SUMMARY
Granulomas, organized aggregates of immune cells, form in response to persistent stimuli and are hallmarks of tuberculosis. Tuberculous granulomas have long been considered host-protective structures formed to contain infection. However, work in zebrafish infected with Mycobacterium marinum suggests that granulomas contribute to early bacterial growth. Here we use quantitative intravital microscopy to reveal distinct steps of granuloma formation and assess their consequence for infection. Intracellular mycobacteria use theESX-1/RD1virulence locus to induce recruitment of new macrophages to, and their rapid movement within, nascent granulomas. This motility enables multiple arriving macrophages to efficiently find and phagocytose infected macrophages undergoing apoptosis, leading to rapid, iterative expansion of infected macrophages and thereby bacterial numbers. The primary granuloma then seeds secondary granulomas via egress of infected macrophages. Our direct observations provide insight into how pathogenic mycobacteria exploit the granuloma during the innate immune phase for local expansion and systemic dissemination.
INTRODUCTION
At the outset of human pulmonary tuberculosis, inhaled Mycobacterium tuberculosis (Mtb) is taken up by phagocytic cells and transported across the alveolar epithelium into the lung. There, infected macrophages recruit additional macrophages and other immune cells to form organized structures called granulomas, pathological hallmarks of tuberculosis (Cosma et al., 2003; Dannenberg, 1993). Granulomas are believed to benefit the host by containing and restricting mycobacteria (Ulrichs and Kaufmann, 2006). One reason for this belief is that granulomas were thought to form only after initiation of adaptive immunity (Saunders and Cooper, 2000), and in animal models of tuberculosis, bacterial growth is rapid for the first 2 weeks of infection and plateaus coincident with the development of adaptive immunity (North and Jung, 2004; Swaim et al., 2006). Hence, according to the classical model, granuloma formation requires adaptive immunity and is critical for restricting bacterial expansion (Andersen, 1997; Saunders and Cooper, 2000).
Studies in transparent zebrafish embryos infected with Mycobacterium marinum (Mm), a system which recapitulates the earliest stages of tuberculosis (Clay et al., 2008; Dannenberg, 1993; Lesley and Ramakrishnan, 2008; Stamm and Brown, 2004; Tobin and Ramakrishnan, 2008), refute the classical model of granuloma initiation as a host-protective event in fundamental ways. First, epithelioid granulomas are found to form within days of infection, well before adaptive immunity is present (Davis et al., 2002). Second, granuloma formation coincides with the accelerated bacterial expansion widely thought to precede it (Volkman et al., 2004). Finally, Mm lacking the ESX-1/RD1 secretion system locus (ΔRD1 Mm) produces attenuated infection (DiGiuseppe Champion and Cox, 2007; Ernst et al., 2007) with poor granuloma formation (Volkman et al., 2004). Together, these findings suggest that granuloma formation actually works as a bacterial tool for expanding infection.
In this study, we sought to determine the mechanisms by which mycobacteria might take advantage of such a widely used host-protective response as granuloma formation (Adams, 1976). The unique visual access to cellular events afforded by the zebrafish embryo has allowed us to investigate the mechanisms and consequences of tuberculous granuloma formation at the whole-animal level. We devised quantitative assays using long-term three-dimensional (3D) differential interference contrast (DIC) and fluorescence in vivo microscopy to isolate the steps of granuloma formation. By comparing virulent and ΔRD1 Mm infection in these assays, we determined the impact of RD1 on each step of pathogenesis. Virulent mycobacteria used RD1 to enhance both macrophage recruitment to and infection within nascent granulomas. This RD1-enhanced infection rate of the arriving macrophages was closely coupled to their continued rapid motility throughout the structure. Thus multiple newly arriving cells found and phagocytosed single infected dead macrophages, creating new bacterial growth niches. Furthermore, we found that the primary granuloma promotes early dissemination of infection via egress of infected macrophages to establish secondary granulomas distally.
RESULTS
Macrophages Infected with Virulent Mycobacteria Promote Chemotactic Recruitment of Uninfected Macrophages in an RD1-Dependent Fashion
We ensured that granuloma formation in the zebrafish embryo model occurred via recruitment of uninfected macrophages to single infected ones, as is likely in human tuberculosis (Dannenberg, 1993). We infected embryos with wild-type (WT) Mm into the hindbrain ventricle (Figure S1 available online) and monitored daily the fate of individual macrophages after they had migrated back into deeper brain tissues (Clay et al., 2007; Davis et al., 2002) (Figure 1A). Infection expanded quickly into multiple macrophages (Figure 1A). These granulomas did not form by aggregation of infected macrophages (Figure S2), suggesting that uninfected macrophages were recruited, then infected. Expansion to new cells did not occur when the inciting macrophage was infected with ΔRD1 Mm (Figure 1B).
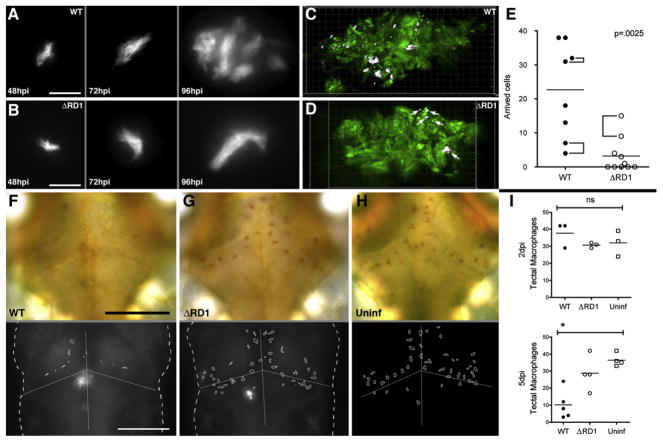
Figure 1. The Early Granuloma Enhances Bacterial Replication by RD1-Dependent Recruitment and Infection of New Cells.
(A and B) Fluorescent images of WT (A) and ΔRD1 (B) infected lesions at 48, 72, and 96 hr post-infection (hpi). Scale bars, 10 μm.
(C and D) Three-dimensional reconstructions of granulomas in WT (C) and ΔRD1 (D) infection, showing Hoechst-positive nuclei rendered white (indicated by white arrows in D) accumulated over 24 hr. Scale grid, 5 μm per square.
(E) Comparison of numbers of Hoechst-positive arrivals in WT and ΔRD1 lesions after 24 hr. Brackets indicate granulomas in the same embryo. Horizontal lines represent means. p value from Mann-Whitney test.
(F–H) Neutral red-stained optic tectum macrophages at 5 days post-hindbrain injection with WT Mm(F), ΔRD1 Mm(G), or no infection (H). Macrophages detected using neutral red dye (upper panels) and their outlines overlayed on images of the fluorescent bacteria present (lower panels—no fluorescent image collected for uninfected embryos). Scale bars, 100 μm.
(I) Numbers of optic tectum macrophages at 2 dpi and 5 dpi. p = 0.47 (2 dpi) and 0.01 (5 dpi) (Kruskal-Wallis test, p < 0.05).
To determine the contributions of macrophage recruitment and retention in RD1-mediated granuloma formation, we developed the Hoechst recruitment assay for macrophage arrival at granulomas; we induced brain granulomas by hindbrain ventricle infection then injected the nuclear dye Hoechst 33342 into the circulation via the caudal vein (Figure S3A). This dye does not penetrate the blood-brain barrier (data not shown) so that any Hoechst-positive cells in brain granulomas represent migrant phagocytes from the circulation. Twenty-four hours after dye injection, Hoechst-positive cells were rarely found in the brains of uninfected embryos (data not shown) whereas they were numerous in the brains of WT Mm-infected embryos (22.6 ± 4.9, n = 8 granulomas in 6 embryos), but only in granulomas (Figures 1C and S3B). Similar-sized ΔRD1 Mm granulomas (induced by infecting with 5-fold more bacteria) attracted 7-fold fewer macrophages than WT (3.2 ± 1.6, n = 10 granulomas in 9 embryos) (Figures 1D and 1E). Again, Hoechst-positive cells were not detected elsewhere in the brain, suggesting that their reduced number in the granulomas was not due to normal recruitment with impaired retention.
The Hoechst recruitment assay quantitates blood macrophage recruitment, a multistep process involving diapedesis across the vascular endothelium, followed by tissue migration to the granuloma (Algood et al., 2003; Imhof and Aurrand-Lions, 2004). To dissect the contributions of macrophage extravasation and tissue migration to recruitment, we assayed recruitment of resident macrophages of the optic tectum of the brain to nearby infected macrophages (Davis et al., 2002; Herbomel et al., 2001). Using neutral red to identify resident macrophages (Davis et al., 2002; Herbomel et al., 2001), we found them reduced in WT infection by 72.1% (±10.4%) from uninfected levels but no significant reduction in ΔRD1-infected embryos (n = 5 WT embryos and 4 each ΔRD1 and uninfected embryos) by 5 days post brain infection (Figures 1F–1I). Therefore, individual infected macrophages promote extravasation and tissue migration of uninfected macrophages to form granulomas in an RD1-dependent fashion.
Macrophages Arriving at Nascent Granulomas Become Infected More Rapidly if the Inciting Macrophages Are Infected with RD1-Competent Bacteria
Macrophages arriving at granulomas appeared to become infected rapidly (Figure 1A). To determine the kinetics of their infection, we adapted the Hoechst recruitment assay to monitor fates of arriving macrophages over 24 hr (Figure S3C). Combining 3D DIC and fluorescence time-lapse microscopy, we could distinguish infected and uninfected cells among the new arrivals (Figures 2A and 2B). Most arrivals at WT Mm granulomas had become infected (89.1% ± 3.1%, n = 6 granulomas in six embryos). We compared infection rates in WT and ΔRD1 granulomas first normalizing for macrophage recruitment by examining subsets of WT and ΔRD1 granulomas with similar numbers of new arrivals (Figure S3C). Only 53.6% (±10.0%) of arrivals to ΔRD1 granulomas had become infected within 24 hr (n = 4 granulomas in four embryos), a 40% reduction versus WT (Figure 2C). Thus, RD1-competent infection substantially increases the rate of macrophage infection in addition to recruitment itself.
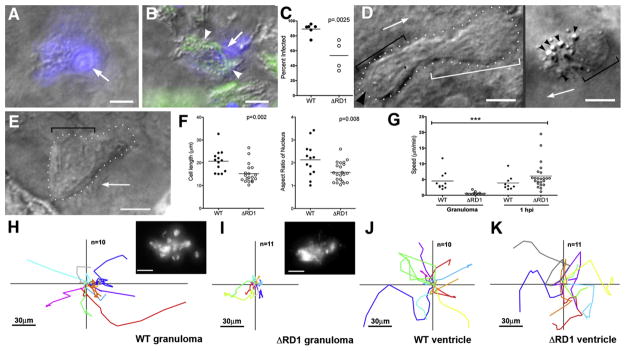
Figure 2. Uninfected Cells at WT Granulomas Show Distinct Morphology and Rapid Motility and Infection.
(A–C) Hoechst-positive nuclei (blue) are distinguishable as uninfected (A) or infected (B). White arrows, Hoechst-positive nuclei; arrowheads, bacteria (green). (C) Infected Hoechst-positive cells in WT versus ΔRD1 lesions over 24 hr. p from Mann-Whitney test.
(D and E) Distinct morphologies of uninfected cells at WT (D) and ΔRD1 (E) lesions. (D) Left: highly motile cell at WT granuloma with lamellipodium (white bracket), elongated nucleus (black bracket), and uropod (large black arrowhead). Arrow indicates direction of travel. Right: distinct appearance in WT granulomas of highly vesicular macrophage shortly before phagocytosis. Small black arrowheads, vesicles. Scale bars, 5 μm. (E) Less motile cell at ΔRD1 granuloma with no lamellipodium and rounded nucleus (black bracket). Scale bar, 5 μm.
(F) Comparison of overall cell length, left, and nuclear aspect ratio, right, of uninfected cells at WT and ΔRD1 granulomas. p by unpaired Student’s t test.
(G) Speeds of uninfected cells at WT or ΔRD1 granulomas, or at the site of injection 1 hr post-infection in the hindbrain ventricle. Bracket above indicates results of 1 way ANOVA (Kruskal-Wallis test)—uninfected cells at ΔRD1 granulomas (***) differed significantly from all others (p < 0.005), other differences not significant.
(H–K) Tracks of uninfected cells in (G). (H and I) Cells at WT and ΔRD1 granulomas—all cells tracked for each strain from one granuloma. Insets, fluorescence view of whole granuloma. Inset scale bar, 30 μm. (J and K) Cells at 1 hr post-injection. 1 hpi tracks from two WT-infected embryos and three ΔRD1-infected embryos. All scale bars = 5 μm unless noted otherwise.
RD1-Competent Infection of Inciting Macrophages in the Nascent Granuloma Is Associated with a Chemotactic Morphology and Rapid Motility of Newly Arriving Macrophages
We next sought to define the mechanism of intercellular bacterial spread in granulomas by examining the appearance and interactions of infected and uninfected cells in WT and ΔRD1 Mm granulomas. Cells attracted to WT granulomas moved rapidly (averaging 4.5 ± 1.0 μm/min, n = 10) and had elongated nuclei with extended lamellipodia and prominent uropods, morphological features of leukocytes undergoing chemotaxis (Figure 2D, left panel; Movie S1) (Sanchez-Madrid and del Pozo, 1999). Some rapidly moving cells were more rounded with extremely prominent cytoplasmic vesicles in their lamellipodia (Figure 2D, right panel; Movie S1). In contrast, uninfected cells in ΔRD1 granulomas moved slowly (averaging 0.6 ± 0.1 μm/min, n = 12) and had rounded shapes and nuclei and fewer cytoplasmic vesicles (Figures 2E and 2F; Movie S2), suggesting that they do not experience the same chemotactic signal as cells at WT granulomas. The speeds and distances covered by uninfected cells in WT granulomas were greater (eight and nine times, respectively) than their counterparts at ΔRD1 granulomas (Figures 2G–2I). In summary, the RD1-mediated chemotactic motility of cells arriving at granulomas was tightly linked to their higher infection rate.
Macrophage Motility in the Context of Extracellular Bacteria at the Initial Site of Infection Is Not RD1 Dependent
Macrophage migration occurs at multiple steps in tuberculous infection, first to the initial site of bacterial infection (Clay et al., 2007; Dannenberg, 1993). Previous work has suggested no difference in cell recruitment to WT or ΔRD1 infection sites (Volkman et al., 2004). We examined cells recruited to the hindbrain ventricle 1 hr after injection of WT or ΔRD1 Mm. In both cases, recruited cells had the chemotactic morphology of those arriving at WT granulomas and in sharp contrast to those at ΔRD1 granulomas (data not shown). Their displacements (Figures 2J and 2K) and speeds (Figure 2G) were also similar to uninfected cells at WT granulomas. Thus, RD1 is dispensable for macrophage chemotaxis to extracellular bacteria at the initial infection site yet required for intracellular bacteria to induce macrophage chemotaxis for granuloma initiation and expansion.
Newly Arriving Macrophages Become Infected by Phagocytosis of Infected Macrophages that Have Recently Undergone Apoptosis
Next, to determine how the rapid movement of uninfected cells at WT Mm-induced granulomas might lead to their higher infection rates, we studied their movements in the context of infected granuloma macrophages (Figure 3A). We were struck by the frequent death of these infected cells, which appeared to be apoptotic based upon morphological hallmarks, i.e., rapid nuclear collapse and fragmentation resulting in spherical remnants (Darzynkiewicz et al., 1997; Tone et al., 2007) (Figures 3B and 3C; Movie S3). The dying cells did not lyse; their membranes appeared intact with the bacteria remaining encased within. In contrast, Salmonella arizonae infection of the embryos featured dying macrophages that developed similar apoptotic nuclear morphology but underwent rapid swelling with bacterial release (Davis et al., 2002; Fink and Cookson, 2005) (Movie S4).
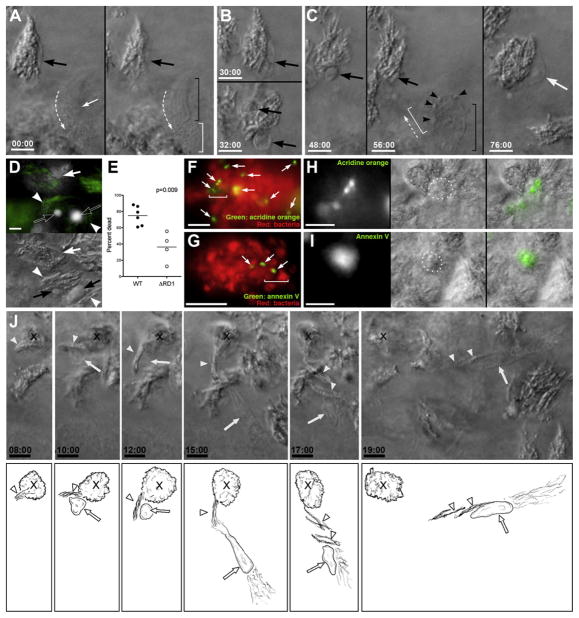
Figure 3. Death and Phagocytosis of Infected Macrophages.
(A–C) DIC time lapse of death and phagocytosis of infected macrophage. (A) two focal planes at time zero showing bacteria in cell with healthy nucleus (large black arrow), with uninfected cell (small white arrow and brackets as in Figure 2D) passing by. Dashed arrow, direction of travel. (B) Same infected cell as in (A), before (top) and after (bottom) nuclear collapse. (C) Same cell after further nuclear collapse into apoptotic sphere and later time points showing complete phagocytosis by new macrophage (black and white brackets and white arrow). Small black arrowheads, vesicles in approaching macrophage.
(D) Hoechst (white) and bacterial GFP (green) overlay above, with matching DIC below, showing compact Hoechst-positive apoptotic nuclei (black arrows) associated with bacteria (white arrowheads), compared to diffuse Hoechst staining of live nuclei (white arrow).
(E) Infected Hoechst-positive apoptotic cells per granuloma in WT and ΔRD1 lesions over 24 hr. p from Mann-Whitney test.
(F–I) Apoptotic cells in WT granulomas detected by acridine orange (F and H) or annexin V (G and I). (F) Granuloma of red fluorescent Mm with acridine orange-positive cells (white arrows). Bracket indicates area shown in (H). (G) Granuloma of red fluorescent Mm with annexin V-positive cells (white arrows). Bracket indicates area shown in (I). (H) Acridine orange signal (left) corresponds with DIC appearance of apoptotic bodies (dotted circle, middle). Overlay, right. Scale bar, 10 μm. (I) Annexin V signal (left) corresponds with DIC appearance of apoptotic bodies (dotted circle, middle). Overlay, right. Scale bar, 10 μm.
(J) DIC time-lapse images of single macrophage (nucleus indicated by white arrow) pulling a small group of bacteria (white arrowheads) from a larger cluster (X). Below, sketches derived from these panels with additional details from other planes at same time point. All scale bars, 5 μm.
The DIC appearance of the nuclear spheres of the mycobacterium-infected dead cells corresponded to distinctive compact fluorescent bodies in cells labeled with Hoechst prior to their death, in contrast to diffuse nuclear staining of living cells (Darzynkiewicz et al., 1997) (Figure 3D). This feature allowed quantitation of the infected immigrant cells that had died over 24 hr using the Hoechst recruitment assay (Figure S3C). Most infected arrivals to WT Mm granulomas had died (74.9% ± 4.8%; n = 6 granulomas) (Figure 3E). That their death was apoptotic was confirmed by staining with acridine orange and Annexin V (Peri and Nusslein-Volhard, 2008) (Figures 3F–3I). Strikingly, their death was followed shortly by phagocytosis of their remains, including the bacteria (Figure 3C; Movie S5). Uninfected cells moved rapidly past living infected cells (Figure 3A), engaging in phagocytosis only upon encountering a dead cell (Figure 3C). The bacteria from a single heavily infected dead macrophage could be phagocytosed by multiple cells, (Figure 3J; Movie S6) accounting for the increase in the number of infected macrophages (Figure 1B).
Dying ΔRD1 Mm-Infected Macrophages Are Not Phagocytosed Rapidly
RD1 has been reported to increase death of infected macrophages in culture (Gao et al., 2004; Guinn et al., 2004) and in zebrafish embryo granulomas (Clay et al., 2008; Volkman et al., 2004). Our analysis of WT Mm granulomas suggested that infection of new macrophages was dependent on death of infected macrophages as well as chemotactic motility of, and phagocytosis by, new macrophages. We sought to determine the contribution of RD1 to each of these. Using the Hoechst recruitment assay and analyzing WT and ΔRD1 granulomas with similar numbers of newly arrived cells (Figure S3C), we found a 40% reduction from WT in dead infected arrivals over 24 hr (n = 6 granulomas) (Figure 3E). This is consistent with previous findings of fewer TUNEL-positive cells in ΔRD1 than WT Mm granulomas (Clay et al., 2008; Volkman et al., 2004). Though fewer, the dead cells in ΔRD1 Mm granulomas showed the same morphology and kinetics of nuclear collapse as those in WT granulomas (Movie S7). Yet their death did not evoke chemotactic morphology or motility even in nearby uninfected macrophages (data not shown).
Phagocytosis of Bacteria from Dying Cells by Multiple Arriving Cells Is Likely a Major Mechanism for Granuloma Expansion
These data suggested that granuloma expansion is driven by the arrival of new cells that phagocytose resident dying cells at a multiplicity of greater than one. First, to test the prediction that granuloma expansion requires new macrophages for niche expansion, we compared the increase in bacterial burden (as judged by an interval increase in fluorescence area of the granuloma) to the arrival of new macrophages during a 24 hr period in eight granulomas from seven embryos. We found a tight correlation (Figure 4A).
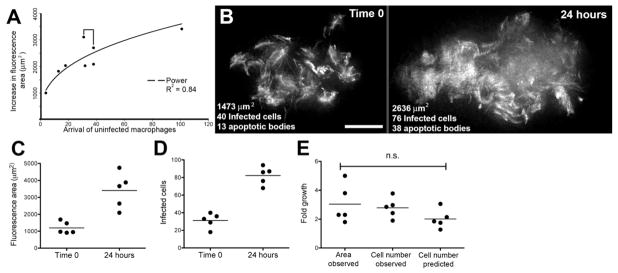
Figure 4. Bacterial Expansion in Early Granulomas as a Function of Macrophage Arrival, Infection, Death, and Rephagocytosis.
(A) Correlation of bacterial growth in granulomas, measured by fluorescence area with arrival of new macrophages. Data from eight granulomas in seven embryos. Bracket indicates pair of granulomas imaged in the same embryo.
(B) Deconvolved fluorescence images of granuloma at time 0 (left) and 24 hr (right). Scale bar, 20 μm.
(C and D) Bacterial expansion in five granulomas over 24 hr as measured by (C) fluorescence area and (D) number of infected cells.
(E) Fold growth as measured in panels (C) and (D) as compared to predicted fold growth based on mathematical modeling. Differences not significant (ANOVA).
Bacterial expansion by intercellular spread could then occur by rephagocytosis of dead infected macrophages, as predicted by our findings, or by bacterial transfer between live cells as previously reported (Davis et al., 2002; Stamm et al., 2003). To assess the likelihood that death and rephagocytosis is a major mechanism for bacterial expansion in granulomas, we constructed a simple mathematical model of granuloma expansion. The model is expressed as
| (1) |
where N0 and N24 are the number of infected macrophages at time zero and 24 hr, respectively, f is the fraction of infected macrophages dying in the 24 hr period, and m is the number of new infected cells resulting from phagocytosis of a single dying infected cell. Rearranged to express proportional growth of the granuloma in terms of infected cells (N24/N0 = G), this becomes
| (2) |
This model and its assumptions are detailed further in the Supplemental Data.
To test this model, we followed five granulomas, in five separate embryos, assessing fluorescence area as a measure of bacterial burdens and counting the numbers of infected macrophages and of apoptotic bodies at 0 and 24 hr (Figure 4B; Table S2). First, we found a 3.0 (±0.6) fold increase in fluorescence area, similar to the 2.8 (±0.3) fold increase in the number of infected macrophages (Figures 4C–4E; Table S3). Second, based on these data, we found f = 0.8 (±0.1) and m = 2.3 (±0.6) (Table S3). These values are in accordance with our previous finding that death of infected macrophages is rapid (Figure 3E) and our observational expectation of multiplicity based on imaging (data not shown). These calculated values for f and m predicted average proportional growth of 2.0 ± 0.5, which was 71.5% (±4.4%) of the observed growth as measured by increase in the number of infected cells (Figure 4E; Table S3) and 75.0% (±25.2%) of growth estimated by increase in fluorescence area. The closeness of these values suggests that the mechanism of continual death and phagocytosis can account for most, if not all, of granuloma expansion.
Some Newly Infected Macrophages Egress the Primary Granuloma
Tracking with 3D DIC microscopy, combined with bacterial volume measurements from fluorescence microscopy, revealed that the motility of infected granuloma macrophages decreased inversely to their bacterial load (Figure 5A; Movies S8–S10). This was confirmed by analysis of long-term 3D confocal microscopy of granulomas where fluorescent bacteria within individual macrophages could be identified as distinct clusters. Following their motion over 18 hr, we discovered that five infected macrophages left the granuloma altogether (Figure 5B; Movie S11). Consistent with our previous observations, these bacterial clusters were among the smallest in volume (Figure 5C). We confirmed by DIC visualization that the departing bacteria were intracellular (Movie S12).
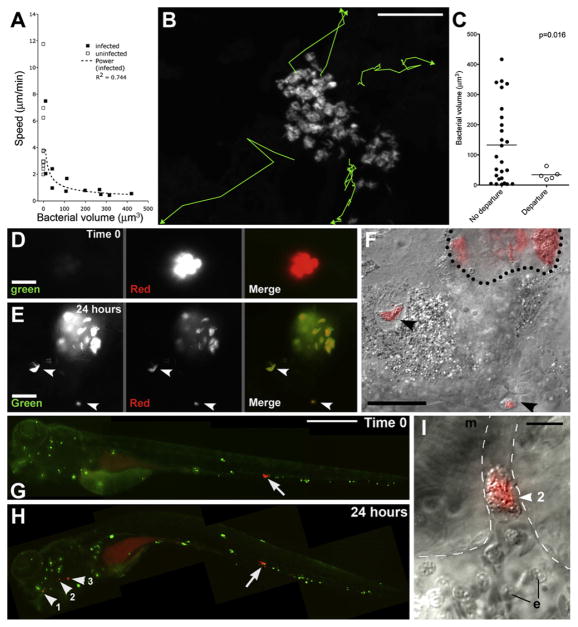
Figure 5. Motility and Departure of Infected Granuloma Macrophages.
(A) Average speeds of infected and uninfected cells versus their bacterial volumes.
(B) Tracks of departing infected macrophages from Movie S11. Scale bar, 50 μm.
(C) Bacterial volumes and average speeds of departing macrophages in (B), compared to those that did not depart. p by Student’s t test.
(D–F) Departure of infected macrophages from brain granuloma. (D) Granuloma immediately after photoactivation (red) and (E) 24 hr later, demonstrating granuloma growth and departure of infected macrophages (arrowheads). (F) DIC/red fluorescence overlay of departing macrophages (arrowheads). Dotted line represents granuloma edge. Scale bars in (D)–(F), 20 μm.
(G–I) Hematogenous dissemination from tail granuloma. (G) A single granuloma (arrow) photoactivated at 3 dpi. Scale bar, 300 μm. (H) At 4 dpi, photoactivated bacteria (arrowheads) seen in gill vasculature. (I) DIC image of cluster #2 from (H). Dashed line: limits of vasculature, e: erythrocyte, m: muscle. Scale bar, 10 μm.
To track departing bacteria, we created MD2, an Mm strain constitutively expressing the Kaede photoactivatable protein (Ando et al., 2002), which switches stably from green to red fluorescence on exposure to ultraviolet light, without damage to the bacteria or their infected cells (Figure S4 and data not shown). Selective photoactivation of single granulomas frequently revealed departure of infected macrophages after 24 hr (Figures 5D–5F). The intracellular MD2 were both red and green fluorescent (Figure 5E), suggesting that they were synthesizing new Kaede protein and thus metabolically active. In addition to departure via tissue migration (Figures 5D–5F), we observed departed infected macrophages lodged in the distant vasculature (Figure 5G–5I and Movie S13). Monitoring 33 granulomas for 24 hr after photoactivation, we recorded one to four departed macrophages in 18 (54.4%) of them and calculated the mean rate of departure at 1.4 (±0.3) per granuloma.
Macrophages Exiting the Primary Granuloma Seed Distal Granulomas to Disseminate Infection
We extended our monitoring to 2 days post-photoactivation and found 18 granulomas with evidence of departed macrophages at day 1. In six cases (33%) the departed bacteria were found in new granulomas (Figures 6A–6C). The bacteria in all departed macrophages remained both red and green fluorescent, showing their prolonged viability after departure and potential to initiate new granulomas.
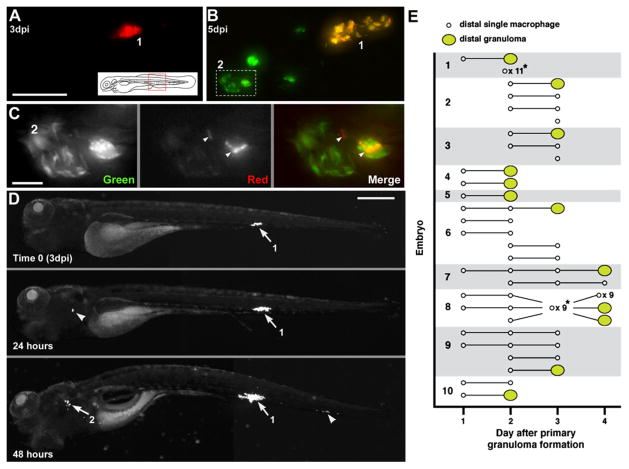
Figure 6. Departing Bacteria form Secondary Granulomas.
(A–C) Bacteria from one granuloma spread to another. (A) Primary granuloma immediately after photoactivation at 3 dpi. Scale bar, 50 μm. (B) At 5 dpi, the original granuloma (1) persists and a new one (2) has appeared nearby. Dashed box, area shown in panel C. (C) Photoactivated bacteria from granuloma 1 spreading in granuloma 2. Scale bar, 10 μm.
(D) Single granuloma at 3 dpi (white arrow, 1) with departure of bacteria to new location 24 hr later (white arrowhead). By 48 hr, the original granuloma (1) continues to grow, the new locus is forming a granuloma (2), and a third locus of infection has appeared (arrowhead). Scale bar, 300 μm.
(E) Dissemination from single primary granulomas in ten embryos. Horizontal lines indicate fate of departing macrophages (remain single or form granuloma) on successive days. *Indicates dissemination of multiple macrophages that could not be tracked definitively between days or that appeared on the last day of tracking. The embryo from (D) is shown as 1.
Mammalian tuberculosis is thought to arise from a single primary lesion (Balasubramanian et al., 1994; Rich, 1946). To determine if egress of infected macrophages could account for this dissemination, we monitored embryos infected with 10–50 CFU of green or red fluorescent bacteria for 3 days and selected ones forming a single granuloma with no other infection foci (Figure 6D). Daily monitoring from the day of granuloma formation (t = 0) showed that primary granulomas were capable of disseminating infection throughout the body by departure of infected macrophages (Figure 6D). All ten embryos monitored had evidence of newly departed infected macrophages by day 2 (Figure 6E), and of secondary granuloma formation by 4 days (Figure 6E). In every case, the appearance of a primary granuloma macrophage preceded secondary granuloma formation, which invariably occurred at the site where the departed macrophage was first observed (Figure 6D and 6E). Over 3 days, the mean rate of macrophage arrival at distal sites was 1.6 (±0.4) per day, similar to the departure rates obtained before. The rate of departure remained relatively constant over the 3 days of observation, with means of 1.4, 2.2, and 1.3 on days 1, 2, and 3 post primary granuloma formation (t = 0), respectively. The rate of new granuloma formation was 0.30 (±0.07) per day, so 19% of newly departed macrophages initiated granuloma formation per day. Together, these data show that infected macrophages frequently depart primary granulomas and migrate both hematogenously and through tissues. The kinetics of departure and secondary granuloma formation suggest that granuloma macrophages constitute the major if not sole mechanism for dissemination.
DISCUSSION
Our results suggest that mycobacterial expansion in early granulomas is driven by a continual cycle of death of infected macrophages and their phagocytosis by multiple newly recruited macrophages (Figure 7). This model is supported by a direct comparison of the kinetics of these events in the presence and absence of the RD1 virulence determinant and their correlation to infection outcome (Figure 7). Furthermore, a simple mathematical model derived from our observed logistics of WT granuloma inception suggests that this mechanism can account for at least 71% and possibly all of granuloma expansion. The small remainder, if any, may be attributable to other mechanisms that may or may not be RD1 dependent. One possible mechanism is direct spread via membrane tethers between individual macrophages as observed in the blood stream (Davis et al., 2002). However, we have not discerned this phenomenon in granulomas during extensive video microscopy. Another possible means of intercellular spread is via actin-based bacterial motility (Stamm et al., 2003; Tobin and Ramakrishnan, 2008). Both Mtb and Mm are reported to escape the phagocytic vacuole into the cytosol in an RD1-dependent fashion (Stamm et al., 2003; van der Wel et al., 2007), and Mtb has also been shown to require RD1 for intercellular spread in vitro (Guinn et al., 2004). In the case of Mm, once cytoplasmic, ~20% of bacteria become motile in cultured macrophage monolayers via host actin polymerization and can spread to adjacent cells (Gao et al., 2004; Stamm et al., 2003, 2005). This motility has not been reported for Mtb and has been proposed to be used for Mm-specific interspecies transmission strategies, rather than the granuloma formation common to Mm and Mtb (Stamm and Brown, 2004; Tobin and Ramakrishnan, 2008). This hypothesis is consistent with our finding that a mechanism distinct from actin-based motility is the major contributor to bacterial expansion in granulomas.
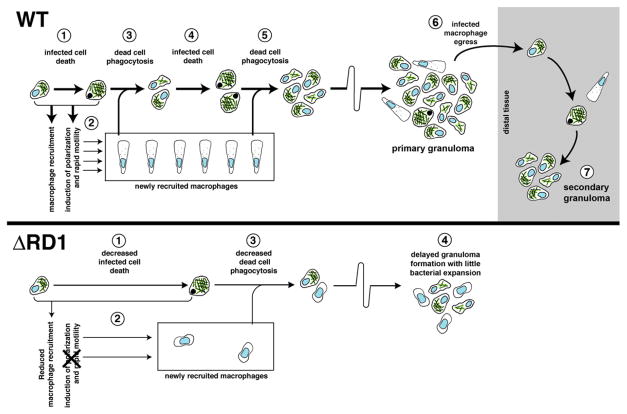
Figure 7. Mechanisms and Consequences of Early Granuloma Formation and the Impact of RD1.
Top: WT pathogenesis. An infected cell (1) recruits new macrophages and induces their rapid motility (2). Upon its death, it is phagocytosed by the recruited cells (3). After more bacterial growth, these infected cells also die (4) and are phagocytosed by more recruited macrophages (5). Infected cells egress the primary granuloma (6) to initiate secondary granulomas (7).
Bottom: The same events altered by the absence of RD1. An infected cell allows intracellular bacterial growth similar to WT. The death of this cell is delayed compared to WT (1), and the recruited macrophages are fewer in number and lack the rapid motility seen in WT (2). Slower infected macrophage death and rephagocytosis of dead cells (3) combine to produce small, delayed granulomas with better containment of infection (4).
The mechanics and dynamics of granuloma formation we observe in response to RD1-deficient infection of zebrafish embryos mirror those found in BCG-induced granulomas in rabbits (Figure 7) (Dannenberg, 2003). BCG produces attenuated infection, similar to ΔRD1 Mm in the zebrafish embryo (Volkman et al., 2004), strongly suggesting that without the faster kinetics of granuloma formation induced by RD1, mycobacteria lose the growth advantage they enjoy specifically in nascent granulomas. Extrapolation of observations from attenuated BCG infection (Dannenberg, 2003) to infections with fully virulent organisms may have led to an incorrect conclusion about the consequence of granuloma formation in response to virulent mycobacteria.
We note that the greatly reduced speeds and displacements of macrophages we observed at ΔRD1 granulomas are in accord with recent work visualizing mouse granulomas formed by the RD1-deficient strain BCG, in which the macrophages are reported to be relatively static (Egen et al., 2008). In this study, in the 2 to 3 weeks after infection, macrophages were recruited to granulomas, consistent with our observations with ΔRD1 Mm granulomas in zebrafish; however, relative macrophage recruitment rates in response to RD1-proficient and -deficient strains were not assessed, precluding a comparison with our findings of the impact of this locus on macrophage recruitment. Strikingly, in that study, while macrophages were recruited to BCG granulomas, once inside they had limited motility in contrast to the highly motile T cells (Egen et al., 2008). This lack of macrophage motility is very possibly due to the lack of RD1 in the inciting bacteria. It is also possible that, independent of RD1, granulomas have distinct macrophage dynamics at this later stage from the very early granulomas we have monitored. While analysis of the effect of RD1 on macrophage dynamics in mature granulomas awaits comparison of isogenic strains in adult animals, there is evidence suggesting that this locus continues to influence granuloma structure after adaptive immunity has been invoked. Adult zebrafish and mice with RD1-deficient Mm and Mtb, respectively, have loose, poorly structured granulomas (Sherman et al., 2004; Swaim et al., 2006). Moreover, ΔRD1 Mm-infected zebrafish embryos continue to have loose noncaseating granulomas even as adults (Volkman et al., 2004). Regardless of its exact role in mature granuloma dynamics, the role of RD1 in nascent granulomas revealed by this study is likely a critical factor in the irrevocable early bacterial expansion that establishes infection (Andersen, 1997; Kaufmann and Ladel, 1994; Saunders and Cooper, 2000).
RD1 has been thought to promote mycobacterial infection by dampening the innate immune response in order to evade its bactericidal mechanisms (Pathak et al., 2007; Stanley et al., 2003), although more recent evidence suggests that RD1 enhances inflammation (Koo et al., 2008). Our findings suggest that at least in the nascent granuloma, bacteria expressing RD1 actually accelerate aspects of the innate immune response, especially macrophage recruitment, motility, and apoptosis (see below). Accelerated granuloma formation in response to Mtb (as compared to BCG) (Adams, 1975) has been assumed to represent the host’s attempt to thwart a more virulent pathogen (Adams, 1975). Our direct observations in vivo suggest that this acceleration is driven by a mycobacterial virulence factor and actually aids bacterial growth.
Our results also shed light on the role of RD1 in granuloma formation (Volkman et al., 2004) and clarify the contributions to this process of the distinct infection phenotypes associated with this virulence locus (DiGiuseppe Champion and Cox, 2007; Ernst et al., 2007). First we show that the presence of intracellular RD1-competent Mm results in enhanced recruitment of uninfected macrophages hematogenously and through tissues to form granulomas. RD1 has been associated with increased death of infected cells in cultured macrophage monolayers. The increased intercellular spread of RD1-competent bacteria that has been observed is thought to result from bacteria being released into the culture medium and becoming accessible for phagocytosis (Gao et al., 2004; Guinn et al., 2004). However, this mechanism seems unlikely within the tight structural restraints of the granuloma. Indeed, our data reveal that while RD1 is associated with increased cell death, this increase alone seems unlikely to account for the increased infection rate. Rather, the motility of uninfected macrophages and their attraction to dead infected cells, both apparently mediated by RD1, appear to be required for intercellular spread. The lack of even a local chemotactic response to cell death in ΔRD1 granulomas suggests that reduced death in these lesions is not the root cause of the overall reduced macrophage motility and spread of bacteria. Alternatively, RD1-competent and -deficient cell death may be molecularly different despite their similar morphology and kinetics, with only the former producing signals that induce macrophage motility. In either case, reduced overall motility of macrophages appears to be responsible for the slower phagocytosis of dead macrophages in ΔRD1 Mm granulomas. This reduced phagocytosis should account substantially for the lower rate of infection of new arrivals, in combination with the decreased death rate itself.
The constant rapid motility of uninfected cells throughout the granuloma is the most profound difference between granulomas with and without RD1. While chemokines have been implicated in cell migration to form granulomas (Algood et al., 2003), we now provide evidence of continued chemotactic migration of macrophages after reaching the granuloma, which in turn correlates with their increased infection. Rapid and continuous migration seems to allow these new arrivals access to the few dying infected cells present at a given moment. These logistics are reminiscent of those described for B and T lymphocytes in lymph nodes as they seek out cognate antigen on other lymphocytes, a process also governed by chemotactic signals (Castellino et al., 2006; Okada et al., 2005)—a comparison made yet more intriguing by reports that the mature tuberculous granuloma has structural and functional similarities to secondary lymph nodes (Kahnert et al., 2007; Ulrichs et al., 2004). The rapidly moving uninfected macrophages in WT granulomas display the classic appearance of leukocytes in the presence of a chemotactic signal (Sanchez-Madrid and del Pozo, 1999). Indeed, their speed (average 4.5 μm/min) is comparable to that reported for mouse macrophages responding to a human recombinant M-CSF-receptor gradient (Webb et al., 1996), as well as those of lymphocytes moving in mouse tuberculous granulomas (Egen et al., 2008) and in lymph nodes (Stoll et al., 2002). While the macrophages we observed at WT granulomas were in constant motion, within a single lesion their movements were not in a unified direction, suggesting that the signal guiding them is uniformly distributed. However, in the midst of this apparently random movement, dying cells are rapidly found and phagocytosed, a sequence that would seem to require specifically directed motion toward newly dead cells. Notably, the macrophages approaching and phagocytosing dead cells have a distinct morphology and more directed motion. To account for this combination of random and directed motion, we propose that a gradient less signal, emanating from infected macrophages throughout the granuloma, induces new cell recruitment and random movement, but a second signal, generated by dying cells, directs nearby macrophages to them for phagocytosis. Both of these movements appear necessary for efficient phagocytosis in nascent granulomas.
The chemokines involved in granuloma formation and the pathways by which they are induced by bacterial RD1 are not yet understood and are likely to be complex (Algood et al., 2003; Peters and Ernst, 2003). Notably, TNF, a key protective cytokine in tuberculosis, was thought to mediate protection by promoting cell migration and granuloma formation, at least in part by modulating the expression of key chemokines and their receptors (Algood et al., 2003; Flynn and Chan, 2001). However we have recently found that TNF signaling is not required for granuloma formation, at least in the innate stages of infection detailed in this study (Clay et al., 2008). Rather its primary role in early protection appears to be to restrict mycobacterial growth within macrophages. Indeed, lack of TNF signaling results in accelerated granuloma formation, presumably because of an increased proportional increase in RD1 within the inciting infected macrophages (Clay et al., 2008; Miller and Ernst, 2008). Infection with ΔRD1 Mm results in attenuated infection with poor granuloma formation even in the absence of TNF signaling, further confirming the lack of involvement of this prime candidate in RD1-mediated granuloma formation.
A striking finding of our study is a very high rate of apoptotic death of infected macrophages, enhanced by RD1, that appears to be a driving factor in granuloma expansion. This finding brings into question our understanding of the role of apoptosis in mycobacterial infection. Macrophage apoptosis has been noted in human tuberculosis granulomas (Cree et al., 1987; Keane et al., 1997) and, based on some in vitro studies, has been regarded as detrimental to the infecting mycobacteria, in contrast to necrosis, which favors bacterial growth (Fratazzi et al., 1997; Gan et al., 2008; Keane et al., 2002; Oddo et al., 1998). However, many of these in vitro studies used additional agents to induce death of the infected macrophages (Fratazzi et al., 1997; Gan et al., 2008; Molloy et al., 1994; Oddo et al., 1998), inducing specific death pathways that may have overridden mycobacterium-induced ones. This point is important for the interpretation of these data given that different molecular inducers of apoptotic death (e.g., Fas ligand versus ATP) have been found to have opposite effects on the viability of the mycobacteria within the dying cell (Lammas et al., 1997). Furthermore, RD1 has been shown to promote death of cultured macrophages infected with Mtb or Mm, and this death shown to be apoptotic in the case of Mm (Gao et al., 2004; Guinn et al., 2004). RD1 is required for intercellular spread in Mtb-infected cultured macrophage monolayers (Guinn et al., 2004; Lesley and Ramakrishnan, 2008), consistent with our in vivo data that apoptotic cell death can play a role in bacterial expansion. In summary, our findings suggest either that apoptotic cell death in vivo is not actually harmful to mycobacteria—perhaps because bacterium-sparing pathways are invoked—or that the rapid phagocytosis after macrophage death, also an RD1-dependent phenomenon, nullifies any bactericidal effects.
Finally, our results shed light on the role of the primary granuloma in the dissemination of tuberculosis. By serial visualization of infection at the whole-organism level from the very earliest events, we have shown how dissemination occurs soon after establishment of the first lesion. Human tuberculosis is increasingly appreciated to be a disseminated infection (Hernandez- Pando et al., 2000) and has been inferred to disseminate hematogenously from the primary granuloma early in infection (Balasubramanian et al., 1994; Chackerian et al., 2002). Our direct imaging of the primary granuloma confirms this model and provides kinetics and a cellular mechanism for this central feature of early pathogenesis. We find that hematogenous dissemination occurs within macrophages that depart established, enlarging granulomas. Macrophages and dendritic cells are required for the initial transport of bacteria to deeper tissues for establishment of infection (Clay et al., 2007; Dannenberg, 2003; Wolf et al., 2007). Our work reveals that macrophages repeat their role as bacterial transporters by disseminating infection from the primary granuloma.
In summary, we propose that the pathway of granuloma formation and subsequent bacterial dissemination is based upon macrophage responses (recruitment, phagocytosis, apoptosis) that are of themselves generally protective (Adams, 1976; Dannenberg, 2003) and that work reasonably well against less virulent (i.e., RD1-deficient) infection. Rather than block these host responses, RD1-competent mycobacteria appear to accelerate them to turn the granuloma response into an effective tool for pathogenesis. The initiation of the adaptive immune response then may halt bacterial expansion not by forming granulomas as suggested by the classical model (Andersen, 1997; Saunders and Cooper, 2000) but by altering the early granuloma into a form of stalemate between host and pathogen.
EXPERIMENTAL PROCEDURES
Animal Care and Strains
Wild-type AB zebrafish embryos were maintained and infected by injection into the caudal vein or hindbrain ventricle at 24–30 hr post-fertilization unless otherwise noted (Cosma et al., 2006; Davis et al., 2002; Volkman et al., 2004).
Bacterial Strains
Fluorescent wild-type Mm strain M (ATCC #BAA-535) and ΔRD1 mutant strains were used (Volkman et al., 2004). MD2 was derived by transformation with plasmid pMD2 expressing a transcriptional msp12::kaede fusion (Chan et al., 2002). See Supplemental Data for details.
Vital Dye Staining of Embryos
Embryos were injected with 200 μg/ml Hoechst 33342 (Invitrogen) via caudal vein or 1/10 dilution Annexin V-AlexaFluor 488 (Invitrogen) via hindbrain ventricle or soaked in 2 μg/ml acridine orange (Sigma-Aldrich) in fish water for 10 min. Neutral red staining was performed as described (Davis et al., 2002).
Microscopy
Widefield microscopy was performed on a Nikon E600 equipped with DIC optics, a Nikon D-FL-E fluorescence unit with 100W Mercury lamp, and MFC-1000 z-step controller (Applied Scientific Instrumentation). Objectives used included 4× Plan Fluor, 0.13 NA, 10× Plan Fluor, 0.3 NA, 20× Plan Fluor, 0.5 NA, 60× Water Fluor, 1.0 NA and 60× Oil Plan Apo, 1.4 NA. Widefield fluorescence and DIC images were captured on a CoolSnap HQ or CoolSnap CF CCD camera (Photometrics) using MetaMorph 7.1 (Molecular Devices). “3D DIC Microscopy” refers to capturing Z stacks of DIC images, for one or multiple time points, and exploring these datasets manually using programs described below. Confocal microscopy was performed on an Olympus Fluoview FV1000 laser scanning confocal microscope equipped with lasers capable of 405 nm, 637 nm, 488 nm, and 561 nm excitation. Objectives used were 10× UplanApo air, 0.4 NA and 20× UplanApo air, 0.75 NA.
Image Processing
Where indicated, Z stacks were deconvolved using AutoDeblur Gold CWF, Version X1.4.1 (Media Cybernetics), with default settings for blind deconvolution. Additional dataset analysis and visualization was performed using Imaris x64 6.0 (Bitplane) and MetaMorph 7.1 (Molecular Devices). Movies were produced either directly from Imaris or from stacks compiled in MetaMorph. Additional movie compilation and formatting was performed in Adobe Premiere 6.0 and QuickTimePro 7.4 (Apple). Figure processing and assembly were performed in Adobe Photoshop CS2.
Statistics
Student’s unpaired t tests and power trendline calculations were performed using Excel 11.4 (Microsoft Corp.). All other statistical tests were performed using Prism 5.0a (GraphPad Software, Inc.).
Supplementary Material
Acknowledgments
We thank M. Troll for mathematical modelling, K. Winglee for data analysis, G. MacDonald, G. Davis, and F. Peri for technical advice, O. Humbert for initial observations, and B. Cormack, P. Edelstein, K. Urdahl, J. Egen, E. Brown, D. Tobin, H. Clay, H. Volkman, R. Lesley, and C. Cosma for discussion and critical comments. J.M.D. is grateful to R. Ahmed and to the late H.K. Ziegler for guidance. This work was supported by NIH RO1 grants AI54503 and AI36396 and the Burroughs Wellcome Foundation (L.R.) and an American Society for Engineering Education predoctoral fellowship (J.M.D.).
Footnotes
Supplemental Data include four figures, Supplemental Experimental Procedures, three tables, and 13 movies and can be found with this article online at http://www.cell.com/supplemental/S0092-8674(08)01443-8.
References
- Adams DO. The structure of mononuclear phagocytes differentiating in vivo. II. The effect of Mycobacterium tuberculosis. Am J Pathol. 1975;80:101–116. [PMC free article] [PubMed] [Google Scholar]
- Adams DO. The granulomatous inflammatory response. A review. Am J Pathol. 1976;84:164–191. [PMC free article] [PubMed] [Google Scholar]
- Algood HM, Chan J, Flynn JL. Chemokines and tuberculosis. Cytokine Growth Factor Rev. 2003;14:467–477. doi: 10.1016/s1359-6101(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Andersen P. Host responses and antigens involved in protective immunity to Mycobacterium tuberculosis. Scand J Immunol. 1997;45:115–131. doi: 10.1046/j.1365-3083.1997.d01-380.x. [DOI] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian V, Wiegeshaus EH, Taylor BT, Smith DW. Pathogenesis of tuberculosis: pathway to apical localization. Tuber Lung Dis. 1994;75:168–178. doi: 10.1016/0962-8479(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440:890–895. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- Chackerian AA, Alt JM, Perera TV, Dascher CC, Behar SM. Dissemination of Mycobacterium tuberculosis is influenced by host factors and precedes the initiation of T-cell immunity. Infect Immun. 2002;70:4501–4509. doi: 10.1128/IAI.70.8.4501-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Knaak T, Satkamp L, Humbert O, Falkow S, Ramakrishnan L. Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc Natl Acad Sci USA. 2002;99:3920–3925. doi: 10.1073/pnas.002024599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay H, Davis JM, Beery D, Huttenlocher A, Lyons SE, Ramakrishnan L. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe. 2007;2:29–39. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay H, Volkman HE, Ramakrishnan L. Tumor necrosis factor signaling mediates resistance to mycobacteria by inhibiting bacterial growth and macrophage death. Immunity. 2008;29:283–294. doi: 10.1016/j.immuni.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma CL, Sherman DR, Ramakrishnan L. The secret lives of the pathogenic mycobacteria. Annu Rev Microbiol. 2003;57:641–676. doi: 10.1146/annurev.micro.57.030502.091033. [DOI] [PubMed] [Google Scholar]
- Cosma CL, Davis JM, Swaim LE, Volkman H, Ramakrishnan L. Animal models of mycobacterium marinum infection. In: Coico CK, et al., editors. Current Protocols in Microbiology. New York: John Wiley & Sons, Inc; 2006. [DOI] [PubMed] [Google Scholar]
- Cree IA, Nurbhai S, Milne G, Beck JS. Cell death in granulomata: the role of apoptosis. J Clin Pathol. 1987;40:1314–1319. doi: 10.1136/jcp.40.11.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg AM., Jr Immunopathogenesis of pulmonary tuberculosis. Hosp Pract. 1993;28:51–58. doi: 10.1080/21548331.1993.11442738. [DOI] [PubMed] [Google Scholar]
- Dannenberg AM., Jr Macrophage turnover, division and activation within developing, peak and “healed” tuberculous lesions produced in rabbits by BCG. Tuberculosis (Edinb) 2003;83:251–260. doi: 10.1016/s1472-9792(03)00048-9. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Juan G, Li X, Gorczyca W, Murakami T, Traganos F. Cytometry in cell necrobiology: analysis of apoptosis and accidental cell death (necrosis) Cytometry. 1997;27:1–20. [PubMed] [Google Scholar]
- Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real-time visualization of Mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693–702. doi: 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- DiGiuseppe Champion PA, Cox JS. Protein secretion systems in Mycobacteria. Cell Microbiol. 2007;9:1376–1384. doi: 10.1111/j.1462-5822.2007.00943.x. [DOI] [PubMed] [Google Scholar]
- Egen JG, Rothfuchs AG, Feng CG, Winter N, Sher A, Germain RN. Macrophage and T cell dynamics during the development and disintegration of mycobacterial granulomas. Immunity. 2008;28:271–284. doi: 10.1016/j.immuni.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JD, Trevejo-Nunez G, Banaiee N. Genomics and the evolution, pathogenesis, and diagnosis of tuberculosis. J Clin Invest. 2007;117:1738–1745. doi: 10.1172/JCI31810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- Fratazzi C, Arbeit RD, Carini C, Remold HG. Programmed cell death of Mycobacterium avium serovar 4-infected human macrophages prevents the mycobacteria from spreading and induces mycobacterial growth inhibition by freshly added, uninfected macrophages. J Immunol. 1997;158:4320–4327. [PubMed] [Google Scholar]
- Gan H, Lee J, Ren F, Chen M, Kornfeld H, Remold HG. Mycobacterium tuberculosis blocks crosslinking of annexin-1 and apoptotic envelope formation on infected macrophages to maintain virulence. Nat Immunol. 2008;9:1189–1197. doi: 10.1038/ni.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol. 2004;53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- Hernandez-Pando R, Jeyanathan M, Mengistu G, Aguilar D, Orozco H, Harboe M, Rook GA, Bjune G. Persistence of DNA from Mycobacterium tuberculosis in superficially normal lung tissue during latent infection. Lancet. 2000;356:2133–2138. doi: 10.1016/s0140-6736(00)03493-0. [DOI] [PubMed] [Google Scholar]
- Imhof BA, Aurrand-Lions M. Adhesion mechanisms regulating the migration of monocytes. Nat Rev Immunol. 2004;4:432–444. doi: 10.1038/nri1375. [DOI] [PubMed] [Google Scholar]
- Kahnert A, Hopken UE, Stein M, Bandermann S, Lipp M, Kaufmann SH. Mycobacterium tuberculosis triggers formation of lymphoid structure in murine lungs. J Infect Dis. 2007;195:46–54. doi: 10.1086/508894. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH, Ladel CH. Role of T cell subsets in immunity against intracellular bacteria: experimental infections of knock-out mice with Listeria monocytogenes and Mycobacterium bovis BCG. Immunobiology. 1994;191:509–519. doi: 10.1016/S0171-2985(11)80457-2. [DOI] [PubMed] [Google Scholar]
- Keane J, Balcewicz-Sablinska MK, Remold HG, Chupp GL, Meek BB, Fenton MJ, Kornfeld H. Infection by Mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect Immun. 1997;65:298–304. doi: 10.1128/iai.65.1.298-304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane J, Shurtleff B, Kornfeld H. TNF-dependent BALB/c murine macrophage apoptosis following Mycobacterium tuberculosis infection inhibits bacillary growth in an IFN-gamma independent manner. Tuberculosis (Edinb) 2002;82:55–61. doi: 10.1054/tube.2002.0322. [DOI] [PubMed] [Google Scholar]
- Koo IC, Wang C, Raghavan S, Morisaki JH, Cox JS, Brown EJ. ESX-1-dependent cytolysis in lysosome secretion and inflammasome activation during mycobacterial infection. Cell Microbiol. 2008;10:1866–1878. doi: 10.1111/j.1462-5822.2008.01177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammas DA, Stober C, Harvey CJ, Kendrick N, Panchalingam S, Kumararatne DS. ATP-induced killing of mycobacteria by human macrophages is mediated by purinergic P2Z(P2X7) receptors. Immunity. 1997;7:433–444. doi: 10.1016/s1074-7613(00)80364-7. [DOI] [PubMed] [Google Scholar]
- Lesley R, Ramakrishnan L. Insights into early mycobacterial pathogenesis from the zebrafish. Curr Opin Microbiol. 2008;11:277–283. doi: 10.1016/j.mib.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Ernst JD. Illuminating the black box of TNF action in tuberculous granulomas. Immunity. 2008;29:175–177. doi: 10.1016/j.immuni.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Molloy A, Laochumroonvorapong P, Kaplan G. Apoptosis, but not necrosis, of infected monocytes is coupled with killing of intracellular bacillus Calmette-Guerin. J Exp Med. 1994;180:1499–1509. doi: 10.1084/jem.180.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North RJ, Jung YJ. Immunity to tuberculosis. Annu Rev Immunol. 2004;22:599–623. doi: 10.1146/annurev.immunol.22.012703.104635. [DOI] [PubMed] [Google Scholar]
- Oddo M, Renno T, Attinger A, Bakker T, MacDonald HR, Meylan PR. Fas ligand-induced apoptosis of infected human macrophages reduces the viability of intracellular Mycobacterium tuberculosis. J Immunol. 1998;160:5448–5454. [PubMed] [Google Scholar]
- Okada T, Miller MJ, Parker I, Krummel MF, Neighbors M, Hartley SB, O’Garra A, Cahalan MD, Cyster JG. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3:e150. doi: 10.1371/journal.pbio.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak SK, Basu S, Basu KK, Banerjee A, Pathak S, Bhattacharyya A, Kaisho T, Kundu M, Basu J. Direct extracellular interaction between the early secreted antigen ESAT-6 of Mycobacterium tuberculosis and TLR2 inhibits TLR signaling in macrophages. Nat Immunol. 2007;8:610–618. doi: 10.1038/ni1468. [DOI] [PubMed] [Google Scholar]
- Peri F, Nusslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- Peters W, Ernst JD. Mechanisms of cell recruitment in the immune response to Mycobacterium tuberculosis. Microbes Infect. 2003;5:151–158. doi: 10.1016/s1286-4579(02)00082-5. [DOI] [PubMed] [Google Scholar]
- Rich AR. Pathogenesis of Tuberculosis. Springfield, IL: Charles C. Thomas; 1946. [Google Scholar]
- Sanchez-Madrid F, del Pozo MA. Leukocyte polarization in cell migration and immune interactions. EMBO J. 1999;18:501–511. doi: 10.1093/emboj/18.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders BM, Cooper AM. Restraining mycobacteria: role of granulomas in mycobacterial infections. Immunol Cell Biol. 2000;78:334–341. doi: 10.1046/j.1440-1711.2000.00933.x. [DOI] [PubMed] [Google Scholar]
- Sherman DR, Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Smith S. Mycobacterium tuberculosis H37Rv: Delta RD1 is more virulent than M. bovis bacille Calmette-Guerin in long-term murine infection. J Infect Dis. 2004;190:123–126. doi: 10.1086/421472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LM, Brown EJ. Mycobacterium marinum: the generalization and specialization of a pathogenic mycobacterium. Microbes Infect. 2004;6:1418–1428. doi: 10.1016/j.micinf.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, Brown EJ. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med. 2003;198:1361–1368. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LM, Pak MA, Morisaki JH, Snapper SB, Rottner K, Lommel S, Brown EJ. Role of the WASP family proteins for Mycobacterium marinum actin tail formation. Proc Natl Acad Sci USA. 2005;102:14837–14842. doi: 10.1073/pnas.0504663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci USA. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll S, Delon J, Brotz TM, Germain RN. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science. 2002;296:1873–1876. doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- Swaim LE, Connolly LE, Volkman HE, Humbert O, Born DE, Ramakrishnan L. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun. 2006;74:6108–6117. doi: 10.1128/IAI.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 2008;10:1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- Tone S, Sugimoto K, Tanda K, Suda T, Uehira K, Kanouchi H, Samejima K, Minatogawa Y, Earnshaw WC. Three distinct stages of apoptotic nuclear condensation revealed by time-lapse imaging, biochemical and electron microscopy analysis of cell-free apoptosis. Exp Cell Res. 2007;313:3635–3644. doi: 10.1016/j.yexcr.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrichs T, Kaufmann SH. New insights into the function of granulomas in human tuberculosis. J Pathol. 2006;208:261–269. doi: 10.1002/path.1906. [DOI] [PubMed] [Google Scholar]
- Ulrichs T, Kosmiadi GA, Trusov V, Jorg S, Pradl L, Titukhina M, Mishenko V, Gushina N, Kaufmann SH. Human tuberculous granulomas induce peripheral lymphoid follicle-like structures to orchestrate local host defence in the lung. J Pathol. 2004;204:217–228. doi: 10.1002/path.1628. [DOI] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004;2:1946–1956. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb SE, Pollard JW, Jones GE. Direct observation and quantification of macrophage chemoattraction to the growth factor CSF-1. J Cell Sci. 1996;109:793–803. doi: 10.1242/jcs.109.4.793. [DOI] [PubMed] [Google Scholar]
- Wolf AJ, Linas B, Trevejo-Nuñez GJ, Kincaid E, Tamura T, Takatsu K, Ernst JD. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol. 2007;179:2509–2519. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.