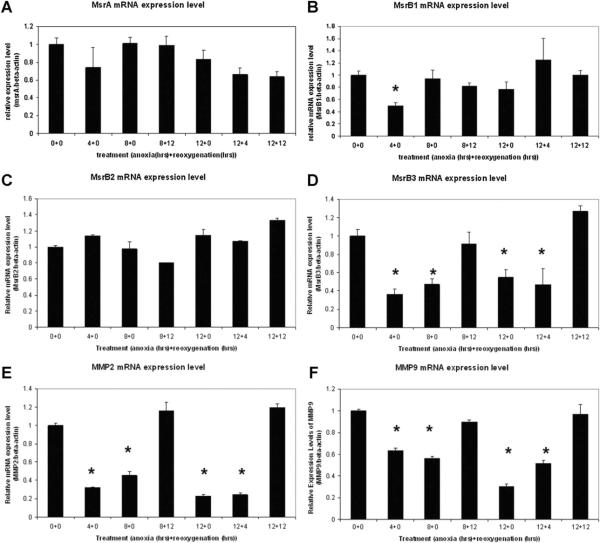
Using real-time RT-PCR, we have compared the regulation of methionine sulfoxide reductase gene expression at the transcription level under oxidative stress. Different levels of oxygen depletion and oxidative stress were induced by treating the cells with increasing time periods of anoxia (in 90% N2, 5% H2, and 5% CO2) and reoxygenation (in air) in combined treatments. Results indicate that MsrA mRNA levels were gradually decreased with prolonged anoxic and reoxygenation treatments (Fig. 2A). Although not statistically significant, possible decreases of MsrA mRNA have been observed as early as 4 h after oxygen depletion, which can be explained by the rapid general response of stem cells to oxygen depletion resulting in a decrease of mRNA transcription for MsrA, and other genes including the Msr family (Fig. 2B–F). From the RT-PCR results of MsrB1 and MsrB2, we have not observed an oxygen-dependent or oxidative stress regulated expression pattern for either, however, there is an oxygen-dependent expression pattern for MsrB3. The sensitivity of MsrB3 expression at the mRNA level to oxygen depletion has been shown clearly in Figure 2D with significantly decreased mRNA concentrations at 4, 8, and 12 h of anoxic conditions to nearly half of the normal levels at normoxia. The mRNA levels return to normal values after prolonged reoxygenation (>4 h). The same oxygen-dependent expression pattern has been observed in matrix metalloprotein 2 and matrix metalloprotein 9 (MMP9) (Fig. 2E,F). MMP2 and MMP9 respond to hypoxia treatment by downregulating their mRNA levels but regain normal expression levels of their mRNA after reoxygenation. Their response to anoxia/reoxygenation treatments also serves as an indicator that the treatments are successful. In all the analyses after real-time RT-PCR experiments, results have been normalized with β-actin levels for comparison between samples.
Fig. 2.
Real-time RT-PCR studies on Msr gene expression under oxidative stress induced by anoxia/reoxygenation. A: MsrA; (B) MsrB1; (C) MsrB2; (D) MsrB3; (E) MMP2; (F) MMP9. MsrB3 shows similar expression responses to MMP2 and MMP9 with their transcription levels downregulated when oxygen is depleted but regaining their normal expression levels quickly after a resupply of oxygen. Numbers show the hours of anoxia and reoxygenation respectively applied to MESCs. For example, 4+0 indicates 4 h of anoxia followed by 0 h of reoxygenation. Results with a statistically significant difference from the 0+0 group were labeled with asterisks (*)(P ≤0.05). Triplets were used in each experiment and two independent experiments were performed. Results were averaged with standard errors of mean (SEM) presented as error bars.
KNOCKDOWN OF MSRA EXPRESSION AT THE MRNA LEVEL HAS NO DIRECT IMPACT ON MSRB1 OR B2 GENE EXPRESSION BUT SIGNIFICANTLY DECREASES MSRB3 EXPRESSION AT THE MRNA LEVEL
Due to previous reports showing that there are potentially gene expression interactions between MsrA and MsrB [Moskovitz, 2007], we have repeated real-time RT-PCR experiments on all three reported MsrB genes (MsrB1, MsrB2, and MsrB3) using cells with MsrA mRNA downregulated by the siRNA transfection approach. Results confirmed that all three MsrBs are expressed in the stem cells. Comparing samples on the same day, neither MsrB1 nor MsrB2 expression at the mRNA level were altered in the cells with MsrA mRNA downregulation (Fig. 3). However, MsrB3 expression was significantly decreased in MsrA specific siRNA transfected cells compared to the cells transfected with negative control siRNA. Our results suggest that there are no direct interactions between MsrA and MsrB1 or MsrB2 gene expression at the mRNA level in mouse embryonic stem cells. On the contrary, for MsrB3, its mRNA expression is shown to be influenced by the levels of MsrA mRNA concentrations in the cells. A nearly 50% reduction of MsrB3 mRNA expression was noted on day 2 after MsrA gene knockdown. It is worth mentioning that MsrA specific siRNA can downregulate MsrA mRNA for days while its protein levels in the cell bounce back quickly after 24 h post-transfection (Fig. 3E,F) [Zhang et al., 2010].
Fig. 3.
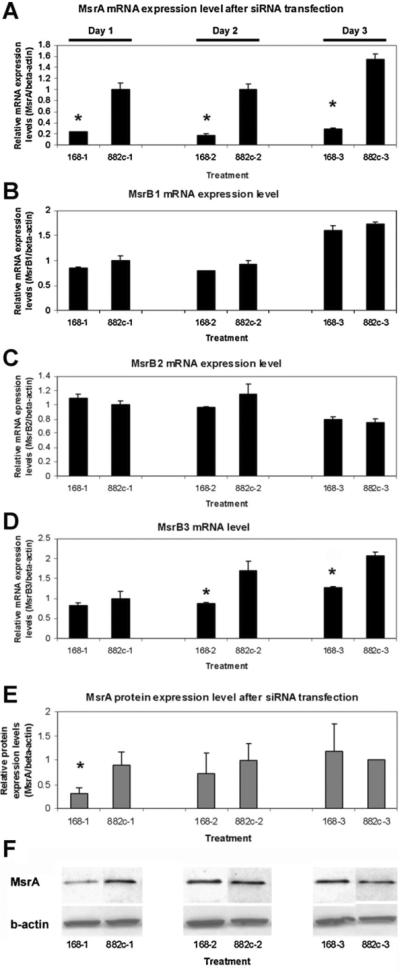
Real-time RT-PCR studies on MsrB gene expression at the mRNA level after downregulation of MsrA expression by siRNA (168) transfection. A: MsrA mRNA levels; (B) MsrB1 mRNA levels; (C) MsrB2 mRNA levels. D: MsrB3 RNA levels. MsrB3 is the only one showing significantly increased mRNA expression after MsrA knockdown. E: MsrA protein levels from densitometry of Western blots shown in F. F: Western blotting assays on MsrA protein and β-actin after treating MESCs with MsrA specific siRNA (168) or negative control siRNA (882) for days 1, 2, and 3. Triplets were used in each experiment. Two independent experiments were performed for real-time RT-PCRs and three Western blotting experiments were done. Results were averaged with standard errors of mean (SEM) presented with error bars. 168-1 represent samples collected on day 1 with siRNA (168) treatment. Results from MsrA specific siRNA (168) treated samples were labeled with asterisks (*) if there are statistically significant differences from negative control siRNA (882) treated groups (P ≤0.05).
PH OF THE CULTURE MEDIUM INFLUENCES MSR GENE EXPRESSION IN CULTURED MOUSE EMBRYONIC STEM CELLS
In our previous work on MsrA, it has been observed that prolonged culture of MES in the same medium, without refreshing, promotes MsrA mRNA expression [Zhang et al., 2010]. To further explore this finding, we cultured the cells in media pre-adjusted to pH 7.5, 6.8, or 6.4 by acetic acid or hydrochloric acid, with both showing similar results. Phosphoric acid was not used in this study because significant growth rate alteration was observed in the cultured stem cells. The media were changed every 24 h for 3 days and cells were collected immediately before the next change. Total RNA and proteins were extracted for real-time RT-PCR and Western blotting. When comparing pH to pH on the same days, we found that MsrA, B1 and B3 genes have significantly increased their mRNA expression in cells cultured in the acidic media on day 3. Significant increases of MsrB3 mRNA were also found in pH 6.8 on day 2. The most significant change was noticed in MsrB3 which had a four- to sixfold increase of its mRNA in either pH 6.4 or 6.8 culture media compared to pH 7.5 on day 3. However, even with a significant increase in MsrA mRNA expression in pH 6.4 medium on day 3, no increase of protein levels in stem cells was observed using MsrA antibody, the only antibody available for Western blotting on mouse Msr family genes (Fig. 4E,F).
Fig. 4.
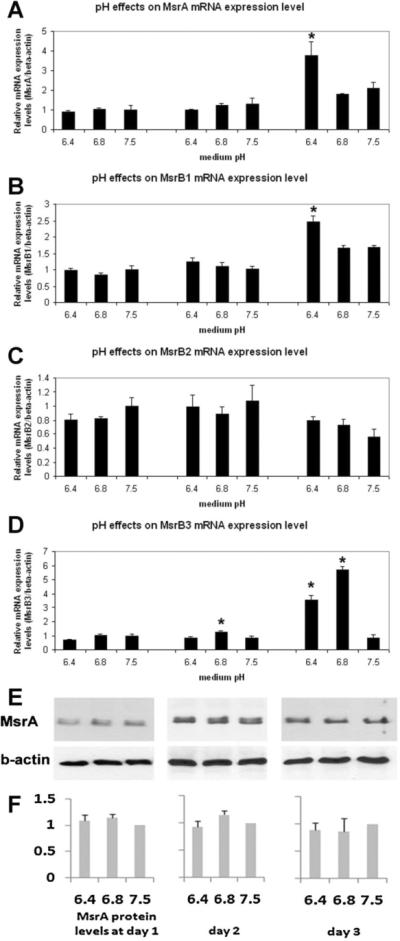
Real-time RT-PCR studies on Msr gene expression in culture media with different pH conditions (pH 6.4, 6.8, and 7.5) at 1, 2, and 3 days in culture. All Msr genes, except for MsrB2, show increased expression at the mRNA level after 3 days of culture in acidic media with MsrB3 showing the most dramatic responses. A: MsrA mRNA levels; (B) MsrB1 mRNA levels; (C) MsrB2 mRNA levels; (D) MsrB3 mRNA levels. E: Western blotting assays on MsrA protein and β-actin after culturing MESCsin media with different pH for days 1, 2, and 3. F: Densitometry of MsrA bands normalized by β-actin after Western blotting shows equal levels of MsrA protein expression in cells cultured in media with different pH on the same days. In all comparisons, data from cells cultured at pH 7.5 are arbitrarily set as unit 1. Results with statistically significant differences from 0+0 group were labeled with asterisks (*)(P ≤0.05). Triplets were used in each experiment and at least two independent experiments were performed. Results were averaged with standard errors of mean (SEM) presented as error bars. Results are labeled with asterisks (*) if there are statistically significant differences (P ≤0.05) comparing samples treated with acidic culture media and cells cultured at pH 7.5.
DISCUSSION
We have previously demonstrated that knockdown of the MsrA gene in mouse embryonic stem cells results in a loss of protection against H2O2-induced oxidative damage [Zhang et al., 2010]. Previous studies from another laboratory suggest a potential intergenic relationship of gene expression between the MsrA and MsrB genes from the MsrA knockout mouse model. MsrA knockout mice show parallel losses at the levels of MsrB1 mRNA and MsrB1 protein [Moskovitz and Stadtman, 2003]. Subsequent studies on MsrA knockout mice on prolonged selenium deficient diets revealed reduced activity of MsrB (which MsrB is not specified) in a tissue specific manner with the cerebellum, liver, and kidney showing dramatic decreases [Moskovitz, 2007]. In the present study, we did not detect a short term (three days) loss of MsrB1 or MsrB2 expression; the long-term effect of MsrA knockdown on MsrB1 and MsrB2 expressions is not known. MsrB3, however, although not showing a difference at day 1 after MsrA siRNA transfection, shows decreased expression at days 2 and 3. Interestingly at day 3, although the mRNA of MsrA is still down 70% compared to negative control siRNA transfected cells, MsrA protein expression has already returned to normal levels. It is possible that the signal to decrease MsrB3 expression is from the level of MsrA mRNA, but not its protein. However, the mechanism by which MsrA regulates MsrB expression remains to be elucidated. It will be a most interesting topic for further studies, considering the fact that MsrB3 and MsrA genes are located on different chromosomes in both human and mouse. Our results, together with previous findings from other laboratories, strongly suggest that MsrA plays a role in MsrB gene expression.
There are only a few studies to date on regulatory factors that influence Msr gene expression. In prokaryotes, studies on H. pylori have shown that certain stress conditions such as treatment with peroxide, peroxynitrite, or iron starvation, can cause approximately a 3- to 3.5-fold transcriptional up-regulation of the Msr gene [Alamuri and Maier, 2006]. H. pylori Msr codes for a 42-kDa protein with fused MsrA- and MsrB-like domains [Weissbach et al., 2002]. The only available evidence that Msr gene expression is influenced by pH stems from studies on Streptococcus gordonii which when entering the blood stream (pH 7.3) from the oral cavity (pH 6.2) promotes MsrA expression and possibly protects and increases survivability of the bacteria [Vriesema et al., 2000]. In Drosophila, ecdysone was found to be effective in promoting MsrA but not MsrB expression [Roesijadi et al., 2007]. In mouse, a selenium deficient diet results in decreased enzymatic activities for both MsrA and MsrB [Uthus and Moskovitz, 2007]. MsrA is also found downregulated in human hepatitis B positive hepatocellular carcinoma (HCC) with metastasis compared to HCCs without metastasis [Lei et al., 2007]. Recent studies on insulin/IGF receptor (IIR)/FOXO pathways indicate that downregulation of signaling in this pathway has been shown to extend lifespan in worms and flies [Murphy, 2006]. FOXO-mediated transcription is required for the long lifespan, thus there is great interest in identifying FOXO target genes. Also, it was reported recently that methionine sulfoxide reductase A expression is regulated by the DAF-16/FOXO pathway in Caenorhabditis elegans [Minniti et al., 2009]. Moreover another study has shown that Spx, a global transcriptional regulator of the disulfide specific oxidative stress response in B. subyilis plays a central role in the paraquat-specific induction of MsrA and MsrB expression [You et al., 2008].
Our studies, for the first time, have demonstrated that Msr genes are responsive to culture condition changes that mimic the environmental changes in pathological situations. We believe that since MsrA is the only enzyme currently known for Met-S-(O) reduction and since MsrB1 has the highest enzymatic activity for Met-R-(O) reduction [Kim and Gladyshev, 2004], and both show only slight responses to oxygen level and media pH changes, MsrA and MsrB1 (and possibly also MsrB2) most likely function as housekeeping enzymes for normalizing oxidative status in the cells. However, they do possess the capability of being regulated in a severely harsh environment. MsrB3, on the other hand, shows the lowest expression level in real-time RT-PCR studies compared to MsrB1 and B2 (data not shown), but the most dramatic response to both oxygen level and media pH among all the Msr genes. Thus, MsrB3 could be a major player responding to increased cellular oxidative stress. It seems also reasonable to hypothesize that an acidic pH is an important signal for MsrB3 expression induction based on current results, while oxygen depletion, although also a tissue damaging signal, shuts down MsrB3 transcription. Both MMP2 and MMP9, the two major matrix metalloproteinases functioning in cellular migration, show the same response as MsrB3, indicating that this could be a normal response of stem cells to their growth niches in tissues which are usually hypoxic [Grayson et al., 2006]. Cells become quiescent and non-motile with all unnecessary cellular functions totally shut down or decreased to minimal levels including MsrB3 gene expression. It is very likely that because both oxidative stress environment and physiological stem cell niche in normal tissues are low in oxygen, stem cells are obliged to use another oxidative stress signal, that is, acidic pH, as the environmental trigger to turn on anti-oxidative stress gene expression. Surprisingly, although media with an acidic pH (pH 6.4) nearly doubles MsrA transcription compared to media with a neutral or alkaline pH, such effects are not seen at the protein level. This could be explained by the short-term follow-up of our studies or, perhaps more likely, by an absence of necessary/essential conditions in our cultures. One of the factors we are considering, for example, is that an appropriate cell density or cell–cell contact level is essential. Relevant to this we have observed increased MsrA protein expression levels along with longer incubation times and increased densities of the cells, even at pH 6.4, arguing against the possibility that failure to detect MsrA protein increases at this pH is due to toxic effects of this physiologically extreme condition to cellular functions. The combined treatments of cells with anoxia and acidosis have been attempted but with difficulty since cells under acidic and anoxic conditions die quickly even though cells cultured under either condition alone can survive and maintain relatively normal morphology for days, indicated by similar cell counts and total amounts of protein extracted from these samples compared to cells from normal culture conditions (data not shown). These results are consistent with the findings of Kubasiak et al. [2002]. Due to the lack of available antibodies for all three of the MsrB proteins, protein level studies are yet to be conducted.
SUMMARY
Our studies using real-time RT-PCR have demonstrated that MsrB3 responds most dramatically among all Msr genes to oxygen deprivation, culture media pH changes and MsrA knockdown. MsrB3 expression decreases significantly under anoxic conditions but increases dramatically after 2 days of culture in acidic medium, suggesting that MsrB3 is a major player in response to changes of tissue oxidative stress in embryonic stem cells. Knockdown of MsrA by siRNA in these cells also has shown a parallel decrease of MsrB3 transcription (30–50%) compared with the negative controls, but not for MsrB1 and B2, indicating different intergenic interactions between MsrA and members of the MsrB group. Our results open up new evidence to examine the expressional regulation of Msr genes. The flexibility of MsrB3 expression levels provides a potential target for future research to improve oxidative stress resistance in therapeutic stem cells or even to reduce this resistance in harmful tissues such as in cancer cells.
ACKNOWLEDGMENTS
This study was supported by NIH grants HL-58435 and HL-061246 and American Heart Association Grant to L. F. L., a grant from the American Heart Association to X. H. and a Summer Research Fellowship from the NIH to C. Z. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung and Blood Institute or the National Institutes of Health. We are grateful to Mrs. Amy Patrick for outstanding secretarial and administrative assistance in the preparation and submission of this manuscript.
Grant sponsor: National Institutes of Health; Grant numbers: HL-58435, HL-061246; Grant sponsor: American Heart Association.
REFERENCES
- Abrams WR, Weinbaum G, Weissbach L, Weissbach H, Brot N. Enzymatic reduction of oxidized alpha-1-proteinase inhibitor restores biological activity. Proc Natl Acad Sci USA. 1981;78(12):7483–7486. doi: 10.1073/pnas.78.12.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamuri P, Maier RJ. Methionine sulfoxide reductase in helicobacter pylori: Interaction with methionine-rich proteins and stress-induced expression. J Bacteriol. 2006;188(16):5839–5850. doi: 10.1128/JB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan LA, Kantorow M. Mitochondrial function and redox control in the aging eye: Role of MsrA and other repair systems in cataract and macular degenerations. Exp Eye Res. 2009;88(2):195–203. doi: 10.1016/j.exer.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F, Picot CR, Perichon M, Castel J, Friguet B, Petropoulos I. Overexpression of mitochondrial methionine sulfoxide reductase B2 protects leukemia cells from oxidative stress-induced cell death and protein damage. J Biol Chem. 2008;283(24):16673–16681. doi: 10.1074/jbc.M708580200. [DOI] [PubMed] [Google Scholar]
- Gabbita SP, Aksenov MY, Lovell MA, Markesbery WR. Decrease in peptide methionine sulfoxide reductase in Alzheimer's disease brain. J Neurochem. 1999;73(4):1660–1666. doi: 10.1046/j.1471-4159.1999.0731660.x. [DOI] [PubMed] [Google Scholar]
- Grayson WL, Zhao F, Izadpanah R, Bunnell B, Ma T. Effects of hypoxia on human mesenchymal stem cell expansion and plasticity in 3D constructs. J Cell Physiol. 2006;207(2):331–339. doi: 10.1002/jcp.20571. [DOI] [PubMed] [Google Scholar]
- Jones EM, Squier TC, Sacksteder CA. An altered mode of calcium coordination in methionine-oxidized calmodulin. Biophys J. 2008;95(11):5268–5280. doi: 10.1529/biophysj.108.139634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: Characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15(3):1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubasiak LA, Hernandez OM, Bishopric NH, Webster KA. Hypoxia and acidosis activate cardiac myocyte death through the bcl-2 family protein BNIP3. Proc Natl Acad Sci USA. 2002;99(20):12825–12830. doi: 10.1073/pnas.202474099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei KF, Wang YF, Zhu XQ, Lu PC, Sun BS, Jia HL, Ren N, Ye QH, Sun HC, Wang L, Tony ZY, Qin LX. Identification of MSRA gene on chromo-some 8p as a candidate metastasis suppressor for human hepatitis B virus-positive hepatocellular carcinoma. BMC Cancer. 2007;7:172. doi: 10.1186/1471-2407-7-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minniti AN, Gataldo R, Trigo C, Vasquez L, Mujica P, Leighton F, Inestrosa NC, Aldunate R. Methionine sulfoxide reductase A expression is regulated by the DAF-16/FOXO pathway in Caenorhabditis elegans. Aging Cell. 2009;8:690–705. doi: 10.1111/j.1474-9726.2009.00521.x. [DOI] [PubMed] [Google Scholar]
- Moskovitz J. Prolonged selenium-deficient diet in MsrA knockout mice causes enhanced oxidative modification to proteins and affects the levels of antioxidant enzymes in a tissue-specific manner. Free Radic Res. 2007;41(2):162–171. doi: 10.1080/10715760600978823. [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Stadtman ER. Selenium-deficient diet enhances protein oxidation and affects methionine sulfoxide reductase (MsrB) protein level in certain mouse tissues. Proc Natl Acad Sci USA. 2003;100(13):7486–7490. doi: 10.1073/pnas.1332607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Fleischmann BK, Selbert S, Ji GJ, Endl E, Middeler G, et al. Selection of ventricular-like cardiomyocytes from ES cells in vitro. FASEB J. 2000;14(15):2540–2548. doi: 10.1096/fj.00-0002com. [DOI] [PubMed] [Google Scholar]
- Murphy CT. The search for DAF-16/FOXO transcriptional targets: Approaches and discoveries. Exp Geronol. 2006;41:910–921. doi: 10.1016/j.exger.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Narayanan R, Higgins KA, Perez JR, Coleman TA, Rosen CA. Evidence for differential functions of the p50 and p65 subunits of NF-kappa B with a cell adhesion model. Mol Cell Biol. 1993;13(6):3802–3810. doi: 10.1128/mcb.13.6.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Oien DB, Ersen FY, Moskovitz J. Elevated levels of brain-pathologies associated with neurodegenerative diseases in the methionine sulfoxide reductase A knockout mouse. Exp Brain Res. 2007;180(4):765–774. doi: 10.1007/s00221-007-0903-6. [DOI] [PubMed] [Google Scholar]
- Roesijadi G, Rezvankhah S, Binninger DM, Weissbach H. Ecdysone induction of MsrA protects against oxidative stress in drosophila. Biochem Biophys Res Commun. 2007;354(2):511–516. doi: 10.1016/j.bbrc.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Shao B, Cavigiolio G, Brot N, Oda MN, Heinecke JW. Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A–I. Proc Natl Acad Sci USA. 2008;105(34):12224–12229. doi: 10.1073/pnas.0802025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart C, Cervantes-Laurean D, Kim G, McElvaney NG, Wehr N, Moss J, et al. Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J Biol Chem. 2000;275(35):27258–27265. doi: 10.1074/jbc.M004850200. [DOI] [PubMed] [Google Scholar]
- Ueda S, Masutani H, Nakamura H, Tanaka T, Ueno M, Yodoi J. Redox control of cell death. Antioxid Redox Signal. 2002;4(3):405–414. doi: 10.1089/15230860260196209. [DOI] [PubMed] [Google Scholar]
- Uthus EO, Moskovitz J. Specific activity of methionine sulfoxide reductase in CD-1 mice is significantly affected by dietary selenium but not zinc. Biol Trace Elem Res. 2007;115(3):265–276. doi: 10.1007/BF02686001. [DOI] [PubMed] [Google Scholar]
- Vriesema AJ, Dankert J, Zaat SA. A shift from oral to blood pH is a stimulus for adaptive gene expression of Streptococcus gordonii CH1 and induces protection against oxidative stress and enhanced bacterial growth by expression of msrA. Infect Immun. 2000;68(3):1061–1068. doi: 10.1128/iai.68.3.1061-1068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach H, Etienne F, Hoshi T, Heinemann SH, Lowther WT, Matthews B, et al. Peptide methionine sulfoxide reductase: Structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;397(2):172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
- You C, Sekowska A, Francetic O, Martin-Verstraete I, Wang Y, Danchin A. Spx mediated oxidative stress regulation of the methionine sulfoxide reductases operon in Bacillus subtilis. BMC Microbiol. 2008;8:128. doi: 10.1186/1471-2180-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Jia P, Jia Y, Weissbach H, Webster KA, Huang X, Lemanski SL, Achary M, Lemanski LF. Methionine sulfoxide reductase A (MsrA) protects cultured mouse embryonic stem cells from H2O2-mediated oxidative stress. J Cell Biochem. 2010;111(1):94–103. doi: 10.1002/jcb.22666. [DOI] [PMC free article] [PubMed] [Google Scholar]